Abstract
Assessing interactions of a glycan-binding protein (GBP) or lectin with glycans on a microarray generates large datasets, making it difficult to identify a glycan structural motif or determinant associated with the highest apparent binding strength of the GBP. We have developed a computational method, termed GlycanMotifMiner, that uses the relative binding of a GBP with glycans within a glycan microarray to automatically reveal the glycan structural motifs recognized by a GBP. We implemented the software with a web-based graphical interface for users to explore and visualize the discovered motifs. The utility of GlycanMotifMiner was determined using five plant lectins, SNA, HPA, PNA, Con A, and UEA-I. Data from the analyses of the lectins at different protein concentrations were processed to rank the glycans based on their relative binding strengths. The motifs, defined as glycan substructures that exist in a large number of the bound glycans and few non-bound glycans, were then discovered by our algorithm and displayed in a web-based graphical user interface (http://glycanmotifminer.emory.edu). The information is used in defining the glycan-binding specificity of GBPs. The results were compared to the known glycan specificities of these lectins generated by manual methods. A more complex analysis was also carried out using glycan microarray data obtained for a recombinant form of human galectin-8. Results for all of these lectins show that GlycanMotifMiner identified the major motifs known in the literature along with some unexpected novel binding motifs.
Introduction
The biological functions of glycans are exerted through their recognition by a wide variety of glycan-binding proteins (GBPs), which include receptors, lectins, and antibodies, as well as GBPs expressed by pathogens (viruses, bacteria, and parasites). However, the glycan determinants recognized by individual GBPs are just beginning to be understood (Cummings, 2009), and is a major quest of modern glycomics research. The exploration of GBP interactions with glycans on microarrays of defined glycan structures represents a high-throughput method for exploring glycan-binding specificities (Blixt et al., 2004; Feizi and Chai, 2004; Fukui et al., 2002; Powell et al., 2009; Rillahan and Paulson, 2011; Song et al., 2008, 2009; Tateno et al., 2008; Willats et al., 2002; Zhi et al., 2006). Such information is important in identifying potential ligands for a GBP and developing hypotheses about their roles in GBP function. For example, the observation that blood group antigens are recognized with relatively high affinity and specificity by certain human galectins led to the discovery of their bactericidal activity and function as innate immune proteins (Stowell et al., 2010).
The utility of defined glycan microarrays is dependent upon the number and diversity of glycans being interrogated with a GBP. Current glycan microarrays, such as the one publicly available from the Consortium for Functional Glycomics (CFG), contain less than a thousand glycans, which is considerably below the estimate of over 7000 glycans/glycan determinants in the human glycomes (Cummings, 2009). Nevertheless, such limited microarrays can provide immense insights into potential ligands recognized by a GBP. However, even these relatively simple analyses with a single GBP typically generate large amounts of data that are difficult to manually or visually analyze to identify glycan structural motifs or determinants required for high-affinity GBP binding, as well as identify glycan substructures that interfere or preclude recognition of the glycan determinant. Thus, there is a clear need to automate the process for motif identification using data from glycan microarrays.
To address this need, we have developed an algorithm termed GlycanMotifMiner, or GLYMMR, which uses frequent subtree mining (Chi et al., 2005), to identify the motifs of a GBP. We used this algorithm to analyze data from the analyses of five biotin-labeled plant lectins and a recombinant form of human galectin-8 (Gal-8), thus representing a spectrum of binding specificities from simple and complex GBPs, respectively. Data were collected using the defined glycan microarray (version 4.0 and 4.2) from the CFG using fluorescent-labeled streptavidin for detection. In this approach the glycan microarray is interrogated with a GBP at multiple concentrations. At each GBP concentration, relative binding strength of each of the glycans on the array, related to the fluorescence intensity measured as relative fluorescence units (RFU), is calculated by normalizing its RFU to a percentage of the maximum RFU for the bound glycans on the array. Non-specifically bound glycans and non-bound glycans are eliminated as binding candidates by a z-score transformation and referred to as non-binding glycans. The percentages of each binding glycan at each GBP concentration are averaged, and the data are sorted, allowing binding glycans to be ranked according to relative binding strengths. The GLYMMR algorithm then finds the common substructures in the binding glycans that exist in none or only a few non-binding glycans to identify the motifs for this GBP. Unlike other existing motif discovery methods (Hashimoto et al., 2008; Maupin et al., 2012; Porter et al., 2010), our method not only considers both binding and non-binding glycans, but it also can reveal unknown motifs automatically with little user interaction.
Materials and Methods
Glycan microarray analysis
The GLYMMR algorithm was developed using the CFG printed glycan microarray, v4.0, comprised of 442 glycan targets, which evolved from earlier versions with ∼200 glycans (Blixt et al., 2004). To determine the utility of the algorithm we used five common biotin-labeled lectins assayed at three different concentrations using the standard protocol for biotinylated proteins that is available on the CFG website, where all raw data are located. Biotinylated concanavalin A (Con A) from the jack bean (Canavalia ensiformis) was purchased from Vector Laboratories (Burlingame, CA; cat. no. B-1005, lot no. T0829), and was assayed at 1.0, 0.1, and 0.001 μg/mL using 5.0 μg/mL of Cy5-labeled streptavidin (Zymed, Carlsbad, CA) for detection. Biotinylated Sambucus nigra agglutinin-I (SNA) was purchased from Vector Laboratories (cat. no. B-1305, lot no. T1204), and was assayed at 1.0, 0.1, and 0.01 μg/mL using 5.0 μg/mL of Cy5-labeled streptavidin for detection. Biotinylated peanut (Arachis hypogaea) agglutinin (PNA) was purchased from Vector Laboratories (cat. no. B-1075, lot no. T0918), and was assayed at 10.0, 1.0, and 0.1 μg/mL using 5.0 μg/mL of Alexa 488-labeled streptavidin (Invitrogen, Carlsbad, CA) for detection. Biotinylated Ulex europaeus agglutinin-I (UEA-I) from the common gorse was purchased from Vector Laboratories (cat. no. B-1065, lot no. U1216), and was assayed at 30.0, 3.0, and 0.3 μg/mL using 5.0 μg/mL of Alexa 488-labeled streptavidin for detection. Biotinylated Helix pomatia agglutinin (HPA) was purchased from Sigma-Aldrich (St. Louis, MO; cat. no. L6512-IMG, lot no. 084k3776), and was assayed at 1.0, 0.1, and 0.05 μg/mL using 5.0 μg/mL of Alexa 488-labeled streptavidin for detection. The glycan microarray slides were processed and data were collected as Excel spreadsheets as previously described (Smith et al., 2010). The application of the algorithm to a more complex protein was done using a biotinylated preparation of Gal-8 prepared as described previously (Stowell et al., 2008), and assayed at 50 and 5.0 μg/mL using Alexa 488-labeled streptavidin at 5.0 μg/mL for detection on version 4.2 of the glycan array containing 511 glycans.
The 442 glycan targets of v4.0 are comprised of 398 unique glycans with one duplicated structure with the same linker, and 43 duplicate glycans with different linkers; each is printed in replicates of six. All glycans are printed at a single concentration from a 100-μM stock solution. Bound GBPs are detected by measuring the RFU at each of the six locations and averaging four of the six RFU values for each after removing the high and low values. Data are reported as average RFU in Supplementary Tables S1–S6 (see online supplementary material at http://www.liebertpub.com), which also include the chemical structure of the linkers associated with each glycan at the bottom of each table. The standard deviations and %CV (100×standard deviation/average) associated with each average value are reported in the original data deposited and available on the CFG website.
Ranking and selecting the binding and non-binding glycans
To reveal relative binding strengths of a GBP for individual glycans, we performed analyses on the glycan array at three different concentrations of each GBP, where the selected concentrations generated signals that are in the linear range of the fluorescence scanner. The signals generated by GBP binding to specific glycans vary with the concentration of the GBP based on the relative affinity of the GBP for a glycan. Before the rank is assigned to individual glycans bound by a GBP, we selected the binding glycans. To avoid using an arbitrary threshold in determining binders and non-binders, we used the z-score as the statistical test for significance of a sample. The z-score transformation is calculated by comparing the value of a sample, relative to the sample mean and standard deviation, with the null hypothesis being that a random sample pulled from the population would be a non-binder. If the converted p value is less than 0.15, the null hypothesis is rejected and the sample is considered a binding glycan. We used a non-conservative p value to allow more glycans in the list of candidate binders as an input to GLYMMR. The z-score transformation is based on the sum of the RFU intensity values for the three different concentrations of the glycan. This statistical test allows the program to discard not only non-binding glycans, but glycans that exhibit non-specific binding, which could distort the motif discovery algorithm. These results of the statistical test are shown in Supplementary Tables S1–S6 (see online supplementary material at http://www.liebertpub.com), where the binding glycans are highlighted in light green. Glycans exhibiting what we define as non-specific binding generate significant binding signals, but the signals do not vary with the concentration of GBP (see Supplementary Table S1, glycan numbers 206, 388, 193, 203, 331, 228, 323, 216, 181, 105, 299, 413, and 427; see online supplementary material at http://www.liebertpub.com). This is in contrast with non-binding glycans, which generate low signals. In this study, we use the term “non-binder” or “non-binding glycans” to denote glycans that are either non-specific binders or non-binders.
