Abstract
Transgene integration into plant genomes is a complex process accompanied by molecular rearrangements. Classic methods that are normally used to study transgenic population genetics are generally inadequate for assessing such integration. Two major characteristics of transgenic populations are that a transgenic genome may harbor many copies of the transgene and that molecular rearrangements can create an unstable transgenic locus. In this work, we examined the segregation of T1, T2 and T3 transgenic tobacco progenies. Since transfer DNA (T-DNA) contains the NptII selectable marker gene that confers resistance to kanamycin, we used this characteristic in developing a method to estimate the number of functional inserts integrated into the genome. This approach was based on calculation of the theoretical segregation ratios in successive generations. Mendelian ratios of 3:1, 15:1 and 63:1 were confirmed for five transformation events whereas six transformation events yielded non-segregating progenies, a finding that raised questions about causal factors. A second approach based on a maximum likelihood method was performed to estimate recombination frequencies between linked inserts. Recombination estimates varied among transformation events and over generations. Some transgenic loci were unstable and evolved continuously to segregate independently in the T3 generation. Recombination and amplification of the transgene and filler DNA yielded additional transformed genotypes.
Keywords: copy number variation, gene rearrangements, genetic inheritance, transgenic
Introduction
Plant transformation mediated by Agrobacterium tumefaciens has become the most used method for introducing foreign genes into plant cells. This method yields a high level of perfect transgenic loci with complete conservation of the host genome (Pawlowski and Somers, 1996). The mechanisms involved in the integration of transfer DNA (T-DNA) are still not well characterized, although integration is considered to occur by illegitimate recombination (Tinland, 1996; Gorbunova and Levy, 1997; Salomon and Puchta, 1998; Britt, 1999; Brunaud et al., 2002; Van Attikum and Hooykaas, 2003). The structure of transgenic loci depends on genomic factors and does not depend on the method how the transgene is transferred to the genome (Somers and Makarevitch, 2004). The complexity of integration mechanisms leads to transgenic loci consisting of two or many copies of the transgene (De Neve et al., 1997; Takano et al., 1997). Frequently, copies of the transgene are arranged in the same direction and separated by filler DNA (Krizkova and Hrouda, 1998). Integration is often associated with complex rearrangements including deletions, filler DNA, inversions and duplication of the original inserted sequence (Jones et al., 1994; Zhu et al., 2010).
Transgenes are inherited sexually as a dominant trait (Christou et al., 1989; Misra, 1989; Pawlowski and Somers, 1996; Theuns et al., 2002), with inheritance conforming to a 3:1 Mendelian ratio (Srivatava et al., 1996; Chen et al., 1998; Perret et al., 2003; Yong et al., 2006; Shrawat et al., 2007). Non-Mendelian segregation occurs at a frequency of 10%–50% through either unstable transmission of the transgene or poor expression (Deroles and Gardner, 1988; Register et al., 1994; McCabe et al., 1999; Limanton-Grevet and Julien, 2001). Many factors influence transgene expression and inheritance, including the transgene itself, the host genome and the interaction between them (Prols and Meyer, 1992; Makarevitch et al., 2003; Yin et al., 2004; Shrawat et al., 2007).
Intra-chromosomal recombination occurs during meiosis and mitosis, with spontaneous recombination normally being a rare event (10−6-10−5 events per cellular division). Embryogenic cells have the highest recombination ability, with an average of 3 × 10−5 recombination events per genome (Yang et al., 2010). The frequency of recombination can be strongly increased by T-DNA integrations, which cause double-strand breaks (Gorbunova and Levy, 1999; Wehrkamp-Richter et al., 2009), or by other factors related to stress (Dong, 2004). Recombination between copies of the transgene has been reported for transgenic loci in various plant species (Jones et al., 1985; Eckes et al., 1986; Christou et al., 1989; Tovar and Lichtenstein, 1992; Choffens et al., 2001).
The objective of this study was to examine the number of functional inserts, the mode of transgene inheritance and the recombination frequencies of linked inserts in the first three generations of transgenic tobacco lines.
Materials and Methods
Transgenic material
Nicotiana tabacum L plants were genetically transformed with Agrobacterium tumefaciens by using the leaf disk method. The T-DNA consisted of a traditional cassette made up of the neomycin phosphotransferase selection marker gene (NptII) driven by the nopaline synthase promoter and the β-glucuronidase reporter gene (GUS) under control of the CaMV 35S promoter (Sanders et al., 1987). Only the expression and inheritance of NptII that conferred resistance to aminoglycosidic antibiotics was studied. Transformed plants were regenerated on selective medium containing kanamycin and were grown in a chamber under carefully controlled conditions. After three weeks, transformed plants were transferred to larger pots and kept in a greenhouse with no pollinating insects or wind in order to avoid pollen dispersal and hybridization. At maturity, seeds from each plant (T0) were collected in a single tube and preserved in bottles with silica gel to avoid humidification and the loss of germination capacity (S Thaminy, unpublished data). Seeds from each plant (line T0) were cultured on medium containing MS salts (Murashige and Skoog, 1962) and 100 mg of kanamycin/L in Petri dishes; this concentration of antibiotic was sufficient to select transformed plants (Klein et al., 1988; Tavazza et al., 1988; Misra, 1989). In this assay, non-transformed seedlings turned brown and died while transformed seedlings survived and grew healthy. T1 transgenic plants (n = 25–30) were transferred to larger pots and kept in a greenhouse. At maturity, seeds from each line were collected and mixed to form the “bulk” for the T2 generation. Subsequently, 50–100 seeds of the bulk were cultured on selective medium and the T2 resistant plants (n = 25–30) were transferred to soil. At maturity, seeds from each transformed plant were harvested and tested for resistance to kanamycin. Table 1 summarizes the experimental protocol.
Table 1.
