Abstract
An important area of genetic research is the identification of functional mechanisms in polymorphisms associated with diseases. A highly relevant functional mechanism is the influence of polymorphisms on gene expression levels (differential allelic expression, DAE). The coding single nucleotide polymorphisms (SNPs) CSF2rs25882 and IL13rs20541 have been associated with asthma. In this work, we investigated whether the mRNA expression levels of CSF2 or IL13 were correlated with these SNPs. Samples were analyzed by mass spectrometry-based quantification of gene expression. Both SNPs influenced gene expression levels (CSF2rs25882: poverall = 0.008 and pDAE samples = 0.00006; IL13rs20541: poverall = 0.059 and pDAE samples = 0.036). For CSF2, the expression level was increased by 27.4% (95% CI: 18.5%–35.4%) in samples with significant DAE in the presence of one copy of risk variant CSF2rs25882-T. The average expression level of IL13 was increased by 29.8% (95% CI: 3.1%–63.4%) in samples with significant DAE in the presence of one copy of risk variant IL13rs20541-A. Enhanced expression of CSF2 could stimulate macrophages and neutrophils during inflammation and may be related to the etiology of asthma. For IL-13, higher expression could enhance the functional activity of the asthma-associated isoform. Overall, the analysis of DAE provides an efficient approach for identifying possible functional mechanisms that link disease-associated variants with altered gene expression levels.
Keywords: candidate gene, gene expression, gene polymorphism, QTL
Introduction
Genetic variation influences the susceptibility to disease. An important mechanism contributing to this effect is the influence of genetic polymorphisms on the expression levels of disease-related genes (differential allelic expression, DAE). DAE affects 20%–50% of genes (Yan et al., 2002; Bray et al., 2003; Lo et al., 2003; Serre et al., 2008) and is involved in gene-dosage compensation of sex chromosomes and imprinting on autosomes (Knight, 2004). Monoallelic expression with random choice between paternal and maternal alleles affects hundreds of autosomal genes and contributes to variability among individual cells (Gimelbrant et al., 2007). To increase our understanding of the role of disease-related polymorphisms in complex disease traits it will be necessary to investigate the effect of these polymorphisms on the transcription levels of disease-related genes (Maniatis et al., 2002; Orphanides and Reinberg, 2002).
In humans, allelic heterogeneity and the polygenic nature of gene expression greatly limit the identification of cis-acting mechanisms by simple comparison of expression levels between individuals carrying different genotypes of the disease-related polymorphism. Genetic heterogeneity can be problematic when polymorphisms interfere with the assay or affect the expression of trans-acting factors such as transcription factors, or when copy number varies among individuals. The comparison of gene expression levels among different genotypes of a polymorphism is further complicated by the occurrence of various regulatory factors and pathways, tissue quality and cDNA preparation, as well as environmental influences (Stamatoyannopoulos, 2004).
These limitations in the identification of cis-directed DAE can be overcome by measuring the influence of disease associated genetic variants on gene expression levels directly in heterozygous individuals (Serre et al., 2008). Since this method relies on comparison of the allelic ratio in DNA (which is expected to be 1:1) and RNA (which may differ from 1:1 if there is DAE) of each heterozygous individual the control for genetic heterogeneity can be obtained with a single sample. This approach markedly reduces confounding by environmental factors, tissue quality and cDNA preparation (Bray et al., 2003; Stamatoyannopoulos, 2004). In the absence of cis-regulatory polymorphisms (or imprinting), both copies of an autosomal gene will contribute equally to mRNA expression levels. However, expression levels will differ if there is DAE. These differences may reflect differences in transcriptional regulation, mRNA stability, processing and/or translational efficiency (Bray et al., 2003).
Allele-specific gene expression levels can be quantified in individual heterozygous cDNA samples by MALDI-TOF (matrix-assisted laser desorption ionization time-of-flight) mass spectrometry. This technique allows accurate measurements and can detect differences as low as 5% in allele expression levels (Knight, 2004).
Interleukin 13 is a cytokine located in the cytokine cluster on 5q31 and plays a crucial role in the etiology of asthma. IL-13 shares a common signaling pathway with IL-4, a key mediator of pro-inflammatory functions in asthma. Like IL-4, IL-13 induces the expression of CD23 and CD72 on B cells, as well as the expression of surface IgM, class II MHC antigens and IgE heavy-chain transcription (Zurawski and de Vries, 1994). The non-synonymous coding SNP IL13rs20541 (R144Q) is associated with increased serum IgE levels and eosinophil counts in asthma (Hunninghake et al., 2007) as well as asthma itself (Heinzmann et al., 2000). IL13rs20541 is also associated with psoriasis (Chang et al., 2008). Detailed molecular modeling has shown that amino acid residue 144 of IL-13 might be important in the internal constitution of the ligand and crucial in ligand-receptor interaction. Vladich et al. (2005) examined the impact of IL13rs20541 (R144Q) on the functional properties of IL-13 by comparing the activity of the glutamine (Q) variant to that of the common R (arginine) variant on primary effector cells of human allergic inflammation. The Q-variant was significantly more active than wild-type IL-13 in multiple effector assays. Based on their findings, these authors suggested that natural variation in the coding region of IL13 might be an important genetic determinant of susceptibility to allergic inflammation. However, there has been no report of DAE in this variant. One of the aims of the present work was therefore to investigate whether IL13rs20541 can influence IL13 mRNA expression levels. Such an interaction could shed light on the contribution of IL13rs20541 to the pathology and development of asthma.