This automatic computational method was developed to provide a systematic and objective process to define binding and non-binding glycans, and to provide an automated method for ranking glycans based on the amount of fluorescence detected, which is related to the relative affinities of the fluorescent-labeled GBP for individual glycan ligands. A rank for each binding glycan is obtained at each GBP concentration using the calculation: rank=100×[RFU bound/highest RFU value in the assay of candidate glycans]. Thus the strongest-binding glycan in each assay at three different concentrations is assigned a maximal rank of 100, and the ranking values decrease with decreased binding strength. Using the three rank values for each glycan, the algorithm calculates an average rank.
As an example of the ranking of glycans bound by a GBP, we analyzed the plant lectin SNA binding to v4.0 of the CFG glycan microarray using biotinylated SNA (Vector Laboratories) at concentrations of 1.0, 0.1, and 0.01 μg/mL (Supplementary Table S1; see online supplementary material at http://www.liebertpub.com). The bound lectin was detected by the fluorescence signal from Cy5-labeled streptavidin (Zymed) at 0.5 μg/mL. Once ranked according to relative binding strengths, the glycans can then be inspected manually (Smith et al., 2010), or with motif analysis algorithms such as the motif discovery tool GLYMMR described here, or by others (Matsumoto and Osawa, 1969; Maupin et al., 2012; Porter et al., 2010), for determining glycan-binding specificity or motifs for a GBP.
Motif discovery using binding and non-binding glycans
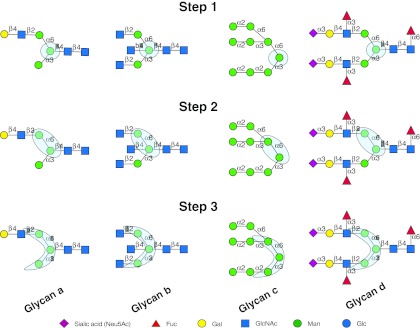
The GLYMMR algorithm functions by automatically finding frequently-occurring patterns in binding glycans that exist in a few or no non-binding glycans. The algorithm then incrementally discovers motifs of larger size and stops when it cannot find any motifs of the next higher size. It accomplishes this by first constructing a two-dimensional tree data structure for all the glycans from their International Union of Pure and Applied Chemistry (IUPAC) notation. The monosaccharides of the glycans form the nodes of the tree, and their connections to other monosaccharides form the edges of the tree. A visual depiction of an example glycan tree is shown in Figure 1, Glycan a, Step 1, where the nodes are represented by the symbols for sialic acid (Neu5Ac), fucose (Fuc), galactose (Gal), N-acetylglucosamine (GlcNAc), and mannose (Man). A monosaccharide, Man, node is highlighted with the light blue circle in Step 1, and the edges are represented with the α or β linkages to the linkage positions 3 or 6 on the neighboring monosaccharides. A subtree is defined as a partial tree with a set of nodes that are attached to each other. GLYMMR initiates a search with trees having a single node. That is, each unique monosaccharide that exists in any of the binding glycans becomes a tree of size one. GLYMMR finds all the nodes that occur in a substantial number (a threshold parameter) of binding glycans. These frequent trees are expanded by adding another node if such a subtree exists in any of the binding glycans. A node (tree of size 1) or in general a subtree that is common to only a few (less than a user-defined threshold, Tb) binding glycans is not expanded, as it is unlikely to be a motif. GLYMMR increases the size of the frequent common subtrees by adding another node until such an addition makes the new subtree infrequent in the binding glycans according to the set threshold. This process is shown in an example in Figure 1, Steps 1 through 3. In Step 1, Man (highlighted by the light blue circle), which is common to all four glycans, is selected. Gal or Neu5Ac are not chosen, as they occur in only 2 of the 4 and 1 of the 4 glycans, respectively. In Step 2, the subtree is expanded to a disaccharide (highlighted in light blue) by adding another Man, and in Step 3 the subtree is further expanded to the trimannosyl core (highlighted in light blue). Subtrees are expanded until they become infrequent and are not expanded further, thus generating a set of possible motifs of different sizes; there is no limit on the size of the motif that GLYMMR can discover. Once the expansion of the common subtrees among the binding glycans is completed, GLYMMR searches if any of these subtrees exist in the non-binding glycans. The subtrees that are present in only binding glycans and none of the non-binding glycans are assumed to represent motifs. The subtrees that exist in many binding glycans, but may also occur in some non-binding glycans, up to a definable limit as described below, are also considered motifs. GLYMMR quickly decreases the possible motifs for a GBP by sorting the common subtrees according to the number of non-binding and binding glycans. Thus the number of motifs discovered can be limited by applying a threshold parameter. The overall algorithm is described in Figure 2.
FIG. 1.
GLYMMR uses frequent subtree mining to discover the glycan-binding motifs of glycan binding proteins (GBPs). Nodes are monosaccharides represented by the symbols defined at the bottom of the figure, and their edges are represented with the α or β linkage to the linkage position on the neighboring monosaccharide. A subtree is a node or a set of nodes (highlighted in light blue). Subtrees are expanded (steps 1–3) until they become infrequent, thus generating a set of possible motifs of different sizes (Fuc, fucose; Gal, galactose; GlcNAc, N-acetylglucosamine; Man, mannose; Glc, glucose).
FIG. 2.
Summary of GLYMMR algorithm. GLYMMR uses a repetitive interrogation of increasingly larger subtrees to discover motifs.
In the application of GLYMMR to determine its utility, we used the parameter values Tb=4, where Tb is the minimum number of binding glycans a subtree must exist in to be a motif; Tn=infinity (essentially no threshold), where Tn is the maximum number of non-binding glycans a subtree can exist in to be a motif; and m=3, where m is a parameter to limit the number of motifs returned by GLYMMR that exist both in binding and non-binding glycans. Note that all motifs that exist only in binding glycans are returned. Also, we sort the motif set in ascending order of the number of non-binding glycans, and then descending order of the number of binding glycans. All of these settings were chosen for CFG array version 4.0 based on evaluating the results of analyses of the selected known lectins after experimenting with different values. We eliminated single nodes as motifs since single nodes (monosaccharides) are not distinguished by their positions in the tree, which would result in an excessive number of motifs. Default parameters were validated based on their ability to generate appropriate motifs for the five well-characterized plant lectins. For a different glycan microarray with larger numbers of defined structures or GBPs with more complex specificities, other parameter values might be more appropriate.
Results
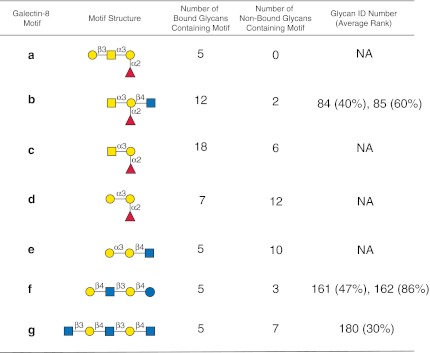
To evaluate the utility of the GLYMMR program, we selected five biotinylated plant lectins, SNA, HPA, PNA, Con A, and UEA-I, whose glycan binding specificities are considered well-defined by a variety of methods over many years, and analyzed their binding on version 4.0 of the CFG mammalian cell glycan microarray. As an example of a more complex GBP we used recombinant biotinylated human Gal-8, which was assayed on v4.2 of the CFG glycan microarray. The glycan microarray data were collected at three concentrations for each biotinylated lectin preparation, and the bound lectins were detected by secondary binding with either Alexa 488- or Cy5-labeled streptavidin. The data were analyzed manually using the concentration-dependent ranking analysis as previously described (Smith et al., 2010), and analyzed automatically with the GLYMMR program. The motifs discovered for each of the five plant lectins are represented in Figures 3 and 6–10 in symbol format where each motif, if it occurs as a glycan on the array, is identified by its glycan number and ranking. In addition, each motif has associated descriptors indicating the number of glycans on the array containing the motif that were binding glycans (binders), and the number of glycans containing the motif on the array that were non-binding glycans (non-binders). Inspection of the structural differences between non-binding and binding glycans possessing the motifs provides information about the specificity of the GBP. The more complex motif analysis of Gal-8, a protein with two unrelated carbohydrate-binding domains, is shown in Figure 11.
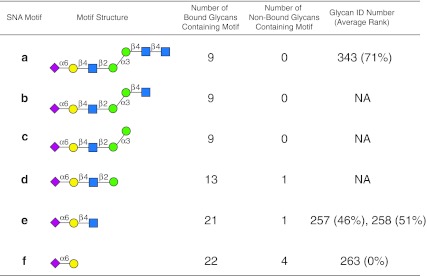
FIG. 3.
Display of motifs for SNA. The structures of motifs (a–f) discovered for SNA over three concentrations are indicated using symbols defined in Figure 1 with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated. The glycan ID number indicating the position on v4.0 of the CFG glycan microarray (Supplementary Table 1; see online supplementary material at http://www.liebertpub.com) is shown for motifs found as glycans on the array with its corresponding average ranking calculated by the algorithm in parentheses. Motifs discovered by the algorithm that are not found as glycans on the array have no glycan ID or ranking and are designated NA (not applicable). The number of bound glycans containing motif is determined by the algorithm and indicates the number of bound glycans found on the glycan array that contain the corresponding motif, while the number of non-bound glycans containing motif indicates the number of glycans found on the glycan array that contain the motif, but are considered non-binding glycans by the algorithm. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S1 (see online supplementary material at http://www.liebertpub.com).