Experimental protocol.
| T0 generation | T0 individuals | Line L1 | |||
|---|---|---|---|---|---|
| T1 generation | T0 seeds |
 50–100 seeds 50–100 seeds |
|||
 resistance test resistance test |
|||||
| T1 individual | 25–30 resistant individuals | ||||
| L1-X | |||||
|
| |||||
| T2 generation | T1 seeds |
 50–100 seeds 50–100 seeds |
|||
 resistance test resistance test |
|||||
| T2 individual | 25–30 resistant individuals | ||||
| L1-X-1,... L1-X-2. L1-X-10 | |||||
|
| |||||
| T3 generation | T2 seeds |
L1-X-1

|
L1-X-2

|
L1-X-10

|
|
| T3 individual | 50–100 seeds

|
50–100 seeds

|
50–100 seeds

|
||
| resistance test | |||||
Segregation analysis
Segregation analysis was done using the χ2 test in which observed values were compared to theoretical values corresponding to the integration of one or more copies of the transgene.
Estimation of recombination frequencies
Recombination frequencies were estimated using a genetic approach derived from the maximum likelihood method. Suppose that there are two linked inserts in the host genome, with both insertions (genes) having two allelic forms (K1R and K1S for the first gene and K2R and K2S for the second gene). KR indicates the presence of a functional insert whereas KS indicates the absence of the insert. The two transgenes are inserted in cis or in trans. The distance separating the physically linked inserts is defined as d and r is the recombination frequency. When r ≥ 50% the two transgenes segregate independently and when r < 50% the two transgenes recombine with a frequency r. Thus, for two linked inserts in cis:
| (1) |
| (2) |
where n1 is the frequency of sensitive seedlings, n2 is the frequency of resistant seedlings and n1 + n2 = 1. The probability of observing n1 or n2 follows multinomial law:
| (3) |
| (4) |
| (5) |
The distance between two inserts is defined as:
| (6) |
For two linked inserts in trans, the recombination frequency was estimated as described above for insertion in cis, but with . Similarly, for cases involving three transgenes with two inserts linked in cis or trans the recombination frequency was estimated based on the principles outlined above; Table 2 summarizes the calculations involved in this analysis.
Table 2.
Estimation of ‘r’ in different cases of linkage.
| Linkage in cis | Linkage in trans | |
|---|---|---|
| Two inserts | ||
| Three inserts of which two are linked |
d: distance between two linked inserts, r: frequency of recombination between two linked inserts, n1: frequency of sensitive seedlings, n2: frequency of resistant seedlings.
Results
Theoretical considerations
Segregation ratios depend on the number of functional inserts integrated into T0 plants. The greater the number of copies of independently segregating inserts in the genome, the greater the probability of obtaining KR gametes and, consequently, the ratio of kanamycin-resistant plants increases. The marker gene NptII is considered a dominant trait. Self-pollinated tobacco plants were grown in a greenhouse to avoid inter-crosses. In this model, there are two variables: generation (T1, T2,... Tn ,Tn+1) and the number of inserts (I1, I2, ..., In, In+1). The following equations were used to calculate theoretical values in transgenic populations:
| (7) |
where XTn is the number of possible genotypes or zygotes in generation Tn, XTn+1 is the number of possible genotypes or zygotes in generation Tn+1 and y is the number of possible gametes that depends on the number of insertions. Generation varies while y is stable.
| (8) |
where yIn is the number of possible gametes in the case of n inserts and yIn+1 is the number of possible gametes in the case of n + 1 inserts.
| (9) |
Equation (9) is applied in the case of one insert where xTn+1 is the number of sensitive seedlings (KS//KS) in generation Tn+1, xT1 is the number of sensitive seedlings (KS//KS) in the case of one insert in T1, xTn is the number of sensitive seedlings (KS//KS) in generation Tn and XT1 is the total number of possible zygotes in T1. This equation can be extended to the case of two, three,... In, In+1 inserts:
| (10) |
where xIn+1 is the number of sensitive seedlings (KS//KS) in the case of n + 1 inserts and xI1 is the number of sensitive seedlings in the case of one insert. When the progenies of each T0 line were tested on selective medium, sensitive homozygous seedlings (KS//KS) died. This selection was taken into account when calculating the theoretical segregation ratios (Table 3).
Table 3.
Genotypic frequencies of self-pollinated plants harboring a single copy of the transgene.
| Absence of selectiona
|
Presence of selectionb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| KR//KR | KR//KS | KS//KS | Segregation ratio | KR//KR | KR//KS | KS//KS | Segregation ratio | |
| T0 | - | 1 | - | 1:0 | - | 1 | - | 1:0 |
| T1 | 1/4 | 1/2 | 1/4 | 3:1 | 1/4 | 1/2 | 1/4 | 3:1 |
| T2 | 3/8 | 2/8 | 3/8 | 5:3 | 3/6 | 2/6 | 1/6 | 5:1 |
T0 individuals were heterozygous for one copy of the transgene.
Plants germinated on non-selective medium without kanamycin,
Plants germinated on selective medium with kanamycin, KR//KR: kanamycin-resistant homozygous genotype, KR//KS: kanamycin-resistant heterozygous genotype and Ks//KS: kanamycin-sensitive homozygous genotype.
When the number of inserts was greater than one copy, Eq. (11) was used to calculate the theoretical number of kanamycin-sensitive homozygous plants in each generation. Sensitive homozygous plants unable to grow on selective medium did not participate in reproduction in the next generation and were excluded from calculations. The populations tended to be homozygous at equilibrium.
| (11) |
where xTn+1 is the number of sensitive seedlings (KS//KS) in generation Tn+1 with selection against (KS//KS) in generation Tn.
Identification of transgenic plants
Eleven self-pollinated transgenic tobacco lines considered as separate transformation events were analyzed (Table 4). Kanamycin-resistant plants had green leaves, were well rooted and developed on selective medium. In contrast, sensitive-kanamycin plants had yellow leaves and weak rooting with delayed growth; these plants died early at the two-leaf stage. The frequency of kanamycin-resistant plants varied among progenies.
Table 4.
Kanamycin-resistance test in T1 and T2 generations.
| T1 generation
|
T2 generation
|
||||
|---|---|---|---|---|---|
| Lines | KR | KS | Lines | KR | KS |
| L1 | 61 | 0 | L1-x | 32 | 0 |
| L7 | 30 | 0 | L7-x | 79 | 0 |
| L2 | 59 | 0 | L2-x | 255 | 3 |
| L3r | 38 | 5 | L3r-x | 105 | 0 |
| L14r | 195 | 31 | L14r-x | 331 | 0 |
| L16r | 68 | 13 | L16r-x | 177 | 0 |
| L4 | 134 | 45 | L4-x | 324 | 96 |
| L6 | 53 | 3 | L6-x | 155 | 11 |
| L17 | 124 | 34 | L17-x | 275 | 24 |
| L17r | 53 | 9 | L17r-x | 317 | 29 |
| L4r | 45 | 21 | L4r-x | 142 | 37 |
KR and KS: kanamycin-resistant and kanamycin-sensitive phenotypes, respectively.