Colony-stimulating factors are proteins necessary for the survival, proliferation and differentiation of hematopoietic progenitor cells. The colony-stimulating factor 2 (CSF2) gene encodes granulocyte/macrophage colony-stimulating factor (GM-CSF). CSF2 regulates macrophage and neutrophil function during inflammatory reactions (Fleetwood et al., 2005) and the blockade of CSF2 signaling reduces airway inflammation and hyper-responsiveness in mouse models of environmentally-induced lung damage and asthma (Ohta et al., 1999; Yamashita et al., 2002). The CSF2 variant I117 (coding SNP CSF2rs25882-T) has been linked to a risk of atopic asthma (Rohrbach et al., 1999) and is associated with increased lung cancer risk (Engels et al., 2007). However, the functional mechanisms by which CSF2 alleles influence the risk of disease are unknown. The aim of this study was therefore to investigate whether the non-synonymous coding variant CSF2rs25882 (I117T) might be related to DAE of the CSF2 gene.
Materials and Methods
For direct measurement of DAE, cell lines derived from Epstein-Barr virus (EBV)-transfected, immortalized B cells of 52 individuals were used. Genomic DNA was extracted (DNeasy Blood & Tissue Kit, Qiagen, Hilden, Germany) and screened for heterozygosity of IL13rs20541 and CSF2rs25882 by mass spectrometry as described elsewhere (Burkhardt et al., 2009). Individual genotyping primers (single base extension or SBE) containing a biotin tag and a UV-cleavable base (L) were used for purification purposes. Total RNA was extracted from heterozygous samples (Nrs20541 = 11; Nrs25882 = 8) and reverse transcribed using standard protocols (RevertAid H Minus first strand cDNA synthesis kit, Fermentas, Germany). PCR primers (Table 1) were designed to span exons 3 to 4 for IL13 and exons 1 to 4 for CSF2, thereby making amplification of genomic DNA (gDNA) improbable. PCR consisted of 40 rounds with an annealing temperature of 58 °C annealing temperature and a 15 s elongation per round. The PCR product size was monitored by agarose gel electrophoresis.
Table 1.
Primer sequences for IL13rs20541 and CSF2rs25882 assays.
| IL13rs20541 | CSF2rs25882 | |
|---|---|---|
| cDNA |
GAGGATGCTGAGCGGATTCTGCCCGCACAAGGTCTCAGC TGGG*CAGTTTTCCAGCTTGCATGTCCGAGACACCAAAAT CGAGGTGGCCCAGTTTGTAAAGGACCTGCTCTTACATTTA AAGAAACTTTTT(C)GCGAGGGAC[A/G]GTTCAACTGAAACTT CGAAAGCATCATTATTTGCAGAGACAGGACCTGACTATTG AAGTTGCAG |
TCCTGAACCTGAGTAGAGACACTGCTGCTGAGATG*AATG AAACAGTAGAAGTCATCTCAGAAATGTTTGACCTCCAG*G AGCCGACCTGCCTACAGACCCGCCTGGAGCTGTACAAGC AGGGCCTGCGGGGCAGCCTCACCAAGCTCAAGGGCCCCT TGACCATGATGGCCAGCCACTACAAGCAGCACTGCCCTCC AACCCCG*GAAACTTCCTGTG(C)AACCCAGA[C/T]TATCACC TTTGAAAGTTTCAAAGAGAACCTGAAGGACTTTCTGCTTG TCATC |
| gDNA |
TCTACTCATGTGCTGACCTCTTTGTCCTGCAGCAGTTTTCC AGCTTGCATGTCCGAGACACCAAAATCGAGGTGGCCCAG TTTGTAAAGGACCTGCTCTTACATTTAAAGAAACTTTTT(C)G CGAGGGAC[A/G]GTTCAACTGAAACTTCGAAAGCATCATT ATTTGCAGAGACAGGACCTGACTATTGAAGTTGCAGATTC ATTTTTCTTTCTGATGTCAAAAATGTCTTGGGTAGGCGGG AAGGAGGGTTAGGGAGGGGTAAAATTC |
TGAGTTCTAAGAGGCAGTAGAGAAACATGCTGGTGCTTCC TTCCCCCACGTTACCCACTTGCCTGGACTCAAGTGTTTTTT ATTTTTCTTTTTTTAAAGGAAACTTCCTGTG(C)AACCCAGA[C /T]TATCACCTTTGAAAGTTTCAAAGAGAACCTGAAGGACT TTCTGCTTGTCATCCCCTTTGACTGCTGGGAGCCAGTCCA GGAGTGAGACCGGCCAGATGAGGCTGGCCAAGCCGGGGA GCTGCTCTCTCATGAAACAAGAGCTAGAAACTCA |
PCR-primer binding sequences are underlined, SNPs are indicated by [], SBE primer (single base extension; genotyping primer) binding sequences are underlined and italicized, SBE bases replaced by a photocleavable linker are indicated by () and exon borders are indicated by *.