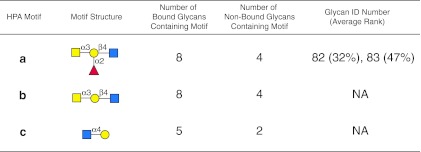
FIG. 6.
Display of motifs for HPA. The structures of motifs (a–c) discovered for HPA over three concentrations are indicated using symbols defined in Figure 1, with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated. The glycan ID number indicating the position on v4.0 of the CFG glycan microarray (Supplementary Table S2; see online supplementary material at http://www.liebertpub.com) is shown for the motif found as a glycan on the array with its corresponding average rank in parentheses. Motifs discovered by the algorithm that are not found as glycans on the array have no ID number or ranking and are designated NA (not applicable). The number of bound glycans containing motif is determined by the algorithm and indicates the number of bound glycans found on the glycan array that contain the corresponding motif; while the number of non-bound glycans containing motif indicates the number of glycans found on the glycan array that contain the motif but are considered non-binding glycans by the algorithm. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S3; see online supplementary material at http://www.liebertpub.com).
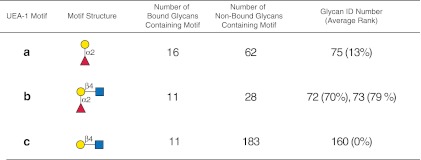
FIG. 10.
Display of motifs for UEA-I. The structures of motifs (a–c) discovered for UEA-I over three concentrations are indicated using symbols defined in Figure 1, with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated. The glycan ID number indicating the position on v4.0 of the CFG glycan microarray (Supplementary Table S5; see online supplementary material at http://www.liebertpub.com) is shown for the motif found as a glycan on the array with its corresponding average rank in parentheses. Motifs discovered by the algorithm that are not found as glycans on the array have no ID number or ranking and are designated NA (not applicable). The number of bound glycans containing motif is determined by the algorithm and indicates the number of bound glycans found on the glycan array that contain the corresponding motif, while the number of non-bound glycans containing motif indicates the number of glycans found on the glycan array that contain the motif but are considered non-binding glycans by the algorithm. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S7 (see online supplementary material at http://www.liebertpub.com).
FIG. 11.
Display of motifs for human recombinant Gal-8. The structures of motifs (a–g) discovered for Gal-8 over two concentrations are indicated using symbols defined in Figure 1, with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated. The glycan ID numbers indicating the position on v4.2 of the CFG glycan microarray (Supplementary Table S6; see online supplementary material at http://www.liebertpub.com) are shown for motifs found as glycans on the array with their corresponding average ranks in parentheses. Motifs discovered by the algorithms that are not found as glycans on the array are designated NA (not applicable). The number of bound glycans containing motif is determined by the algorithm and indicates the number of bound glycans found on the glycan array that contain the corresponding motif, while the number of non-bound glycans containing motif indicates the number of glycans found on the glycan array that contain the motif but are considered non-binding glycans by the algorithm. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S8, panel c (see online supplementary material at http://www.liebertpub.com).
Analysis of SNA
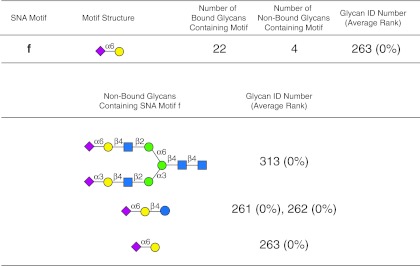
The results of the motif analysis generated for SNA using data in Supplementary Table S1 (see online supplementary material at http://www.liebertpub.com), and with the default algorithm settings (no filter, m=3, and normal sorting as described in Fig. 2) is shown in Figure 3. Results with different parameter settings are shown in Supplementary Figure S1 (see online supplementary material at http://www.liebertpub.com), which provides the display of the web-based graphical user interface generated by GLYMMR. The motifs a, b, and c are the primary motifs that occur in 9 binding glycans, but do not exist in any non-binding glycans. Motifs d, e, and f exist in some non-binding glycans along with many binding glycans. According to our parameter, m, only three motifs are shown that exist in both binding and non-binding glycans. Note that GLYMMR discovered motifs of different sizes, as it searched frequent subtrees of increasing size until the subtrees are infrequent in binding glycans. All of the motifs discovered by the algorithm are related in that they possess the minimal motif f (Neu5Acα2-6Gal). The web-based graphical user interface allows the investigator to inspect the structures of the non-binding glycans and binding glycans possessing a motif by clicking on the underlined number next to binding glycans (binders) and non-binding glycans (non-binders) associated with the motif (Supplementary Figure S1a; see online supplementary material at http://www.liebertpub.com).
For example, there are 22 binding glycans (not shown) that contain motif f (Neu5Acα2-6Gal), and 4 non-binding glycans (Fig. 3 and Supplementary Figure S1a; see online supplementary material at http://www.liebertpub.com). The structures of the 4 non-binding glycans possessing the minimal motif f (glycan no. 263), are displayed by clicking on the number 4 associated with the “non-binders” for that structure and viewing the resulting graphical user interface (Supplementary Figure S2; see online supplementary material at http://www.liebertpub.com), which is summarized in Figure 4. The results indicate that the lack of SNA binding by these glycans is consistent with the results of manual inspection analysis of SNA using the identical data (Smith et al., 2010). Inspection of the motifs discovered by GLYMMR indicate that the lectin prefers the determinant Neu5Acα2-6Galβ1-4GlcNAc (motif e, no. 257 and 258), at rankings of 46% and 51% (Fig. 3), compared to the determinants Neu5Acα2-6Galβ1-4Glc (no. 261 and 262), at a ranking of 0% (Fig. 4), or Neu5Acα2-6Galβ (motif f, no. 263), also at 0% (Figs. 3 and 4). The non-binding of N-glycan no. 313 (Fig. 4), containing the preferred trisaccharide Neu5Acα2-6Galβ1-4GlcNAc (motif e) at a ranking of 0%, is because this trisaccharide determinant is located on the six-branched mannose of the biantennary glycan, while Neu5Acα2-3Galβ1-4GlcNAc, a non-determinant, is on the three-branched mannose of no. 313. This observation is consistent with our manual analysis that led to the conclusions that SNA binds to the trisaccharide determinant Neu5Acα2-6Galβ1-4GlcNAc when it is present on the three-branched mannose of the biantennary structure (Smith et al., 2010), as found in glycan no. 343 and in motifs a, b, and c of Figure 3, but will not bind to the trisaccharide determinant when it is present on the six-branched mannose of the biantennary structure. The motif, Neu5Acα2-6Galβ1-4GlcNAc (no. 257 and 258), is also a component of the four larger motifs and is found in 21 binding glycans (not shown), and in only one non-binding glycan (no. 313, Fig. 4), where it is found on the six-branched mannose of the biantennary structure mentioned above. Thus, motif e (Fig. 3), which is found in motifs a–d, represents a minimal motif, since glycan no. 263 (motif f in Fig. 3), and Neu5Acα2-6Galβ1-4Glc (no. 261 and 262, Fig. 4) are not bound (0%). Interestingly, the disaccharide Neu5Acα2-6Gal (no.263), in spite of being a commonly recognized determinant for SNA and being necessary for binding, is not sufficient alone for SNA binding. The specificity of SNA determined by the GLYMMR was consistent with the conclusions based on manual inspection of the data in Supplementary Table S1 (see online supplementary material at http://www.liebertpub.com), as described previously (Smith et al., 2010), but using this program to decipher this specificity can be accomplished in a fraction of the time.
FIG. 4.
Motif f of SNA discovered in Figure 3 occurs in bound and non-binding glycans on v4.0 of the microarray. Motif f (Neu5Acα2-6Gal) is found in 22 bound glycans and in only 4 non-binding glycans. The 4 non-binding glycans that contain motif f are indicated using symbols defined in Figure 1, with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated, and with the glycan ID number indicating the positions on v4.0 of the CFG glycan microarray (Supplementary Table S1; see online supplementary material at http://www.liebertpub.com), and their corresponding average rankings. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S2; see online supplementary material at http://www.liebertpub.com).