Lines L1 and L7 had homogenous progenies with a kanamycin-resistant phenotype in T1 and T2 generations. There was no segregation in either generation, possibly because the parent plants were homozygous for one or more copies of the transgene.
Line L2 progeny was homogenous for kanamycin-resistance in T1 and heterogeneous in T2. The segregation in T2 did not reflect the hypothesis of a homozygous locus. This finding indicated that the genome harbored many copies of the transgene since the frequency of resistant plants depends on number of expressed inserts.
Lines L3r, L14r and L16r had both kanamycin-resistant and kanamycin-sensitive plants in T1, but in T2 all progenies were kanamycin-resistant. The increase in transgene expression may have resulted from the amplification of one or more copies of the original insert or, alternatively, the transgenic locus may have become homozygous in T2.
Lines L4, L4r, L6, L17 and L17r had heterogeneous progenies in both generations and the respective parents may have been heterozygous for one or more copies of the transgene. Segregating progenies are the primary material for studying transgene inheritance and recombination.
Inter-transformant variability is attributable to the fact that each transgenic line is the result of a single, separate transformation event, with one or more NpTII transgenes being inserted at a single locus, or inserted independently at segregating loci. Consequently, screening a transgenic line based only on transgene expression is insufficient; it is more convenient to screen for plants with a single copy of the transgene.
Segregation analysis
Hypotheses of segregation can be tested only for heterogeneous progenies (Table 5). The progenies of L2 were homogenous with regard to the resistant phenotype in T1. The number of individuals screened was much greater in T2, in which a sensitive phenotype was observed. The χ2 test was significant for one, two and three inserts, suggesting that this line may harbor more than three copies of the transgene.
Table 5.
χ2 test results for transgene segregation in the T1 and T2 generations.
| Lines | Number of inserts in T1 generation
|
Number of inserts in T2 generation
|
||||
|---|---|---|---|---|---|---|
| Onea 3:1b | Twoa 15:1b | Threea 63:1b | Onea 5:1b | Twoa 55:5b | Threea 485:19b | |
| L3r | 0.74 (ns) | 61.17 (hs) | 393.3 (hs) | - | - | - |
| L16r | 3.46 (ns) | 13.27 (hs) | 110.5 (hs) | - | - | - |
| L14r | 15.35 (hs) | 21.50 (hs) | 217 (hs) | - | - | - |
| L2 | - | - | - | 44.05 (hs) | 17.10 (hs) | 4.73 (s) |
| L4 | 0.00 (ns) | 109 (hs) | 646.9 (hs) | 11.59 (hs) | 115.9 (hs) | 421.8 (hs) |
| L6 | 11.52 (hs) | 0.08 (ns) | 5.24 (s) | 12.05 (hs) | 0.63 (ns) | 3.73 (ns) |
| L17 | 1.02 (ns) | 62.87 (hs) | 409.1 (hs) | 16.07 (hs) | 0.04 (ns) | 14.94 (hs) |
| L17r | 3.63 (ns) | 7.23 (hs) | 67.64 (hs) | 17.10 (hs) | 0.00 (ns) | 20.28 (hs) |
| L4r | 1.64 (ns) | 73.64 (hs) | 392.8 (hs) | 2.07 (ns) | 35.67 (hs) | 140.9 (hs) |
number of inserts,
theoretical segregation ratios, ns: not significant, s: significant, hs: highly significant, (-) homogenous resistant progenies were not tested because no segregation was observed.
Lines L3r, L14r and L16r had heterogeneous progenies in T1. The χ2 test suggested the presence of more than one insert for lines L3r and L16r, whereas for L14r, this test was significant for one, two and three inserts. The corresponding progenies became homogenous and had a kanamycin-resistant phenotype in T2, making it impossible to test the hypothesis of segregation in this generation. The instability of these transgenic lines may be attributable to amplification of the original transgenic loci or other complex rearrangements.
Lines L4, L4r, L6, L17 and L17r had heterogeneous progenies in T1 and T2. Line L4r had a stable 3:1 segregation ratio in both generations, in agreement with the presence of a single functional insert. For line L4, the hypothesis of a single insert was confirmed in T1 but not verified in T2. A 15:1 segregation ratio was confirmed for L6 in T1 and T2 and the hypothesis of three inserts was also confirmed in T2. For lines L17 and L17r, the χ2 test was not significant for a single insert in T1 and not significant for two inserts in T2. The frequency of resistant individuals increased in T2, possibly as a result of recombination and amplification.
Genetic recombination analysis
When inserts were physically linked (Figure 1), the distance between the two inserts and their position (cis or trans) need to be considered. Only distances between 0 and 0.5 Morgans were considered (Table 6). This analysis in lines L3r and L16r confirmed the hypothesis of a single insert. The estimated recombination frequencies between two linked inserts in cis were 32% for L3r and 20% for L16r. Linked inserts were not sufficiently far apart to segregate independently, which explains why the χ2 test was not significant for a single copy of the transgene. For L14r, the χ2 test was not significant for one, two and three transgene copies. The estimated recombination frequency between two linked inserts in cis was 26%.
