Abstract
Cellular microtubules are marked by abundant and evolutionarily conserved post-translational modifications that have the potential to tune their functions. This review focuses on the astonishing chemical complexity introduced in the tubulin heterodimer at the post-translational level and summarizes the recent advances in identifying the enzymes responsible for these modifications and deciphering the consequences of tubulin’s chemical diversity on the function of molecular motors and microtubule associated proteins.
Introduction
Microtubules are dynamic polymers essential for cell morphogenesis, cell division, and intracellular transport. Microtubules perform their diverse cellular functions by forming suprastructures with highly distinctive geometries and dynamic behaviours: the radial cytoplasmic array, the stable short parallel axonemal array, the dynamic bipolar spindle array, the long tiled axonal array or the highly complex dendritic array. Microtubules are hollow cylindrical polymers of ~ 25 nm diameter of highly variable lengths. Their building block is the α/β heterodimer. Repeating α/β-tubulin heterodimers form a protofilament and typically thirteen protofilaments associate laterally to form the microtubule.
How does a single building block, the α/β-tubulin heterodimer, create the diverse microtubule functions observed in cells? The answer lies in the myriad of cytoskeletal regulators that act on microtubules as well as the genetic and chemical diversity of tubulin itself. In many eukaryotes, particularly higher vertebrates, both α- and β-tubulin are encoded by multigene families—comprising, for instance, six and seven genes, respectively, in humans [Sullivan 1988]. While tubulin isotypes are restricted to specialized cells, most tissues express several, thus possibly creating many combinations of α/β heterodimers. Most microtubules are made of mixtures of isotypes, while some contain predominantly a single isotype [Miller et al. 2010]. Different isotypes can have specific functions, and one isotype is not always interchangeable with another [Nielsen et al. 2001]. For example, the divergent β-tubulin found in platelets localizes to the marginal band and is important for giving platelets their discoid shape [Schwer et al. 2001]. Presently we still know very little about the effects of tubulin isotype composition on polymer assembly and dynamics, although several studies support the idea that microtubule dynamics can be modulated by titrating tubulin isotype levels [Lu and Luduena 1994; Panda et al. 1994; Newton et al. 2002] and that microtubule associated proteins can differentially interact with different tubulin isotypes [Lu et al. 2004]. A second layer of complexity is added to the tubulin dimer by highly diverse post-translational modifications. This review focuses primarily on the types of post-translational modifications found on tubulin, the enzymes responsible for these modifications and the consequences of this chemical diversity on the function of molecular motors and microtubule associated proteins (MAPs).
The chemical complexity of the microtubule
Tubulins and microtubules are subject to evolutionarily conserved and developmentally regulated post-translational modifications, including the removal and subsequent addition of the C-terminal tyrosine in α-tubulin [Barra et al. 1973], the non-reversible removal of the conserved penultimate glutamate residue of α-tubulin [Paturle-Lafanechere et al. 1991], acetylation of α-tubulin [L'Hernault and Rosenbaum 1983; L'Hernault and Rosenbaum 1985], poly-glutamylation of α- and β-tubulin [Edde et al. 1990; Alexander et al. 1991; Redeker et al. 1992; Rudiger et al. 1992], poly-glycylation of α- and β-tubulin [Redeker et al. 1994], phosphorylation of β-tubulin [Eipper 1972] and palmitoylation of α-tubulin [Caron 1997; Ozols and Caron 1997; Caron et al. 2001]. Acetylation of β-tubulin has also been reported recently [Chu et al. 2011] and this addition most likely signals that we do not yet have a complete compendium of tubulin post-translational modifications. Expansion of tubulin’s post-translational repertoire coincides with increased metazoan complexity and the highest density and variety of post-translational modifications is found in especially complex microtubule arrays like those of neurons or cilia.
The diversity of the tubulin repertoire is evident both at the cellular and subcellular levels. At the subcellular level, post-translational modifications mark distinct subpopulations of microtubules in the cell. These marks may serve to locally adapt microtubules for specific functions: thus, microtubules oriented towards a wound in a confluent monolayer of cells are enriched in detyrosination [Bre et al. 1991; Nagasaki and Gundersen 1996], microtubules in a newly extended neurite that is destined to become the axon are enriched in acetylation and poly-glutamylation [Hammond et al. 2010], astral microtubules are tyrosinated, spindle microtubules are enriched in detyrosination [Gundersen et al. 1984; Gundersen and Bulinski 1986], while microtubules in cilia, flagella or basal bodies are acetylated [Piperno and Fuller 1985] as well as heavily poly-glutamylated [Edde et al. 1990] and poly-glycylated [Levilliers et al. 1995; Verhey and Gaertig 2007; Fukushima et al. 2009; Wloga and Gaertig 2010].
The tubulin C-terminal tails: hotspots for chemical diversity
The well defined globular domain of tubulins (the “tubulin body”) contributes a large molecular surface to the protomer-protomer interface in the microtubule lattice, while the outside of the microtubule shaft (Figure 1) is decorated by a predominantly unstructured, highly negatively-charged C-terminal region (the “tubulin tail”; [Nogales et al. 1998]). Interestingly, the C-terminal tails of α- and β-tubulin are the sites of most sequence variation among isotypes [Sullivan 1988; Tischfield and Engle 2010] as well as the site of the majority of known post-translational modifications: detyrosination/tyrosination [Arce et al. 1975], poly-glutamylation [Edde et al. 1990; Alexander et al. 1991; Redeker et al. 1992; Rudiger et al. 1992], poly-glycylation [Redeker et al. 1994] and phosphorylation [Eipper 1972] (Figures 1 and 2). Thus the outside of the microtubule shaft, the surface “seen” by most MAPs and motors, is decorated by highly diverse tubulin tails that can influence their behaviour. One notable exception seems to be acetylation at position Lys40 on α-tubulin [L'Hernault and Rosenbaum 1983], a residue that is predicted to reside inside the microtubule lumen (Figure 1) and thus likely “invisible” to the regulators that bind to the microtubule surface. From an evolutionarily standpoint, it makes perfect sense that for an essential polymer additional regulatory controls were mainly added to regions of tubulin that do not participate in lattice interactions and where modifications would be less likely to result in loss of viability. This situation is analogous to the introduction of diverse modifications to the N-terminal tails of histones [Jenuwein and Allis 2001; Verhey and Gaertig 2007].
Figure 1. Tubulin post-translational modifications.
Ribbons representation of the tubulin dimer (PDB 1TUB [Nogales et al. 1998]) with α- and β-tubulin colored green and blue, respectively. The unstructured C-terminal tails were modeled to illustrate their span and are colored red. The location and type of known post-translational modifications is indicated on the structure. Residues colored grey are conserved only in a subset of tubulin isoforms. Phosphorylation occurs at Tyr437 and Ser444 of βIII-tubulin and Ser441 of βVI-tubulin; glycylation occurs on Glu445 of αIIIA/B-tubulin and Glu437 of βIV-tubulin; glutamylation occurs on Glu443 and Glu445 of αIVA-tubulin, and Glu445 of αIA/B-tubulin, Glu435 of βII-tubulin, Glu438 of βIII-tubulin, and Glu433 of βIVa-tubulin. For a more complete list of modifications, see [Redeker 2010].
Figure 2. Chemical structures of poly-Glu and poly-Gly chains added to tubulin.
A. Tyrosination B. Branch point created by the addition of a glutamate C. Branching and elongation of a poly-Glu chain D. Branch point created by the addition of a glycine E. Branching and elongation of a poly-Gly chain.
In addition to being chemically diverse, tubulin post-translational modifications have the potential to be informationally highly complex, in effect functioning as a “tubulin code”. Poly-glutamylation and poly-glycylation are not simple ON/OFF signals like tyrosination/detyrosination, phosphorylation or acetylation. The specific length of the added chain shows large variation: glutamic acid chains can be as long as 20 residues, although most are between one and six [Edde et al. 1992; Geimer et al. 1997; Redeker et al. 1998; Schneider et al. 1998; Redeker 2010]. Glycine chains of up to 34 residues have been identified [Redeker et al. 1994]. This variation substantially increases the heterogeneity of the microtubule lattice and can potentially act as a rheostat for the recruitment of regulators. To complicate matters even further, multiple post-translational modifications often occur on the same microtubule [Edde et al. 1991; Edde et al. 1992], giving rise to a staggering number of possible combinations that could tune the recruitment and behaviour of microtubule regulators.
Until recently, the majority of the work on tubulin post-translational modifications was made possible by use of antibodies that could distinguish differently modified microtubules in the cell. However, with the exception of tubulin tyrosine ligase, the enzyme that catalyzes the addition of the terminal Tyr to α-tubulin (Figures 1 and 2A), the identity of the enzymes responsible for the various tubulin modifications remained elusive for many years. A major breakthrough came in 2005 with the identification of an enzyme complex from brain extracts that poly-glutamylates tubulin [Janke et al. 2005]. This initial breakthrough was followed in the last five years by multiple studies identifying the enzymes responsible for both the addition [Janke et al. 2005; Ikegami et al. 2006; van Dijk et al. 2007; Ikegami et al. 2008; Rogowski et al. 2009; Wloga et al. 2009; Akella et al. 2010; Shida et al. 2010] (Tables I, II) and the removal of various tubulin post-translational modifications [Kalinina et al. 2007; Rogowski et al. 2010; Lalle et al. 2011] (Table III), as well as several knockout studies demonstrating the importance of these enzymes to organismal development [Ikegami et al. 2007; Akella et al. 2010; Rogowski et al. 2010; Shida et al. 2010; Vogel et al. 2010].
Table I.
Enzymes involved in the addition of mono-modifications to tubulin
| Modification | Enzyme | Substrate preference (MT vs. tubulin) |
Substrate preference (α- vs. β - tubulin) |
Activity (autonomous vs. complex) |
Function | Disease involvement |
|---|---|---|---|---|---|---|
| Tyrosination | TTL | tubulin dimer | α-tubulin | autonomous | Recruitment of +TIPs [Peris et al. 2006]; Regulation of kinesin-13 MCAK [Konishi and Setou 2009; Peris et al. 2009]; TTL inhibits tubulin polymerization [Szyk et al. 2011] | Decreased levels of TTL are associated with various cancers [Mialhe et al. 2001; Mas et al. 2002; Kato et al. 2004; Soucek et al. 2006; Whipple et al. 2010] |
| Acetylation | MEC17/αTAT1 | MT | Lys40 of α–tubulin [Akella et al. 2010; Shida et al. 2010] | autonomous | Guidance cue for Kinesin-1 in neurons [Reed et al. 2006] Interacts with and regulates Na+/K+-ATPase pump [Santander et al. 2006] | Huntington’s disease [Dompierre et al. 2007] Alzheimer’s disease [Perez et al. 2009] |
Table II.
Enzymes involved in the addition of poly-modifications to tubulin
| Modification | Enzyme | Substrate preference (MT vs. tubulin) |
Substrate preference (α vs. β -tubulin) |
Specificity (initiase vs. elongase) |
Activity (autono- mous vs. complex) |
Function | Disease involvement |
|---|---|---|---|---|---|---|---|
| Glutamylation | TTLL1 | MT | α– and β-tubulin [Janke et al. 2005; van Dijk et al. 2007] | initiase | complex | Main poly-glutamylase in neurons [Janke et al. 2005]; glutamylation of ciliary axonemes [Ikegami et al. 2010]; glutamylation of basal body MTs [Wloga et al. 2008] | |
| TTLL2 | MT | unknown | unknown | complex? | unknown | ||
| TTLL4 | MT | β-tubulin [van Dijk et al. 2007] | initiase | autonomous | Increased levels of TTLL4 correlate with pancreatic cancer [Kashiwaya et al. 2010] | ||
| TTLL5 | MT | α-tubulin [van Dijk et al. 2007] | initiase | autonomous | unknown | ||
| TTLL6 | MT | α-tubulin [van Dijk et al. 2007]; β-tubulin [Janke et al. 2005; Suryavanshi et al. 2010] | elongase | autonomous | Poly-glutamylation of axonemal MTs [Janke et al. 2005; Pathak et al. 2011]; Regulation of MT severing enzymes [Lacroix et al. 2010]; Ciliary motility, regulation of inner-arm dynein activity [Suryavanshi et al. 2010] | ||
| TTLL7 | MT | β-tubulin [Ikegami et al. 2006; van Dijk et al. 2007; Mukai et al. 2009] | initiase/ elongase | autonomous | Required for growth of MAP-2 positive neurites [Ikegami et al. 2006] | ||
| TTLL9 | MT | α-tubulin [Wloga et al. 2008; Kubo et al. 2010] | elongase | complex? | Poly-glutamylation of axonemal MTs [Wloga et al. 2008; Kubo et al. 2010] | ||
| TTLL11 | MT | α-tubulin [van Dijk et al. 2007] | elongase | autonomous | Regulation of the MT severing enzyme spastin [Lacroix et al. 2010] | ||
| TTLL12 | unknown | unknown | unknown | unknown | unknown | Increased levels of TTLL12 correlate with prostate cancer [Wasylyk et al. 2010] | |
| TTLL13 | MT | α-tubulin [van Dijk et al. 2007] | elongase | autonomous | unknown | ||
| Glycylation | TTLL3 | MT Additional substrates | α- and β-tubulin [Rogowski et al. 2009; Wloga et al. 2009] | initiase/elongase | autonomous | Glycylation of axonemes [Wloga et al. 2009; Pathak et al. 2011]; maintenance of axonemal structure; knockdown causes male sterility in D. melanogaster [Rogowski et al. 2009] | Inactivating mutations associated with increased cancer risk [Sjoblom et al. 2006; Rogowski et al. 2009] |
| TTLL8 | MT Additional substrates | α- and β-tubulin [Rogowski et al. 2009] | initiase | autonomous | unknown | ||
| TTLL10 | MT Additional substrates | α- and β-tubulin, preference for α–tubulin [Ikegami et al. 2008; Ikegami and Setou 2009; Rogowski et al. 2009] | elongase | autonomous | Note: Human TTLL10 is catalytically inactive due to two point mutations [Rogowski et al. 2009] |
Table III.