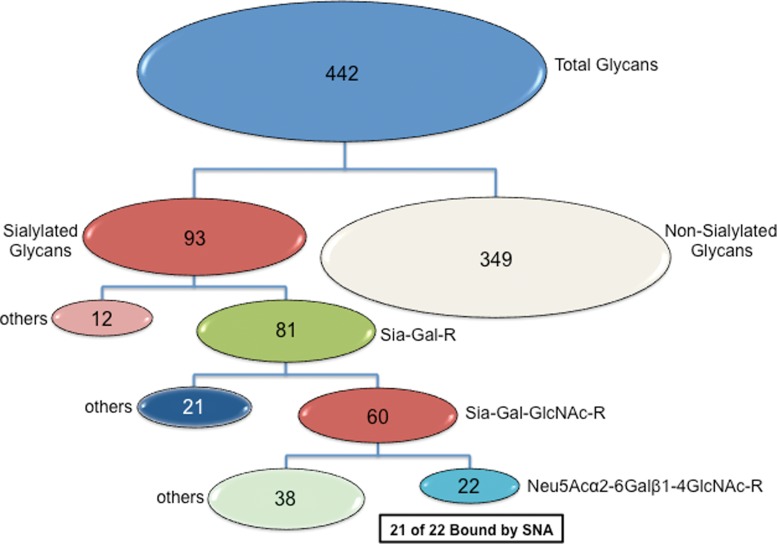
A graphical description of the discriminatory capability of GLYMMR is illustrated in Figure 5, in which the subsets of all glycans are exhibited relative to their recognition by SNA. Twenty-two glycans of the total 442 in the glycan microarray possess the determinant Neu5Acα2-6Galβ1-4GlcNAc. GLYMMR reported that 21 of them that possessed that motif were bound by SNA and identified one as a non-binding glycan. Similar types of graphical illustrations showing the selectivity of each lectin for subsets of glycans can be easily generated from the datasets described below.
FIG. 5.
Discriminatory capability of GLYMMR. The subsets of all glycans are displayed based on their structural attributes relative to their recognition by SNA. Twenty-two glycans of the total 442 on v4.0 of the CFG microarray are recognized as binding glycans by SNA, and all of those glycans contain the sequence Neu5Acα2-6Galβ1-4GlcNAc.
Analysis of HPA
HPA is a lectin isolated from the albumin gland of the edible snail Helix pomatia, and specifically agglutinates human blood group A, but not B and O erythrocytes (Hammarstrom and Kabat, 1971; Uhlenbruck and Prokop, 1966). This historically-defined specificity results from its binding a group of defined glycans with the following order of preference: Forssman antigen (GalNAcα1-3GalNAc-R), blood group A substance (GalNAcα1-3(Fucα1-2)Galβ1-R), Tn antigen (GalNAcα-Ser/Thr), GalNAc, and GlcNAc (Wu, 1991). For this reason, HPA has been classified as an α-GalNAc-binding lectin and used extensively to define the presence of this sugar residue at the non-reducing termini of mammalian oligosaccharides. The results of the motif analysis generated for HPA using glycan array data on v4.0 of the CFG array (Supplementary Table S2; see online supplementary material at http://www.liebertpub.com), with the algorithm set with no filter, m=3, and normal sorting (see the web-based graphical user interface in Supplementary Figure S3; see online supplementary material at http://www.liebertpub.com), are summarized in Figure 6, where two very different motifs were discovered. Motifs a and b are related and clearly consistent with the known specificity for terminal α-GalNAc, but motif c terminates in α-GlcNAc. This second glycan recognition by HPA has largely been overlooked, since α-GlcNAc occurs rarely in mammals, and has so far only been found in O-glycans of human gastric mucins (Nakayama et al., 1999; Zhang et al., 2001). HPA was recently described as a member of a group of invertebrate lectins that are involved in innate immunity in the snail, where it participates in the protection of fertilized eggs by agglutinating a variety of microorganisms (Sanchez et al., 2006). Unlike mammalian cells, microorganisms may be more likely to present α-GlcNAc on their surfaces.
Inspection of the non-binding and binding glycans associated with the α-GalNAc terminating sequences (motifs a and b in Fig. 6) indicates that binding by HPA is associated with terminal α-GalNAc residues, as previously described with one exception. That exception is the presence of a GalNAc at the reducing end of a glycan expressing a difucosylated blood group A-like sequence as seen in GalNAcα1-3(Fucα1-2)Galβ1-4(Fucα1-3)GlcNAcβ1-3GalNAc (no. 415), as shown in Supplementary Table S2 (see online supplementary material at http://www.liebertpub.com). HPA does not recognize this glycan, although it binds well to glycan no. 80 with the simpler sequence GalNAcα1-3(Fucα1-2)Galβ1-4(Fucα1-3)GlcNAc. The structural difference between no. 80 and no. 415 that confers such a remarkable difference in HPA recognition is not known, but was easily revealed using the motif discovery algorithm.
Inspection of the non-binding and binding determinants associated with the other HPA binding motif c, GlcNAcα1-4Gal (Fig. 6), shows that this determinant is present in the five binding glycans (glycans no. 333, 334, 335, 336, and 338 in red type in Supplementary Table S2; see online supplementary material at http://www.liebertpub.com), and in two non-binding glycans (no. 337 and 339 at 21% and 12%, respectively, in Supplementary Table S2; see online supplementary material at http://www.liebertpub.com). The non-binding glycan no.337 is closely related to the five binding glycans sharing the terminal sequence GlcNAcα1-4Galβ1-4GlcNAcβ1-3Gal, with the binding glycan numbers 333, 336, and 338. This is the result of not meeting the criteria for statistical significance after performing the z-score transformation due to the lower-than-anticipated binding at the highest HPA concentration. While the data in Supplementary Table S2 (see online supplementary material at http://www.liebertpub.com) show a ranking value of 21 for glycan 337, the automated calculation considered it a non-binding glycan. Glycan no. 339 at 12% also fell below the cutoff, but this reflects a significant difference in structure, which possesses a reducing terminal GalNAc (no. 339, GlcNAcα1-4Galβ1-3GalNAc). Simple inspection of the motif analysis results provides subtle specificity information indicating that HPA must be affected in its binding by more than the presence of the terminal monosaccharide, since a modification of the structure GlcNAcα1-4Galβ1-4GlcNAc- (no. 335) to GlcNAcα1-4Galβ1-3GalNAc- (no. 339) has a significant negative impact on HPA recognition (Supplementary Table S2; see online supplementary material at http://www.liebertpub.com).
Analysis of PNA
The lectin from the peanut (Arachis hypogaea) agglutinates neuraminidase-treated human erythrocytes (Bird, 1964; Uhlenbruck et al., 1969), and was originally designated “anti-T agglutinin” because, like the mammalian anti-T antibody, it induced T-polyagglutination that was associated with certain bacterial and viral infections (Lotan et al., 1975). The T-antigen was shown to have the structure Galβ1-3GalNAc-R (Springer and Desai, 1974), and purified peanut agglutinin (PNA) was shown to be very specific for this disaccharide sequence, which is typically found within core 1 and core 2 O-glycans. The proposed binding site on the protein was restricted to the non-reducing terminal β-linked Gal of the T-antigen (Lotan et al., 1975).
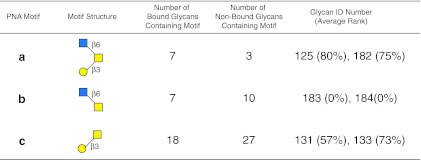
The results of the motif analysis generated for PNA using glycan array data on v4.0 of the CFG array (Supplementary Table S3; see online supplementary material at http://www.liebertpub.com), and with the algorithm set with no filter, m=3, and normal sorting (see the web-based graphical user interface in Supplementary Figure S4; see online supplementary material at http://www.liebertpub.com) is summarized in Figure 7. The disaccharide Galβ1-3GalNAc-R (motif c) was among the three motifs identified by this algorithm. The largest motif was GlcNAcβ1-6(Galβ1-3)GalNAc (motif a), which is a composite of the other two disaccharide motifs, GlcNAcβ1-6GalNAc (motif b) and Galβ1-3GalNAc (motif c). Thus, the motif analysis discovered the T-antigen (glycans no. 131 and 133 in Supplementary Table S3, and motif c in Fig. 7; see online supplementary material at http://www.liebertpub.com) among the motifs for this lectin, but the trisaccharide GlcNAcβ1-6(Galβ1-3)GalNAc (motif a; glycans no. 125 and 182 ranked 80% and 75%, respectively, in Supplementary Table S3; see online supplementary material at http://www.liebertpub.com) was a stronger binder (higher ranking) than the simple T-antigen disaccharide alone. Although motif b (glycans no. 183 and 184 in Supplementary Table S3; see online supplementary material at http://www.liebertpub.com) itself is non-binding glycan, the algorithm includes this motif since it appears in the binding glycans. Inspection of the ranking of the glycans bound by PNA (Supplementary Table S3; see online supplementary material at http://www.liebertpub.com) indicated that the core 2 O-glycans tetrasaccharide Galβ1-4GlcNAcβ1-6(Galβ1-3)GalNAc (glycans no. 157 and 159, ranked 98% and 89%, respectively), pentasaccharide Galβ1-3GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAc (glycan no. 377 ranked 85%), and trisaccharide GlcNAcβ1-6(Galβ1-3)GalNAc (motif a; glycans no. 125 and 182, ranked 80% and 75%, respectively), all are ranked higher than the disaccharide Galβ1-3GalNAc (glycans no. 131 and 133, ranked 57% and 73%, respectively). This is another example of how increased numbers of glycans available on glycan microarrays provide new paradigms for lectin specificities. The apparently strict specificity and the proposed restricted binding site on PNA for the T-antigen disaccharide must clearly be revised based on the fact that many diverse glycans exist that bind PNA stronger than the simple T-antigen disaccharide found within O-glycans. It is noteworthy that PNA did not recognize any glycan in which the disaccharide motif Galβ1-3GalNAc was sialylated or fucosylated.