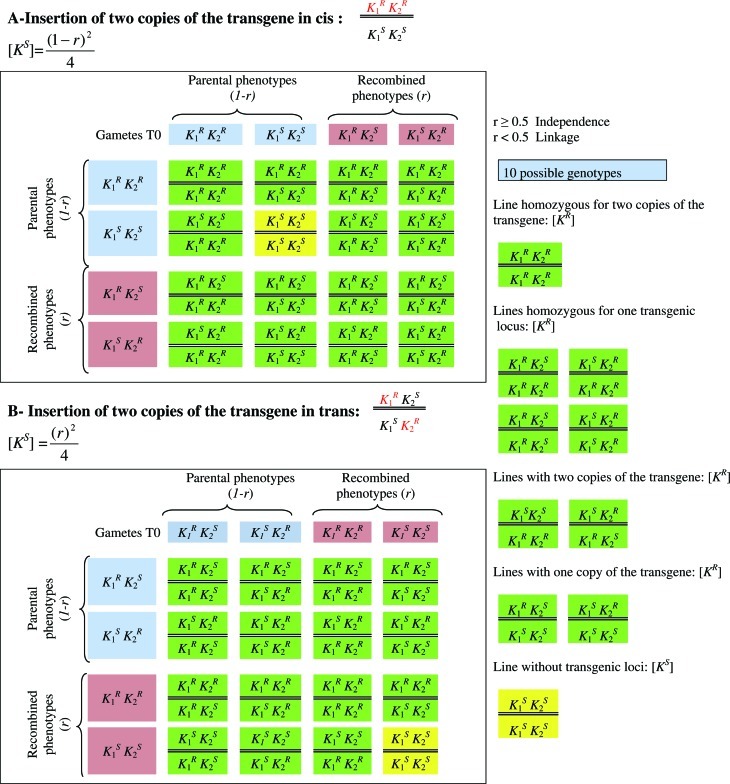
Figure 1.
Segregation of two copies of the transgene linked in cis (A) or trans (B). K1R and K2R indicate transgenic loci whereas K1S and K2S indicate absence of the transgene on the chromosome. [KR] and [KS] are the kanamycin-resistant and kanamycin-sensitive phenotypes, respectively. r - recombination frequency between the linked inserts. Gametes T0 are gametes produced by T0 transformed plants. When r ≥ 0.5, the two transgenes segregate independently while for r < 0.5 the two transgenes recombined with a frequency (r) equal to the distance separating the physically linked inserts.
Table 6.
Estimated distances between linked inserts (Morgan) in the T1 and T2 generations.
| T1 generation | T2 generation | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Lines | Two linked inserts
|
Three inserts of which two are linked
|
Two linked inserts
|
Three inserts of which two are linked
|
||||
| Cis | Trans | Cis | Trans | Cis | Trans | Cis | Trans | |
| L3r | 0.32 | 0.68 | −0.36 | 1.36 | 1 | 0 | 1 | 0 |
| L16r | 0.2 | 0.8 | 0.6 | 0.4 | 1 | 0 | 1 | 0 |
| L14r | 0.26 | 0.74 | −0.48 | 1.48 | 1 | 0 | 1 | 0 |
| L2 | 1 | 0 | 1 | 0 | 0.8 | 0.2 | 0.6 | 0.4 |
| L4 | 0 | 1 | −1.01 | 2.01 | 0.04 | 0.96 | −0.91 | 1.91 |
| L6 | 0.54 | 0.46 | 0.07 | 0.93 | 0.49 | 0.51 | −0.03 | 1.03 |
| L17 | 0.07 | 0.97 | −0.86 | 1.86 | 0.43 | 0.57 | −0.13 | 1.13 |
| L17r | 0.24 | 0.76 | −0.52 | 1.52 | 0.42 | 0.58 | −0.16 | 1.16 |
| L4r | −0.13 | 1.13 | −1.26 | 2.26 | 0.09 | 0.91 | −0.82 | 1.82 |
For L2 in T2, the χ2 test was highly significant for the presence of two copies and significant for three inserts. Based on estimated recombination frequencies, we concluded that L2 harbored three inserts, of which two were linked in trans and recombined with a frequency of 40%.
T1 of line L4 confirmed the hypothesis of one copy, with a recombination frequency of 0%. Hypotheses of one and two independent inserts were not confirmed in T2 and the estimated recombination frequency between two linked inserts in cis was very low (4%). For line L4r, the χ2 test confirmed the hypothesis of a single insert in T1 and T2 and the recombination frequencies were not considered in these cases.
For line L6, hypothesis of two inserts was confirmed in T1 and T2, both of which had high recombination frequencies (46% in T1 and 49% in T2), indicating that the two inserts segregated independently.
For line L17, in which the hypothesis of a single insert was confirmed in T1, the recombination frequency was low (7%). In T2, hypothesis of two inserts was confirmed with a recombination frequency of 43%. These results suggested the presence of two tightly linked inserts in cis in T1, with a low recombination frequency. The transgenic locus evolved in T2 and inserts were far enough apart to recombine with a high frequency.
For line L17r in T1, two linked inserts in cis recombined with a frequency of 24%, which explained why the χ2 test was not significant for the presence of a single insert. The recombination frequency increased sufficiently (to 42%) to allow independent segregation in T2.
The findings described above indicated that L4, L17 and L17r carried unstable transgenic loci. Since filler DNA is a property of complex integration and amplification was detected at sites of insertion, we hypothesized that linked inserts were far apart because filler DNA may have been amplified. Similarly, the original copy of the transgene could have been amplified or duplicated. Molecular approaches could be used to confirm these hypotheses.
T3 Generation analysis and screening good events of transformation
For each line with heterogeneous progenies in T1 and T2 we separately analyzed six T3 progenies obtained by self-pollination of T2 individuals (Table 7). Lines L4-x-1, L4-x-2 and L4-x-3 had heterogeneous progenies with a ratio of 15:1, indicating the segregation of two independent inserts. The corresponding recombination frequencies were high (44%, 40% and 46%, respectively). These lines might be heterozygous with two copies of the transgene. Two progenies originated from L4-x-5 and L4-x-6 progenies were homogenous for the kanamycin-resistant phenotype, indicating that the corresponding parents might be homozygous for one or two copies of the transgene. For line L4-x-4, the presence of one, two and three copies of the transgene was not confirmed and the recombination frequency between two linked inserts in cis was 27%. Line L4 was possibly heterozygous for two tightly linked inserts in cis in T1, with recombination frequencies varying between 27% and 46%. The original parent may also have been heterozygous for a single insert which was amplified in T3.
Table 7.
