Abstract
The prevalence of metabolic disorders varies among ethnic populations and these disorders represent a critical health care issue for elderly women. This study investigated the correlation between genetic ancestry and body composition, metabolic traits and clinical status in a sample of elderly women. Clinical, nutritional and anthropometric data were collected from 176 volunteers. Genetic ancestry was estimated using 23 ancestry-informative markers. Pearsons correlation test was used to examine the relationship between continuous variables and an independent samples t-test was used to compare the means of continuous traits within categorical variables. Overall ancestry was a combination of European (57.49%), Native American (25.78%) and African (16.73%). Significant correlations were found for European ancestry with body mass index (r = 0.165; p = 0.037) and obesity (mean difference (MD) = 5.3%; p = 0.042). African ancestry showed a significant correlation with LDL (r = 0.159, p = 0.035), VLDL (r = −0.185; p = 0.014), hypertriglyceridemia (MD = 6.4%; p = 0.003) and hyperlipidemia (MD = 4.8%; p = 0.026). Amerindian ancestry showed a significant correlation with triglyceride levels (r = 0.150; p = 0.047) and hypertriglyceridemia (MD = 4.5%; p = 0.039). These findings suggest that genetic admixture may influence the etiology of lipid metabolism-related diseases and obesity in elderly women.
Keywords: dyslipidemia, genetic ancestry, lipid profile, obesity, postmenopausal women
Introduction
Obesity and lifestyle factors are the main contributors to an increased risk of chronic non-communicable (non-transmissible) diseases, especially type 2 diabetes (T2D), lipid disorders, hypertension and other metabolic-related comorbidities (Sullivan et al., 2005). Many of these conditions may ultimately lead to cardiovascular disease. In the elderly, the risk of these disorders increases progressively with age (Wilson and Kannel, 2002; Cabrera et al., 2007). As ageing progresses, overweight increases the risk for cardiovascular disease, mostly in women with high-risk LDL and HDL cholesterol profiles (Perissinotto et al., 2002). As the incidence of these diseases increases worldwide there is an ever greater need to study the underlying genetic factors.
Recent findings from genome-wide association studies have identified several loci associated with chronic metabolic and cardiovascular diseases. However, the allelic architecture that can explain the heritability accounting for these complex phenotypes is not well understood. Informative single nucleotide polymorphisms (SNPs) have been used to characterize the structure of the main continental populations, and the data indicate that ancestry estimates are the most important correction factors in genetic association studies, especially in studies using admixed populations such as those of Latin America (Bonilla et al., 2004a; Tsai et al., 2005; Gentil et al., 2007; Moreno Lima et al., 2007; Gentil et al., 2009; Lai et al., 2009; Ruiz-Narvaez et al., 2010). Consequently, the clinical application of genetic risk markers may not be straightforward across different ethnic populations and may be vulnerable to spurious allelic association in genetic studies (Tsai et al., 2005).
The risk factors for metabolic syndrome and other phenotypes related to postmenopausal biology have distinct frequencies among populations of different ethnic backgrounds (Deurenberg et al., 1998; Wulan et al., 2010). Genetic studies of admixed populations have substantially increased our knowledge of various genetic diseases through the detection and correction for population stratification (Bonilla et al., 2004a; Lai et al., 2009; Ruiz-Narvaez et al., 2010).
Most genetic studies of metabolic phenotypes have investigated adult and middle-aged populations, with studies on elderly women being underrepresented. This group is more susceptible to chronic illnesses (Perissinotto et al., 2002; Wilson and Kannel, 2002; Cabrera et al., 2007) and is the fastest growing population group in developing countries (Nobrega et al., 2009). The aim of the present study was therefore to investigate in a sample of elderly Brazilian women the contribution of admixture to several body composition measurements, metabolic traits and parameters of clinical status based on their correlation with genetic ancestry, estimated by ancestry-informative markers.
Subjects and Methods
Subjects
This cross-sectional study was done using data obtained from apparently healthy female outpatients from the low-income suburbs of Brasília, Brazil, who volunteered to undergo health screenings and medical, nutritional and/or pharmacological interventions as part of a larger research project entitled the Elderly Health Promotion Project (EHPP). Additional characterization of the subjects is presented elsewhere (Paula et al., 2010; Nobrega et al., 2011). Project eligibility criteria consisted of being female, aged 60 years or older, completing the clinical course of assessment for hypertension, diabetes and dyslipidemia, and recording and informing their dietary profile. Since clinical data and laboratory results were obtained more than once, only the most recent data collected prior to any health intervention (especially blood pressure therapy) were considered. All volunteers provided written informed consent and the research protocol was approved by the Catholic University of Brasília Ethics Committee.