FIG. 7.
Display of motifs for PNA. The structures of motifs (a–c) discovered for PNA over three concentrations are indicated using symbols defined in Figure 1, with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated. The glycan ID number indicating the position on v4.0 of the CFG glycan microarray (Supplementary Table S3; see online supplementary material at http://www.liebertpub.com) is shown for the motif found as a glycan on the array with its corresponding average rank in parentheses. Motifs discovered by the algorithm that are not found as glycans on the array have no ID number or ranking and are designated NA (not applicable). The number of bound glycans containing motif is determined by the algorithm, and indicates the number of bound glycans found on the glycan array that contain the corresponding motif, while the number of non-bound glycans containing motif indicates the number of glycans found on the glycan array that contain the motif, but are considered non-binding glycans by the algorithm. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S4; see online supplementary material at http://www.liebertpub.com).
Analysis of Con A
The GLYMMR analyses of the lectins described above were based on data generated after the glycans were ranked and averaged over three concentrations. During this process each dataset is submitted to analysis by the ranking program utilizing the z-score transformation, and the ranking of each glycan at three concentrations is averaged and the glycans are sorted by rank from high to low for motif discovery analysis. The ranking program and GLYMMR were designed to analyze three concentrations of GBP; however, these analyses can also be performed on one or two datasets. If multiple concentrations are selected for analysis they should be in a linear range of the fluorescence scanner used for the analysis (between 1000 and 40,000 to 60,000 RFU). If data are included that are not in this linear range, the motif pattern will be altered based on the bias generated by the non-linear data. This effect can be demonstrated in the analysis of Con A from the jack bean (Canavalia ensiformis), known to be specific for either terminal α-linked mannose or glucose residues on glycans (Goldstein, 1975; Goldstein et al., 1965), and for the internal trimannosyl core structure of N-glycans (Baenziger and Fiete, 1979; Cummings, 1994; Merkle and Cummings, 1987). The biotinylated lectin was assayed at 1.0, 0.01, and 0.001 μg/mL, and detected with Cy5-labeled streptavidin. The data are shown in Supplementary Table S4b (see online supplementary material at http://www.liebertpub.com), where the RFU values for the 50 strongest binding glycans at 1.0 μg/mL were between 30,000 and 60,000 RFU, and z-score transformation resulted in 57 binding glycans that were statistically significant. At the highest concentration of the lectin, the ranking is difficult due to the inability to discriminate between binding and non-bound glycans.
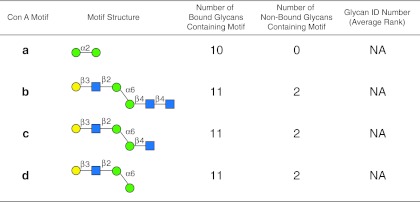
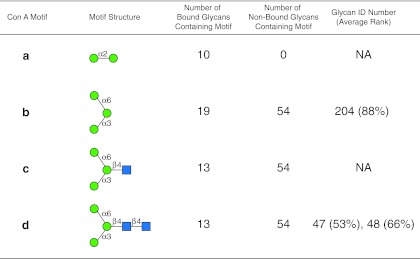
The motif analysis generated for Con A with the algorithm set with no filter, m=3, and normal sorting (see the web-based graphical user interface in Supplementary Figure S5; see online supplementary material at http://www.liebertpub.com), based on all three concentrations for which the high concentration is well above the linear range, is summarized in Figure 8. The motif analysis using these data reveals the well-characterized terminal α-mannose specificity (Goldstein et al., 1965) of Con A in motif a (Fig. 8), and a portion of a biantennary N-glycan (motifs b–d), which is known to be a motif of Con A (Baenziger and Fiete, 1979; Merkle and Cummings, 1987), but none of the motifs discovered occur as an individual glycan on the array. However, if the motif analysis is carried out using only the two low concentrations of Con A, which are clearly in the linear range of the instrument, the motif with the same algorithm settings (see the web-based graphical user interface in Supplementary Figure S6; see online supplementary material at http://www.liebertpub.com) is more representative of what is known for this lectin and is summarized in Figure 9. Con A was initially characterized as a mannan-binding lectin with specificity for terminal Manα1-2 (Goldstein, 1975; Goldstein et al., 1965), found in motif a of both Figures 8 and 9. This motif a does not appear on the array as a glycan, but it is present in the 10 highest-ranked mannans shown in Supplementary Tables S4a and S4b (see online supplementary material at http://www.liebertpub.com), while no non-binding glycans possess this determinant. As mentioned above, Con A is also known to bind biantennary N-glycans that possess the trimannosyl core structure shown in motif b, which appears as glycan no. 204 on the array ranked at 88%. Motifs c and d (Fig. 9) both contain motif b, and this trimannosyl core with the core GlcNAc (motif c), or core chitobiose (motif d; glycans no. 47 and 48 at rankings of 53% and 66%, respectively), and are found in 13 binding glycans and 54 non-binding glycans. Of the 54 non-binding glycans, many are triantennary, bisected, and derivatized N-glycans that possess this chitobiose, trimannosyl core structure, and are known to be poorly bound by Con A (Baenziger and Fiete, 1979). These observations clearly demonstrate the need to work with data that are in the linear range of the analysis to obtain representative motifs.
FIG. 8.
Display of motifs for concanavalin A (Con A) based on three concentrations of the lectin. The structures of motifs discovered for Con A (a–d) are indicated using symbols defined in Figure 1, with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated. The glycan ID number indicating the position on v4.0 of the CFG glycan microarray (Supplementary Table S4b; see online supplementary material at http://www.liebertpub.com), is shown for the motif found as a glycan on the array with its corresponding average rank in parentheses. Motifs discovered by the algorithm that are not found as glycans on the array have no ID number or ranking and are designated NA (not applicable). The number of bound glycans containing motif is determined by the algorithm and indicates the number of bound glycans found on the glycan array that contain the corresponding motif, while the number of non-bound glycans containing motif indicates the number of glycans found on the glycan array that contain the motif, but are considered non-binding glycans by the algorithm. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S5; see online supplementary material at http://www.liebertpub.com).
FIG. 9.
Display of motifs for Con A based on two concentrations of the lectin. The structures of motifs discovered for Con A (a–d) are indicated using symbols defined in Figure 1, with α and β anomeric carbons and linkage positions to the adjacent monosaccharides indicated. The glycan ID number indicating the position on v4.0 of the CFG glycan microarray (Supplementary Table S4a; see online supplementary material at http://www.liebertpub.com), is shown for the motif found as a glycan on the array with its corresponding average rank in parentheses. Motifs discovered by the algorithm that are not found as glycans on the array have no ID number or ranking and are designated NA (not applicable). The number of bound glycans containing motif is determined by the algorithm and indicates the number of bound glycans found on the glycan array that contain the corresponding motif, while the number of non-bound glycans containing motif indicates the number of glycans found on the glycan array that contain the motif, but are considered non-binding glycans by the algorithm. The display of the graphical user interface used to generate this summary is shown in Supplementary Figure S6; see online supplementary material at http://www.liebertpub.com).
Analysis of UEA-I
UEA-I is a lectin isolated from Ulex europaeus (common gorse), and specifically binds H (type 2) glycans that represent the O blood group antigen (Matsumoto and Osawa, 1969). The known specificity of UEA-I for blood group H (type 2) is clearly presented as the motif b (Fig. 10) discovered by the GLYMMR program using data from three concentrations of this lectin as shown in Supplementary Table S5 with the algorithm set with no filter, m=3, and normal sorting (see the web-based graphical user interface in Supplementary Figure S7; see online supplementary material at http://www.liebertpub.com). The H (type 2) structure Fucα1-2Galβ1-4GlcNAc (motif b) includes the other discovered motifs a and c, both of which are presented as glycan targets on the array (glycans no. 75 and 160, respectively). Consistent with early observations of this lectin (Baldus et al., 1996; Debray et al., 1981; Petryniak and Goldstein, 1986; Sughii et al., 1982), the α1-2-linked fucose is an absolute requirement for UEA-I binding. This is based on the observations that the non-fucosylated Galβ1-4GlcNAc (LacNAc), which is a component of most of the binding glycans (Supplementary Table S5; see online supplementary material at http://www.liebertpub.com), is found in over 180 non-binding glycans, lacks fucose addition, and is not bound by UEA-I.