To quantitatively compare the expression of both alleles of an SNP in a heterozygous cDNA sample we compared the signal ratios of both alleles in cDNA with their corresponding signal ratios in simultaneously processed gDNA. Ratios of SNP allele expression within the range of gDNA allelic fluctuation (mean ± 1.96 SD), were indicative of no regulation.
To quantify allelic expression the MALDI-TOF-based genotyping system genoSNIP marketed by Bruker Daltonik GmbH (Bremen, Germany) was used, as described elsewhere (Burkhardt et al., 2009), with minor modifications. Measurements were done at least in triplicate. The natural logarithm of the signal-to-noise ratios (intensity of allele-specific mass peaks divided by background noise) reported by Flexcontrol 3.0 (Bruker Daltonik GmbH, Bremen, Germany) was used for statistical analysis. Quality control ensured appropriate spectrum quality by excluding low-intensity spectra (signal to noise ration < 5) and spectra with abnormal ion-peaks.
We screened for the presence of DAE in general (averaged across all samples) as well as for DAE in individual samples. For general screening, we determined whether the natural logarithms of allelic signal ratios for both transcripts differed between cDNA and gDNA by using a standard linear model (R Development Core Team, 2010). In this case, allelic ratios that were averaged across 3–4 measurements were used. For individual samples, we assessed whether an observed allelic ratio in cDNA was more extreme than expected by chance given the variance of allelic ratios observed within gDNA samples. For this we initially estimated the distribution of allelic ratios within gDNA samples by calculating the mean and standard deviation. Subsequently, p-values indicating the presence of DAE in cDNA samples were determined by calculating the quantile of the distribution that corresponded to the measured allelic ratios of DAE in the samples. To avoid batch effects, the cDNA and gDNA analyses were always done simultaneously.
To facilitate the comparison of DAE between different studies we applied a method based on a one sample t-test, as described previously for DAE analysis (Antoun et al., 1991). This method differs from the analysis described above as it does not take into account fluctuation of gDNA measurements. Briefly, in this method, significant deviation (based on a one sample Student’s t-test) from one of the allelic ratios between cDNA and gDNA samples indicated DAE. A similar direction of altered allelic ratios across all heterozygous samples of a SNP indicated cis-regulation.
To verify the occurrence of cis-directed DAE purified PCR products (PCR purifying kit, Seqlab, Göttingen, Germany) from cDNA and genomic DNA of selected samples were analyzed by eurofins-MWG/operon (Ebersberg, Germany) using fluorescence-based sequencing (cycle sequencing technology with dideoxy chain termination in an Applied Biosystems ABI 3730XL-sequencer).
The software Genevar 3.5.0 was used to locate expression quantitative trait loci (eQTL) related to IL13 or CSF2 within published genome-wide datasets (Dimas et al., 2009; Nica et al., 2011). The Gene Expression Atlas maintained by the European Bioinformatics Institute v. 2.0.5 05/11 was searched for published mRNA expression of IL13 and CSF2 in asthma relevant tissues or animal models.
Results
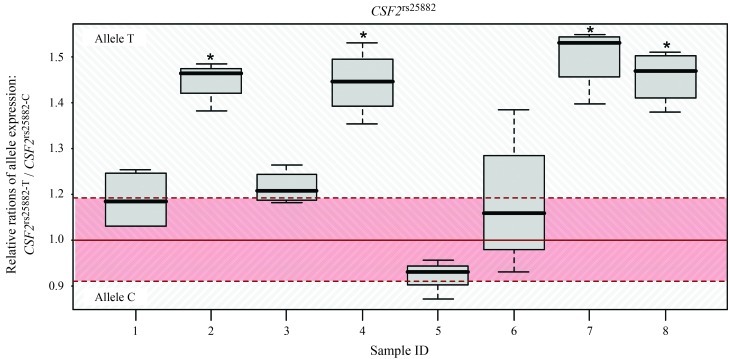
We found cis-directed DAE for CSF2rs25882 and IL13rs20541 (Figures 1 and 2). For CSF2rs25882 a significant effect of DAE (p = 0.008) was found across all samples with allele CSF2rs25882-T (corresponding to isoleucine) and was overexpressed by an average of 17.7% (95% CI: 5.7%–28.2%). The one sample t-test method showed similar significant results (p = 0.008). Of the eight heterozygous samples analyzed, four showed individual DAE, all in the same direction (cis-effect). When only those samples with a significant DAE were analyzed the p value was 0.00006, with CSF2rs25882-T being overexpressed by an average of 27.4% (95% CI: 18.5%–35.4%) compared to allele A. The results obtained by sequencing agreed with those of mass spectrometry (Table 2).
