Abstract
Context:
Hand, foot, and mouth disease (HFMD) remains a common problem in India, yet its etiology is largely unknown as diagnosis is based on clinical characteristics. There are very few laboratory-based molecular studies on HFMD outbreaks.
Aim:
The aim of this study was to characterize HFMD-related isolates by molecular techniques.
Settings and Design:
Between 2005 and 2008, during two documented HFMD outbreaks, 30 suspected HFMD cases presented at the Outpatient Unit of the Department of Dermatology, Christian Medical College (CMC), Vellore. Seventy-eight clinical specimens (swabs from throat, mouth, rectum, anus, buttocks, tongue, forearm, sole, and foot) were received from these patients at the Department of Clinical Virology, CMC, for routine diagnosis of hand, foot, and mouth disease.
Materials and Methods:
Samples from these patients were cultured in Vero and rhabdomyosarcoma (RD) cell lines. Isolates producing enterovirus-like cytopathogenic effect (CPE) in cell culture were identified by a nested reverse transcription–based polymerase chain reaction (RT-PCR) and sequenced. The nucleotide sequences were analyzed using the BioEdit sequence program. Homology searches were performed using the Basic Local Alignment Search Tool (BLAST) algorithm.
Statistical Analysis used:
The statistical analysis was performed using Epi Info version 6.04b and Microsoft Excel 2002 (Microsoft Office XP).
Results:
Of the 30 suspected HFMD cases, only 17 (57%) were laboratory confirmed and Coxsackievirus A16 (CVA16) was identified as the etiological agent in all these cases.
Conclusions:
Coxsackievirus A16 (CVA16) was identified as the virus that caused the HFMD outbreaks in Vellore between 2005 and 2008. Early confirmation of HFMD helps to initiate control measures to interrupt virus transmission. In the laboratory, classical diagnostic methods, culture and serological tests are being replaced by molecular techniques. Routine surveillance systems will help understand the epidemiology of HFMD in India.
Keywords: Coxsackie virus, Hand, foot and mouth disease, South India
INTRODUCTION
Hand, foot and mouth disease (HFMD) is a mild, self-limiting, exanthematous disease of infants and children below 10 years of age. Human enterovirus 71 (HEV-71) and Coxsackievirus A16 (CV-A16) are common etiological agents causing HFMD epidemics. Other Enteroviruses (EV) causing sporadic HFMD cases include CV-A4 to CV-A7, CV-A9, CV-A10, CV-B1 to CV-B3, and CV- B5. The characteristic distribution of the vesicle gives the disease its name. HFMD usually spreads from person-to-person through contact with nose or throat discharges, feces or vesicular fluid.[1]
The etiological agents of HFMD belong to the Enterovirus genus, family Picornaviridae. They are small, non-enveloped, single-stranded, positive-sense RNA viruses. HEV-71-related HFMD epidemics in Singapore,[2] Sarawak,[3] and Taiwan[4] have reported serious complications like encephalitis, myocarditis, and death. Comparatively, CV-A16 causes milder infections than HEV-71.[5]
The genome of enteroviruses is about 7.5 kb in length; the open reading frame is preceded by a long untranslated (UTR) 5′ region, followed by a shorter 3′ UTR. Enteroviruses possess four viral structural proteins: VP1, VP2, VP3, and VP4.[6] The highly conserved 5′ UTR is frequently employed for characterization and the more variable VP1 region, for genotyping of enteroviruses.[4,7,8]
Enterovirus infections are common in children under four years of age and peak incidence is seen in one-year-olds.[2,3] Moreover, infants younger than six months of age had lower mortality compared to those 0.5 to <1 year of age.[9] Highly susceptible children congregate in childcare centers, kindergartens, and preschools. These institutions are excellent reservoirs for the rapid spread of HFMD, which is then transmitted to their families and the rest of the population.[10]
There are very few reports of HFMD from India. As laboratory testing of HFMD is not readily available in India, diagnosis is often based on clinical characteristics alone. Laboratory-confirmed HFMD outbreaks have been reported from Calicut and Nagpur. The microneutralization test helped to identify HEV-71, the etiological agent of the Calicut outbreak. In Nagpur, CVA16 was detected from the serum sample of an HFMD patient by RT-PCR.[11,12] In 2005 and 2008, there were HFMD outbreaks in Vellore, Tamil Nadu. Here we report the laboratory diagnosis of HFMD cases that presented at the Department of Dermatology, CMC, Vellore. Samples from multiple sites were cultured in cell lines and isolates subjected to nested PCR followed by sequencing. To the best of our knowledge, this is the first complete report on the molecular characterization of HFMD-related isolates from India.
MATERIALS AND METHODS
Thirty suspected HFMD pediatric cases, who presented to the Outpatient Unit of the Department of Dermatology, CMC, Vellore, with clinical evidence of hand, foot and mouth disease were taken for the study (male to female ratio was 1 : 1). Most of the samples were collected during two documented outbreaks in November and December of 2005 and in January and February of 2008. The significant clinical features seen were fever; ulcers with erythematous halo in the oral mucosa, predominantly on the tongue and buccal mucosa; elliptical vesicles on the hands and feet, and erythematous papular eruptions on the buttocks, elbows, and knees. The average age of the male and female patients was 3.8 and 2.7 years respectively. Whenever possible, samples from multiple sites were collected. Seventy eight (n=78) clinical specimens, (which included swabs from throat, mouth, rectum, anus, buttocks, mouth, tongue, forearm, sole, and foot) were received at the Department of Clinical Virology, CMC, for routine diagnosis of hand, foot, and mouth disease.