Segregation ratios, χ2 test results and distances between linked inserts (Morgan) in the T3 generation.
| Number of inserts
|
Two linked inserts
|
Three inserts of which two are linked
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Lines | KRa | KSa | Oneb | Twob | Threeb | Cisc | Transc | Cisc | Transc |
| L4-x-1 | 82 | 7 | 13.94 (hs) | 0.40 (ns) | 22.99 (hs) | 0.44 | 0.56 | −0.12 | 1.12 |
| L4-x-2 | 10 | 1 | 1.48 (ns) | 0.15 (ns) | 4.05 (s) | 0.40 | 0.60 | −0.21 | 1.21 |
| L4-x-3 | 75 | 6 | 13.37 (hs) | 0.19 (ns) | 17.99 (hs) | 0.46 | 0.54 | −0.09 | 1.09 |
| L4-x-4 | 91 | 14 | 7.62 (hs) | 8.99 (hs) | 94.59 (hs) | 0.27 | 0.73 | −0.46 | 1.46 |
| L4-x-5 | 199 | 0 | - | - | - | - | - | - | - |
| L4-x-6 | 95 | 0 | - | - | - | - | - | - | - |
| L6-x-1 | 41 | 1 | 11.46 (hs) | 1.07 (ns) | 0.18 (ns) | 0.69 | 0.31 | 0.38 | 0.62 |
| L6-x-2 | 61 | 0 | - | - | - | - | - | - | - |
| L6-x-3 | 74 | 2 | 20.28 (hs) | 1.70 (ns) | 0.56 (ns) | 0.68 | 0.32 | 0.35 | 0.65 |
| L6-x-4 | 53 | 0 | - | - | - | - | - | - | - |
| L6-x-5 | 29 | 0 | - | - | - | - | - | - | - |
| L6-x-6 | 97 | 0 | - | - | - | - | - | - | - |
| L4r-x-1 | 86 | 34 | 1.43 (ns) | 92.96 (hs) | 516.9 (hs) | −0.06 | 1.06 | −1.13 | 2.13 |
| L4r-x-2 | 66 | 21 | 0.03 (ns) | 47.51 (hs) | 288.3 (hs) | 0.02 | 0.98 | −0.97 | 1.97 |
| L4r-x-3 | 84 | 0 | - | - | - | - | - | - | - |
| L4r-x-4 | 76 | 0 | - | - | - | - | - | - | - |
| L4r-x-5 | 84 | 28 | 0.00 (ns) | 67.2 (hs) | 400 (hs) | 0.00 | 1.00 | −1.00 | 2.00 |
| L4r-x-6 | 61 | 0 | - | - | - | - | - | - | - |
| L17-x-1 | 47 | 6 | 5.29 (s) | 2.33 (ns) | 32.81 (hs) | 0.33 | 0.65 | −0.35 | 1.35 |
| L17-x-2 | 75 | 0 | - | - | - | - | - | - | - |
| L17-x-3 | 67 | 0 | - | - | - | - | - | - | - |
| L17-x-4 | 50 | 0 | - | - | - | - | - | - | - |
| L17-x-5 | 92 | 8 | 15.41 (hs) | 0.52 (ns) | 26.94 (hs) | 0.43 | 0.57 | −0.13 | 1.13 |
| L17-x-6 | 62 | 0 | - | - | - | - | - | - | - |
| L17r-x-1 | 98 | 0 | - | - | - | - | - | - | - |
| L17r-x-2 | 37 | 0 | - | - | - | - | - | - | - |
| L17r-x-3 | 0 | 65 | 195.0 (hs) | 975 (hs) | 4095 (hs) | - | - | - | - |
| L17r-x-4 | 56 | 0 | - | - | - | - | - | - | - |
| L17r-x-5 | 48 | 2 | 4.51 (s) | 2.82 (ns) | 35.41 (hs) | 0.31 | 0.69 | −0.39 | 1.39 |
| L17r-x-6 | 63 | 0 | - | - | - | - | - | - | - |
segregation ratios,
chi square values,
genetic distances, ns: non-significant, s: significant and hs: highly significant χ2 test result, KR and KS: kanamycin-resistant and kanamycin-sensitive phenotypes, respectively.
Two lines (L6-x-1, and L6-x-3) had heterogeneous progenies, which confirmed the hypothesis of two independently segregating transgenic loci. We rejected the possibility of two linked inserts in trans because the hypothesis of two independent inserts was confirmed in T1 and T2. The hypothesis of three inserts, two of which were linked in cis, was also confirmed; in this case, the recombination frequencies were 38% (L6-x-1) and 35% (L6-x-3). Four lines (L6-x-2, L6-x-4, L6-x-5 and L6-x-6) had homogenous progenies with a kanamycin-resistant phenotype; these lines may be homozygous for one, two or three copies of the transgene.
T3 confirmed the stability of line L4r; three lines (L4r-x-1, L4r-x-2, L4r-x-1) had segregation ratios indicative of the presence of a single functional transgenic locus. Lines L4r-x-3, L4r-x-4 and L4r-x-6 were homozygous for one copy of the transgene. From these three progenies, we screened homozygous lines for a single copy of the transgene with stable and acceptable transgene expression. T3 progenies showed Mendelian inheritance of the transgene and confirmed the hypotheses for T1 and T2.
Lines L17-x-1 and L17-x-5 had two inserts each, both of which were linked in T1 and were sufficiently far apart in T3 to allow detectable crossing-over, with frequencies of 33% (L17-x-1) and 43% (L17-x-5). Lines L17-x-2, L17x-3, L17-x-4 and L17-x-6, which had homogenous kanamycin-resistant progenies, were possibly homozygous for two copies of the transgene.
Line L17r-x had only one heterogeneous progeny (L17r-x-5) and the estimated recombination frequency between two linked inserts in cis was 31%. Lines L17r-x-1, L17r-x-2, L17r-x-4 and L17r-x-6 were probably homozygous for two copies of the transgene because their progenies were homogenous for kanamycin-resistance. Interestingly, for the kanamycin-sensitive progeny of L17r-x-3 the genotype was probably homozygous (KS//KS). Sensitive progeny would be expected with a probability of 1:4 in the case of one copy and 1:16 in the case of two copies.
For the genetic engineer who desires excessive expression of the transgene, the best approach would be to screen homozygous lines for two copies of the transgene in homogenous kanamycin-resistant progenies of lines L4, L6, L17 and L17r. In addition, lines that yielded homogenous resistant progenies in T1 and/or T2 (L1, L7, L3r, L14r and L16r) may have more than three copies of the transgene.