Figure 1.
Differential cis-directed expression in samples heterozygous for CSF2rs25882. Boxplots show the allelic expression in individual samples; horizontal lines indicate the median and whiskers indicate the 1.5 x interquartile range. The minor allele is C (T, threonine) and the major allele is T (I, isoleucine). The gDNA fluctuation range (mean ± 1.96 SD) is indicated in red and represents the range (defined in the Methods) in which there is no differential allelic expression. Allelic ratios were calibrated relative to the mean for gDNA samples, which corresponded to an allelic ratio of 1. *p < 0.05. SNR – signal-to-noise-ratio.
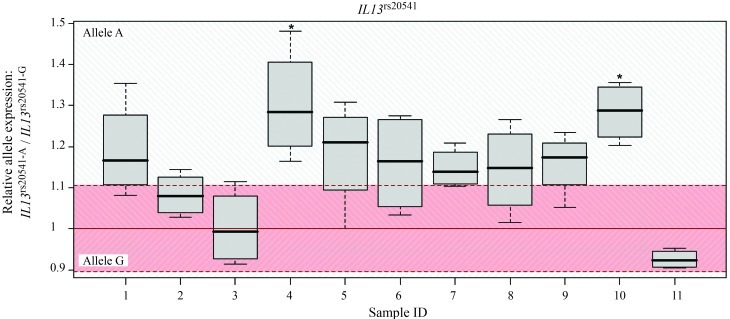
Figure 2.
Differential cis-directed expression in samples heterozygous for IL13rs20541. Boxplots show the allelic expression in individual samples; horizontal lines indicate the median and whiskers indicate the 1.5 x interquartile range. The minor allele is A (Q, glutamine) and the major allele is G (R, arginine). The gDNA fluctuation range (mean ± 1.96 SD) is indicated in red and represents the range (defined in the Methods) in which there is no differential allelic expression. Allelic ratios were calibrated relative to the mean for gDNA samples, which corresponded to an allelic ratio of 1. *p < 0.05. SNR – signal-to-noise-ratio.
Table 2.
Differential expression of the allelic ratio CSF2rs25882-T/CSF2rs25882-C in individual heterozygous B cell line samples. Two techniques were used to determine the allelic expression ratios, with sequencing being applied to selected samples. Allelic ratios that differed significantly from a cDNA/gDNA ratio of 1 are indicated in bold.
| Sample | p value | cDNA/gDNA ratio
|
|
|---|---|---|---|
| Genotyping | Sequencing | ||
| 1 | 0.34 | 1.10 | - |
| 2 | 0.002 | 1.37 | - |
| 3 | 0.23 | 0.89 | |
| 4 | 0.002 | 1.12 | 1.41 |
| 5 | 1 | 0.93 | - |
| 6 | 0.19 | 1.14 | 1.05 |
| 7 | 0.0005 | 1.41 | 1.35 |
| 8 | 0.002 | 1.37 | - |
We also found evidence for cis-directed DAE of IL13rs20541. We found DAE across all samples to be marginally significant (p = 0.059) with higher expression of allele A; the one sample t-test method showed p = 0.001. Two out of 11 samples were individually significant for DAE, with both showing similar direction (cis-effect; p = 0.036). Within these samples, allele A was overexpressed by an average of 29.8% (95% CI: 3.1%–63.4%). The results obtained by sequencing selected samples agreed with these findings (Table 3).
Table 3.
Differential expression of the allelic ratio IL13rs20541-A / IL13rs20541-G in individual heterozygous B cell line samples. Two techniques were used to determine the allelic expression ratios, with sequencing being applied to selected samples. Allelic ratios that differed significantly from a cDNA/gDNA ratio of 1 are indicated in bold.