Data collection
Venous blood samples were collected in EDTA-containing tubes after 12 h overnight fast. Laboratory tests involved routine clinical analyses with reagents from Boehringer-Mannheim (Germany). Low-density lipoprotein (LDL) was quantified by using Friedewald’s formula. During consultation, blood pressure was measured after at least a 10 min rest in a sitting position. The blood pressure value of each subject was the mean of three physician-obtained measurements recorded 3 min apart. The body mass index (BMI = weight (kg)/height2 (m2)) of each patient was calculated by measuring body weight (kg) while the subject was wearing light clothing, and the subject’s height (m), without shoes, was assessed to the nearest millimeter. Obesity was defined as a BMI = 30 kg/m2. Body composition (fat and fat-free soft tissue) was measured using a dual-energy X-ray absorptiometer (DXA; Lunar DPX-IQ model, software version 4.7e, Lunar Radiation Corp., Madison, WI, USA) according to standard procedures provided by the manufacturer.
Each subject was classified based on the consensus of Brazilian Medical Societies, particularly the 5th Brazilian Guidelines in Arterial Hypertension (Diretrizes, 2007) and the 4th Brazilian Guidelines for Dyslipidemia and Atherosclerosis Prevention (Sposito et al., 2007). Briefly, hypertension was defined as a systolic blood pressure ≥ 130 mm Hg or a diastolic blood pressure ≥ 85 mm Hg, or the current use of antihypertensive medication. Type 2 diabetes mellitus (DM) was characterized by fasting blood glucose ≥ 126 mg/dL or the current use of insulin or oral anti-diabetic drugs. Glucose intolerance included DM cases and any cases with a fasting blood glucose ≥ 110 mg/dL. Hypertriglyceridemia was defined as triglyceride levels ≥ 150 mg/dL, and hypercholesterolemia was defined as total cholesterol ≥ 200 mg/dL or LDL-cholesterol ≥ 130 mg/dL. The current use of any anti-lipemic medication was considered to represent the presence of both entities. Hyperlipidemia was defined as isolated or combined hypertriglyceridemia and hypercholesterolemia, whereas dyslipidemia encompassed all cases of hyperlipidemia with or without a HDL-cholesterol < 50 mg/dL.
The study design included an estimation of fat consumption based on each subject’s natural, ad libitum food intake. Dietary analysis was performed based on an average 3-day food intake (two weekdays and one weekend day) recorded by the patient at home. Clinical dietitians and assistants instructed each patient on how to record food intake by defining the size and number of each serving. The forms were returned at a clinical interview during which the amounts and qualities of food were checked for completion and missing data were collected. Diet composition was calculated using Diet-Pro® software, version 4.0 (A.S. Sistemas, Viçosa, MG, Brazil). Software was set for all food tables and completed with Philippi’s composition table for local food (Philippi, 2002). Total lipid was expressed as the percentage of total energy. To construct categorical variables of fat consumption, intakes were classified into two groups: those with a normal lipid diet (10%–35%) and those with a hyperlipid diet (> 35%).
Marker selection and genotyping
For individual genetic ancestry estimation, we selected 23 ancestry-informative markers (AIMs) that displayed differential allele frequencies among European, African and Amerindian parental populations (Fernandez et al., 2003; Bonilla e et al., 2004b; Smith et al., 2004; Shriver et al., 2005). Some SNPs were previously used in cross-sectional studies to correct for population structure (Gentil et al., 2007, 2009; Moreno Lima et al., 2007) and the informativeness of these SNPs for a three hybrid population panel has been evaluated in a Brazilian population (Lins et al., 2010). The AIMs were genotyped using a modified, single-base extension protocol, as previously described (Lins et al., 2007). Briefly, genomic DNA was extracted using a modified salting-out method and amplified by the polymerase chain reaction (PCR). Products were treated with Exo I/SAP enzymes and genotyping was done by single-base extension using the SNaPshot Multiplex System (Applied Biosystems, Foster City, CA), followed by capillary electrophoresis on an ABI Prism 3130XL genetic analyzer. The data were analyzed using GeneMapper software (Applied Biosystems).
Statistical analysis
Individual genetic ancestry was estimated using an algorithm based on maximum likelihood estimation (MLE) (Tsai et al., 2005) that uses allele frequencies from the parental populations (European – EUR, African – AFR and Native American – AMR). Allelic frequencies were retrieved via genomic mapping in multiple panels of unrelated ethnic samples (Fernandez et al., 2003; Bonilla et al., 2004b; Shriver et al., 2005).