Discussion
The analysis of transgenic segregating progenies based on the two approaches described here provided additional information concerning the transgenic population. The major findings of this study agreed with those of previous reports. Genetic analysis confirmed high inter-transformant variability. Indeed, expression levels can vary considerably among plants transformed with the same construct (Hobbs et al., 1990; Peach and Velten, 1991) and in most cases, this expression does not correlate with the copy number (Mlynarova et al., 1991; Hobbs et al., 1993). The copy numbers of transgenic and rearranged fragments are often highly variable, possibly because one or more transgenes can occur at any site.
The two approaches described here were useful for confirming hypotheses regarding the number of insert copies (one insert, two independent or linked inserts, three independent inserts or three inserts of which two were linked) but were unsuitable for non-segregating progenies. Lines with homogenous kanamycin-resistant progenies in T1, T2 and T3 may harbor many copies of the transgene. The best hypothesis for explaining non-segregating progenies is that each chromatid sister possesses a functional transgene. Kohli et al. (1998) reported that the first integrated site acts as a hot spot to integrate more copies of the transgene. This can result in multiple T-DNA insertions (De Neve et al., 1997), with single transgene insertions occurring at a low frequency (Huang et al., 2001). T-DNA acts as an endogenous stimulus that activates the cellular machinery (Fagard and Vaucheret, 2000). As a result, a previously stable genome can become particularly reactive in response to newly inserted transgenes, depending on the extent of inter-genic reactions (Jones et al., 1985; Gheysen et al., 1987; Mayerhofer et al., 1991; Petrov, 1997; Drews and Yadegari, 2002; Brunaud et al., 2002; Van Attikum and Hooykaas, 2003).
Inter-transformant variability was accompanied by variation within the transformed line; transgene expression in most of the lines was unstable and increased across generations. Enhanced transgene expression can be explained by amplification or duplication of the original transgene loci. Since amplification and duplication are frequent events during the repair of double-strand breaks (Spencer et al., 1992; Cannell et al., 1999; Cucu et al., 2002) the number of transgene copies increases in the host genome. This explains why the progenies of lines L3r, L14r and L16r became homogenous for the kanamycin-resistant phenotype in T2 and T3. This observation agrees with the finding of Yong et al. (2006) who reported that homozygous transgenic progeny plants were obtained in T2. In meiotic cells, a copy of the transgene on one chromatid can be passed to the allelic position on the opposite homologue so that the transformed line becomes homozygous for the transgene. Moreover, for self-pollinating species, all loci become homozygous at equilibrium. Upon selfing, the epigenetically silenced loci may segregate, thereby restoring expression of the trans-silenced locus (Khaitová et al., 2011).
The modified maximum likelihood method used here showed that there was frequent crossing-over between linked inserts. Crossing-over occurs naturally in plants and its major role is to generate new genetic combinations; this phenomenon is observed at meiosis and during mitosis between sister-chromatids (Gal et al., 1991; Gorbunova and Levy, 1999). The frequency of crossing-over increases in response to endogenous and exogenous stimuli such as transgenes newly integrated into the genome. Filler DNA, which has been observed in complex transgenic loci (Gheysen et al., 1987; Krizkova and Hrouda, 1998; Brunaud et al., 2002; Theuns et al., 2002; Somers and Makarevitch, 2004), may be amplified such that inserts that were previously tightly linked at the same transgenic site, now become sufficiently separated from each other physically to allow detectable crossing-over.
Several studies have shown that transgene integration sites exhibit different levels of structural complexity ranging from the simple integration of two apparently contiguous transgene copies to tightly linked clusters of multiple copies of transgenes interspersed with host DNA (Svitashev et al., 2000). Epistatic interaction between different loci and/or allelic interaction within a single locus also occur (Matzke and Matzke, 1995; Nap et al., 1997). In the present study, only line L4r was stable, with a 3:1 segregation ratio, indicating the presence of a single functional transgenic locus. This line represented a good transformation event since the stability of transgene expression is a challenge for genetic engineering. However, such analyses should not be limited to the first or second generation.
The results described here showed that transgene inheritance followed Mendelian laws. Mendelian segregation has not been verified for most transformed lines because of transgene instability. The instability of transgenic loci may reflect complex rearrangements, especially amplification of the transgene and filler DNA. Amplification can increase the recombination frequencies, leading to more transformed genotypes. The scenario of transgene introduction may reflect what happened in the history of gene movement among relatives in land races or through horizontal gene transfer (Parrott, 2010).
The genetic approaches developed in this work were efficient because they allowed us to address fundamental and practical issues: (1) they allowed us to screen for stable genetic transformation events that are desirable for breeding programs, (2) they provided insights into the evolution and variation of transgenic loci in early generations (T1, T2 and T3) and (3) they facilitated the study of transgene inheritance. However, future investigations should use molecular analyses such as quantitative PCR to quantify transgenes in the host genome.
Acknowledgments
The present research was funded by the Center of Biotechnology of Borj Cedria. We thank the reviewers for their relevant comments and suggestions to the original paper. We also wish to thank Karima Jandoubi and Fakhri Boukhari for their technical support.