| Sample | p value | cDNA/gDNA ratio
|
|
|---|---|---|---|
| Genotyping | Sequencing | ||
| 1 | 0.07 | 1.20 | - |
| 2 | 0.38 | 1.09 | - |
| 3 | 0.95 | 1.01 | - |
| 4 | 0.006 | 1.31 | 1.11 |
| 5 | 0.08 | 1.18 | - |
| 6 | 0.12 | 1.16 | - |
| 7 | 0.14 | 1.15 | - |
| 8 | 0.15 | 1.15 | - |
| 9 | 0.12 | 1.16 | - |
| 10 | 0.009 | 1.29 | 1.52 |
| 11 | 1 | 0.93 | - |
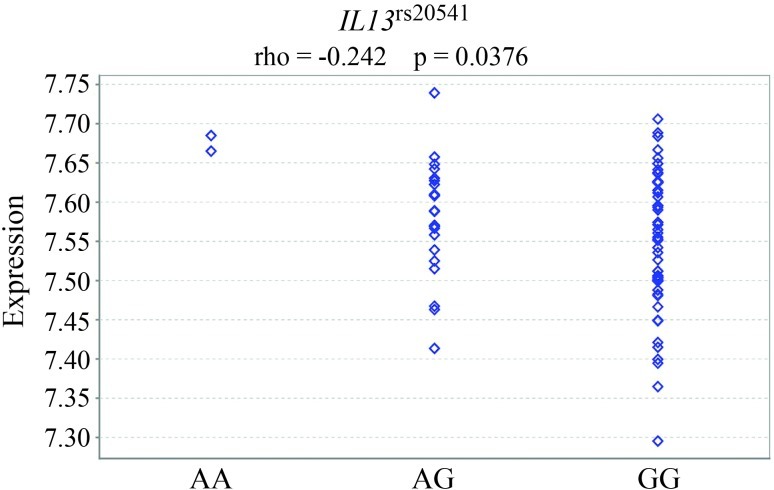
The findings described above were confirmed by Genevar, which revealed a nominally significant cis-directed DAE for IL13 in the MuTHER pilot project (Figure 3). The expression of IL13 mRNA was increased in carriers of IL13rs20541-A (p = 0.04). All other SNPs showing evidence for cis-directed eQTL in IL13 (< 5 kbp distance) were in close linkage disequilibrium based on HapMap (release #27) data (D’: 1.0 and r2 > 0.88 with rs847, rs848, rs1295685, rs1295686 and rs1881457).
Figure 3.
Genevar eQTL analysis for IL13rs20541 in which the rare allele A was significantly associated with elevated IL13 mRNA expression (Spearman’s rank correlation coefficient, rho = −0.242 and p = 0.0376). Analysis of expression quantitative trait loci (eQTL) was based on the genotyping data and mRNA expression data available from the MuTHER (multiple tissue human expression resource) project, lymphocyte twin set 1 (N = 156; Nica et al., 2011).
Discussion
The results of this study demonstrate an association between two previously identified asthma-related SNPs (CSF2rs25882 and IL13rs20541) and the expression levels of the genes CSF2 and IL13; both of these genes are important candidates for involvement in the etiology of asthma and other diseases. Overall, asthma-related CSF2rs25882-T and IL13rs20541-A were associated with higher gene expression levels of CSF2 and IL13, respectively. One copy of the CSF2rs25882-T allele increased expression by an average of 17.7% in all samples and by 27.4% in samples that showed individually significant DAE of the two alleles. However, not all of the samples displayed DAE. This finding may reflect (1) the presence of a different, possibly unknown causative variant in linkage disequilibrium with CSF2rs25882-T, (2) complex modulation by various transcription factors or other means of gene-gene interaction or (3) interaction with extrinsic environmental factors.
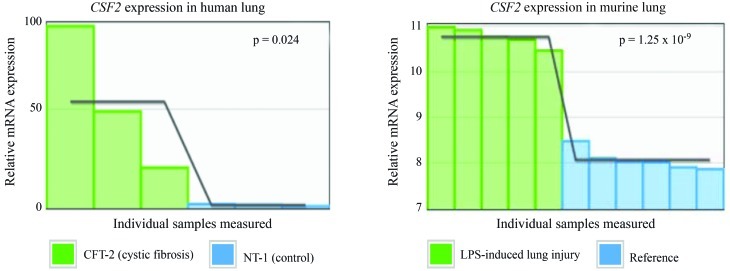
Data from the Gene Expression Atlas (experiment E-MEXP-980; Verhaeghe et al., 2007) showed increased expression of CSF2 in a human cystic fibrosis cell line (CFT-2) compared to a control cell line (NT-1) (p = 0.024). The CFT-2 cell line shares similar characteristics with inflamed asthmatic bronchoepithelial tissue. CSF2 was also up-regulated after LPS-induced lung injury in mice (E-GEOD-1871; Jacobson et al., 2005), a model for airway inflammation similar to that in asthmatic diseases (p = 1.25 x 10−9) (Figure 4). Therefore, our finding that known risk variant CSF2rs25882-T and not CSF2rs25882-C increased the expression of CSF2 agreed with published results.
Figure 4.
CSF2 expression in human and murine lung. Left panel – CSF2 expression in the human cystic fibrosis cell line CFT-2 and human NT-1 cells (control); right panel – CSF2 expression in murine lung before and after LPS-induced lung injury. The relative CSF2 mRNA expression is shown based on data derived from the Gene Expression Atlas, experiments E-MEXP-980 (Verhaeghe et al., 2007) and E-GEOD-1871 (Jacobson et al., 2005). Each bar represents an individual sample, the moving average is shown as solid line.