The Kolmogorov-Smirnov test was used to check for normal distribution and Pearsons correlation test was applied to each continuous variable: age, weight, height, body mass index (BMI), fat mass (FM), fat-free mass (FFM), triglycerides (TG), total lipids (TL), total low-density lipoprotein (LDL), very-low density lipoprotein (VLDL), high-density lipoprotein (HDL), fasting plasma glucose (GLU), systolic blood pressure (SBP), diastolic blood pressure (DBP), dietary intake of total lipids (LIP), European ancestry proportion (EUR), African ancestry proportion (AFR) and Native American, or Amerindian, ancestry proportion (AMR). Cases were labeled as positive (affected) or negative (unaffected) with regard to metabolic and clinical features, including medication status determined from medical records, to assemble categorical variables for each metabolic disorder or trait (obesity, hypertriglyceridemia, hypercholesterolemia, hyperlipidemia, dyslipidemia, glucose intolerance, type 2 diabetes, hypertension and normolipid diet). An independent samples t-test was used to compare the means of continuous traits across carriers and non-carriers of the aforementioned metabolic disorders. Linear regression analysis was used to assess possible covariance for ancestry estimates and the variable rendered as interferential was later included in a covariance analysis with Bonferroni adjustments to correct for its effects. A value of p < 0.05 was considered statistically significant for all tests. The software SPSS, version 13 (SPSS Inc., Chicago, IL) was used for statistical calculations.
Results
The Kolmogorov-Smirnov test showed that the data for all continuous traits were normally distributed. The quantitative and categorical traits of the subjects are summarized in Table 1 and Table 2, respectively.
Table 1.
Continuous variables in the studied population (n = 176).
| Variable | Mean | SD | Min | Max |
|---|---|---|---|---|
| Age (years) | 68.31 | 5.77 | 58.00 | 83.00 |
| Weight (kg) | 63.21 | 9.58 | 32.00 | 89.00 |
| Height (m) | 1.52 | 0.06 | 1.38 | 1.72 |
| Body mass index (kg/m2) | 27.28 | 4.04 | 14.61 | 37.69 |
| Fat mass (%) | 37.28 | 6.84 | 8.05 | 57.74 |
| Fat free mass (%) | 58.88 | 6.41 | 39.80 | 87.00 |
| Triglycerides (mg/dL) | 157.24 | 97.11 | 38.00 | 967.00 |
| Total lipids (mg/dL) | 744.73 | 143.26 | 218.00 | 1117.00 |
| Total cholesterol (mg/dL) | 233.24 | 43.07 | 126.00 | 343.00 |
| LDL (mg/dL) | 139.93 | 39.58 | 43.00 | 250.00 |
| VLDL (mg/dL) | 29.84 | 13.12 | 7.60 | 76.00 |
| HDL (mg/dL) | 61.65 | 10.07 | 35.00 | 92.00 |
| Fasting glucose (mg/dL) | 104.33 | 29.67 | 70.00 | 324.00 |
| SBP (mmHg) | 136.42 | 26.54 | 70.00 | 210.00 |
| DBP (mmHg) | 81.16 | 15.74 | 40.00 | 120.00 |
| LIP (%) | 34.31 | 5.15 | 19.09 | 65.52 |
| EUR (%) | 57.49 | 17.21 | 2.00 | 100.00 |
| AFR (%) | 16.73 | 14.73 | 0.00 | 80.00 |
| AMR (%) | 25.78 | 14.50 | 0.00 | 65.00 |
AFR = African ancestry; AMR = Amerindian ancestry; DBP = diastolic blood pressure; EUR = European ancestry; HDL = high density lipoprotein; LDL = low density lipoprotein; LIP = lipid dietary intake; Max = maximum; Min = minimum; SD = standard deviation; SBP = systolic blood pressure; VLDL = very low density lipoprotein.
Table 2.
Categorical variables of the clinical and nutritional assessments in the sampled population.
| Trait | Unaffected
|
Affected
|
||
|---|---|---|---|---|
| n | % | n | % | |
| Obesity | 85 | 48.3 | 91 | 51.7 |
| Hypertriglyceridemia | 92 | 52.3 | 84 | 47.7 |
| Hypercholesterolemia | 41 | 23.3 | 135 | 76.7 |
| Hyperlipidemia | 103 | 58.5 | 73 | 41.5 |
| Dyslipidemia | 23 | 13.1 | 153 | 86.9 |
| Normal lipid diet | 59 | 33.5 | 117 | 66.5 |
| Glucose intolerance | 134 | 76.1 | 42 | 23.9 |
| Type 2 diabetes | 152 | 86.4 | 24 | 13.6 |
| Hypertension | 29 | 16.5 | 147 | 83.5 |
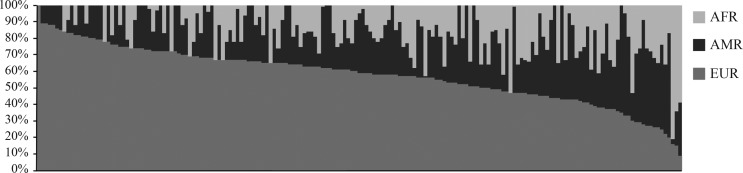
The allelic frequencies of the AIMs in the sample population differed from those reported for reference ancestral populations, thereby indicating the admixture structure (Table 3). The distribution of the individual ancestry estimates (IAE) comprised a wide range of ancestry proportions in a diverse three-hybrid pattern of variation (Figure 1). The sample population displayed a genetic constitution with a marked contribution from European ancestry (57.49%) followed by Native American (25.78%) and African (16.73%) ancestries. Fewer individuals displayed a di-hybrid ancestral arrangement, e.g., EUR-AFR or EUR-AMR.