Enzymes involved in the removal of post-translational modifications on tubulin
| Modification | Enzyme | Substrate preference (MT vs. tubulin) |
Substrate preference (α vs. β -tubulin) |
Specificity (branch vs. extended chain) |
Activity (autonomous vs. complex) |
Function | Disease involvement |
|---|---|---|---|---|---|---|---|
| Detyrosination | unknown | MT | α-tubulin | - | unknown | preferential association of KIF5 [Konishi and Setou 2009] | |
| Deacetylation | HDAC6, SIRT2 | MT | Lys40 of α-tubulin [Hubbert et al. 2002; North et al. 2003] | - | Autonomous May function as part of a complex in vivo [North et al. 2003] | Regulation of chemotactic cell motility [Hubbert et al. 2002] | Huntington’s disease [Dompierre et al. 2007] Alzheimer’s disease [Perez et al. 2009] |
| Deglutmaylation | CCP1 | MT Additional substrates: Telokin, MLCK1 | α- and β-tubulin; Generates Δ2-tubulin [Rogowski et al. 2010] | Extended Glu chains | autonomous | Regulation of polyglutamylation in neuronal cells during development [Rogowski et al. 2010]; Ciliary ultrastructure maintenance, localization of kinesin-3 KLP-6, regulation of kinesin-2 OSM-3 speed [O'Hagan et al. 2011] | Purkinje cell degeneration, mutated in pcd mice [Rogowski et al. 2010] |
| CCP4 | MT | α- and β-tubulin; Generates Δ2-tubulin [Rogowski et al. 2010] | Extended Glu chains | autonomous | unknown | ||
| CCP5 | MT | α- and β-tubulin [Rogowski et al. 2010] | Branching-point Glu | autonomous | Regulation of polyglutamylation in neuronal cells during development [Rogowski et al. 2010] | ||
| CCP6 | MT | α- and β-tubulin; Generates Δ2 tubulin [Rogowski et al. 2010] | Extended Glu chains | autonomous | Deglutamylation of MTs in sensory cilia [Kimura et al. 2010] | ||
| Deglycylation | unknown | unknown | unknown | unknown | unknown | unknown |
Tubulin monomodifications - the detyrosination /tyrosination cycle
It is rather fitting to write this review about tubulin post-translational modifications in a special issue dedicated to a conference focused on the neuronal cytoskeleton held in South America since the first tubulin specific post-translational modification was discovered by a team of Argentinian scientists. In 1973, Hector Barra and colleagues reported an RNA-independent addition of a tyrosine to an abundant protein present in brain extracts that they later identified as tubulin [Barra et al. 1973; Barra et al. 1974; Arce et al. 1975]. Subsequent studies have shown that most isoforms of α-tubulin contain a C-terminal genome-encoded tyrosine residue [Valenzuela et al. 1981] that undergoes a cycle of detyrosination and tyrosination [Russell et al. 1984; Sherwin et al. 1987; Warn et al. 1990]. The product of the detyrosination reaction is known as “Glu-tubulin” since the penultimate C-terminal residue of α-tubulin is a glutamate. This form of tubulin is prevalent in stable/long-lasting microtubules that display little dynamicity (t1/2 ~ 16 hrs). Tyrosinated tubulin (known as “Tyr-tubulin”) is found mainly in dynamic microtubules (t1/2 ~ 3–5 min; [Webster et al. 1987]). Recent studies have shown that this difference in stability between Tyr- and Glu-tubulin is not due to intrinsic changes in the properties of the microtubule polymer itself, but the modification-dependent recruitment of microtubule dynamics regulators [Peris et al. 2006; Peris et al. 2009].
The enzyme responsible for the addition of the terminal tyrosine to α-tubulin, tubulin tyrosine ligase (TTL), was isolated from brain extracts in a stable complex with tubulin and shown to require ATP and magnesium ions for the tyrosination reaction [Raybin and Flavin 1977; Murofushi 1980; Schroder et al. 1985]. The TTL gene was cloned after intense efforts in 1993 [Ersfeld et al. 1993], more than ten years after the initial isolation of the enzyme. Subsequent studies by Weber and coworkers [Rudiger et al. 1994] established the exquisite specificity that TTL has for the C-terminal tail of tubulin, showing that any substitutions in the last six residues of the α-tubulin C-terminal tail had drastic effects on TTL activity. Despite the fact that the detyrosination reaction was identified more than 30 years ago [Hallak et al. 1977], the identity of the enzyme that catalyzes the removal of the C-terminal tyrosine on α-tubulin has proven to be elusive. The product of the detyrosination reaction, Glu-tubulin, can be further modified by the removal of the penultimate glutamate, forming Δ2-tubulin [Paturle-Lafanechere et al. 1991]. Microtubules made of Δ2-tubulin are abundant in neuronal cells [Paturle-Lafanechere et al. 1994] and this tubulin form cannot be reconverted to Glu- and subsequently Tyr-tubulin and is thus removed from the detyrosination/tyrosination cycle. The enzymes that are capable of catalyzing the formation of Δ2-tubulin were recently identified and belong to a family of cytosolic carboxypeptidases [Rogowski et al. 2010] (Table III).
Tubulin tyrosination – an ON/OFF signal for motors and +TIPs
Studies from several labs have shown that the C-terminal tyrosine on α-tubulin can act as a binary ON/OFF switch for the recruitment of microtubule dynamics regulators (Figure 3A, B). The kinesin-13 MCAK preferentially depolymerizes Tyr-microtubules over Glu-microtubules, thus contributing to the shorter lifetime of tyrosinated microtubules in cells [Peris et al. 2009]. Tyrosination status also affects microtubule plus-end dynamics as the C-terminal tyrosine in α-tubulin is required for the recruitment of microtubule plus-end interacting proteins such as cytoplasmic linker protein-170 (CLIP170) and p150Glued. CLIP170 tracks the microtubule plus-end by using its CAP-Gly domains to interact with a composite binding site formed by the C-terminal EEY motif of the α-tubulin tail and the end-binding protein EB1 [Honnappa et al. 2006; Hayashi et al. 2007; Weisbrich et al. 2007; Bieling et al. 2008; Dixit et al. 2009; Gupta et al. 2010]. In Saccharaomyces cerevisiae, the tyrosination/detyrosination cycle is absent, however, both α-tubulin isoforms in this organism contain an EEF sequence at their C termini, similar to the EEY C-terminal sequence of mammalian α-tubulins. The yeast ortholog of CLIP170, Bik1p, is unable to track the growing microtubule plus-ends in an engineered strain of S. cerevisiae that lacks the C-terminal Phe residue of α-tubulin and this strain also shows a decrease in spindle dynamics as well as a dampening in nuclear oscillations during budding [Badin-Larcon et al. 2004].
Figure 3. Tubulin post-translation modifications affect the recruitment of motors and MAPs.
A. Tyrosinated tubulin is enriched at the microtubule plus-end where it recruits +TIP CAP-Gly domain-containing protein CLIP170 and the depolymerizing kinesin MCAK B. Microtubules in the somatodendritic compartment are tyrosinated while axonal microtubules are detyrosinated. Kinesin-1 KIF5 prefers detyrosinated microtubules to tyrosinated microtubules and is therefore sequestered within the axon. C. Microtubule severing enzyme spastin acts preferentially on poly-glutamylated microtubules D. Poly-glutamylation regulates the interaction between inner-arm dynein and the axonemal B tubule. Left, a cross-sectional view of a “9+2” axoneme. Microtubules are colored blue, inner-arm dyneins, green, outer-arm dyneins, orange, and tubulin tails, red. Right, close up view of the boxed region. Inner-arm dyneins project away from the A-tubule of one microtubule doublet and interact with the B-tubule of an adjacent microtubule doublet using their microtubule interacting domains (colored yellow). Tubulin tails are colored red.
In neurons, tyrosination acts as a negative cue by preventing the entry of KIF5, a kinesin-1 family member, into dendrites that are rich in Tyr-microtubules, thus targeting the motor only to the axon that is rich in Glu-microtubules [Konishi and Setou 2009] (Figure 3B). Setou and co-workers elegantly showed that kinesin-1 uses a structural element in its motor domain to sense the presence of the C-terminal tyrosine on microtubules. Mutation of this structural element or depletion of TTL by RNAi breaks the asymmetric distribution of kinesin-1 and allows the motor to distribute both to the axonal and dendritic compartments [Konishi and Setou 2009]. Kinesin-1 preferential targeting to detyrosinated microtubules has been observed also in non-neuronal cells [Kreitzer et al. 1999; Dunn et al. 2008; Konishi and Setou 2009] and in vitro studies have in fact shown that kinesin-1 binds with higher affinity to Glu-microtubules [Liao and Gundersen 1998]. Consistent with its tighter binding, kinesin-1 moves slower on Glu-tubulin enriched microtubules than Tyr-tubulin enriched microtubules and it was proposed that this velocity change could ensure a more consistent delivery of cargo throughout the cell. Detyrosination could also serve as a targeting cue for the asymmetric accumulation of kinesin-1 cargo [Dunn et al. 2008]. For example, stable Glu-tubulin rich microtubules are prevalent at the leading edge of migrating cells and kinesin’s preference for the detyrosinated track could result in an asymmetric distribution of cargo at the leading edge [Gundersen and Bulinski 1988].
Given the importance of the tyrosination status to the recruitment of microtubule regulators, it is not surprising that TTL activity is essential for normal development. Mice that lack TTL die shortly after birth primarily due to abnormal neuritic and axonal projections [Erck et al. 2005]. The microtubule plus-end tracking protein CLIP170 is absent in the neurite extensions and growth cones of these mice and TTL null mouse embryonic fibroblasts display cell shape abnormalities and defects in mitotic spindle positioning [Peris et al. 2006]. Reduction in TTL levels has also been linked to several cancers of poor prognosis [Mialhe et al. 2001; Kato et al. 2004] and a reduction in TTL levels accompanied by an increase in Glu- and Δ2-tubulin is a frequent marker for tumors that are resistant to drug treatments [Mialhe et al. 2001; Soucek et al. 2006].
Tubulin acetylation, a modification predicted to be in the microtubule lumen
Acetylation of α-tubulin is one of the earliest tubulin post-translational modifications discovered [L'Hernault and Rosenbaum 1983; L'Hernault and Rosenbaum 1985]. Acetylation has been associated with long-lived microtubules like those found in ciliary axonemes [Gaertig et al. 1995] and axons [Hammond et al. 2010]. Elegant early studies by Borisy and colleagues established that acetylation is most likely a consequence of microtubule stabilization and not a cause [Webster and Borisy 1989]. Supporting these results, acetylated and deacetylated microtubules have similar stabilities in vitro [Maruta et al. 1986], however the microtubules used in these experiments carried additional post-translational modifications and it cannot be excluded that an effect of acetylation would be observed on an otherwise “naked” microtubule.
Unlike the majority of tubulin post-translational modifications that decorate C-terminal tails on the outside of the microtubule, acetylation occurs on Lys40 of α-tubulin on a loop that is largely disordered and predicted to be inside the microtubule lumen (Figure 1) based on the electron crystal structure of the αβ-tubulin dimer obtained from Zn sheets [Nogales et al. 1998]. However, unlike microtubules, Zn sheets are flat and assembled from protofilaments of random polarity. Thus, the exact disposition of the acetylation loop in the context of the microtubule is presently not clear. It is conceivable that this loop can rearrange in an assembled microtubule to present the acetylated Lys on the outside of the microtubule shaft. Alternatively, the acetylation loop is close to the protofilament-protofilament interface in the microtubule and thus could affect the packing of the heterodimers in the microtubule and affect lattice dynamics or the conformation of MAP or motor binding sites. The predicted lumenal position of the acetylated Lys makes it puzzling to understand its reported effects on motor function. For example, in vitro assays using axonemes purified from wild-type Tetrahymena thermophila and strains carrying tubulin mutants that cannot be acetylated show that kinesin-1 microtubule binding is sensitive to acetylation and that the speed of this motor is reduced on axonemes that have non-acetylated microtubules [Reed et al. 2006]. Kinesin-1 has also been shown to target preferentially to acetylated microtubules in neurons, although both detyrosination and poly-glutamylation also affect the differential recruitment of this motor [Reed et al. 2006; Hammond et al. 2010].
There have been multiple reports of histone acetyltransferases that are able to acetylate tubulin [Marmorstein and Trievel 2009]; however, a bona fide α-tubulin acetylase, MEC17/αTAT, has been just recently identified as a gene required for the function of touch receptor neurons in C. elegans [Akella et al. 2010; Shida et al. 2010]. This tubulin acetyltransferase shows homology to the Gcn5 family histone acetyltransferase [Steczkiewicz et al. 2006] and has homologs in several species including man. Importantly, recombinant MEC17/αTAT acetylates α-tubulin exclusively on Lys-40 in vitro [Akella et al. 2010]. Interestingly, Akella et al. show that the acetylation of an axoneme proceeds in vitro from the ends, supporting the idea that the enzyme diffuses through the microtubule lumen. Intriguingly, early experiments have shown that the acetylation reaction is not dependent on the length distribution of the microtubules which would be expected if the enzyme were to diffuse through the lumen [Maruta et al. 1986]. Alternatively, Shida et al. proposed that the acetyltransferase is able to reach the hidden acetylation site by accessing the microtubule lumen through transient holes that exist in the microtubule lattice [Shida et al. 2010]. Clearly there are still many facets of the microtubule acetylation reaction that we do not understand. Higher resolution models of the microtubule as well as better insight into substrate recognition for the tubulin acetyltransferase should shed light on some of these fascinating questions.