We also used MatInspector and Genomatix to assess whether transcription factor binding sites were created or lost in the presence of CSF2rs25882-T. Interestingly, a probable binding site for Smad3 was lost when the T allele was present (core similarity = 1; matrix similarity = 0.997). Smad3 is a negative regulator of the normal TGF-β response (Zhang et al., 1996) and a direct mediator of transcriptional activation by the TGF-β receptor. This effect is mediated either by induction of cyclin-dependent kinase inhibitors or by repression of other growth controlling genes such as MYC, a key oncogene (Chen et al., 2002). In asthma, airway remodeling following immune driven inflammation is thought to contribute to airway dysfunction and there is evidence that TGF-β, expressed mainly by eosinophils in asthmatic lung tissue, is a major mediator in airway remodeling (Fattouh and Jordana, 2008). In summary, the loss of a Smad3 binding site through the presence of CSF2rs25882-T may be a pathologically relevant event for the development of asthma.
IL13rs20541-A was associated with higher expression in some of the samples investigated. This association may have arisen for reasons similar to those stated for CSF2rs25882, i.e., the influence of different variants and transcription factors, imprinting, gene-gene interactions or extrinsic factors. In samples with evidence of DAE allele A was overexpressed by an average of 29.8%.
Based on in silico analyses using Genomatix and MatInspector (Tasheva et al., 2004) the presence of the asthma risk allele IL13rs20541-A was predicted to create two transcription factor binding sites: EN1 and MYT1. However, a role for these two transcription factors in asthma is not immediately obvious as they are both commonly expressed in neuronal, but not peripheral, tissues (Kim et al., 1997; Logan et al., 1992). Since SNPs in linkage disequilibrium with IL13rs20541 could be responsible for the observed effect we repeated this analysis with all SNPs in linkage disequilibrium with IL13rs20541, according to the HapMap CEU population (r2 > 0.8), and additionally displaying cis-directed eQTL in the Genevar database. Our findings revealed several new or lost transcription factor binding sites in the presence of these linked SNPs (Table 4).
Table 4.
Loss or gain of transcription factor (TF) binding sites as predicted by Genomatix for IL13rs20541 or SNPs in linkage disequilibrium. The loss or gain was predicted to occur in carriers of the rare allele compared to the common allele (common allele → rare allele). The similarity scores for the core and matrix sequences represent the similarity between expected TF binding sites (the core TF binding sequence and the TF family binding sequence) and the corresponding sequences altered by the indicated SNPs.
| SNP rs # | Allele | Loss/gain | TF family/matrix | Core similarity | Matrix similarity |
|---|---|---|---|---|---|
| rs1881457 | A → C | gain | MIF1 | 0.8 | 0.772 |
| rs847 | A → G | loss | EN1 | 1 | 0.775 |
| rs1881457 | A → C | loss | CTCF | 0.787 | 0.781 |
| rs848 | G → T | loss | ZNF76_143 | 1 | 0.789 |
| rs1881457 | A → C | loss | DICE | 0.783 | 0.806 |
| rs20541 | C → T | gain | EN1 | 0.782 | 0.816 |
| rs20541 | C → T | loss | MYBL1 | 1 | 0.837 |
| rs1295685 | C → T | loss | AML3 | 1 | 0.84 |
| rs1295685 | C → T | loss | MYOD | 1 | 0.881 |
| rs1295686 | A → G | gain | XBP1 | 1 | 0.887 |
| rs1295686 | A → G | loss | PARAXIS | 0.882 | 0.914 |
| rs1295685 | C → T | loss | USF | 0.851 | 0.927 |
| rs20541 | C → T | gain | MYT1L | 1 | 0.93 |
| rs1295686 | A → G | gain | BHLHB2 | 1 | 0.933 |
| rs1295686 | A → G | loss | MIT | 1 | 0.956 |
| rs1295686 | A → G | gain | NMYC | 1 | 0.976 |
| rs1295686 | A → G | gain | HRE | 1 | 0.984 |
| rs1295686 | A → G | gain | HIF1 | 1 | 0.991 |
The increased expression of IL13 in IL13rs20541-A carriers agreed with a previous report showing that variant IL13rs20541-A was significantly more active in effector assays (Vladich et al., 2005). The latter authors suggested that the amino acid switch from R to Q increased the activity of the IL-13 Q144 protein variant. Our data provide evidence for an additional effect, namely that the enhanced activity of IL-13 in carriers of Q144 may be amplified by increased expression via DAE.
Increased IL-13 levels may contribute to the development of asthma by inducing the expression of CD23 and CD72 in B cells, both of which are key molecules in B cell activation and differentiation. IL-13 also enhances the expression of surface IgM and class-II MHC antigen and induces germline IgE heavy-chain gene transcription (Zurawski and de Vries, 1994), indicating a possible role for IL-13 in the pathogenesis of immune disorders such as asthma. The finding that risk variant IL13rs20541-A was related to increased IL-13 levels therefore agrees with our current understanding of the role of IL-13 in asthma.