Table 3.
Ancestry informative markers with chromosomal positions and allele frequencies in Europeans (EUR), Africans (AFR), Amerindians (AMR) and the sampled population.
| Locus | Position | Allele | EUR | AFR | AMR | Sample | Reference |
|---|---|---|---|---|---|---|---|
| FY-null (rs2814778) | 1q23 | C | 0.006 | 0.983 | 0.018 | 0.209 | Smith et al. (2004) |
| rs1129038 | 15q13 | C | 0.224 | 0.996 | 0.983 | 0.783 | Smith et al. (2004) |
| rs3796384 | 3p14 | C | 0.154 | 0.783 | 0.875 | 0.626 | Smith et al. (2004) |
| rs1480642 | 6q23 | C | 0.993 | 0.121 | 0.621 | 0.791 | Smith et al. (2004) |
| rs1871534 | 8q24.3 | C | 0.981 | 0.071 | 1.000 | 1.000 | Smith et al. (2004) |
| rs267071 | 5q22 | C | 0.654 | 0.088 | 1.000 | 0.421 | Smith et al. (2004) |
| rs4280128 | 13q22 | A | 0.357 | 0.941 | 0.034 | 0.494 | Smith et al. (2004) |
| rs6034866 | 20p12 | A | 0.083 | 0.954 | 0.143 | 0.100 | Smith et al. (2004) |
| rs7349 | 10p11.2 | A | 0.067 | 0.969 | 0.000 | 0.221 | Smith et al. (2004) |
| rs803733 | 9q33 | C | 0.880 | 0.013 | 0.410 | 0.636 | Smith et al. (2004) |
| rs1426654 | 15q21 | C | 0.013 | 0.967 | 0.931 | 0.291 | Smith et al. (2004) |
| TSC1102055 (rs2065160) | 1q32 | C | 0.088 | 0.487 | 0.875 | 0.231 | Smith et al. (2004) |
| rs730570 | 14q32 | A | 0.896 | 0.197 | 0.054 | 0.646 | Smith et al. (2004) |
| rs1240709 | 1p36.3 | A | 0.766 | 0.050 | 0.103 | 0.526 | Smith et al. (2004) |
| rs2278354 | 5p15.2 | T | 0.120 | 0.703 | 0.839 | 0.713 | Smith et al. (2004) |
| rs803733 | 9q33 | C | 0.880 | 0.013 | 0.410 | 0.636 | Smith et al. (2004) |
| rs1980888 | 9q22 | T | 0.100 | 0.143 | 0.950 | 0.204 | Shriver et al. (2005) |
| rs1415878 | Xq26 | A | 0.917 | 1.000 | 0.025 | 0.708 | Shriver et al. (2005) |
| CYP3A4 (rs2740574) | 7q22 | G | 0.040 | 0.800 | 0.040 | 0.211 | Fernandez et al. (2003) |
| AT3 (rs3138521) | 1q25 | Insertion | 0.282 | 0.858 | 0.061 | 0.429 | Bonilla et al. (2004a) |
| WI-11153 (rs17203) | 3p12.1 | G | 0.171 | 0.785 | 0.805 | 0.621 | Bonilla et al. (2004a) |
| MID-93 (rs16383) | 22q13.2 | A | 0.220 | 0.739 | 0.895 | 0.391 | Bonilla et al. (2004a) |
| RB (rs2252544) | 13q14 | C | 0.320 | 0.930 | 0.180 | 0.253 | Bonilla et al. (2004a) |
Figure 1.
Individual ancestry estimates for the sampled, admixed population. Each bar represents an individual subject and each color represents the corresponding relative ancestry proportion in relation to a parental population (light gray = African, dark gray = Amerindian and gray = European).
When ancestry proportions were used as continuous traits, significant results were found for the European ancestry proportion, which was positively correlated with weight (r = 0.157, p = 0.037) and BMI (r = 0.165, p = 0.028), and negatively correlated with DBP (r = −0.187, p = 0.013). For the African ancestry proportion, there was a positive significant correlation with LDL (r = 0.159, p = 0.035) and a negative correlation with VLDL (r = −0.185; p = 0.014). For the Native American ancestry proportion, there was a positive correlation only with triglyceride levels (r = 0.150; p = 0.047). The correlation between variants other than ancestry also yielded significant associations and are shown in Table S1 (Supplementary Material).