Tubulin acetylation is reversible and several enzymes that are able to deacetylate tubulin have been identified so far. In addition to their roles in epigenetic silencing, histone deacetylase 6 (HDAC6) and the NAD-dependent deacetylase sirtuin-2 (SIRT2) colocalize with microtubules and can deacetylate tubulin in vivo and in vitro [Hubbert et al. 2002; North et al. 2003]. Even though they seem to function as part of a complex [North et al. 2003], each is capable of deacetylating α-tubulin on its own [Hubbert et al. 2002; North et al. 2003]. SIRT2 can use both soluble tubulin as well as microtubules as substrates [North et al. 2003]. The substrate preference for HDAC6 is less clear as one study shows that it preferentially deacetylates α-tubulin already incorporated in the microtubule lattice and not soluble tubulin [Hubbert et al. 2002], while another shows that it can deacetylate both [North et al. 2003].
Tubulin acetylation and its effects on cellular function
Even though acetylation was one of the earliest tubulin post-translational modifications identified, its effects on cellular physiology have been slow to emerge. Substitution of wild-type α-tubulin with non-acetylatable mutants in T. thermophila produces no morphological abnormalities and has no effect on growth [Kozminski et al. 1993; Gaertig et al. 1995]. Cilia dependent functions were also the same as those observed in wild-type cells. Likewise, knock-down of MEC17/αTAT1 in RPE-hTERT cells does not cause visible morphological or proliferation defects, however it does delay cilia assembly upon serum starvation [Shida et al. 2010]. At the organismal level, zebrafish mec17 morphants display gross anatomical defects, such as a curved body shape, short body axis, small head and eyes and they also have a drastically impaired startle response, consistent with neuromuscular defects [Akella et al. 2010]. However, it is not yet clear what the cellular mechanisms responsible for these phenotypes are and whether they are all related to the cilia assembly defects observed in cells.
Several studies have now linked microtubule acetylation to membrane dynamics. Interestingly, a direct interaction between acetylated tubulin and a membrane protein, the Na+/K+ pump was one of the first reported molecular functions for tubulin acetylation [Santander et al. 2006; Zampar et al. 2009] and this interaction is responsible for inhibiting the ATPase activity of the pump [Casale et al. 2001; Casale et al. 2003]. Elegant imaging studies have shown that the ER associates and slides along the microtubule shaft [Lee and Chen 1988; Waterman-Storer and Salmon 1998] and recent work established that this sliding occurs primarily on acetylated microtubules [Friedman et al. 2010]. The identity of the ER receptor for the acetylated microtubule is still unknown. It is possible that ER sliding is mediated by molecular motors [Wozniak et al. 2009], consistent with the bidirectional nature of the sliding motion and the observation that kinesin-1 shows a preference for acetylated microtubules. Like the ER, mitochondria also move preferentially on acetylated microtubules and a recent tomographic study revealed that mitochondrial fission was triggered by the gradual constriction of the ER tubule around the mitochondria and subsequent recruitment of dynamin to the constriction site [Friedman et al. 2011]. It is tempting to speculate whether the location of the patches of acetylation on microtubules influences the site of the fission event.
Our understanding of the cellular consequences of tubulin acetylation has lagged behind because the identity of the principal tubulin acetyltransferase was unknown until very recently and thus the cellular effects of tubulin acetylation were mostly explored by manipulating the levels of tubulin deacetylases either by siRNA or using chemical inhibitors. However, it is important to keep in mind that these deacetylases have other substrates in addition to tubulin that could contribute to the observed phenotypes. For example, knock-down of HDAC6 in NIH-3T3 cells increases their motility [Hubbert et al. 2002] and since HDAC6 localizes to the leading edge of fibroblasts, a highly dynamic environment without microtubules enriched in acetylation, HDAC6 was proposed to regulate cell motility by controlling microtubule acetylation at the leading edge [Hubbert et al. 2002]. However, HDAC6 has also been shown to have an effect on actin-dependent cell motility by deacetylating cortactin [Zhang et al. 2007]. The recent identification of the tubulin acetylase finally allows the mechanistic dissection of the consequences of tubulin acetylation for cells and the organism.
Tubulin poly-modifications – poly-glutamylation and poly-glycylation
Poly-glutamylation and poly-glycylation are abundant and evolutionarily conserved post-translational modifications that lead to the addition of glutamate and glycine chains of variable lengths to the C-terminal tails of either α- or β-tubulin. The reaction involves the ATP-dependent formation of an isopeptide bond between the α-amino group of the first newly added residue (either Glu or Gly) and the side-chain γ-carboxyl group of a glutamate (Figures 2B–E). Additional glutamates or glycines are then added to the growing chain via standard peptide bonds [Redeker et al. 1991; Redeker et al. 1996; Redeker 2010] (Figures 2C, E). Glutamylation and glycylation occur on microtubules [Audebert et al. 1993; Regnard et al. 1998], as opposed to tyrosination that occurs primarily on soluble tubulin [Arce et al. 1975; Raybin and Flavin 1975; Szyk et al. 2011]. Tubulin glutamylation was first identified in cultured mouse brain neurons more than 20 years ago [Edde et al. 1990], while tubulin glycylation was first observed in paramecium axonemes [Redeker et al. 1994]. Like Glu- and Δ2-tubulin, poly-glutamylation is prevalent on neuronal microtubules [Audebert et al. 1993], as well as in stable microtubule assemblies like those found in axonemes [Bre et al. 1994], centrioles, basal bodies and parts of the mitotic spindle [Bobinnec et al. 1998]. The distribution of glycylated microtubules is much more limited and this modification is found predominantly on the stable microtubules of cilia and flagella [Redeker et al. 1994; Bre et al. 1996].
Poly-glutamylases and poly-glycylases – the TTLL family of enzymes
Despite the fact that tubulin poly-glutamylation and poly-glycylation have been known for decades, the enzymes responsible for these modifications were identified only recently [Janke et al. 2005; Ikegami et al. 2006; van Dijk et al. 2007; Rogowski et al. 2009; Wloga et al. 2009; Rogowski et al. 2010; Lalle et al. 2011] and are just beginning to be characterized (Table II). The first tubulin poly-glutamylation enzyme identified showed homology to TTL [Janke et al. 2005] and based on this homology an extended family of poly-glutamylases and poly-glycylases was later identified [Ikegami et al. 2006; van Dijk et al. 2007; Ikegami and Setou 2009; Rogowski et al. 2009; Wloga et al. 2009]. Consequently, all tubulin glutamylases and glycylases form the TTL-like (TTLL) family of tubulin modifying enzymes. Mammals have 13 distinct TTLL enzymes (TTLL1-13) [van Dijk et al. 2007], and several TTLL family members have been identified in simpler eukaryotic organisms, including Chlamydomonas reinhardtii [Kubo et al. 2010], T. thermophila [Janke et al. 2005; Wloga et al. 2010], Caenorhabditis elegans [Kimura et al. 2010], and Giardia duodenalis [Lalle et al. 2011]. TTLL enzymes can have preference for either α- or β-tubulin, although many are able to modify either protomer efficiently when overexpressed [Janke et al. 2005; Ikegami et al. 2006; van Dijk et al. 2007]. The initial characterization of the mouse TTLL family showed that TTLL5, 6, 11, and 13 preferentially poly-glutamylate α-tubulin, while TTLL4 and 7 prefer β-tubulin. TTLL1 has a preference for α-tubulin tails [Janke et al. 2005]. Interestingly, a TTLL1 knockout mouse displayed decreased glutamylation levels on both α- and β-tubulin [Ikegami et al. 2010]. Ttll6Ap, one of six TTLL6 orthologs present in T. thermophila, preferentially modifies β-tubulin. Interestingly, mouse TTLL6 shows a preference for α-tubulin [Janke et al. 2005]. It is important to point out that given the divergence of TTLL family members as well as our very limited understanding of the molecular determinants for their specificity, sequence conservation can sometimes be misleading in classifying the various TTLLs from different organisms. Further structural and functional studies should aid in their classification.
TTLL enzymes are also specialized to either initiate the poly-glutamylation or poly-glycylation reaction by catalyzing the synthesis of the first branched peptide bond (and thus are called “initiases”), or elongate the amino acid chain starting from the branch site (and thus are called “elongases”). TTLL4, 5, and 7 function as initiases adding a branched glutamic acid to the tubulin tail (Figure 2B), while TTLL6, 11, and 13 function as elongases and add poly-Glu chains of variable lengths to the branched glutamic acid (Figure 2C). TTLL7 has been reported to also have an elongase function [Mukai et al. 2009]. The specificity of mammalian TTLL2, 9, and TTLL12 are unknown. However, TTLL9 orthologs from C. reinhardtii and T. thermophila appear to function primarily as α-tubulin elongases [Wloga et al. 2008; Kubo et al. 2010], and recent evidence indicates that TTLL12 might be inactive yet function as a regulator of other poly-glutamylase TTLLs in vivo [Wasylyk et al. 2010]. TTLL3, 8, and 10 are glycylases [Ikegami et al. 2008; Ikegami and Setou, 2009; Rogowski et al. 2009; Wloga et al. 2009], with TTLL3 and 8 serving as initiases, and TTLL10 serving as an elongase.
Intriguingly, several studies show that some TTLL enzymes can function both as initiases and elongases. Recombinant mouse TTLL7 can both initiate the addition as well as the elongation of glutamate chains at least 16 residues long on the C-terminal tail of β-tubulin in vitro [Mukai et al. 2009]. Drosophila melanogaster lacks a functional copy of TTLL10, an elongating glycylase [Rogowski et al. 2009]. However, it produces two TTLL3 isoforms (TLLL3A and TTLL3B) capable of initiating and elongating poly-glycine chains. While no direct structural data is currently available, optimized reversed-phase separations indicate that, in the case of glutamylation, the first residue joined to the tubulin C-terminal tail is attached via a γ-peptide bond between the α-amide group of the incoming residue and the side-chain γ-carboxyl group of the main-chain glutamate residue in the tubulin tail, while the second and third (and presumably all additional) glutamates are attached to the newly adjoined residue via standard α-peptide bonds [Redeker et al. 1991; Redeker et al. 1996; Redeker 2010] (Figure 2C). From a mechanistic standpoint it is intriguing that some TTLL enzymes are capable of catalyzing the formation of both peptide and isopeptide bonds. This bi-functionality implies a remarkable plasticity of the enzyme active site. This is even more surprising for poly-glycylases that have been reported to perform both initiating and elongating reactions, since this would require them to first accommodate a glutamate side chain in their active site during the initiating reaction, followed by a glycine during chain elongation (Figure 2E). The side chain of a glutamate is quite different from the linear glycine peptide chain because in the case of the glutamate, the carboxylate is preceded by two methylene groups while in the linear glycine peptide chain, the carboxylate is preceded by one methylene which in turn is preceded by a polar conjugated amide that is distinctly different from the hydrophobic methylene (Figure 2E). High-resolution structural information on the various TTLL enzymes of different specificities will hopefully shed light on the mechanisms they use for chain initiation and elongation and the accommodation the active site needs to undergo in order to support these two different reactions.
TTL structure – a scaffold upon which the diversity of the TTLL family is built
TTLL family members are highly variable in length, ranging from ~ 400 amino acids (TTLL1) to ~1300 (TTLL5; [van Dijk et al. 2007]). Despite this variation, TTLL enzymes contain a conserved core that is homologous to TTL. The X-ray crystal structure of TTL was reported recently [Szyk et al. 2011] and revealed an elongated molecule consisting of three domains: a N-terminal, a central, and C-terminal domain (Figure 4) with the active site supported at the interface between the central and C-terminal domains. On the basis of multiple sequence analyses, van Dijk et al. defined a "core TTL domain" shared between all TTLL family members [van Dijk et al. 2007]. Their definition is in all likelihood too restrictive. The TTL crystal structure shows an intimate association between the N-, central and C-terminal domains of TTL [Szyk et al. 2011] and revealed that the original definition covers only part of the central and the C-terminal domains that are unlikely to fold and function on their own. Thus the "TTL core" most likely extends beyond the one identified initially based on primary structure and comprises the entire TTL three-domain structure that is likely shared by all TTLL family members.