Footnotes
Associate Editor: Adriana Hemerly
References
- Britt AB. Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 1999;4:20–25. doi: 10.1016/s1360-1385(98)01355-7. [DOI] [PubMed] [Google Scholar]
- Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, et al. T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertions sites. EMBO J. 2002;3:1152–1157. doi: 10.1093/embo-reports/kvf237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell ME, Doherty A, Lazzeri PA, Barcelo P. A population of wheat and tritordeum transformants showing a high degree of marker gene stability and heritability. Theor Appl Genet. 1999;99:772–784. [Google Scholar]
- Chen L, Marmey P, Taylor JN, Brizard JP, Espinoza C, D’Cruz P, Huet H, Zhan S, Kochko A, Beachy RN, et al. Expression and inheritance of multiple transgenes in rice plants. Nat Biotechnol. 1998;16:1060–1064. doi: 10.1038/3511. [DOI] [PubMed] [Google Scholar]
- Choffnes DS, Philip R, Vodkin LO. A transgenic locus in soybean exhibits a high level of recombination. In Vitro Cell Dev Biol Plant. 2001;37:756–762. [Google Scholar]
- Christou P, Swain FW, Yang NS, McCabe DE. Inheritance and expression of foreign genes in transgenic soybean plants. Proc Natl Acad Sci USA. 1989;86:7500–7504. doi: 10.1073/pnas.86.19.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucu N, Tenea G, Gavrila L. Genetically modified medicinal plants: II Transfer and expression of a marker kanamycin resistance gene in Atropa belladonna plants. Roum Biotechnol. 2002;7:869–874. [Google Scholar]
- De Neve N, De Buck S, Jacobs A, Van Montagu M, Depicker A. T-DNA integration patterns in cotrans formed plant cells suggest that T-DNA repeats originate from cointegration of separate T-DNAs. Plant J. 1997;11:15–29. doi: 10.1046/j.1365-313x.1997.11010015.x. [DOI] [PubMed] [Google Scholar]
- Deroles SC, Gardner RC. Expression and inheritance of kanamycin resistance in a large number of transgenic petunias generated by Agrobacterium-mediated transformation. Plant Mol Biol. 1988;11:355–364. doi: 10.1007/BF00027392. [DOI] [PubMed] [Google Scholar]
- Dong X. Pathogen-induced systemic DNA rearrangement in plants. Trends Plant Sci. 2004;9:60–61. doi: 10.1016/j.tplants.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Drews GN, Yadegari R. Development and function of the Angiosperm female gametophyte. Annu Rev Genet. 2002;36:99–124. doi: 10.1146/annurev.genet.36.040102.131941. [DOI] [PubMed] [Google Scholar]
- Eckes P, Rosahl S, Schell J, Willmitzer L. Isolation and characterization of a light inducible, organ specific gene from potato and its analysis of expression after tagging and transfer into tobacco and potato schoots. Mol Gen Genet. 1986;205:14–22. [Google Scholar]
- Fagard M, Vaucheret H. (Trans)Gene silencing in plants: How many mechanisms? Annu Rev Plant Physiol. 2000;51:167–194. doi: 10.1146/annurev.arplant.51.1.167. [DOI] [PubMed] [Google Scholar]
- Gal S, Pisan B, Hohn T, Grimsley N, Hohn B. Genomic homologous recombination in plants. EMBO J. 1991;10:1571–1578. doi: 10.1002/j.1460-2075.1991.tb07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen G, Van Montagu M, Zambryski P. Integration of Agrobacterium tumefaciens: Transfer DNA involves rearrangements of target plant DNA sequences. Proc Natl Acad Sci USA. 1987;84:61–69. doi: 10.1073/pnas.84.17.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Levy AA. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 1997;25:4650–4657. doi: 10.1093/nar/25.22.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Levy AA. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 1999;4:263–268. doi: 10.1016/s1360-1385(99)01430-2. [DOI] [PubMed] [Google Scholar]
- Hobbs SLA, Kpodar P, Delong CMO. The effect of T-DNA number and methylation on reporter gene expression in tobacco transformants. Plant Mol Biol. 1990;15:851–864. doi: 10.1007/BF00039425. [DOI] [PubMed] [Google Scholar]
- Hobbs SLA, Warkentin TD, Delong CMO. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol. 1993;21:17–26. doi: 10.1007/BF00039614. [DOI] [PubMed] [Google Scholar]
- Huang JQ, Wei ZM, An HL, Zhu YX. Agrobacterium tumefaciens mediated transformation of rice with the spider insecticidal gene conferring resistance to leaf folder and striped stem borer. Cell Res. 2001;11:149–155. doi: 10.1038/sj.cr.7290080. [DOI] [PubMed] [Google Scholar]
- Jones JD, Weller SC, Goldsbrough PB. Selection for kanamycin resistance in transformed Petunia cells leads to the co-amplification of a linked gene. Plant Mol Biol. 1994;24:505–514. doi: 10.1007/BF00024118. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dunsmuir P, Bedbrook J. High level of expression of introduced chimeric genes in regenerated transformed plants. EMBO J. 1985;4:2411–2418. doi: 10.1002/j.1460-2075.1985.tb03949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitová LC, Fojtová M, Krízová K, Lunerová J, Fulnecek J, Depicker A, Kovaík A. Paramutation of tobacco transgenes by small RNA-mediated transcriptional gene silencing. Epigenetics. 2011;6:650–660. doi: 10.4161/epi.6.5.15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TM, Harper EC, Sanford JC, Fromm ME, Maliga P. Stable genetic transformation of intact Nicotiana cells by the particle bombardment process. Proc Natl Acad Sci USA. 1988;85:8502–8505. doi: 10.1073/pnas.85.22.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A, Leech M, Vain P, Laurice DA, Christou P. Transgene organization in rice engineered through direct DNA transfer supports a two-phase integration mechanism mediated by the establishment of integration hot spots. Proc Natl Acad Sci USA. 1998;95:7203–7208. doi: 10.1073/pnas.95.12.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizkova L, Hrouda M. Direct repeats of T-DNA integrated in tobacco chromosome: Characterization of junction regions. Plant J. 1998;16:673–680. doi: 10.1046/j.1365-313x.1998.00330.x. [DOI] [PubMed] [Google Scholar]
- Limanton-Grevet A, Jullien M. Agrobacterium-mediated transformation of Asparagus officinalis L., molecular and genetic analysis of transgenic plants. Mol Breed. 2001;7:141–150. [Google Scholar]