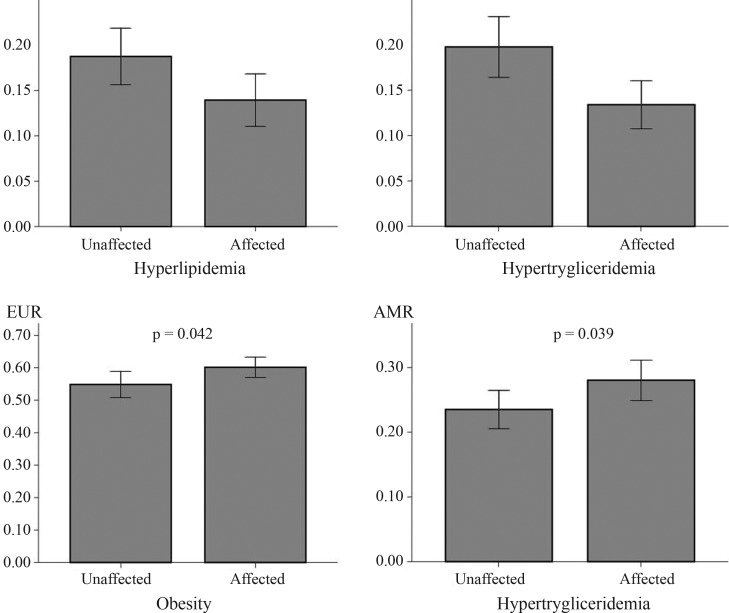
Regarding the affected status, the mean difference (MD) of the African ancestry proportion was significantly different for hypertriglyceridemia (MD = 6.4%, p = 0.004) and hyperlipidemia (MD = 4.8% p = 0.026) (Figure 2). The contribution of European ancestry was significantly different for obesity (MD = 5.3%, p = 0.042) and the Native American ancestry for hypertriglyceridemia (MD = 4.5%, p = 0.039). The mean differences for traits other than those previously described also yielded significant associations and are shown in Table S2 (Supplementary Material). Given that lipid- and obesity-related variables yielded a significant association with ancestry, linear regression analysis used to select these phenotypes as possible covariates for African, Native American and European associations. These analyses revealed that SBP and DBP could be included as covariates for the African ancestry proportion (SBP β coefficient = −0.319, p = 0.014 and DBP β coefficient = 0.389, p = 0.003) and for the European ancestry proportion (SBP β coefficient = 0.256, p = 0.042 and DBP β coefficient = −0.404, p = 0.002). No potential predictors were identified for the Native American ancestry.
Figure 2.
Mean difference in the ancestry coefficients of significant traits. AFR = African ancestry; AMR = Amerindian ancestry; EUR = European ancestry. Error bars indicate the 95% confidence intervals.
There was no divergence in the mean difference of the African ancestry proportion in the category of hypertriglyceridemia for the corrected model with SBP and DBP as covariates. For hyperlipidemia, an increase of 0.1% occurred in the corrected model for the African ancestry proportion (MD from 4.8% to 4.9%, p = 0.034). Using blood pressures as covariates for obesity status, the mean difference of the European ancestry levels among groups increased from 5.3% (p = 0.042) to 6.5% (p = 0.011). No other association was found for covariate-corrected models.
Discussion
The present study identified significant correlations between genetic ancestry and lipid profiles and a fluctuation in the metabolic parameters in a heterogeneous and admixed group of elderly Brazilian women. Interestingly, population ancestry estimates were relatively higher for the Native American and slightly lower for the African and European ancestry proportions than those in the general Brazilian population (Lins et al., 2010) or in other elderly samples (Gentil et al., 2007, 2009; Moreno Lima et al., 2007), however, no significant differences in standard deviations or variance were found between the present sample population and other populations from the same region (data not shown) (Lins et al., 2010, 2011). Although self-reported ancestry is socially correlated with income in most parts of Brazil, income and education status were not assessed in the present study. Notwithstanding, total lipid intake was considered as a covariate that correlates with increased income in Brazilian women (Bonomo et al., 2003) and may represent an additional risk for cholesterol and lipid disorders.
Genetic and environmental factors, such as age, diet and lifestyle are known to contribute to phenotypic traits such as those evaluated in this study. Genetic ancestry has an inherent influence on the etiology of lipid and metabolism-related diseases, as demonstrated in cases such as African ancestry associations with triglyceride, LDL-cholesterol and HDL-cholesterol levels in admixed African-Americans (Deo et al., 2009), or an inverse association in individuals with European ancestry (Basu et al., 2009). In the present study, increased African ancestry was significantly associated with decreased levels of triglyceride-rich serum components (TG and VLDL) and augmented levels of cholesterol-rich lipoproteins (LDL), as has been reported by others (Deo et al., 2009). No correlation was found for HDL levels, as previously reported (Basu et al., 2009; Deo et al., 2009). Although the effect of European ancestry observed here was modest, it was nevertheless significant; based on this finding, we suggest that European ancestry is an additional risk factor and that higher levels of African ancestry account for a more beneficial lipid profile in elderly women with admixed ethnicity.