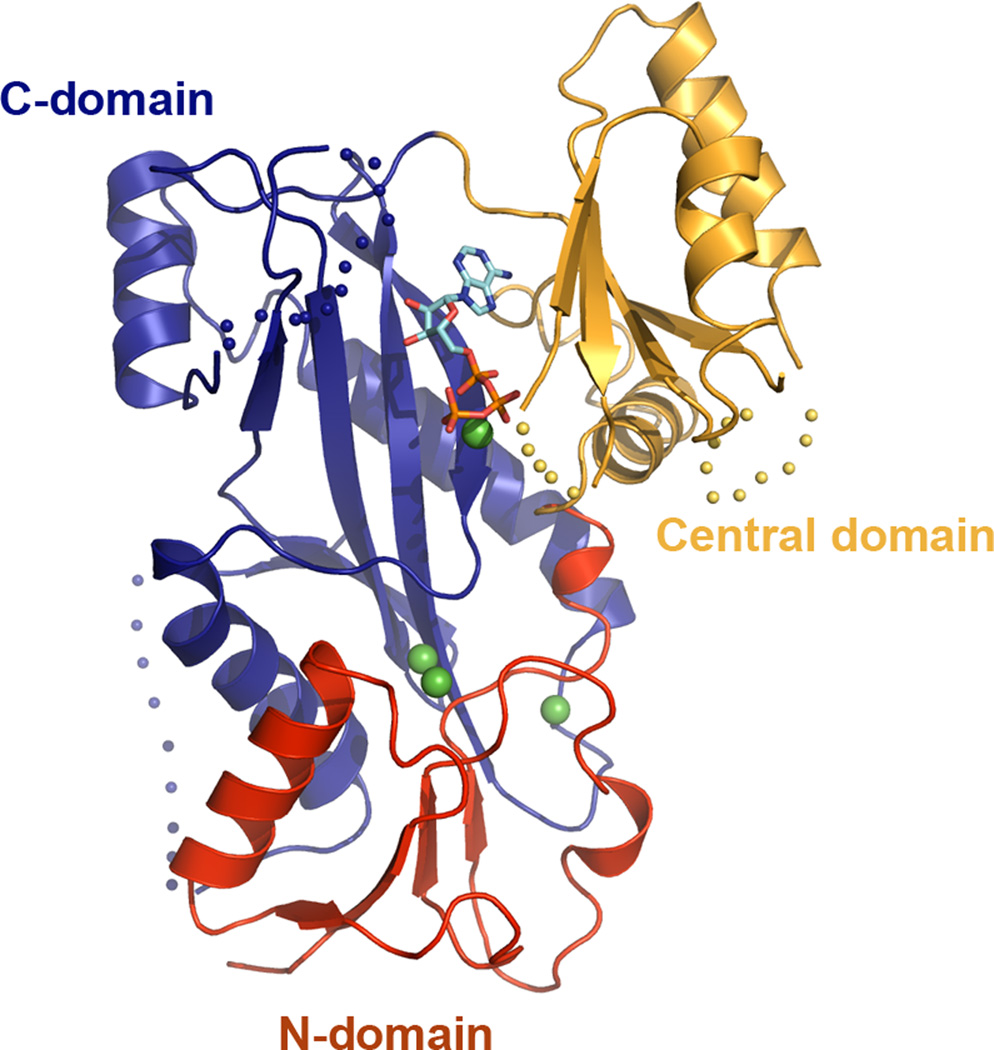
Figure 4. Crystal structure of tubulin tyrosine ligase.
Ribbons representation of the TTL crystal structure bound to ATP [Szyk et al. 2011]; N-terminal domain, red; central domain, gold; C-domain, dark blue; ATP shown in ball-and-stick representation.
Structure-based functional studies showed that TTL uses a conserved positively-charged surface adjacent to the active site to interact with the negatively-charged C-terminal tail of α-tubulin [Szyk et al. 2011]. Structure-based sequence alignments revealed that while active site residues are invariant between all TTL and TTLL family members, consistent with a common reaction mechanism for these enzymes, the identities of residues that form the C-terminal peptide binding surface were reshuffled in order to accommodate the different substrate specificities of the various TTLLs [Szyk et al. 2011]. However, unlike the TTLL enzymes characterized so far, TTL prefers monomeric tubulin and is inefficient at modifying tubulin already incorporated in the microtubule lattice [Arce et al. 1975; Raybin and Flavin 1975]. Small angle X-ray scattering coupled with analytical ultracentrifugation revealed that the enzyme forms an elongated 1:1 complex with the tubulin heterodimer by capping the longitudinal interface of the α/β-heterodimer on the surface that would otherwise be engaged in microtubule lattice interactions. Consequently, TTL inhibits tubulin polymerization in vitro and decreases microtubule growth rates when overexpressed in vivo. The tubulin polymerization inhibition activity is independent of the tyrosination activity of the enzyme [Szyk et al. 2011]. Interestingly, the surface employed by TTL to recognize the tubulin dimer is used as a homo-oligomerization interface in structurally related enzymes that belong to the ATP-grasp superfamily of ATP-dependent carboxylate-amine/thiol ligases, such as glutathione S-transferase or D-Ala:D-Ala ligase to which TTL is distantly related [Szyk et al. 2011] (Figure 5). Thus, TTL has preserved the active site architecture that was refined evolutionarily to support the carboxylate-amine/thiol ligation reaction for small ligands (i.e. glutathione or D-Ala) but evolved to convert a homo-oligomerization interface into a hetero-oligomerization interface with its large ligand, tubulin.
Figure 5.
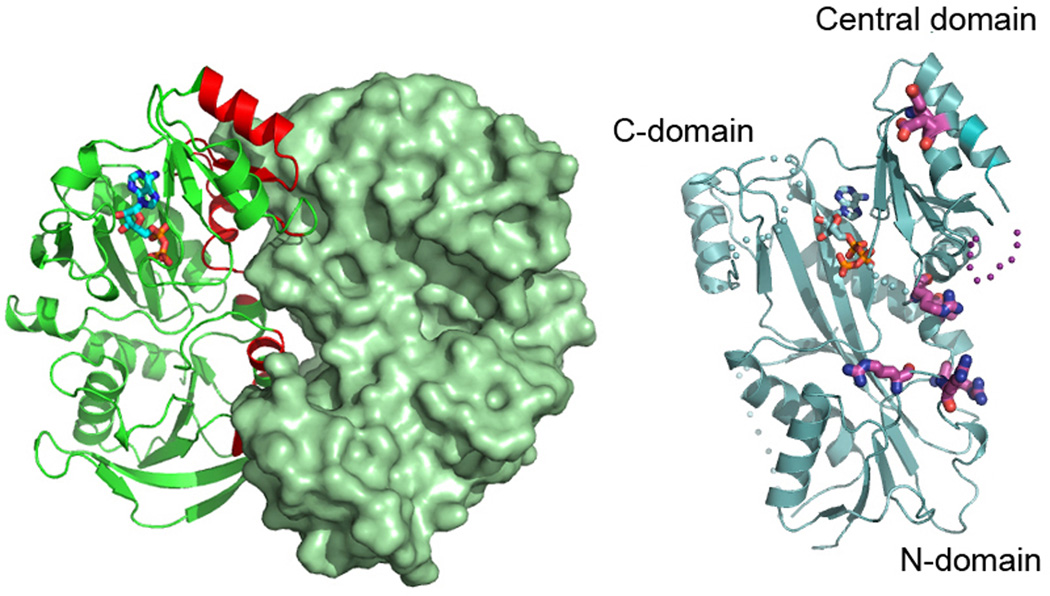
Monomeric TTL exploits a homo-oligomerization interface common to ATP-grasp enzymes to form a hetero-oligomeric complex with tubulin. Right, structure of the distantly related glutathione S-transferase dimer ([Ji et al. 1992] 1GST.pdb). Left, structure of TTL [Szyk et al. 2011] in the same orientation as the GST monomer with residues delineated as important for tubulin binding from functional studies [Szyk et al. 2011] shown in ball and stick representation.
In addition to the TTL core, several of the TTLL enzymes contain additional domains that target them to particular subcellular structures. In T. thermophila, full-length Ttll6Ap localizes only to a subset of assembling cilia, while C-terminal Ttll6Ap truncations are retained in the cell body [Wloga et al. 2010]. Likewise, the C-terminal 117 residues of mouse TTLL6 are essential for its targeting to cilia [van Dijk et al. 2007] and the N-terminal non-catalytic domain of TTLL3A, one of the six TTLL3 orthologs found in T. thermophila, is required for the ciliary targeting of this enzyme [Wloga et al. 2009]. It is not yet clear whether these accessory domains enhance catalysis and specificity for certain substrates or whether the substrate specificity in vivo is a result of the sequestration of the enzyme to the correct subcellular compartment, like the cilia for example. Determining how these accessory domains fold and are arranged in relation to the TTL catalytic core as well as what targets they recognize and how they engage them is crucial to our understanding of the regulation of TTLL enzymes in vivo.
Tubulin poly-glutamylation tunes the activity of motors and MAPs
Microtubules decorated with poly-glutamate tails of different lengths are spatially segregated in the cell, with the longest poly-glutamate chains found in cilia. The poly-glutamate modified tails are in close proximity to the binding sites of motors and MAPs where they can influence their activity and possibly act as a “cellular GPS” for their localization. Consistent with this, proteolytic removal of the tubulin C-terminal tails decreases the processivity of the molecular motors dynein and kinesin [Wang and Sheetz 2000] and early studies using antibodies have shown that the presence and length of the poly-Glu tail can affect the recruitment of cytoskeletal regulators to the microtubule. For example, qualitative studies using blot overlay assays have shown that rat kinesin-1, MAP1B, MAP2 and Tau interact preferentially with microtubules displaying moderately long (up to three) poly-Glu tails [Boucher et al. 1994; Larcher et al. 1996; Bonnet et al. 2001], while MAP1A prefers microtubules with longer tails (up to six; [Bonnet et al. 2001]). Quantitative biophysical studies are needed to reveal the mechanistic basis for these differences.
A recent study revealed that tubulin poly-glutamylation modulates synaptic transmission by affecting the efficient transport of synaptic vesicles by kinesin-2. Mice lacking functional PGs1, a subunit of the TTLL1-containing α-tubulin poly-glutamylase complex, display a drastic loss of poly-glutamylated α-tubulin in neurons and a decrease in the levels of the kinesin-2 KIF1A in neurites. This results in a drop in the density of synaptic vesicles transported by KIF1A to synaptic terminals, which are as a result more easily depleted during continuous stimulation [Ikegami et al. 2007]. The decreased α-tubulin poly-glutamylation lowers the microtubule binding affinity of kinesin-2 (KIF1A) as well as those of kinesin-1 (KIF5), dynein and MAP1A, but has no effect on the affinity of MAP2. Interestingly, despite the fact that kinesin-1, dynein and MAP1A all displayed lower microtubule binding affinity in response to lower poly-glutamylation levels, only kinesin-2 showed changes in its subcellular targeting [Ikegami et al. 2007] indicating that the motor localization is more than just a simple function of differential binding affinities.
The poly-glutamate chain on the tubulin tail also regulates the activity of microtubule severing enzymes. Both spastin and katanin prefer microtubules decorated with poly-glutamate chains as their substrates [Sharma et al. 2007; Roll-Mecak and Vale 2008; Lacroix et al. 2010; Roll-Mecak and McNally 2010] and spastin is in fact extremely inefficient at severing microtubules that are not poly-glutamylated [Roll-Mecak and Vale 2008; Lacroix et al. 2010; Roll-Mecak and McNally 2010]. Structural studies have shown that spastin forms a hexamer with a positively-charged central pore [Roll-Mecak and Vale 2008] and removal of the negatively-charged C-terminal tails of tubulin by subtilisin treatment inhibits severing by spastin and katanin [McNally and Vale 1993; Roll-Mecak and Vale 2005]. Based on structural and functional data, it was proposed that spastin and katanin sever the microtubule by engaging the negatively-charged C-terminal tails of tubulin through their positively-charged central pore and exerting repeated “tugs” on the tubulin dimer until the fabric of the microtubule weakens and leads to catastrophic breakdown of the microtubule lattice [Roll-Mecak and Vale 2008; Roll-Mecak and McNally 2010]. The microtubule severing reaction can be inhibited by antibodies that recognize the glutamylated tails of tubulin and presumably occlude access of the enzyme to these tails, but not by antibodies recognizing the Tyr-tubulin tail [Roll-Mecak and Vale 2008]. Overexpression of either mouse TTLL6 or TTLL11 (α-tubulin glutamylases that add long poly-Glu chains) in HeLa cells that have very low levels of poly-glutamylation reduced interphase microtubule mass by 70% and this effect could be reversed almost completely by knocking down spastin [Lacroix et al. 2010]. Moreover, microtubules with long poly-glutamate chains that were purified from HeLa cells overexpressing TTLL6 are effectively severed by spastin, those with shorter poly-Glu chains added by TTLL4 less so, while non-glutamylated microtubules are severed extremely inefficiently [Lacroix et al. 2010] (Figure 3C). It is possible that the very weak activity observed in the latter case could be due to the small amount of fluorescently labeled heavily glutamylated brain tubulin that was used in their assay to assemble the microtubules. It is presently unclear how increased glutamylation of the C-terminal tail of tubulin stimulates microtubule severing, however the proposed mechanism for microtubule severing enzymes suggests a possibly tighter interaction between the positive pore of spastin and the more negatively-charged poly-glutamylated tails. Recent work shows that katanin prefers to sever also acetylated microtubules [Sudo and Baas 2010], however the mechanism behind this preference is not clear at the moment. One possibility is that acetylated microtubules display slightly different packing of their protofilaments that makes it energetically less costly to disrupt lateral interactions in the microtubule lattice.
Poly-glutamylation has more recently been implicated in the microtubule association of the septin family member, SEPT2 [Spiliotis et al. 2008]. The accumulation of septins on poly-glutamylated microtubules precludes the binding of MAP4 [Spiliotis et al. 2008] which acts as an inhibitor of vesicle transport [Bulinski et al. 1997] and this is thought to create a poly-glutamylated microtubule/septin “fast track” for the efficient transport of vesicles from the Golgi to the plasma membrane that is required for apical-basolateral polarization.