- Makarevitch I, Svitashev SK, Somers DA. Complete sequence analysis of transgene loci from plants transformed via microprojectile bombardment. Plant Mol Biol. 2003;52:421–432. doi: 10.1023/a:1023968920830. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Matzke AJM. How and why do plants inactivate homologous (trans) genes? Plant Physiol. 1995;107:679–685. doi: 10.1104/pp.107.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C. T-DNA integration: A mode of illegitimate recombination in plants. EMBO J. 1991;10:697–704. doi: 10.1002/j.1460-2075.1991.tb07999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MS, Mohapatra UB, Debnath SC, Power JB, Davey MR. Integration, expression and inheritance of two linked T-DNA marker genes in transgenic lettuce. Mol Breed. 1999;5:329–344. [Google Scholar]
- Misra S. Transformation of Brassica napus L. with a disarmed-octopine plasmid of Agrobacterium tumefaciens: Molecular analysis and inheritance of the transformed phenotype. J Exp Bot. 1989;41:269–275. [Google Scholar]
- Mlynrov L, Loonen A, Heldens J, Jansen RC, Keizer P, Stiekema WJ, Nap JP. Reduced position effect in mature transgenic plants conferred by the chicken lysozyme matrix associated region. Plant Cell. 1994;6:417–426. doi: 10.1105/tpc.6.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nap JP, Conner AJ, Mlynarova L, Stiekema WJ, Jansen RC. Dissection of a synthesized quantitative trait to characterize transgene interactions. Genetics. 1997;147:315–320. doi: 10.1093/genetics/147.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott W. Genetically modified myths and realities. N Biotechnol. 2010;27:545–551. doi: 10.1016/j.nbt.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Somers DA. Transgenic inheritance in plants genetically enginerated using microprojectile bombardment. Mol Biotechnol. 1996;6:17–30. doi: 10.1007/BF02762320. [DOI] [PubMed] [Google Scholar]
- Peach C, Velten J. Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol Biol. 1991;17:49–60. doi: 10.1007/BF00036805. [DOI] [PubMed] [Google Scholar]
- Perret SJ, Valentine J, Leggett JM, Morris P. Integration, expression and inheritance of transgenes in hexaploid oat (Avena sativa L.) J Plant Physiol. 2003;160:931–943. doi: 10.1078/0176-1617-00880. [DOI] [PubMed] [Google Scholar]
- Petrov D. Slow but steady: Reduction of genome size through biased mutation. Plant Cell. 1997;9:1900–1901. doi: 10.1105/tpc.9.11.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prols F, Meyer P. The methylation patterns of chromosomal integration regions influence gene activity of transferred DNA in Petunia hybrida. Plant J. 1992;2:465–475. doi: 10.1046/j.1365-313x.1992.t01-20-00999.x. [DOI] [PubMed] [Google Scholar]
- Register JC, Peterson DJ, Bell PJ, Bullock WP, Evans IJ, Frame B, Greenland AJ, Higgs NS, Jepson I, Jiao S, et al. Structure and function of selectable and non-selectable transgenes in maize after introduction by particle bombardment. Plant Mol Biol. 1994;25:951–961. doi: 10.1007/BF00014669. [DOI] [PubMed] [Google Scholar]
- Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PR, Winter JA, Bamason AR, Rogers SG, Fraley RT. Comparison of cauliflower mosaic virus 35S and nopaline synthase promoters in transgenic plants. Nucleic Acids Res. 1987;15:1543–1558. doi: 10.1093/nar/15.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrawat AK, Becker D, Lörz H. Agrobacterium tumefaciens-mediated genetic transformation of barley (Hordeum vulgare L) Plant Sci. 2007;172:281–290. doi: 10.1111/j.1467-7652.2005.00178.x. [DOI] [PubMed] [Google Scholar]
- Somers DA, Makarevitch I. Transgene integration in plants poking or patching holes in promiscuous genomes? Curr Opin Biotechnol. 2004;15:126–131. doi: 10.1016/j.copbio.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Spencer TM, O’Brien JV, Start WG, Adams TR, Gordon-Kamm WJ, Lemaux PG. Segregation of transgenes in maize. Plant Mol Biol. 1992;18:201–210. doi: 10.1007/BF00034949. [DOI] [PubMed] [Google Scholar]
- Srivastava V, Vasil V, Vasil IK. Molecular characterization of the fate of transgenes in transformed wheat Triticum aesitivum L. Theor Appl Genet. 1996;92:1031–1037. doi: 10.1007/BF00224045. [DOI] [PubMed] [Google Scholar]
- Svitashev S, Ananiev E, Pawlowski WP, Somers DA. Association of transgene integration sites with chromosome rearrangements in hexaploid oat. Theor Appl Genet. 2000;100:872–880. [Google Scholar]
- Takano M, Egawa H, Ikeda JE, Wakasa K. The structures of integration sites in transgenic rice: Three structures of integration sites in transgenic rice. Plant J. 1997;11:353–361. doi: 10.1046/j.1365-313x.1997.11030353.x. [DOI] [PubMed] [Google Scholar]
- Tavazza R, Ordas RJ, Tavazza M, Ancora G, Benvenuto E. Genetic transformation of Nicotiana clevelendi using a Ti-plasmid derived vector. Plant Physiol. 1988;133:640–644. [Google Scholar]
- Theuns I, Windels P, Debuck S, Depicker A, Van Bockstale E, Deloose M. Identification and characterization of T-DNA inserts by T-DNA finger printing. Euphytica. 2002;123:75–84. [Google Scholar]
- Tinland B. The integration of T-DNA into plant genome. Trends Plant Sc. 1996;1:178–184. [Google Scholar]
- Tovar J, Lichtenstein C. Somatic and meiotic chromosomal recombination between inverted duplications in transgenic tobacco plants. Plant Cell. 1992;4:319–322. doi: 10.1105/tpc.4.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Attikum H, Hooykaas PJJ. Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:826–832. doi: 10.1093/nar/gkg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrkamp-Richter S, Degroote F, Laffaire JB, Wyatt P, Perez P, Picard G. Characterisation of a new reporter system allowing high throughput in planta screening for recombination events before and after controlled DNA double strand break induction. Plant Physiol Biochem. 2009;47:248–255. doi: 10.1016/j.plaphy.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tang L, Li M, Chen L, Xu J, Wu G, Li H. Monitoring homologous recombination in rice (Oryza sativa L.) Mutat Res. 2010;691:55–63. doi: 10.1016/j.mrfmmm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Yin Z, Plader W, Malepszy S. Transgene inheritance in plants. J Appl Genet. 2004;45:127–144. [PubMed] [Google Scholar]
- Yong Z, Bao-Yu Y, Shi-Yun S. Inheritance and analysis of herbicide-resistant transgenic soybean lines. Acta Genet Sin. 2006;33:1105–1111. doi: 10.1016/S0379-4172(06)60148-0. [DOI] [PubMed] [Google Scholar]
- Zhu C, Wu J, He C. Induction of chromosomal inversion by integration of T-DNA in the rice genome. J Genet Genomics. 2010;37:189–196. doi: 10.1016/S1673-8527(09)60037-0. [DOI] [PubMed] [Google Scholar]