The levels of LDL expression in African-American women are often described as being higher than those in European-American women. Conversely, increments of triglycerides in lean African-Americans are described as lower than in lean European-American women (Bower et al., 2002). Here, African ancestry exerted a protective effect against hyperlipidemia and hypertriglyceridemia since it was correlated with higher levels of LDL and lower levels of VLDL. Therefore, in this sample population, African ancestry influenced lipid profiles, but, in contrast to previous studies (Fernandez et al., 2003; Lai et al., 2009), was not correlated to obesity, diabetes or hypertension. This lack of association may reflect a difference in genetic stratification (Wulan et al., 2010), and the levels and range of admixture (Lins et al., 2011) may confound results because of pronounced genetic effects on phenotype expression. For this reason, association studies should not be labeled as analyzing deterministic, causal relationships, but rather as an elucidative contribution to the pathophysiology of these complex phenotypes.
The prevalence of obesity varies significantly among ethnic groups, especially African- and European-Americans (Deurenberg et al., 1998). In addition, some studies have demonstrated associations between European ancestry and body composition traits in admixed populations, especially bone mineral density (BMD) (Bonilla et al., 2004b) and BMI (Basu et al., 2009). Others have reported an association between African ancestry and BMI, FM, FFM and BMD in African-American women (Fernandez et al., 2003). In the present admixed cohort, European ancestry was significantly correlated with BMI and with obesity, but in a previous study (Gentil et al., 2009) this association was not detected. Fat mass and fat-free mass were not correlated with any ancestry estimate, neither in the present nor in a previous study of elderly Brazilian women (Moreno Lima et al., 2007). Such associations are particularly important for understanding the role of genetic ancestry in metabolic-related traits and how interacting variables modulate these phenotypes.
Although lipid intake had no effect on overall traits, we do not advocate excluding this variable in future assessments in view of possible environmental influences on the risk of obesity and metabolic syndrome (Sichieri, 2002) or heart disease (Nobrega et al., 2011). Notably, Brazilian women display significant differences in their pattern of lipid macronutrient consumption, with a trend toward increased consumption with increased income (Bonomo et al., 2003) and a suggestive risk for heart disease (Nobrega et al., 2011). With regard to ethnicity, in a study of the dietary pattern of a Brazilian-urban population, a correlation was identified between categories of skin color (defined by the interviewer) and dietary patterns, in which lighter skin was associated with a more varied diet (Sichieri et al., 2003). However, this study was not income-adjusted, and categorical ethnicity groups might comprise a wide degree of admixture (Lins et al., 2011). Consequently, other factors relating ancestry and dietary patterns to obesity and comorbidities should be investigated.
Other associations found in this study are of interest to the clinical status of elderly women. However, they are merely well-established descriptions of metabolic health risk conditions, such as the association between lipid profile, diabetes and/or hypertension, but do not represent a novel addition to our current knowledge. The supplemental tables, available as part of the online article, contain all the association tests and their significance. Although a body of evidence advocates that the overall ancestry background influences the lipid profile of individuals, the novelty of the present paper is the approach used to address this matter. The Brazilian population displays an extensive degree of admixture that it renders suitable for estimating a variety of ancestries under standardized socio-economic conditions (Lins et al., 2011).
The relatively small sample size used may be a limitation of this study. However, the sample size was sufficiently large to significantly corroborate previous reports in other populations. This lends plausibility to the present data since the replication of positive findings is an important requirement for genetic studies. This scenario might help identify the relationship between ancestry backgrounds and specific phenotypes. The present study may also have been subject to other unmeasured confounders, such as the important contribution of individual interactions with environmental factors. Lifestyle behaviors in the elderly population that may be used as a covariant, such as smoking, alcohol intake, stress or depression, were not assessed here. Since this study used a cohort of elderly women, these results cannot be extended to another population strata or gender.
In conclusion, our results support the role of genetic ancestry in chronic non-communicable diseases prevalent in elderly women and suggest that caution should be taken when performing gene-based association studies in admixed populations because of the genetic heterogeneity. Our results raise the possibility of undertaking association studies based on admixture linkage disequilibrium; such studies may identify genomic loci that could explain correlations between lipid metabolic traits and ancestry.
Acknowledgments
The authors thank Cristiane Urcina Lima, Fernanda Colares-Bento and Sarah Peres da Silveira for gathering the nutritional data and for ensuring adherence to the food registration procedure. This research was supported by CNPq (grants 484318/2006-3 and 402699/2007-6) and FAPDF (grants 193.000.309/2007 and 193.000.449/2008). R.S. Paula was supported by a studentship from CAPES (PROSUP), A.S. Pires was supported by a student scholarship from CNPq (grant 550315/2008-0) and R.W. Pereira was supported by a research fellowship from CNPq.
Footnotes
Associate Editor: Carlos F.M. Menck
Supplementary Material
The following online material is available for this article:
- Table S1 - Pearson correlation for continuous traits.