Poly-glutamylases and poly-glycylases – regulators of axonemal function
Several of the TTLL knockout mice characterized to date show defects in axonemal structure and function. For example, ROSA22 mice lacking a functional copy of PGs1, a non-catalytic subunit of the multi-protein TTLL1 poly-glutamylase complex, are sterile due to defects in flagellar assembly [Campbell et al. 2002; Regnard et al. 2003; Janke et al. 2005]. Interestingly, these mice also display abnormal behavioral traits, including reduced aggressiveness. The underlying mechanisms for these behavioral phenotypes are presently unclear. Axonemes isolated from the tracheal epithelial cilia of TTLL1 knockout mice are often completely straight and have a decrease in the average bend compared to wild-type, leading to an increase in cilia beat frequency and a loss of ciliary beating asymmetry that is important for directed fluid flow [Ikegami et al. 2010]. Like the ROSA22 mice, they are also sterile due to defects in sperm morphology and motility [Ikegami et al. 2010; Vogel et al. 2010]. While the exact mechanism behind the loss in curvature and beating asymmetry is unclear, it was proposed that the asymmetric distribution of glutamylation in the axoneme [Fouquet et al. 1996] contributes to the asymmetry of beating in wild-type cilia, and that the down regulation in the levels of poly-glutamylation in the axonemes of the TTLL1 knock-out mice eliminates axonemal beating asymmetry.
Two additional studies also demonstrate a link between tubulin poly-glutamylation and axonemal function. In T. thermophila, the double knockout Ttll6Ap/Ttll6Fp (Ttll6A and Ttll6F are two of the six TTLL6 paralogs present in that organism) had significantly reduced motility and much straighter locomotory cilia compared to wild-type [Suryavanshi et al. 2010]. Surprisingly, despite these differences, isolated axonemal microtubules from wild-type and double mutant strains displayed similar sliding velocities. However, a triple knockout of Ttll6Ap/Ttll6Fp and outer arm dynein displayed a 2-fold increase in axonemal microtubule sliding when compared to the single outer-arm dynein knockout. Knockout of TTLL9 in C. reinhardtii (one of ten TTLL genes found in this organism) results in a significant decrease in α-tubulin poly-glutamylation on the B-tubule of outer doublet axonemal microtubules, but does not cause any structural defects in the axonemes and the sliding velocities of isolated axonemal microtubules are unaffected [Kubo et al. 2010]. Interestingly, however, double knockout strains of TTLL9 and either outer-arm or inner-arm dynein have dramatically reduced swimming velocities and increased microtubule sliding velocities when compared to wild-type. Both studies proposed that tubulin poly-glutamylation could affect the strength of the interaction between the dynein stalk and microtubules (Figure 3D) and that a decrease in glutamylation could thus increase sliding velocity [Kubo et al., 2010; Suryavanshi et al., 2010; Vogel et al. 2010].
Poly-glycylation is the tubulin post-translation modification most strongly associated with axonemes. Consistent with an axonemal association, the poly-glycylase TTLL3 is abundantly expressed in testes and its knockdown in Drosophila sperm causes defects in sperm tail axonemes as well as sterility [Rogowski et al. 2009]. In T. thermophila, knockdown of two of the TTLL3 genes found in this organism accelerated the growth of cilia as well as overall cell growth, the latter possibly due to the alteration in microtubule dynamics observed in these cells. Overexpression of a dominant negative mutant of TTLL3 resulted in very short cilia [Wloga et al. 2009]. In zebra fish, TTLL3 knockdown results in defective motile and primary cilia in the olfactory placode and axonemes that are shorter or completely missing [Wloga et al. 2009]. However, a more recent study found only minor effects on axonemal structure and motility [Pathak et al. 2011], but dramatic phenotypes both in axonemal structure and cilia motility in the double TTLL3/TTLL6 knockdown. Interestingly, the double knockdown phenotype is reminiscent of that obtained for the fleer gene, possibly implicating it as a regulator of both poly-glutamylation and poly-glycylation [Pathak et al. 2007; Pathak et al. 2011].
The B-tubule of axonemal microtubules carries both long poly-Gly as well as poly-Glu chains and the positions on the tubulin chain at which these modifications are added are in close proximity, raising the possibility of whether these different modifications can regulate each other. Interestingly, knockdown of the poly-glycylase TTLL3 and the accompanying decrease in poly-glycylation levels in axonemes is accompanied also by an increase in poly-glutamylation [Rogowski et al. 2009; Wloga et al. 2009], supporting the idea that poly-glutamylases and poly-glycylases compete for the tubulin tails and the length of the poly-Glu or poly-Gly chains on microtubules is reached by balancing the activities of these enzymes.
TTLL substrates - more than just tubulin
The identification of the various TTLL enzymes also revealed that their substrate repertoires are more diverse than initially suspected and extend beyond tubulin [van Dijk et al. 2007; van Dijk et al. 2008]. Mammalian TTLL4 and 5 can poly-glutamylate several other proteins, including nucleosome assembly proteins 1 and 2 (NAP1 and NAP2), nuclear factor 45 (NF45), SET, nucleophosmin B23, and PELP1 [Regnard et al. 2000; van Dijk et al. 2007; van Dijk et al. 2008; Kashiwaya et al. 2010], while TTLL10 poly-glycylates NAP1 [Ikegami et al. 2008]. While the sequences of these proteins all contain glutamate stretches, there is no easily identifiable poly-glutamylation/poly-glycylation consensus sequence. Many of the non-tubulin substrates are involved in chromatin remodeling and it was proposed that poly-glutamylation of both microtubules and chromatin-binding proteins could facilitate an intimate coordination between spindle dynamics and changes in chromatin structure during mitosis [van Dijk et al. 2008]. Interestingly, the histone chaperones NAP1 and NAP2 are more heavily poly-glutamytated in HeLa cells than tubulin: while only a few percent of tubulin is estimated to be poly-glutamyltated in these cells, more than 50% of the NAPs seem to be modified with as many as nine glutamates [Regnard et al. 2000]. NAP1 is involved in the deposition of histones onto DNA and interacts with both histones H3▪H4 as well as H2A▪H2B dimers. It is tempting to speculate that the affinity of NAP1 for intermediates in the chromatin fiber assembly pathway is differentially regulated by poly-glutamylation. At the present time, it is not at all clear how the TTLL modification enzymes are able to coordinate their activities between tubulin and histone chaperones and how they access these substrates in postmitotic cells such as neurons. Recent data is pointing more and more towards the fact the TTLLs (and not only TLLL1 that was initially identified as a five-subunit complex) are part of larger complexes that possess adapter proteins that govern their localization [Lee et al. 2012]. The mechanism of targeting TTLL activities to histone chaperones and their roles in tuning chaperone functions will likely be a fertile area of future exploration.
The existence of many additional substrates certainly complicates the in vivo dissection of the effects of tubulin post-translational modifications on the microtubule cytoskeleton. Thus, the effects of tubulin post-translational modifications on microtubules cannot be inferred from enzyme depletion studies alone and will need to be accompanied by a gradual reconstitution in vitro of microtubule arrays with different modifications to dissect the effects of these modifications in tuning the behaviour of motors and MAPs.
What goes up must come down - enzymes that remove tubulin poly-modifications
The existence of enzymes that remove the poly-glutamate or poly-glycine chains from tubulin was hypothesized for a long time, however their identity proved elusive until recently when a family of enzymes that catalyze the removal of glutamate residues from the C-terminal tails of both α- and β-tubulin was identified in multiple organisms [Kalinina et al. 2007; Rodriguez de la Vega et al. 2007; Kimura et al. 2010; Rogowski et al. 2010]. These enzymes, termed cytosolic carboxypeptidases (CCPs), are members of the MC clan, M14 family, subfamily M14D of metallopeptidases [Kalinina et al. 2007]. The various CCPs identified to date are specific for either long glutamate chains or branching-point glutamate residues (Table III). Mouse CCP1, 4, and 6 are functionally homologous and remove long glutamate chains from tubulin, while CCP5 specifically removes branching-point glutamates [Rogowski et al. 2010]. Interestingly, Purkinje cell degeneration (pcd) mice have defects in the gene encoding for CCP1 [Mullen et al. 1976; Fernandez-Gonzalez et al. 2002]. These mice have an altered gait due to degeneration of their Purkinje cells. They are also sterile and have abnormal sperm shape, phenotypes consistent with defects in axonemal function [Mullen et al. 1976; Landis and Mullen 1978]. The Purkinje cell degeneration could be rescued by expression of wild-type CCP1 but not a mutant predicted to be defective in substrate binding [Wang et al. 2006], thus establishing the carboxypeptidase activity as important for the function of CCP1. A recent re-examination of the pcd mice revealed that they have high levels of tubulin poly-glutamylation in the areas of the brain that are affected by neurodegeneration. Most importantly, knockdown in the cerebellum of young pcd mice of TTLL1, the enzyme responsible for most of the tubulin poly-glutamylation activity in the brain, prevented Purkinje cell death and resulted in improved motor coordination [Rogowski et al. 2010]. This new finding establishes a direct link between tubulin hyperglutamylation and neurodegeneration.
C. elegans contains only two CCP orthologs, CeCCP1 and CeCCP6 [Kimura et al. 2010]. CeCCP6 is homologous to mouse CCP5 and like its mammalian counterpart is also responsible for removing branching-point glutamates. CeCCP1 is unable to catalyze the removal of glutamates and does not seem to function as a tubulin deglutamylase. Interestingly, the ability of mouse CCP1 (and presumably CCP4 and 6) to remove glutamates from the C-terminal tails of tubulin is dependent upon the specific TTLL enzyme that added them initially. CCP1 can remove both long-chain and branching-point glutamates on microtubules modified by TTLL6, however it cannot remove branching-point glutamates from microtubules modified by TTLL4 [Rogowski et al. 2010]. Mouse TTLL6 functions primarily as an α-tubulin elongase while TTLL4 functions primarily as a β-tubulin initiase. These results strongly suggest that sequence context and the length of the poly-glutamate tails are important determinants for CCP specificity. Like the TTLL family, CCPs also act on non-tubulin targets. For example, CCP1 removes the genetically encoded glutamate chains in the C-terminal tails of myosin light chain kinase 1 and telokin, two regulators of myosin function [Rogowski et al. 2010]. It is interesting to note that none of the many non-tubulin targets known to be glutamylated by TTLL enzymes (i.e. NAP1, NAP2) were identified as substrates for the known CCP enzymes, although it is likely that future proteomic studies will uncover additional substrates for the CCP enzymes that overlap with those found for TTLLs.
Deglycylating enzymes that remove glycine chains have yet to be identified. Interestingly, two enzymes with deglycylating activity have been identified in the flagellated protozoan Giardia duodenalis [Lalle et al. 2011]. These enzymes, termed giardial dipeptidase 1 and 2 (gDIP1 and gDIP2) are aminoacylhistidine dipeptidases that belong to the MH clan, M20 family of metallopeptidases, a different enzyme family from the CCP deglutamylases. gDIP1 and gDIP2 are capable of removing most, but not all glycine residues from the C-terminal tail of protein 14-3-3. The inability to remove all glycine moieties is likely due to the activity of gTTLL3, a G. duodenalis TTLL glycylase. Based on the multi-substrate specificity of the TTLL family and CCP family of deglutamylating enzymes, it is possible that gDIP1 and gDIP2, and possibly other members of the MH clan, M20 family of metallopeptidases function as tubulin deglycylases. It is interesting to note that none of the CCP enzymes identified so far are capable of detyrosinating α-tubulin. Thus, more than thirty years since the identification of the detyrosination/tyrosination cycle, the search for the tubulin detyrosinase continues.
Tubulin post-translational modifications and human disease
Changes in tubulin post-translational modifications have been linked to several human disease states ranging from cancers and neurodegenerative disorders to stroke. Low TTL levels have long been associated with aggressive tumors that are resistant to chemotherapy [Mialhe et al. 2001; Kato et al. 2004; Soucek et al. 2006] and a decrease in TTL levels is associated with the formation of tentacles rich in detyrosinated tubulin that facilitate penetration into the endothelial layer of circulating tumor cells, thus contributing to metastasis [Whipple et al. 2010]. Likewise, an increase in TTLL12 levels and consequently in glutamylated tubulin has been associated with metastatic progression in prostate cancer [Wasylyk et al. 2010]. In a mouse model of Charcot-Marie-Tooth disease, the levels of acetylated tubulin were repressed and the axonal transport defects observed in these mice were corrected by the administration of HDAC6 inhibitors [d'Ydewalle et al. 2011] that also partially recovered the acetylated tubulin levels. Similar observations were made in neurodegenerative disorders such as Huntington’s and Alzheimer’s [Dompierre et al., 2007; Kazantsev and Thompson 2008]. The mechanisms behind these changes in tubulin post-translational modification profiles are largely unclear. A clearer connection between a tubulin modification enzyme and a human disease was demonstrated in a very recent study that identifies the ciliary protein CEP41 as important for targeting the poly-glutamylation activity of TTLL6 to cilia and establishes that its mutation is associated with Joubert syndrome, a neurological disorder characterized by ataxia, abnormal breathing patterns and low muscle tone [Lee et al. 2012]. A better understanding of the cellular mechanisms behind the observed changes in tubulin post-translational modifications will likely lead to a better understanding of these disease states as well as aid in the identification of novel drug targets. The recent study showing that TTLL1 downregulation can reverse the neurodegenerative phenotype in pcd mice [Rogowski et al. 2010] makes a strong case for inhibitors of TTLL polyglutamylases as having therapeutic potential.