A limitation of this study is that we used immortalized B-cell lines that did not originate from affected individuals. Further studies are therefore necessary to validate our findings in other tissues, e.g., diseased primary tissue. Serre et al. (2008) examined the possible influence of lymphoblastoid culture conditions and passages on DAE but found very little evidence for a bias attributable to technical aspects in a genome-wide study. In addition, data obtained from the MuTHER pilot project (Nica et al., 2011) indicated DAE for IL13rs20541, as also observed here; unfortunately, CSF2rs25882 was not genotyped in that dataset. The IL13 mRNA expression reported in the MuTHER project was based on fresh blood lymphocytes, thereby eliminating any influence of immortalization. Together, these findings indicate that changes in gene expression seen with DAE-based techniques tend to reflect true biological responses rather than technical artifacts.
The effects of both SNPs on gene expression can be considered moderate. However, common diseases are expected to be influenced by many risk factors and it is the sum of the effects of these factors that accounts for a substantial part of phenotypic variation (Morley et al., 2004). In general, observed effect sizes are similar to known sizes of effects of genetic variants on gene expression. Thus, in a genome-wide study of DAE, only 18.1% of 349 SNPs with DAE (excluding X-linked SNPs) showed fold changes in DAE ≥ 1.5 and only 5.4% showed fold changes ≥ 3 (Serre et al., 2008).
Acknowledgments
This work was funded by the German Federal Ministry of Education and Research (grants 0315883 to JB and HK, and 01KN0702 to PA). HK and PA are supported by LIFE – Leipzig Research Center for Civilization Diseases, Universität Leipzig. LIFE is funded by the European Union, the European Regional Development Fund (ERDF) and the Free State of Saxony within the framework of an excellence initiative.
Footnotes
Associate Editor: Mara Hutz
References
- Antoun GR, Re GG, Terry NH, Zipf TF. Molecular genetic evidence for a differentiation-proliferation coupling during DMSO-induced myeloid maturation of HL-60 cells: Role of the transcription elongation block in the c-myc gene. Leuk Res. 1991;15:1029–1036. doi: 10.1016/0145-2126(91)90108-6. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Buckland PR, Owen MJ, O’Donovan MC. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet. 2003;113:149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- Burkhardt J, Petit-Teixeira E, Teixeira VH, Kirsten H, Garnier S, Ruehle S, Oeser C, Wolfram G, Scholz M, Migliorini P, et al. Association of the X-chromosomal genes TIMP1 and IL9R with rheumatoid arthritis. J Rheumatol. 2009;36:2149–2157. doi: 10.3899/jrheum.090059. [DOI] [PubMed] [Google Scholar]
- Chang M, Li Y, Yan C, Callis-Duffin KP, Matsunami N, Garcia VE, Cargill M, Civello D, Bui N, Catanese JJ, et al. Variants in the 5q31 cytokine gene cluster are associated with psoriasis. Genes Immun. 2008;9:176–181. doi: 10.1038/sj.gene.6364451. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massagué J. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez AM, Sekowska M, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels EA, Wu X, Gu J, Dong Q, Liu J, Spitz MR. Systematic evaluation of genetic variants in the inflammation pathway and risk of lung cancer. Cancer Res. 2007;67:6520–6527. doi: 10.1158/0008-5472.CAN-07-0370. [DOI] [PubMed] [Google Scholar]
- Fattouh R, Jordana M. TGF-β, eosinophils and IL-13 in allergic airway remodeling: A critical appraisal with therapeutic considerations. Inflamm Allergy Drug Targets. 2008;7:224–236. doi: 10.2174/187152808786848388. [DOI] [PubMed] [Google Scholar]
- Fleetwood AJ, Cook AD, Hamilton JA. Functions of granulocyte-macrophage colony-stimulating factor. Crit Rev Immunol. 2005;25:405–428. doi: 10.1615/critrevimmunol.v25.i5.50. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Heinzmann A, Mao XQ, Akaiwa M, Kreomer RT, Gao PS, Ohshima K, Umeshita R, Abe Y, Braun S, Yamashita T, et al. Genetic variants of IL-13 signalling and human asthma and atopy. Hum Mol Genet. 2000;9:549–559. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- Hunninghake GM, Soto-Quiros ME, Avila L, Su J, Murphy A, Demeo DL, Ly NP, Liang C, Sylvia JS, Klanderman BJ, et al. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol. 2007;120:84–90. doi: 10.1016/j.jaci.2007.04.032. [DOI] [PubMed] [Google Scholar]
- Jacobson JR, Barnard JW, Grigoryev DN, Ma SF, Tuder RM, Garcia JG. Simvastatin attenuates vascular leak and inflammation in murine inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;288:L1026–L1032. doi: 10.1152/ajplung.00354.2004. [DOI] [PubMed] [Google Scholar]