Motif b is present at two locations on the array with slightly different spacers as glycans no. 72 and 73 ranked at 70% and 79%, respectively. Higher-ranking glycans were mono- and di-sulfated derivatives of fucosylated LacNAc or lactose (Supplementary Table S5; see online supplementary material at http://www.liebertpub.com), which have not been previously reported as inhibitors of UEA-I binding, and were not identified as a motif since there were only three sulfated structures on the array, and the algorithm was set for Tb=4. Interestingly, the Lewis y (Ley) antigenic tetrasaccharide determinant [Fucα1-2Galβ1-4(Fucα1-3)GlcNAc] was found in two of the 11 binding glycans containing motif b as glycans no. 68 and 69 ranked at 39% and 41%, respectively. The lower ranking relative to motif b suggests that the additional fucose on the Ley-related structure destabilizes its binding to UEA-I, which is consistent with previous studies on the inhibition of blood group substance precipitation by UEA-I (Sughii et al., 1982). Again, although the Ley-containing glycans are found among the binding glycans, GLYMMR does not identify this as a motif since it exists in only two bound glycans, and the threshold, Tb, in our algorithm was set to 4. Motif b also occurs in 28 non-binding glycans, of which 13 were related to blood group A, GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAc, and six were related to blood group B, Galα1-3(Fucα1-2)Galβ1-4GlcNAc, indicating that substituting the Gal of motif b with anything at its 3-position destroyed UEA-I binding, which is consistent with the H(O) specificity (Matsumoto and Osawa, 1969) of UEA-I; however, substitution of this motif at the 4-position (glycans no. 90 with terminal α4GalNAc and no. 112 with α4Gal, ranked at 58% and 74%, respectively; Supplementary Table S5; see online supplementary material at http://www.liebertpub.com) did not significantly affect binding. The weaker-binding Ley tetrasaccharide is ranked even lower when it is linked β1-3 to GalNAc at the reducing end (no. 413), β1-2 to Man in di- and triantennary N-glycans (no. 358 and 442), or β1-3 to Gal in longer fucosylated polylactosamines (no. 66 and 67). Thus, the binding pattern of UEA-I on v4.0 of the CFG array demonstrates exquisite specificity for H (type 2) glycans, with complete lack of binding to blood group A, B, and H (type 1) glycans, as previously described (Petryniak and Goldstein, 1986; Sughii et al., 1982), and differential ranking of the known glycan structures provides a definition of the UEA-I specificity. The GLYMMR program permits rapid identification of the binding motif, and facilitates deciphering the specificity by providing the structures of the binding and non-binding glycans containing each motif.
Analysis of recombinant human galectin-8 (Gal-8)
Human Gal-8, a member of the galectin family of galactose-binding proteins, was selected for analysis using GLYMMR as an example of a biologically relevant and complex glycan-binding protein. Gal-8, a tandem repeat galectin whose single polypeptide has two carbohydrate recognition domains (CRDs) that bind different sets of glycans (Carlsson et al., 2007; Stowell et al., 2008), was assayed at two different concentrations and analyzed using GLYMMR. As expected for a GBP with mixed specificity, Gal-8 binds strongly to 46 glycans (Supplementary Table S6; see online supplementary material at http://www.liebertpub.com) on the CFG glycan array v4.2 with the strongest being no. 162, LNnT (Galβ1-4GlcNAcβ1-3Galβ1-4Glc) at a rank of 86%, while 20 binding glycans express human blood group A or B. Glycans containing poly-N-acetyllactosamine sequences (Galβ1-4GlcNAc)n comprised a significant number of the binding glycans, and several sialylated and sulfated glycans were among this group. These data are consistent with previous analyses of Gal-8 on the CFG array (Stowell et al., 2008). When the data from the two concentrations (50 μg/mL and 5.0 μg/mL) were analyzed using GLYMMR with the default settings of m=3, normal sorting, and no filtering, 5 motifs were discovered as shown in the web-based graphical user interface shown in Supplementary Figure S8a (see online supplementary material at http://www.liebertpub.com). With these settings the algorithm discovered an extended blood group A determinant [Galβ1-3GalNAcα1-3(Fucα1-2)Gal], the blood group A [GalNAcα1-3(Fucα1-2)Gal], and LNnT (Galβ1-4GlcNAcβ1-3Galβ1-4Glc), but did not discover the blood group B motif [Galα1-3(Fucα1-2)Gal]. By increasing the value of m to 9, we were able to discover the blood group B motif as shown in Supplementary Figure S8b (see online supplementary material at http://www.liebertpub.com), which shows 11 motifs, that can be filtered to remove subsets of identical structures to show the 7 motifs in Supplementary Figure S8c (see online supplementary material at http://www.liebertpub.com), which are summarized in Figure 11. The inability of the algorithm to detect blood group B motifs is due to the fact that the parameter setting of m=3 restricted the number of motifs that exist both in binding and non-binding glycans to 3. Increasing this value resulted in the discovery of the human blood group B motifs. In addition, the array contains only four Gal-8 binding glycans containing the blood group B tetrasaccharide determinant Galβ1-3(Fucα1-2)Galβ1-4GlcNAc, while there are seven Gal-8 binding glycans containing the blood group A tetrasaccharide determinant GalNAcβ1-3(Fucα1-2)Galβ1-4GlcNAc. Thus, GLYMMR discovered both the human blood group motif, as well as the poly-N-acetyllactosamine motif of Gal-8, which is known to possess different specificities because it has two different CRDs, and by modifying the parameters of the algorithm it is possible to accommodate the discovery of multiple motifs in a single sample.
Discussion
Our results show that GLYMMR is a glycan binding motif discovery algorithm that first ranks and classifies glycans interrogated with a GBP into binding and non-binding glycans, and then identifies the motifs based on the glycan structures. The motif discovery results are shown for five representative and well-characterized lectins: SNA, HPA, PNA, Con A, and UEA-I. Each motif discovered by GLYMMR was consistent with the published and accepted specificities of these well known plant lectins that have been used for many years for detecting specific glycan structures, although our findings expand the knowledge about the glycan motifs recognized by several of these lectins. The results of GLYMMR can be presented to the user in a web-based graphical user interface (Supplementary Figures S1–S8; see online supplementary material at http://www.liebertpub.com) for quick interpretation in a web-based format.
GLYMMR offers several advantages for finding glycan-binding motifs using microarray approaches. In contrast to the algorithm described here, other methods for motif discovery either limit the motifs to a predefined list, or focus only on binding glycans, while ignoring the presence of the motif in non-binding glycans. The work by Porter and associates (Porter et al., 2010) demonstrated an approach that searches for the existence of a motif from pre-defined set of 63 motifs using an intensity segregation method. In this method, motif segregation is based on fluorescence intensity, in which glycans are segregated according to the presence or absence of a given motif, and then the statistical difference in fluorescence signal is calculated between the glycans in the two groups. A drawback of this method is that it can find only a predetermined set of known motifs. This approach was recently refined (Maupin et al., 2012) to permit the manual identification of new determinants to be included in subsequent motif segregation analysis. The other available method for motif discovery (Hashimoto et al., 2008) provides a definition of the common subtrees among a list of glycans without evaluating binding intensities.
The GLYMMR algorithm coupled with a web-based graphical user interface to evaluate results addresses the limitations in prior methodologies. GLYMMR is a relatively simple system for exploring the CFG glycan microarray for the purpose of identifying the glycan-binding specificity of GBPs. It incorporates a non-biased selection of candidate glycans, based on their binding strength, with a simple approach to searching for glycan determinants or motifs, and reports both the binding and non-binding glycans in which the determinants or motifs occur. While the algorithm itself does not generate the specificity for a GBP, it provides the researcher the ability to quickly find and compare the structural features of the binding and non-binding glycans so that specificity determinations can quickly be made.
In evaluating the utility of GLYMMR by comparing the known specificities of a selection of plant and animal lectins, we found the algorithm to be useful and accurate if applied to sound datasets with an understanding of the limitation of the program based on the threshold parameters used. We suggest a default setting for the threshold parameters for our initial analyses so that the variables would be associated with the data used for input as we developed the application with the 442 glycan targets of v4.0 of the CFG array. As we discovered motifs of known lectins, we found that modifying the method for the value of m was useful in revealing all of the known motifs. The modifications were useful, as the complement of structures on the array can bias the sorting parameters. Filtering the results so that subsets are combined into the largest glycan limited the number of motifs. It may be necessary to make further modifications of parameters such as the sorting method as the application is applied to later versions of the array (i.e., v5.0 comprised of 611 glycan targets). The application to newer versions of the array requires only the addition of the structures to the library of glycans, and it is possible to apply the algorithm to any array of defined structures.
This algorithm permits a rapid and accurate method for defining subsets of glycans when it is necessary to work with large numbers of structures that have to be represented with a relatively complex nomenclature. For example the data generated to support the analysis of the lectin SNA can be described in subsets according to the diagram in Figure 5. There are 442 glycan targets printed on v4.0 of the CFG array. For SNA, which is considered to be a sialic acid-specific lectin, there are two major subsets of glycans: sialylated structures comprising a subset of 93 glycans, and non-sialylated glycans making up a subset of the remaining 349. The subsets within the sialylated glycans can be subdivided first into those with a terminal sialic acid coupled to Gal by all possible linkages (Sia-Gal-R), which make up a subset of 81 glycans, which is further reduced to a subset of 60 by eliminating all except those with the terminal trisaccharide Sia-Gal-GlcNAc in all possible linkages, which would include both type 1 (Galβ1-3GlcNAc) and type 2 (Galβ1-4GlcNAc) glycans, as well as some glycans having modified sialic acids. Restricting that subset to those containing only Neu5Acα2-6Galβ1-4GlcNAc results in 22 glycans. GLYMMR reported 21 glycans containing Neu5Acα2-6Galβ1-4GlcNAc or motif a in Figure 3. The other glycan on the array that contains motif a is glycan 313, which was considered a non-binder by the algorithm as shown in Figure 4. Similar subset distributions are easily carried out for all of the lectins analyzed.