Future directions and challenges
Despite the recent and significant advances in identifying the plethora of tubulin post-translational modification enzymes, we are just beginning to understand their cellular mechanism of action as well as the effects of tubulin post-translational modifications on microtubule structure and dynamics. A future challenge will be to mechanistically dissect the observed phenotypes in cells or organisms in which tubulin post-translational modification enzymes have been knocked-down. Are the effects seen due to changes in tubulin modifications or/and modifications of other non-tubulin substrates? How complete is our list of bona fide non-tubulin substrates of poly-glutamylases and poly-glycylases? What are the effects of the newly discovered tubulin modification enzymes on microtubule dynamics? In vitro microtubule dynamics studies with homogenously modified microtubules have been missing so far. What are the base dynamics of non-modified microtubules and how do they change as a function of tubulin modifications? While challenging, these types of studies are now within reach, as the identification of tubulin modification enzymes that both add and remove the various modifications opens up the possibility of systematically manipulating biochemically the modification state of the microtubule to produce homogenous preparations. In addition, the recent resurgence of S. cerevisiae [Barnes et al. 1992; Bode et al. 2003; Johnson et al. 2011] and S. pombe [Drummond et al. 2011] as systems suitable for the expression and purification of biochemical amounts of tubulin that is "naive" (i.e. not post-translationally modified) opens the exciting possibility of a systematic mechanistic dissection of the consequences of chemical and genetic variability in the tubulin pool on microtubule dynamics and the activity of effectors that interact with tubulin and microtubules.
Antibodies recognizing various forms of modified tubulin have been instrumental in revealing their cellular distribution as well as in the identification of the TTLL and CCP enzyme families. However, antibodies are blunt tools for probing the precise chemical structure of the amino acid chains added by these enzymes. In addition, it is not clear how the existence of additional post-translational modifications at neighboring sites affects the specificity of these antibodies. This phenomenon was just recently recognized for modification-specific antibodies against histones [Egelhofer et al. 2011; Fuchs et al. 2011]. In order to better understand the mechanism of action of tubulin post-translational modification enzymes, future biochemical and biophysical methods will be needed to characterize the products of the reactions they catalyze [Redeker 2010] and understand how their activity and specificity can be tuned. The specificity of the TTLL enzymes is not very well characterized and many of them seem to be rather promiscuous. It is possible that this promiscuity is due to overexpression in some cases or because the enzymes might be part of larger complexes that serve to tune their functions. While TTLL1 was biochemically isolated from brain extracts as part of a five subunit complex [Regnard et al. 2003; Janke et al. 2005], many of the TTLL enzymes were later identified through genomic searches based on their homology to TTL [van Dijk et al. 2007], and thus it is possible that some of their functional partners remain to be discovered.
Enzymatic assays for TTL and TTLL enzymes have been performed so far on brain tubulin that is abundantly modified [Zambito et al. 2002], or tubulin isolated from cell lines that are known to have different levels of post-translational modifications. For example, microtubules from HeLa cells display low levels of poly-glutamylation, and were therefore used as a substrate to test poly-glutamylase initiase activity, while microtubules from adult neurons are highly poly-glutamylated, and were used as a substrate to test elongase activity [Bobinnec et al. 1998; Wloga et al. 2008]. Thus, there is a need to examine the enzymatic activity and specificity of the various TTLLs using homogenous tubulin preparations where the exact post-translational modification landscape of the starting microtubule is known. How much does the presence and type of a specific post-translational modification adjacent to the new modification site affect the activity of an enzyme? Do some elongases show preference for the length of the poly-Glu chain? Can they distinguish between chains that are three versus five or ten amino acids long? Are some enzymes specific only for certain tubulin isoforms? An attractive hypothesis is that part of the tubulin genetic complexity is there to ensure differential interactions and modifications by various post-translational modification enzymes.
We also need better reporters of tubulin post-translational modifications in cells. Recent advances in recombinant antibody technology offer the opportunity for generating a second generation of antibodies that are more specific and cover a larger selection of post-translational modified tubulins. Recombinant antibodies can be produced in large quantities, as well as rationally engineered and easily labeled for imaging studies. High-resolution structural studies of the various TTLL enzymes should also aid in the design of amino acid analogs (fluorescent or for click chemistry [Kolb et al. 2001; Banerjee et al. 2010]) that could be used as reporters in cells to monitor live the dynamics of tubulin post-translational modifications and couple those with the dynamics of the microtubule network. Structural studies will also shed light on the molecular determinants of substrate specificity of TTLL enzymes: α- versus β-tubulin recognition, glutamylation versus glycylation, and chain initiation versus chain elongation.
Knockout mice for the tubulin modification enzymes TTL [Erck et al. 2005], TTLL1 [Campbell et al. 2002; Ikegami et al. 2010; Vogel et al. 2010] and CCP1 [Mullen et al., 1976; Fernandez-Gonzalez et al., 2002; Rogowski et al. 2010] have proven very useful in understanding the roles of these enzymes in normal development and disease. Mouse knockout studies of additional modification enzymes will prove equally informative and will hopefully be pursued by various groups. More than thirty years after the discovery of the first tubulin specific post-translational modification, we are close to having a complete list of the principal molecular players involved and have the tools to embark on a systematic dissection of the molecular underpinnings of the chemical complexity of tubulin in cells.
Acknowledgements
We would like to apologize to the many colleagues whose work we were unable to cover in our review because of space limitations. We thank our anonymous reviewers for their careful reading of our manuscript and their useful suggestions. A. R-M. is a Searle Scholar and is supported by the intramural programs of the National Institute of Neurological Disorder and Stroke (NINDS) and the National Heart, Lung and Blood Institute (NHLBI).
Abbreviations
- CLIP170
cytoplasmic linker protein-170
- CCP
cytosolic carboxypeptidase
- MAP
microtubule associated protein
- MCAK
mitotic centromere associated kinesin
- +TIP
plus-end tracking protein
- TTL
tubulin tyrosine ligase
- TTLL
tubulin tyrosine ligase-like
References
- Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467(7312):218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JE, Hunt DF, Lee MK, Shabanowitz J, Michel H, Berlin SC, MacDonald TL, Sundberg RJ, Rebhun LI, Frankfurter A. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc Natl Acad Sci U S A. 1991;88(11):4685–4689. doi: 10.1073/pnas.88.11.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce CA, Rodriguez JA, Barra HS, Caputo R. Incorporation of L-tyrosine, L-phenylalanine and L-3,4-dihydroxyphenylalanine as single units into rat brain tubulin. Eur J Biochem. 1975;59(1):145–149. doi: 10.1111/j.1432-1033.1975.tb02435.x. [DOI] [PubMed] [Google Scholar]
- Audebert S, Desbruyeres E, Gruszczynski C, Koulakoff A, Gros F, Denoulet P, Edde B. Reversible polyglutamylation of alpha- and beta-tubulin and microtubule dynamics in mouse brain neurons. Mol Biol Cell. 1993;4(6):615–626. doi: 10.1091/mbc.4.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badin-Larcon AC, Boscheron C, Soleilhac JM, Piel M, Mann C, Denarier E, Fourest-Lieuvin A, Lafanechere L, Bornens M, Job D. Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc Natl Acad Sci U S A. 2004;101(15):5577–5582. doi: 10.1073/pnas.0307917101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Panosian TD, Mukherjee K, Ravindra R, Gal S, Sackett DL, Bane S. Site-specific orthogonal labeling of the carboxy terminus of alpha-tubulin. ACS Chem Biol. 2010;5(8):777–785. doi: 10.1021/cb100060v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes G, Louie KA, Botstein D. Yeast proteins associated with microtubules in vitro and in vivo. Mol Biol Cell. 1992;3(1):29–47. doi: 10.1091/mbc.3.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra HS, Arce CA, Rodriguez JA, Caputto R. Some common properties of the protein that incorporates tyrosine as a single unit and the microtubule proteins. Biochem Biophys Res Commun. 1974;60(4):1384–1390. doi: 10.1016/0006-291x(74)90351-9. [DOI] [PubMed] [Google Scholar]
- Barra HS, Rodriguez JA, Arce CA, Caputto R. A soluble preparation from rat brain that incorporates into its own proteins (14 C)arginine by a ribonuclease-sensitive system and (14 C)tyrosine by a ribonuclease-insensitive system. J Neurochem. 1973;20(1):97–108. doi: 10.1111/j.1471-4159.1973.tb12108.x. [DOI] [PubMed] [Google Scholar]
- Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol. 2008;183(7):1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y, Moudjou M, Fouquet JP, Desbruyeres E, Edde B, Bornens M. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton. 1998;39(3):223–232. doi: 10.1002/(SICI)1097-0169(1998)39:3<223::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Bode CJ, Gupta ML, Suprenant KA, Himes RH. The two alpha-tubulin isotypes in budding yeast have opposing effects on microtubule dynamics in vitro. EMBO Rep. 2003;4(1):94–99. doi: 10.1038/sj.embor.embor716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet C, Boucher D, Lazereg S, Pedrotti B, Islam K, Denoulet P, Larcher JC. Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J Biol Chem. 2001;276(16):12839–12848. doi: 10.1074/jbc.M011380200. [DOI] [PubMed] [Google Scholar]
- Boucher D, Larcher JC, Gros F, Denoulet P. Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry. 1994;33(41):12471–12477. doi: 10.1021/bi00207a014. [DOI] [PubMed] [Google Scholar]
- Bre MH, de Nechaud B, Wolff A, Fleury A. Glutamylated tubulin probed in ciliates with the monoclonal antibody GT335. Cell Motil Cytoskeleton. 1994;27(4):337–349. doi: 10.1002/cm.970270406. [DOI] [PubMed] [Google Scholar]
- Bre MH, Pepperkok R, Kreis TE, Karsenti E. Cellular interactions and tubulin detyrosination in fibroblastic and epithelial cells. Biol Cell. 1991;71(1–2):149–160. doi: 10.1016/0248-4900(91)90061-q. [DOI] [PubMed] [Google Scholar]
- Bre MH, Redeker V, Quibell M, Darmanaden-Delorme J, Bressac C, Cosson J, Huitorel P, Schmitter JM, Rossler J, Johnson T, et al. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J Cell Sci. 1996;109(Pt 4):727–738. doi: 10.1242/jcs.109.4.727. [DOI] [PubMed] [Google Scholar]
- Bulinski JC, McGraw TE, Gruber D, Nguyen HL, Sheetz MP. Overexpression of MAP4 inhibits organelle motility and trafficking in vivo. J Cell Sci. 1997;110(Pt 24):3055–3064. doi: 10.1242/jcs.110.24.3055. [DOI] [PubMed] [Google Scholar]
- Campbell PK, Waymire KG, Heier RL, Sharer C, Day DE, Reimann H, Jaje JM, Friedrich GA, Burmeister M, Bartness TJ, et al. Mutation of a novel gene results in abnormal development of spermatid flagella, loss of intermale aggression and reduced body fat in mice. Genetics. 2002;162(1):307–320. doi: 10.1093/genetics/162.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron JM. Posttranslational modification of tubulin by palmitoylation: I. In vivo and cell-free studies. Mol Biol Cell. 1997;8(4):621–636. doi: 10.1091/mbc.8.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron JM, Vega LR, Fleming J, Bishop R, Solomon F. Single site alpha-tubulin mutation affects astral microtubules and nuclear positioning during anaphase in Saccharomyces cerevisiae: possible role for palmitoylation of alpha-tubulin. Mol Biol Cell. 2001;12(9):2672–2687. doi: 10.1091/mbc.12.9.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casale CH, Alonso AD, Barra HS. Brain plasma membrane Na+,K+-ATPase is inhibited by acetylated tubulin. Mol Cell Biochem. 2001;216(1–2):85–92. doi: 10.1023/a:1011029125228. [DOI] [PubMed] [Google Scholar]
- Casale CH, Previtali G, Barra HS. Involvement of acetylated tubulin in the regulation of Na+,K+-ATPase activity in cultured astrocytes. FEBS Lett. 2003;534(1–3):115–118. doi: 10.1016/s0014-5793(02)03802-4. [DOI] [PubMed] [Google Scholar]
- Chu CW, Hou F, Zhang J, Phu L, Loktev AV, Kirkpatrick DS, Jackson PK, Zhao Y, Zou H. A novel acetylation of beta-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol Biol Cell. 2011;22(4):448–456. doi: 10.1091/mbc.E10-03-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Ydewalle C, Krishnan J, Chiheb DM, Van Damme P, Irobi J, Kozikowski AP, Vanden Berghe P, Timmerman V, Robberecht W, Van Den Bosch L. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat Med. 2011;17(8):968–974. doi: 10.1038/nm.2396. [DOI] [PubMed] [Google Scholar]
- Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur EL. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci U S A. 2009;106(2):492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Kain S, Newcombe A, Hoey C, Katsuki M, Cross RA. Purification of tubulin from the fission yeast Schizosaccharomyces pombe. Methods Mol Biol. 2011;777:29–55. doi: 10.1007/978-1-61779-252-6_3. [DOI] [PubMed] [Google Scholar]