- Kim JG, Armstrong RC, Agoston D, Robinsky A, Wiese C, Nagle J, Hudson LD. Myelin transcription factor 1 (Myt1) of the oligodendrocyte lineage, along with a closely related CCHC zinc finger, is expressed in developing neurons in the mammalian central nervous system. J Neurosci Res. 1997;50:272–290. doi: 10.1002/(SICI)1097-4547(19971015)50:2<272::AID-JNR16>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20:113–116. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, Lee MP. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C, Hanks MC, Noble-Topham S, Nallainathan D, Provart NJ, Joyner AL. Cloning and sequence comparison of the mouse, human, and chicken engrailed genes reveal potential functional domains and regulatory regions. Dev Genet. 1992;13:345–358. doi: 10.1002/dvg.1020130505. [DOI] [PubMed] [Google Scholar]
- Maniatis N, Collins A, Xu CF, McCarthy LC, Hewett DR, Tapper W, Ennis S, Ke X, Morton NE. The first linkage disequilibrium (LD) maps: Delineation of hot and cold blocks by diplotype analysis. Proc Natl Acad Sci USA. 2002;99:2228–2233. doi: 10.1073/pnas.042680999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M, Travers M, Potter S, Grundberg E, Small K, et al. The architecture of gene regulatory variation across multiple human tissues: The MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K, Yamashita N, Tajima M, Miyasaka T, Nakano J, Nakajima M, Ishii A, Horiuchi T, Mano K, Miyamoto T. Diesel exhaust particulate induces airway hyper-responsiveness in a murine model: Essential role of GM-CSF. J Allergy Clin Immunol. 1999;104:1024–1030. doi: 10.1016/s0091-6749(99)70084-9. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Rohrbach M, Frey U, Kraemer R, Liechti-Gallati S. A variant in the gene for GM-CSF, I117T, is associated with atopic asthma in a Swiss population of asthmatic children. J Allergy Clin Immunol. 1999;104:247–248. doi: 10.1016/s0091-6749(99)70147-8. [DOI] [PubMed] [Google Scholar]
- Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, Bibikova M, Chudin E, Barker DL, Dickinson T, et al. Differential allelic expression in the human genome: A robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA. The genomics of gene expression. Genomics. 2004;84:449–457. doi: 10.1016/j.ygeno.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Tasheva ES, Klocke B, Conrad GW. Analysis of transcriptional regulation of the small leucine rich proteoglycans. Mol Vis. 2004;10:758–772. [PubMed] [Google Scholar]
- Verhaeghe C, Remouchamps C, Hennuy B, Vanderplasschen A, Chariot A, Tabruyn SP, Oury C, Bours V. Role of IKK and ERK pathways in intrinsic inflammation of cystic fibrosis airways. Biochem Pharmacol. 2007;73:1982–1994. doi: 10.1016/j.bcp.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Vladich FD, Brazille SM, Stern D, Peck ML, Ghittoni R, Vercelli D. IL-13 R130Q, a common variant associated with allergy and asthma, enhances effector mechanisms essential for human allergic inflammation. J Clin Invest. 2005;115:747–754. doi: 10.1172/JCI22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Tashimo H, Ishida H, Kaneko F, Nakano J, Kato H, Hirai K, Horiuchi T, Ohta K. Attenuation of airway hyperresponsiveness in a murine asthma model by neutralization of granulocyte-macrophage colony-stimulating factor (GM-CSF) Cell Immunol. 2002;219:92–97. doi: 10.1016/s0008-8749(02)00565-8. [DOI] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- Zurawski G, de Vries JE. Interleukin 13 elicits a subset of the activities of its close relative interleukin 4. Stem Cells. 1994;12:169–174. doi: 10.1002/stem.5530120204. [DOI] [PubMed] [Google Scholar]
Internet Resources
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2010. http://www:R-project.org/ (August 29, 2011)
- R package lme4: Linear mixed-effects models using S4 classes, http://CRAN.R-project.org/package=lme4 (August 29, 2011).
- Analysis of genetic linkage disequilibrium, http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap24_B36/ (September 14, 2011)
- Genevar: Genome-wide expression quantitative trait loci database and analysis, http://www.sanger.ac.uk/resources/software/genevar/ (September 14, 2011)
- Genomatix and MatInspector Software Suite for transcription factor binding site analysis, http://www.genomatix.de/on-line_help/help_matinspector/matinspector_help.html (August 29, 2011)
- Gene expression atlas database and analysis: Experiment E-MEXP-980, http://www.ebi.ac.uk/gxa/experiment/E-MEXP-980 (September 14, 2011); Experiment E-GEOD-1871, http://www.ebi.ac.uk/gxa/experiment/E-GEOD-1871 (September 14, 2011)