GLYMMR successfully defined motifs of all of the well-characterized lectins that we selected for analysis. We had previously shown by manual inspection of data from SNA binding in the CFG glycan array that this lectin, which was generally thought to be specific for Neu5Acα2-6Gal, prefers that this disaccharide be on a type 2 glycan sequence (Neu5Acα2-6Galβ1-4GlcNAc), for which it exhibits stronger binding than to sialylated lactose, which lacks the GlcNAc residue and contains Glc instead (Smith et al., 2010). These data are consistent with the original studies of Goldstein and colleagues showing that SNA binds glycans with the sequence Neu5Acα2-6Gal(NAc)-R (Shibuya et al., 1987). Interestingly, GLYMMR revealed that SNA binds well to the Neu5Acα2-6Galβ1-4GlcNAc determinant within N-glycans, except when it is linked to the α6-branched mannose of an N-glycan (Smith et al., 2010); the motifs discovered by GLYMMR were all related to the heptasaccharide (motif a shown in Fig. 3). The ability to compare the structural features of binding and non-binding motif-containing glycans, provided a rapid method for confirming what aspects of a glycan structure destabilize binding. For example, the presence of a non-binding determinant on the 3-branch of glycan no. 313 (Fig. 4) strongly supported the conclusion that SNA will recognize the determinant Neu5Acα2-6Galβ1-4GlcNAc on the 3-branch as a binding motif, but not if the determinant is on the 6-branched mannose.
The analysis of HPA resulted in the discovery of two extremely different motifs, terminal α-linked-GalNAc and α-linked-GlcNAc, and provided an interesting example of how the numbers of glycans on a microarray and the threshold parameters of GLYMMR interact. As shown in Figure 6, for HPA binding there are eight binding glycans that possess terminal α-linked-GalNAc, which is consistent with prior studies of its binding specificity (Wu, 1991). However, GLYMMR also identified five binding glycans possessing terminal α-linked-GlcNAc, which is a relatively rare glycan in animals (Nakayama et al., 1999; Zhang et al., 2001), and was not previously described to be well recognized by HPA. If glycans with this latter structural feature had not been on the array, this motif would not have been discovered. In fact, if it had been represented fewer than four times on the array, it would have been identified as a binding glycan, but not as a motif, since the threshold parameter Ts was set at 4. Thus when using GLYMMR, it is important to initiate the specificity determination by listing all of the binding glycans sorted from high to low ranking in order to identify any rare glycan that is a binding glycan for the GBP in question. For example, if one inspects the ranking of glycans that are bound by SNA (Supplementary Table S1; see online supplementary material at http://www.liebertpub.com), glycans no. 353 (Kdnα2-6Galβ1-4GlcNAc) and 256 [Neu5Acα2-6Galβ1-4(6OSO3)GlcNAc] are the highest ranking glycans; this suggests the possibility that SNA might prefer sulfated glycans or other sialic acid derivatives, but these motifs do not arise from analysis by GLYMMR, since they are represented fewer than four times on the array. Interestingly, in other work on arrays comprised of derivatives of sialic acid, we observed that SNA preferred Kdn and its acetylated derivatives to Neu5Ac and Neu5Gc, the more common mammalian sialic acids (Song et al., 2011). These results also illustrate the need in glycan microarray studies for presenting glycan determinants in a variety of formats and backbones on multiple glycans to facilitate identification of motifs.
GLYMMR revealed new motifs associated with PNA binding that had not been previously reported. The discovery of new motifs for well-defined GBPs will probably become common as GLYMMR and other algorithms are applied to additional well-defined lectins. This will occur largely because glycan microarrays allow us to evaluate hundreds of glycans simultaneously, as opposed to the classical method of hapten inhibition of lectin binding that was generally accomplished using limited numbers of structures and a single oligosaccharide per experiment. With hundreds of glycans on a single array, unexpected discoveries become common. The original studies defining the specificity of PNA as being directed against Galβ1-3GalNAc are certainly valid and the definition of PNA as an anti-T agglutinin is appropriate (Lotan et al., 1975), but the fact that more extended structures are also equivalently recognized, such as the non-sialylated core 2 O-glycan Galβ1-4GlcNAcβ1-6(Galβ1-3)GalNAc, should now be taken into account. The utility of using multiple concentrations for analysis of GBP specificities is demonstrated in data obtained for PNA, as shown in Supplementary Table S3 (see online supplementary material at http://www.liebertpub.com), where the glycans 206, 299, 203, and 150 are in bold type. These glycans received a low but significant ranking of 3–7% in this analysis; however, these glycans, which obviously do not contain a PNA motif, received high rankings when lower concentrations of lectin were analyzed due to the lower value of the maximum RFU. When averaged, this high ranking at low lectin concentration resulted in an elevated average ranking. These glycans are examples of non-specific binders, since the RFU values do not change as a function of concentration, and they are eliminated from the candidate glycans by the z-score transformation described above. Because GLYMMR eliminates the non-binding glycans (non-specifically bound and non-bound) prior to calculating their rank, the average rankings generated by the algorithm (Figs. 3, 4, and 6–11) may be different from the average rankings generated manually in Supplementary Tables S1–S6 (see online supplementary material at http://www.liebertpub.com).
We purposely evaluated Con A, which binds mannose-containing N-glycans (Goldstein et al., 1965), at a concentration outside the range of linearity to demonstrate the effects of such data on binding discrimination using GLYMMR. Using non-linear data results in a bias in which weaker binding glycans receive a higher-than-appropriate rank, which causes the algorithm, with the parameters used, to miss the trimannosyl core as a motif (Fig. 8). When we eliminated the non-linear data, the analysis by GLYMMR discovered the trimannosyl core as a motif (Fig. 9), demonstrating the requirement of high-quality data, and the ability of the algorithm to use fewer than three datasets for analysis. In fact, the analysis can be used with a single data set if the data are in a linear range; however, non-specific binding glycans, which do not vary with concentration of GBP, would not be identified.
The analysis of UEA-I revealed the well-known specificity of this lectin for the H type 2 structure (Matsumoto and Osawa, 1969), but some unexpected observations were made. In this case the inspection of the ranking of glycans (Supplementary Table S5; see online supplementary material at http://www.liebertpub.com) indicates that the three highest-ranking glycans are sulfated derivatives of the UEA-I determinant (glycans 251, 213, and 212), which would not be detected as motifs because they were not found on a sufficient number of glycans. These data suggest that sulfated glycans may be important in natural UEA-I-glycan interactions.
We included the analysis of Gal-8 on a different version of the array (Fig. 11), to demonstrate the limitations of a single set of parameters and the versatility of the algorithm to perform well using any glycan array that is entered into the database with diverse and complex lectins. While the initial set of parameters worked well for GBPs with a single carbohydrate recognition domain (CRD), like Con A, SNA, HPA, PNA, and UEA-I, the evaluation of GBPs with multiple specificities or multiple CRDs required an expanded display of motifs and the ability to filter out motifs that are substructures of larger motifs. The user can change easily these parameters using the web-based interface for any lectin analysis without the need to modify the parameters in the algorithm itself. This would give users the flexibility to investigate both simple and complex lectins in even larger arrays, while adjusting the number of motif structures they want to view depending on if they want to do a quick or thorough analysis. As future work, we will add the ability to change other parameters such as Tb (the minimum number of binding glycans a subtree must exist in to be a motif), and Tn (the maximum number of non-binding glycans a subtree can exist in to be a motif), from the user interface. We will also develop more filtering options like eliminating all the smaller motifs if they are part of a larger motif, regardless of their binding glycans.
In summary, GLYMMR is a useful motif discovery algorithm incorporated into a web-based application. It has been tested and is publically available at http://glycanmotifminer.emory.edu for versions 4.0 and 4.2 of the CFG mammalian cell glycan microarray using the data presented in this article, and it will eventually be expanded for use on all versions of the CFG glycan microarray. Participating investigators of the CFG and the public can use it to define the specificity of GBPs, which is difficult to accomplish by manual inspection of hundreds of different glycan structures.