- Dunn S, Morrison EE, Liverpool TB, Molina-Paris C, Cross RA, Alonso MC, Peckham M. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121(Pt 7):1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- Edde B, Rossier J, Le Caer JP, Berwald-Netter Y, Koulakoff A, Gros F, Denoulet P. A combination of posttranslational modifications is responsible for the production of neuronal alpha-tubulin heterogeneity. J Cell Biochem. 1991;46(2):134–142. doi: 10.1002/jcb.240460207. [DOI] [PubMed] [Google Scholar]
- Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247(4938):83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- Edde B, Rossier J, Le Caer JP, Prome JC, Desbruyeres E, Gros F, Denoulet P. Polyglutamylated alpha-tubulin can enter the tyrosination/detyrosination cycle. Biochemistry. 1992;31(2):403–410. doi: 10.1021/bi00117a014. [DOI] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18(1):91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper BA. Rat brain microtubule protein: purification and determination of covalently bound phosphate and carbohydrate. Proc Natl Acad Sci U S A. 1972;69(8):2283–2287. doi: 10.1073/pnas.69.8.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erck C, Peris L, Andrieux A, Meissirel C, Gruber AD, Vernet M, Schweitzer A, Saoudi Y, Pointu H, Bosc C, et al. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc Natl Acad Sci U S A. 2005;102(22):7853–7858. doi: 10.1073/pnas.0409626102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld K, Wehland J, Plessmann U, Dodemont H, Gerke V, Weber K. Characterization of the tubulin-tyrosine ligase. J Cell Biol. 1993;120(3):725–732. doi: 10.1083/jcb.120.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez A, La Spada AR, Treadaway J, Higdon JC, Harris BS, Sidman RL, Morgan JI, Zuo J. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 2002;295(5561):1904–1906. doi: 10.1126/science.1068912. [DOI] [PubMed] [Google Scholar]
- Fouquet JP, Prigent Y, Kann ML. Comparative immunogold analysis of tubulin isoforms in the mouse sperm flagellum: unique distribution of glutamylated tubulin. Mol Reprod Dev. 1996;43(3):358–365. doi: 10.1002/(SICI)1098-2795(199603)43:3<358::AID-MRD10>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190(3):363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2011;21(1):53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima N, Furuta D, Hidaka Y, Moriyama R, Tsujiuchi T. Post-translational modifications of tubulin in the nervous system. J Neurochem. 2009;109(3):683–693. doi: 10.1111/j.1471-4159.2009.06013.x. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Cruz MA, Bowen J, Gu L, Pennock DG, Gorovsky MA. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J Cell Biol. 1995;129(5):1301–1310. doi: 10.1083/jcb.129.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimer S, Teltenkotter A, Plessmann U, Weber K, Lechtreck KF. Purification and characterization of basal apparatuses from a flagellate green alga. Cell Motil Cytoskeleton. 1997;37(1):72–85. doi: 10.1002/(SICI)1097-0169(1997)37:1<72::AID-CM7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Bulinski JC. Distribution of tyrosinated and nontyrosinated alpha-tubulin during mitosis. J Cell Biol. 1986;102(3):1118–1126. doi: 10.1083/jcb.102.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Bulinski JC. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc Natl Acad Sci U S A. 1988;85(16):5946–5950. doi: 10.1073/pnas.85.16.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 1984;38(3):779–789. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- Gupta KK, Joyce MV, Slabbekoorn AR, Zhu ZC, Paulson BA, Boggess B, Goodson HV. Probing interactions between CLIP-170, EB1, and microtubules. J Mol Biol. 2010;395(5):1049–1062. doi: 10.1016/j.jmb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallak ME, Rodriguez JA, Barra HS, Caputto R. Release of tyrosine from tyrosinated tubulin. Some common factors that affect this process and the assembly of tubulin. FEBS Lett. 1977;73(2):147–150. doi: 10.1016/0014-5793(77)80968-x. [DOI] [PubMed] [Google Scholar]
- Hammond JW, Huang CF, Kaech S, Jacobson C, Banker G, Verhey KJ. Posttranslational modifications of tubulin and the polarized transport of kinesin-1 in neurons. Mol Biol Cell. 2010;21(4):572–583. doi: 10.1091/mbc.E09-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I, Plevin MJ, Ikura M. CLIP170 autoinhibition mimics intermolecular interactions with p150Glued or EB1. Nat Struct Mol Biol. 2007;14(10):980–981. doi: 10.1038/nsmb1299. [DOI] [PubMed] [Google Scholar]
- Honnappa S, Okhrimenko O, Jaussi R, Jawhari H, Jelesarov I, Winkler FK, Steinmetz MO. Key interaction modes of dynamic +TIP networks. Mol Cell. 2006;23(5):663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Heier RL, Taruishi M, Takagi H, Mukai M, Shimma S, Taira S, Hatanaka K, Morone N, Yao I, et al. Loss of alpha-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc Natl Acad Sci U S A. 2007;104(9):3213–3218. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Horigome D, Mukai M, Livnat I, MacGregor GR, Setou M. TTLL10 is a protein polyglycylase that can modify nucleosome assembly protein 1. FEBS Lett. 2008;582(7):1129–1134. doi: 10.1016/j.febslet.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Mukai M, Tsuchida J, Heier RL, Macgregor GR, Setou M. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J Biol Chem. 2006;281(41):30707–30716. doi: 10.1074/jbc.M603984200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Sato S, Nakamura K, Ostrowski LE, Setou M. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc Natl Acad Sci U S A. 2010;107(23):10490–10495. doi: 10.1073/pnas.1002128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Setou M. TTLL10 can perform tubulin glycylation when co-expressed with TTLL8. FEBS Lett. 2009;583(12):1957–1963. doi: 10.1016/j.febslet.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308(5729):1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Ji X, Zhang P, Armstrong RN, Gilliland GL. The three-dimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2-A resolution. Biochemistry. 1992;31(42):10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- Johnson V, Ayaz P, Huddleston P, Rice LM. Design, overexpression, and purification of polymerization-blocked yeast alphabeta-tubulin mutants. Biochemistry. 2011;50(40):8636–8644. doi: 10.1021/bi2005174. [DOI] [PubMed] [Google Scholar]
- Kalinina E, Biswas R, Berezniuk I, Hermoso A, Aviles FX, Fricker LD. A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 2007;21(3):836–850. doi: 10.1096/fj.06-7329com. [DOI] [PubMed] [Google Scholar]
- Kashiwaya K, Nakagawa H, Hosokawa M, Mochizuki Y, Ueda K, Piao L, Chung S, Hamamoto R, Eguchi H, Ohigashi H, et al. Involvement of the tubulin tyrosine ligase-like family member 4 polyglutamylase in PELP1 polyglutamylation and chromatin remodeling in pancreatic cancer cells. Cancer Res. 2010;70(10):4024–4033. doi: 10.1158/0008-5472.CAN-09-4444. [DOI] [PubMed] [Google Scholar]
- Kato C, Miyazaki K, Nakagawa A, Ohira M, Nakamura Y, Ozaki T, Imai T, Nakagawara A. Low expression of human tubulin tyrosine ligase and suppressed tubulin tyrosination/detyrosination cycle are associated with impaired neuronal differentiation in neuroblastomas with poor prognosis. Int J Cancer. 2004;112(3):365–375. doi: 10.1002/ijc.20431. [DOI] [PubMed] [Google Scholar]
- Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7(10):854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Kurabe N, Ikegami K, Tsutsumi K, Konishi Y, Kaplan OI, Kunitomo H, Iino Y, Blacque OE, Setou M. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs) J Biol Chem. 2010;285(30):22936–22941. doi: 10.1074/jbc.C110.128280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12(5):559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- Kozminski KG, Diener DR, Rosenbaum JL. High level expression of nonacetylatable alpha-tubulin in Chlamydomonas reinhardtii. Cell Motil Cytoskeleton. 1993;25(2):158–170. doi: 10.1002/cm.970250205. [DOI] [PubMed] [Google Scholar]
- Kreitzer G, Liao G, Gundersen GG. Detyrosination of tubulin regulates the interaction of intermediate filaments with microtubules in vivo via a kinesin-dependent mechanism. Mol Biol Cell. 1999;10(4):1105–1118. doi: 10.1091/mbc.10.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Yanagisawa HA, Yagi T, Hirono M, Kamiya R. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr Biol. 2010;20(5):441–445. doi: 10.1016/j.cub.2009.12.058. [DOI] [PubMed] [Google Scholar]
- L'Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified in the flagella during flagellar assembly. J Cell Biol. 1983;97(1):258–263. doi: 10.1083/jcb.97.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24(2):473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- Lacroix B, van Dijk J, Gold ND, Guizetti J, Aldrian-Herrada G, Rogowski K, Gerlich DW, Janke C. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J Cell Biol. 2010;189(6):945–954. doi: 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalle M, Camerini S, Cecchetti S, Fantauzzi CB, Crescenzi M, Pozio E. Giardia duodenalis 14-3-3 protein is polyglycylated by a tubulin tyrosine ligase-like member and deglycylated by two metallocarboxypeptidases. J Biol Chem. 2011;286(6):4471–4484. doi: 10.1074/jbc.M110.181511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis SC, Mullen RJ. The development and degeneration of Purkinje cells in pcd mutant mice. J Comp Neurol. 1978;177(1):125–143. doi: 10.1002/cne.901770109. [DOI] [PubMed] [Google Scholar]
- Larcher JC, Boucher D, Lazereg S, Gros F, Denoulet P. Interaction of kinesin motor domains with alpha- and beta-tubulin subunits at a tau-independent binding site. Regulation by polyglutamylation. J Biol Chem. 1996;271(36):22117–22124. doi: 10.1074/jbc.271.36.22117. [DOI] [PubMed] [Google Scholar]
- Lee C, Chen LB. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54(1):37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, Marsh SE, Olvera J, Brancati F, Iannicelli M, Ikegami K, et al. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat Genet. 2012;44(2):193–199. doi: 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levilliers N, Fleury A, Hill AM. Monoclonal and polyclonal antibodies detect a new type of post-translational modification of axonemal tubulin. J Cell Sci. 1995;108(Pt 9):3013–3028. doi: 10.1242/jcs.108.9.3013. [DOI] [PubMed] [Google Scholar]
- Liao G, Gundersen GG. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J Biol Chem. 1998;273(16):9797–9803. doi: 10.1074/jbc.273.16.9797. [DOI] [PubMed] [Google Scholar]
- Lu C, Srayko M, Mains PE. The Caenorhabditis elegans microtubule-severing complex MEI-1/MEI-2 katanin interacts differently with two superficially redundant beta-tubulin isotypes. Mol Biol Cell. 2004;15(1):142–150. doi: 10.1091/mbc.E03-06-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Luduena RF. In vitro analysis of microtubule assembly of isotypically pure tubulin dimers. Intrinsic differences in the assembly properties of alpha beta II, alpha beta III, and alpha beta IV tubulin dimers in the absence of microtubule-associated proteins. J Biol Chem. 1994;269(3):2041–2047. [PubMed] [Google Scholar]
- Marmorstein R, Trievel RC. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim Biophys Acta. 2009;1789(1):58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta H, Greer K, Rosenbaum JL. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J Cell Biol. 1986;103(2):571–579. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75(3):419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- Mialhe A, Lafanechere L, Treilleux I, Peloux N, Dumontet C, Bremond A, Panh MH, Payan R, Wehland J, Margolis RL, et al. Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Res. 2001;61(13):5024–5027. [PubMed] [Google Scholar]
- Miller LM, Xiao H, Burd B, Horwitz SB, Angeletti RH, Verdier-Pinard P. Methods in tubulin proteomics. Methods Cell Biol. 2010;95:105–126. doi: 10.1016/S0091-679X(10)95007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M, Ikegami K, Sugiura Y, Takeshita K, Nakagawa A, Setou M. Recombinant mammalian tubulin polyglutamylase TTLL7 performs both initiation and elongation of polyglutamylation on beta-tubulin through a random sequential pathway. Biochemistry. 2009;48(5):1084–1093. doi: 10.1021/bi802047y. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Eicher EM, Sidman RL. Purkinje cell degeneration, a new neurological mutation in the mouse. Proc Natl Acad Sci U S A. 1976;73(1):208–212. doi: 10.1073/pnas.73.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murofushi H. Purification and characterization of tubulin-tyrosine ligase from porcine brain. J Biochem. 1980;87(3):979–984. doi: 10.1093/oxfordjournals.jbchem.a132828. [DOI] [PubMed] [Google Scholar]
- Nagasaki T, Gundersen GG. Depletion of lysophosphatidic acid triggers a loss of oriented detyrosinated microtubules in motile fibroblasts. J Cell Sci. 1996;109(Pt 10):2461–2469. doi: 10.1242/jcs.109.10.2461. [DOI] [PubMed] [Google Scholar]
- Newton CN, DeLuca JG, Himes RH, Miller HP, Jordan MA, Wilson L. Intrinsically slow dynamic instability of HeLa cell microtubules in vitro. J Biol Chem. 2002;277(45):42456–42462. doi: 10.1074/jbc.M207134200. [DOI] [PubMed] [Google Scholar]
- Nielsen MG, Turner FR, Hutchens JA, Raff EC. Axoneme-specific beta-tubulin specialization: a conserved C-terminal motif specifies the central pair. Curr Biol. 2001;11(7):529–533. doi: 10.1016/s0960-9822(01)00150-6. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391(6663):199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Ozols J, Caron JM. Posttranslational modification of tubulin by palmitoylation: II. Identification of sites of palmitoylation. Mol Biol Cell. 1997;8(4):637–645. doi: 10.1091/mbc.8.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D, Miller HP, Banerjee A, Luduena RF, Wilson L. Microtubule dynamics in vitro are regulated by the tubulin isotype composition. Proc Natl Acad Sci U S A. 1994;91(24):11358–11362. doi: 10.1073/pnas.91.24.11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N, Austin CA, Drummond IA. Tubulin tyrosine ligase-like genes ttll3 and ttll6 maintain zebrafish cilia structure and motility. J Biol Chem. 2011;286(13):11685–11695. doi: 10.1074/jbc.M110.209817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N, Obara T, Mangos S, Liu Y, Drummond IA. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol Biol Cell. 2007;18(11):4353–4364. doi: 10.1091/mbc.E07-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paturle-Lafanechere L, Edde B, Denoulet P, Van Dorsselaer A, Mazarguil H, Le Caer JP, Wehland J, Job D. Characterization of a major brain tubulin variant which cannot be tyrosinated. Biochemistry. 1991;30(43):10523–10528. doi: 10.1021/bi00107a022. [DOI] [PubMed] [Google Scholar]