- Table S2 - Independent-samples t-test comparing the means of continuous traits among categorical groups.
This material is available as part of the online article from http://www.scielo.br/gmb.
References
- Basu A, Tang H, Arnett D, Gu CC, Mosley T, Kardia S, Luke A, Tayo B, Cooper R, Zhu X, et al. Admixture mapping of quantitative trait loci for BMI in African Americans: Evidence for loci on chromosomes 3q, 5q, and 15q. Obesity. 2009;17:1226–1231. doi: 10.1038/oby.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla C, Shriver MD, Parra EJ, Jones A, Fernandez JR. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York city. Hum Genet. 2004a;115:57–68. doi: 10.1007/s00439-004-1125-7. [DOI] [PubMed] [Google Scholar]
- Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet. 2004b;68:139–153. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- Bonomo E, Caiaffa WT, Cesar CC, Lopes AC, Lima-Costa MF. Food intake according to socioeconomic and demographic profile: The Bambui Project. Cad Saude Publica. 2003;19:1461–1471. doi: 10.1590/s0102-311x2003000500025. (in Portuguese, Abstract in English). [DOI] [PubMed] [Google Scholar]
- Bower JF, Deshaies Y, Pfeifer M, Tanenberg RJ, Barakat HA. Ethnic differences in postprandial triglyceride response to a fatty meal and lipoprotein lipase in lean and obese African American and Caucasian women. Metabolism. 2002;51:211–217. doi: 10.1053/meta.2002.29991. [DOI] [PubMed] [Google Scholar]
- Cabrera MA, Gebara OC, Diament J, Nussbacher A, Rosano G, Wajngarten M. Metabolic syndrome, abdominal obesity, and cardiovascular risk in elderly women. Int J Cardiol. 2007;114:224–229. doi: 10.1016/j.ijcard.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Deo RC, Reich D, Tandon A, Akylbekova E, Patterson N, Waliszewska A, Kathiresan S, Sarpong D, Taylor HA, Jr, Wilson JG. Genetic differences between the determinants of lipid profile phenotypes in African and European Americans: The Jackson Heart Study. PLoS Genetics. 2009;5:e1000342. doi: 10.1371/journal.pgen.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg P, Yap M, van Staveren WA. Body mass index and percent body fat: A meta analysis among different ethnic groups. Int J Obes Relat Metab Disord. 1998;22:1164–1171. doi: 10.1038/sj.ijo.0800741. [DOI] [PubMed] [Google Scholar]
- Diretrizes V. Brazilian guidelines in arterial hypertension. Arq Bras Cardiol. 2007;89:e24–79. (in Portuguese). [PubMed] [Google Scholar]
- Fernandez JR, Shriver MD, Beasley TM, Rafla-Demetrious N, Parra E, Albu J, Nicklas B, Ryan AS, McKeigue PM, Hoggart CL, et al. Association of African genetic admixture with resting metabolic rate and obesity among women. Obes Res. 2003;11:904–911. doi: 10.1038/oby.2003.124. [DOI] [PubMed] [Google Scholar]
- Gentil P, Lima RM, Lins TC, Abreu BS, Pereira RW, Oliveira RJ. Physical activity, Cdx-2 genotype, and BMD. Int J Sports Med. 2007;28:1065–1069. doi: 10.1055/s-2007-965130. [DOI] [PubMed] [Google Scholar]
- Gentil P, de Lima Lins TC, Lima RM, de Abreu BS, Grattapaglia D, Bottaro M, de Oliveira RJ, Pereira RW. Vitamin-D-receptor genotypes and bone-mineral density in post-menopausal women: Interaction with physical activity. J Aging Phys Act. 2009;17:31–45. doi: 10.1123/japa.17.1.31. [DOI] [PubMed] [Google Scholar]
- Lai CQ, Tucker KL, Choudhry S, Parnell LD, Mattei J, Garcia-Bailo B, Beckman K, Burchard EG, Ordovas JM. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum Genet. 2009;125:199–209. doi: 10.1007/s00439-008-0612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins TC, Nogueira LR, Lima RM, Gentil P, Oliveira RJ, Pereira RW. A multiplex single-base extension protocol for genotyping Cdx2, FokI, BsmI, ApaI, and TaqI polymorphisms of the vitamin D receptor gene. Genet Mol Res. 2007;6:216–224. [PubMed] [Google Scholar]
- Lins TC, Vieira RG, Abreu BS, Grattapaglia D, Pereira RW. Genetic composition of Brazilian population samples based on a set of twenty-eight ancestry informative SNPs. Am J Hum Biol. 2010;22:187–192. doi: 10.1002/ajhb.20976. [DOI] [PubMed] [Google Scholar]