Supplementary Material
Acknowledgments
The authors acknowledge the use of the Consortium for Functional Glycomics' Glycan Microarray, and the efforts of Core D of the CFG for maintaining the glycan library and manufacturing the printed array, and Core H of the CFG for analysis of the lectins, all supported by National Institutes of Health grant GM62116. A CFG Bridging Grant (GM62116 to D.F.S.), and a EUREKA Grant (GM85448 to D.F.S.) also supported this work. The authors also thank Jamie Heimburg-Molinaro for manuscript editing and review, and Keri Bencuya for coordinating parts of bioinformatics components of the project, and appreciate the support from grant UL1RR025008 (NCRR CTSA) to J.H.S.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Baenziger J.U. Fiete D. Structural determinants of concanavalin A specificity for oligosaccharides. J Biol Chem. 1979;254:2400–2407. [PubMed] [Google Scholar]
- Baldus S.E. Thiele J. Park Y.O. Hanisch F.G. Bara J. Fischer R. Characterization of the binding specificity of Anguilla anguilla agglutinin (AAA) in comparison to Ulex europaeus agglutinin I (UEA-I) Glycoconj J. 1996;13:585–590. doi: 10.1007/BF00731446. [DOI] [PubMed] [Google Scholar]
- Bird G.W. Anti-T in Peanuts. Vox Sang. 1964;9:748–749. doi: 10.1111/j.1423-0410.1964.tb04072.x. [DOI] [PubMed] [Google Scholar]
- Blixt O. Head S. Mondala T., et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson S. Oberg C.T. Carlsson M.C., et al. Affinity of galectin-8 and its carbohydrate recognition domains for ligands in solution and at the cell surface. Glycobiology. 2007;17:663–676. doi: 10.1093/glycob/cwm026. [DOI] [PubMed] [Google Scholar]
- Chi Y. Muntz R.R.S. Nijssen S. Kok J.N. Frequent subtree mining—An overview. Fundamenta Informaticae. 2005;66:161–198. [Google Scholar]
- Cummings R.D. Use of lectins in analysis of glycoconjugates. Methods Enzymol. 1994;230:66–86. doi: 10.1016/0076-6879(94)30008-9. [DOI] [PubMed] [Google Scholar]
- Cummings R.D. The repertoire of glycan determinants in the human glycome. Mol Biosyst. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- Debray H. Decout D. Strecker G. Spik G. Montreuil J. Specificity of twelve lectins towards oligosaccharides and glycopeptides related to N-glycosylproteins. Eur J Biochem. 1981;117:41–55. doi: 10.1111/j.1432-1033.1981.tb06300.x. [DOI] [PubMed] [Google Scholar]
- Feizi T. CHAI W. Oligosaccharide microarrays to decipher the glyco code. Nat Rev Mol Cell Biol. 2004;5:582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- Fukui S. Feizi T. Galustian C. Lawson A.M. Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- Goldstein I.J. Hollerman C.E. Smith E.E. Protein-carbohydrate interaction. Ii. Inhibition studies on the interaction of concanavalin a with polysaccharides. Biochemistry. 1965;4:876–883. doi: 10.1021/bi00881a013. [DOI] [PubMed] [Google Scholar]
- Goldstein I.J. Studies on the combining sites of concanavalin A. Adv Exp Med Biol. 1975;55:35–53. doi: 10.1007/978-1-4684-0949-9_3. [DOI] [PubMed] [Google Scholar]
- Hammarstrom S. Kabat E.A. Studies on specificity and binding properties of the blood group A reactive hemagglutinin from Helix pomatia. Biochemistry. 1971;10:1684–1692. doi: 10.1021/bi00785a028. [DOI] [PubMed] [Google Scholar]
- Hashimoto K. Takigawa I. Shiga M. Kanehisa M. Mamitsuka H. Mining significant tree patterns in carbohydrate sugar chains. Bioinformatics. 2008;24:i167–i173. doi: 10.1093/bioinformatics/btn293. [DOI] [PubMed] [Google Scholar]
- Lotan R. Skutelsky E. Danon D. Sharon N. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea) J Biol Chem. 1975;250:8518–8523. [PubMed] [Google Scholar]
- Matsumoto I. Osawa T. Purification and characterization of an anti-H(O) phytohemagglutinin of Ulex europeus. Biochim Biophys Acta. 1969;194:180–189. doi: 10.1016/0005-2795(69)90193-7. [DOI] [PubMed] [Google Scholar]
- Maupin K.A. Liden D. Haab B.B. The fine specificity of mannose-binding and galactose-binding lectins revealed using outlier motif analysis of glycan array data. Glycobiology. 2012;22:160–169. doi: 10.1093/glycob/cwr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle R.K. Cummings R.D. Lectin affinity chromatography of glycopeptides. Methods Enzymol. 1987;138:232–259. doi: 10.1016/0076-6879(87)38020-6. [DOI] [PubMed] [Google Scholar]
- Nakayama J. Yeh J.C. Misra A.K. Ito S. Katsuyama T. Fukuda M. Expression cloning of a human alpha1, 4–N-acetylglucosaminyltransferase that forms GlcNAcalpha1-–>4Galbeta–>R, a glycan specifically expressed in the gastric gland mucous cell-type mucin. Proc Natl Acad Sci USA. 1999;96:8991–8996. doi: 10.1073/pnas.96.16.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak J. Goldstein I.J. Immunochemical studies on the interaction between synthetic glycoconjugates and alpha-L-fucosyl binding lectins. Biochemistry. 1986;25:2829–2838. doi: 10.1021/bi00358a014. [DOI] [PubMed] [Google Scholar]
- Porter A. Yue T. Heeringa L. Day S. Suh E. Haab B.B. A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology. 2010;20:369–380. doi: 10.1093/glycob/cwp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A.K. Zhi Z.L. Turnbull J.E. Saccharide microarrays for high-throughput interrogation of glycan-protein binding interactions. Methods Mol Biol. 2009;534:313–329. doi: 10.1007/978-1-59745-022-5_22. [DOI] [PubMed] [Google Scholar]
- Rillahan C.D. Paulson J.C. Glycan microarrays for decoding the glycome. Annu Rev Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J.F. Lescar J. Chazalet V., et al. Biochemical and structural analysis of Helix pomatia agglutinin. A hexameric lectin with a novel fold. J Biol Chem. 2006;281:20171–20180. doi: 10.1074/jbc.M603452200. [DOI] [PubMed] [Google Scholar]
- Shibuya N. Goldstein I.J. Broekaert W.F. Nsimba-Lubaki M. Peeters B. Peumans W.J. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2-6)Gal/GalNAc sequence. J Biological Chem. 1987;262:1596–1601. [PubMed] [Google Scholar]
- Smith D.F. Song X. Cummings R.D. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 2010;480:417–444. doi: 10.1016/S0076-6879(10)80033-3. [DOI] [PubMed] [Google Scholar]
- Song X. Xia B. Lasanajak Y. Smith D.F. Cummings R.D. Quantifiable fluorescent glycan microarrays. Glycoconj J. 2008;25:15–25. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- Song X. Xia B. Stowell S.R. Lasanajak Y. Smith D.F. Cummings R.D. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X. Yu H. Chen X., et al. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J Biol Chem. 2011;286:31610–31622. doi: 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer G.F. Desai P.R. Common precursors of human blood group MN specificities. Biochem Biophys Res Commun. 1974;61:470–475. doi: 10.1016/0006-291x(74)90980-2. [DOI] [PubMed] [Google Scholar]
- Stowell S.R. Arthur C.M. Dias-Baruffi M., et al. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell S.R. Arthur C.M. Slanina K.A. Horton J.R. Smith D.F. Cummings R.D. Dimeric Galectin-8 induces phosphatidylserine exposure in leukocytes through polylactosamine recognition by the C-terminal domain. J Biol Chem. 2008;283:20547–20559. doi: 10.1074/jbc.M802495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sughii S. Kabat E.A. Baer H.H. Further immunochemical studies on the combining sites of Lotus tetragonolobus and Ulex europaeus I and II lectins. Carbohydr Res. 1982;99:99–101. doi: 10.1016/s0008-6215(00)80982-9. [DOI] [PubMed] [Google Scholar]
- Tateno H. Mori A. Uchiyama N., et al. Glycoconjugate microarray based on an evanescent-field fluorescence-assisted detection principle for investigation of glycan-binding proteins. Glycobiology. 2008;18:789–798. doi: 10.1093/glycob/cwn068. [DOI] [PubMed] [Google Scholar]
- Uhlenbruck G. Pardoe G.I. Bird G.W. On the specificity of lectins with a broad agglutination spectrum. II. Studies on the nature of the T-antigen and the specific receptors for the lectin of Arachis hypogoea (ground-nut) Z Immunitatsforsch Allerg Klin Immunol. 1969;138:423–433. [PubMed] [Google Scholar]
- Uhlenbruck G. Prokop O. An agglutinin from Helix pomatia, which reacts with terminal N-acetyl-D-galactosamine. Vox Sang. 1966;11:519–520. doi: 10.1111/j.1423-0410.1966.tb04250.x. [DOI] [PubMed] [Google Scholar]
- Willats W.G. Rasmussen S.E. Kristensen T. Mikkelsen J.D. Knox J.P. Sugar-coated microarrays: a novel slide surface for the high-throughput analysis of glycans. Proteomics. 2002;2:1666–1671. doi: 10.1002/1615-9861(200212)2:12<1666::AID-PROT1666>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Wu A.M.A.S. Coding and classification of D-galactose, N-acetyl-D-galactosamine, and B-D-Gal[I-3(4)]-B-D-GlcNAc, specificities of applied lectins. Carbohydrate Res. 1991;213:17. [Google Scholar]
- Zhang M.X. Nakayama J. Hidaka E., et al. Immunohistochemical demonstration of alpha1,4-N-acetylglucosaminyltransferase that forms GlcNAcalpha1,4Galbeta residues in human gastrointestinal mucosa. J Histochem Cytochem. 2001;49:587–596. doi: 10.1177/002215540104900505. [DOI] [PubMed] [Google Scholar]
- Zhi Z.L. Powell A.K. Turnbull J.E. Fabrication of carbohydrate microarrays on gold surfaces: direct attachment of nonderivatized oligosaccharides to hydrazide-modified self-assembled monolayers. Anal Chem. 2006;78:4786–4793. doi: 10.1021/ac060084f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.