- Paturle-Lafanechere L, Manier M, Trigault N, Pirollet F, Mazarguil H, Job D. Accumulation of delta 2-tubulin, a major tubulin variant that cannot be tyrosinated, in neuronal tissues and in stable microtubule assemblies. J Cell Sci. 1994;107(Pt 6):1529–1543. doi: 10.1242/jcs.107.6.1529. [DOI] [PubMed] [Google Scholar]
- Peris L, Thery M, Faure J, Saoudi Y, Lafanechere L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, Wordeman L, et al. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol. 2006;174(6):839–849. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L, Wagenbach M, Lafanechere L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, Andrieux A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol. 2009;185(7):1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101(6):2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybin D, Flavin M. An enzyme tyrosylating alpha-tubulin and its role in microtubule assembly. Biochem Biophys Res Commun. 1975;65(3):1088–1095. doi: 10.1016/s0006-291x(75)80497-9. [DOI] [PubMed] [Google Scholar]
- Raybin D, Flavin M. Enzyme which specifically adds tyrosine to the alpha chain of tubulin. Biochemistry. 1977;16(10):2189–2194. doi: 10.1021/bi00629a023. [DOI] [PubMed] [Google Scholar]
- Redeker V. Mass spectrometry analysis of C-terminal posttranslational modifications of tubulins. Methods Cell Biol. 2010;95:77–103. doi: 10.1016/S0091-679X(10)95006-1. [DOI] [PubMed] [Google Scholar]
- Redeker V, Le Caer JP, Rossier J, Prome JC. Structure of the polyglutamyl side chain posttranslationally added to alpha-tubulin. J Biol Chem. 1991;266(34):23461–23466. [PubMed] [Google Scholar]
- Redeker V, Levilliers N, Schmitter JM, Le Caer JP, Rossier J, Adoutte A, Bre MH. Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science. 1994;266(5191):1688–1691. doi: 10.1126/science.7992051. [DOI] [PubMed] [Google Scholar]
- Redeker V, Melki R, Prome D, Le Caer JP, Rossier J. Structure of tubulin C-terminal domain obtained by subtilisin treatment. The major alpha and beta tubulin isotypes from pig brain are glutamylated. FEBS Lett. 1992;313(2):185–192. doi: 10.1016/0014-5793(92)81441-n. [DOI] [PubMed] [Google Scholar]
- Redeker V, Rossier J, Frankfurter A. Posttranslational modifications of the C-terminus of alpha-tubulin in adult rat brain: alpha 4 is glutamylated at two residues. Biochemistry. 1998;37(42):14838–14844. doi: 10.1021/bi981335k. [DOI] [PubMed] [Google Scholar]
- Redeker V, Rusconi F, Mary J, Prome D, Rossier J. Structure of the C-terminal tail of alpha-tubulin: increase of heterogeneity from newborn to adult. J Neurochem. 1996;67(5):2104–2114. doi: 10.1046/j.1471-4159.1996.67052104.x. [DOI] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16(21):2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Regnard C, Audebert S, Desbruyeres, Denoulet P, Edde B. Tubulin polyglutamylase: partial purification and enzymatic properties. Biochemistry. 1998;37(23):8395–8404. doi: 10.1021/bi9804131. [DOI] [PubMed] [Google Scholar]
- Regnard C, Desbruyeres E, Huet JC, Beauvallet C, Pernollet JC, Edde B. Polyglutamylation of nucleosome assembly proteins. J Biol Chem. 2000;275(21):15969–15976. doi: 10.1074/jbc.M000045200. [DOI] [PubMed] [Google Scholar]
- Regnard C, Fesquet D, Janke C, Boucher D, Desbruyeres E, Koulakoff A, Insina C, Travo P, Edde B. Characterisation of PGs1, a subunit of a protein complex co-purifying with tubulin polyglutamylase. J Cell Sci. 2003;116(Pt 20):4181–4190. doi: 10.1242/jcs.00743. [DOI] [PubMed] [Google Scholar]
- Rodriguez de la Vega M, Sevilla RG, Hermoso A, Lorenzo J, Tanco S, Diez A, Fricker LD, Bautista JM, Aviles FX. Nna1-like proteins are active metallocarboxypeptidases of a new and diverse M14 subfamily. FASEB J. 2007;21(3):851–865. doi: 10.1096/fj.06-7330com. [DOI] [PubMed] [Google Scholar]
- Rogowski K, Juge F, van Dijk J, Wloga D, Strub JM, Levilliers N, Thomas D, Bre MH, Van Dorsselaer A, Gaertig J, et al. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 2009;137(6):1076–1087. doi: 10.1016/j.cell.2009.05.020. [DOI] [PubMed] [Google Scholar]
- Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A, Peris L, Gold ND, Lacroix B, Grau MB, et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143(4):564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22(1):96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale RD. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr Biol. 2005;15(7):650–655. doi: 10.1016/j.cub.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451(7176):363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger M, Plessman U, Kloppel KD, Wehland J, Weber K. Class II tubulin, the major brain beta tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 1992;308(1):101–105. doi: 10.1016/0014-5793(92)81061-p. [DOI] [PubMed] [Google Scholar]
- Rudiger M, Wehland J, Weber K. The carboxy-terminal peptide of detyrosinated alpha tubulin provides a minimal system to study the substrate specificity of tubulin-tyrosine ligase. Eur J Biochem. 1994;220(2):309–320. doi: 10.1111/j.1432-1033.1994.tb18627.x. [DOI] [PubMed] [Google Scholar]
- Russell DG, Miller D, Gull K. Tubulin heterogeneity in the trypanosome Crithidia fasciculata. Mol Cell Biol. 1984;4(4):779–790. doi: 10.1128/mcb.4.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santander VS, Bisig CG, Purro SA, Casale CH, Arce CA, Barra HS. Tubulin must be acetylated in order to form a complex with membrane Na(+),K (+)-ATPase and to inhibit its enzyme activity. Mol Cell Biochem. 2006;291(1–2):167–174. doi: 10.1007/s11010-006-9212-9. [DOI] [PubMed] [Google Scholar]
- Schneider A, Plessmann U, Felleisen R, Weber K. Posttranslational modifications of trichomonad tubulins; identification of multiple glutamylation sites. FEBS Lett. 1998;429(3):399–402. doi: 10.1016/s0014-5793(98)00644-9. [DOI] [PubMed] [Google Scholar]
- Schroder HC, Wehland J, Weber K. Purification of brain tubulin-tyrosine ligase by biochemical and immunological methods. J Cell Biol. 1985;100(1):276–281. doi: 10.1083/jcb.100.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer HD, Lecine P, Tiwari S, Italiano JE, Jr, Hartwig JH, Shivdasani RA. A lineage-restricted and divergent beta-tubulin isoform is essential for the biogenesis, structure and function of blood platelets. Curr Biol. 2001;11(8):579–586. doi: 10.1016/s0960-9822(01)00153-1. [DOI] [PubMed] [Google Scholar]
- Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, Jerka-Dziadosz M, Gaertig J. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol. 2007;178(6):1065–1079. doi: 10.1083/jcb.200704021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J Cell Biol. 1987;104(3):439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci U S A. 2010;107(50):21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek K, Kamaid A, Phung AD, Kubala L, Bulinski JC, Harper RW, Eiserich JP. Normal and prostate cancer cells display distinct molecular profiles of alpha-tubulin posttranslational modifications. Prostate. 2006;66(9):954–965. doi: 10.1002/pros.20416. [DOI] [PubMed] [Google Scholar]
- Spiliotis ET, Hunt SJ, Hu Q, Kinoshita M, Nelson WJ. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J Cell Biol. 2008;180(2):295–303. doi: 10.1083/jcb.200710039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steczkiewicz K, Kinch L, Grishin NV, Rychlewski L, Ginalski K. Eukaryotic domain of unknown function DUF738 belongs to Gcn5-related N-acetyltransferase superfamily. Cell Cycle. 2006;5(24):2927–2930. doi: 10.4161/cc.5.24.3572. [DOI] [PubMed] [Google Scholar]
- Sudo H, Baas PW. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J Neurosci. 2010;30(21):7215–7226. doi: 10.1523/JNEUROSCI.0048-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF. Structure and utilization of tubulin isotypes. Annu Rev Cell Biol. 1988;4:687–716. doi: 10.1146/annurev.cb.04.110188.003351. [DOI] [PubMed] [Google Scholar]
- Suryavanshi S, Edde B, Fox LA, Guerrero S, Hard R, Hennessey T, Kabi A, Malison D, Pennock D, Sale WS, et al. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr Biol. 2010;20(5):435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Deaconescu AM, Piszczek G, Roll-Mecak A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat Struct Mol Biol. 2011;18(11):1250–1258. doi: 10.1038/nsmb.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Engle EC. Distinct alpha- and beta-tubulin isotypes are required for the positioning, differentiation and survival of neurons: new support for the 'multi-tubulin' hypothesis. Biosci Rep. 2010;30(5):319–330. doi: 10.1042/BSR20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P, Quiroga M, Zaldivar J, Rutter WJ, Kirschner MW, Cleveland DW. Nucleotide and corresponding amino acid sequences encoded by alpha and beta tubulin mRNAs. Nature. 1981;289(5799):650–655. doi: 10.1038/289650a0. [DOI] [PubMed] [Google Scholar]
- van Dijk J, Miro J, Strub JM, Lacroix B, van Dorsselaer A, Edde B, Janke C. Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem. 2008;283(7):3915–3922. doi: 10.1074/jbc.M705813200. [DOI] [PubMed] [Google Scholar]
- van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell. 2007;26(3):437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6(17):2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- Vogel P, Hansen G, Fontenot G, Read R. Tubulin tyrosine ligase-like 1 deficiency results in chronic rhinosinusitis and abnormal development of spermatid flagella in mice. Vet Pathol. 2010;47(4):703–712. doi: 10.1177/0300985810363485. [DOI] [PubMed] [Google Scholar]
- Wang T, Parris J, Li L, Morgan JI. The carboxypeptidase-like substrate-binding site in Nna1 is essential for the rescue of the Purkinje cell degeneration (pcd) phenotype. Mol Cell Neurosci. 2006;33(2):200–213. doi: 10.1016/j.mcn.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Wang Z, Sheetz MP. The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys J. 2000;78(4):1955–1964. doi: 10.1016/S0006-3495(00)76743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warn RM, Harrison A, Planques V, Robert-Nicoud N, Wehland J. Distribution of microtubules containing post-translationally modified alpha-tubulin during Drosophila embryogenesis. Cell Motil Cytoskeleton. 1990;17(1):34–45. doi: 10.1002/cm.970170106. [DOI] [PubMed] [Google Scholar]
- Wasylyk C, Zambrano A, Zhao C, Brants J, Abecassis J, Schalken JA, Rogatsch H, Schaefer G, Pycha A, Klocker H, et al. Tubulin tyrosine ligase like 12 links to prostate cancer through tubulin posttranslational modification and chromosome ploidy. Int J Cancer. 2010;127(11):2542–2553. doi: 10.1002/ijc.25261. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol. 1998;8(14):798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- Webster DR, Borisy GG. Microtubules are acetylated in domains that turn over slowly. J Cell Sci. 1989;92(Pt 1):57–65. doi: 10.1242/jcs.92.1.57. [DOI] [PubMed] [Google Scholar]
- Webster DR, Gundersen GG, Bulinski JC, Borisy GG. Differential turnover of tyrosinated and detyrosinated microtubules. Proc Natl Acad Sci U S A. 1987;84(24):9040–9044. doi: 10.1073/pnas.84.24.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrich A, Honnappa S, Jaussi R, Okhrimenko O, Frey D, Jelesarov I, Akhmanova A, Steinmetz MO. Structure-function relationship of CAP-Gly domains. Nat Struct Mol Biol. 2007;14(10):959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- Whipple RA, Matrone MA, Cho EH, Balzer EM, Vitolo MI, Yoon JR, Ioffe OB, Tuttle KC, Yang J, Martin SS. Epithelial-to-mesenchymal transition promotes tubulin detyrosination and microtentacles that enhance endothelial engagement. Cancer Res. 2010;70(20):8127–8137. doi: 10.1158/0008-5472.CAN-09-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Dave D, Meagley J, Rogowski K, Jerka-Dziadosz M, Gaertig J. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot Cell. 2010;9(1):184–193. doi: 10.1128/EC.00176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Gaertig J. Post-translational modifications of microtubules. J Cell Sci. 2010;123(Pt 20):3447–3455. doi: 10.1242/jcs.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Rogowski K, Sharma N, Van Dijk J, Janke C, Edde B, Bre MH, Levilliers N, Redeker V, Duan J, et al. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot Cell. 2008;7(8):1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Webster DM, Rogowski K, Bre MH, Levilliers N, Jerka-Dziadosz M, Janke C, Dougan ST, Gaertig J. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev Cell. 2009;16(6):867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Wozniak MJ, Bola B, Brownhill K, Yang YC, Levakova V, Allan VJ. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci. 2009;122(Pt 12):1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambito AM, Knipling L, Wolff J. Charge variants of tubulin, tubulin S, membrane-bound and palmitoylated tubulin from brain and pheochromocytoma cells. Biochim Biophys Acta. 2002;1601(2):200–207. doi: 10.1016/s1570-9639(02)00472-7. [DOI] [PubMed] [Google Scholar]
- Zampar GG, Chesta ME, Carbajal A, Chanaday NL, Diaz NM, Casale CH, Arce CA. Acetylated tubulin associates with the fifth cytoplasmic domain of Na(+)/K(+)-ATPase: possible anchorage site of microtubules to the plasma membrane. Biochem J. 2009;422(1):129–137. doi: 10.1042/BJ20082410. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27(2):197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]