- Lins TC, Vieira RG, Abreu BS, Gentil P, Moreno-Lima R, Oliveira RJ, Pereira RW. Genetic heterogeneity of self-reported ancestry groups in an admixed Brazilian population. J Epidemiol. 2011;21:240–245. doi: 10.2188/jea.JE20100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Lima R, Silva de Abreu B, Gentil P, Cesar de Lima Lins T, Grattapaglia D, Pereira RW, Jaco de Oliveira R. Lack of association between vitamin D receptor genotypes and haplotypes with fat-free mass in postmenopausal Brazilian women. J Gerontol A Biol Sci Med Sci. 2007;62:966–972. doi: 10.1093/gerona/62.9.966. [DOI] [PubMed] [Google Scholar]
- Nobrega OT, Faleiros VP, Telles JL. Gerontology in the developing Brazil: Achievements and challenges in public policies. Geriatr Gerontol Int. 2009;9:135–139. doi: 10.1111/j.1447-0594.2008.00499.x. [DOI] [PubMed] [Google Scholar]
- Nobrega OT, Paula RS, Silveira SR, Pires AS, Toledo JO, Moraes CF, Cordova C. Usual dietary intake and cardiovascular risk factors in older Brazilian women. Aging Clin Exp Res. 2011 doi: 10.3275/7674. [DOI] [PubMed] [Google Scholar]
- Paula RS, Souza VC, Benedet AL, Souza ER, Toledo JO, Moraes CF, Gomes L, Alho CS, Cordova C, Nobrega OT. Dietary fat and apolipoprotein genotypes modulate plasma lipoprotein levels in Brazilian elderly women. Mol Cell Biochem. 2010;337:307–315. doi: 10.1007/s11010-009-0313-0. [DOI] [PubMed] [Google Scholar]
- Perissinotto E, Pisent C, Sergi G, Grigoletto F. Anthropometric measurements in the elderly: Age and gender differences. Br J Nutr. 2002;87:177–186. doi: 10.1079/bjn2001487. [DOI] [PubMed] [Google Scholar]
- Philippi ST. Tabela de Composição de Alimentos: Suporte para Decisão Nutricional. Coronário, São Paulo. 2002:135. [Google Scholar]
- Ruiz-Narvaez EA, Bare L, Arellano A, Catanese J, Campos H. West African and Amerindian ancestry and risk of myocardial infarction and metabolic syndrome in the Central Valley population of Costa Rica. Hum Genet. 2010;127:629–638. doi: 10.1007/s00439-010-0803-x. [DOI] [PubMed] [Google Scholar]
- Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, Schurr TG, Zhadanov SI, Osipova LP, Brutsaert TD, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichieri R. Dietary patterns and their associations with obesity in the Brazilian city of Rio de Janeiro. Obes Res. 2002;10:42–48. doi: 10.1038/oby.2002.6. [DOI] [PubMed] [Google Scholar]
- Sichieri R, Castro JF, Moura AS. Factors associated with dietary patterns in the urban Brazilian population. Cad Saude Publica. 2003;19(Suppl 1):S47–53. doi: 10.1590/s0102-311x2003000700006. (in Portuguese, Abstract in English). [DOI] [PubMed] [Google Scholar]
- Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sposito AC, Caramelli B, Fonseca FA, Bertolami MC, Afiune Neto A, Souza AD, Lottenberg AM, Chacra AP, Faludi AA, Loures-Vale AA, et al. IV Brazilian Guideline for Dyslipidemia and Atherosclerosis Prevention: Department of Atherosclerosis of the Brazilian Society of Cardiology. Arq Bras Cardiol. 2007;88(Suppl 1):2–19. doi: 10.1590/s0066-782x2007000700002. (in Portuguese, Abstract in English). [DOI] [PubMed] [Google Scholar]
- Sullivan PW, Morrato EH, Ghushchyan V, Wyatt HR, Hill JO. Obesity, inactivity, and the prevalence of diabetes and diabetes-related cardiovascular comorbidities in the U.S., 2000–2002. Diabetes Care. 2005;28:1599–1603. doi: 10.2337/diacare.28.7.1599. [DOI] [PubMed] [Google Scholar]
- Tsai H-J, Choudhry S, Naqvi M, Rodriguez-Cintron W, Burchard E, Ziv E. Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Hum Genet. 2005;118:424–433. doi: 10.1007/s00439-005-0067-z. [DOI] [PubMed] [Google Scholar]
- Wilson PW, Kannel WB. Obesity, diabetes, and risk of cardiovascular disease in the elderly. Am J Geriatr Cardiol. 2002;11:119–125. doi: 10.1111/j.1076-7460.2002.00998.x. [DOI] [PubMed] [Google Scholar]
- Wulan SN, Westerterp KR, Plasqui G. Ethnic differences in body composition and the associated metabolic profile: A comparative study between Asians and Caucasians. Maturitas. 2010;65:315–319. doi: 10.1016/j.maturitas.2009.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
- Table S1 - Pearson correlation for continuous traits.
- Table S2 - Independent-samples t-test comparing the means of continuous traits among categorical groups.