Virus culture and characterization of isolates
All samples were processed within an hour of receipt and inoculated into flasks of Vero cells. The inoculated cell lines were incubated at 37°C and examined daily for cytopathic effect (CPE). CPE suggestive of EV infection consisted of rounding, shrinking, nuclear pyknosis, refractility, and monolayer degeneration.[1] If virus-induced CPE was observed, the infected cells were frozen and thawed, and the cell culture supernatant was used for RNA extraction.[13] Negative cultures were incubated for 10 days.
Viral RNA was extracted from 140 μl of cell culture supernatant using the Qiamp Viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Reverse transcription was carried out with the Moloney murine leukemia virus (MMLV) enzyme (Invitrogen Corp., Carlsbad, CA, USA) and random hexamers. The cDNA obtained was then subjected to a nested polymerase chain reaction (PCR).[13] The outer primers were 5’-CGGTACCTT TGTACGCCTGTT- 3’ and 5’-CCGCATTCAGGGGCCGGAGGACT-3’, while the inner primers were 5’-GCACTTCTGTTTCCCC-3’ and 5’-CATTCAGGGGCCGGAGGA-3’.
Appropriate positive and negative controls were included with every PCR assay. The nested PCR yielded a 304 bp product. Furthermore, EV isolates (one per patient) were subjected to typing with consensus EV sequencing primers, which spanned the VP1-2A region of the EV genome.[13] The typing primers used included 5’-ATGTAYGTXCCXCCXGGXGG-3’, 5’-ATGTAYRTXCCXMCXGGXGC-3’, and 5’-GCXCCXGAYTGXTGXCCRAA-3’. All the primers used in our study were from published sources.
The PCR products were purified and sequenced on an automated DNA sequencer (ABI 310, PE Applied Biosystems). The nucleotide sequences were analyzed using the BioEdit sequence program. Homology searches were performed using the Basic Local Alignment Search Tool (BLAST) algorithm. Enterovirus sequences AM292476.1 (Human Coxsackievirus A16 partial VP1 gene), AY895110.1 (Human Coxsackievirus A16 strain), AM292435.1 (Human Coxsackievirus A16 partial VP1 gene), AM292460.1 (Human Coxsackievirus A16 partial VP1 gene), and AM292447.1 (Human Coxsackievirus A16 partial VP1 gene) of reference strains from the GenBank were used for phylogenetic analysis. The neighbor-joining method of phylogenetic analysis from the MEGA 4 program package, version 4.0, was used [Figure 1].
Figure 1.
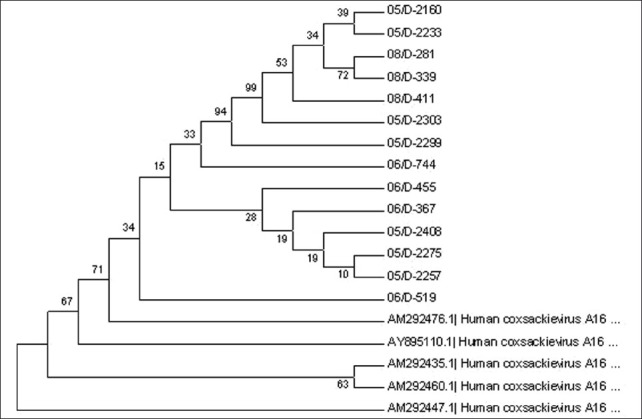
Dendrogram of the 14 strains, along with Genbank reference strains is presented. The percentage of bootstrap frequency of each branch in the tree is indicated
RESULTS
Of the 78 samples received from 30 patients, 28 showed CPE suggestive of enterovirus infection. Of the 30 suspected HFMD cases, only 17 (57%) were laboratory confirmed. On an average, CPE appeared after five days of inoculation. All suspected EV isolates, when re-passaged, showed evidence of virus growth within 24 to 48 hours. The results of virus isolation from multiple sample sites have been represented in Table 1. All patients recovered without any complications.
Table 1.
Details of enterovirus isolates from different samples
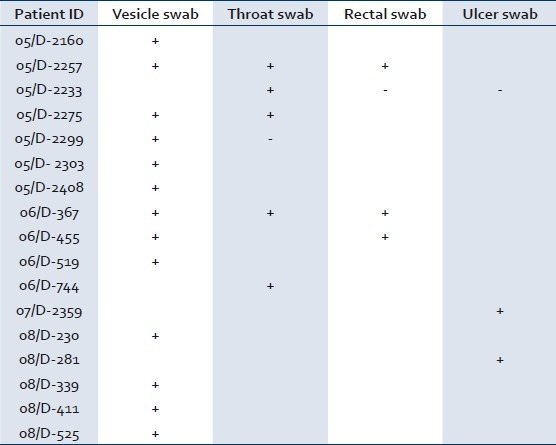
All isolates were characterized as enterovirus by nested RT-PCR. One isolate per patient (17 isolates) was subjected to sequencing PCR. Three isolates (18%) failed to amplify in sequencing PCR. The other 14 (82%) isolates were sequenced. The sequences when subjected to BLAST analysis were identified as serotype CVA16.

DISCUSSION
The emergence of non-polio enteroviruses has assumed great clinical importance as polioviruses are nearly eradicated and there are no effective antivirals or vaccines currently available.
Unlike the benign type of HFMD caused by CVA 16 and other Coxsackieviruses, HEV-71 can cause large epidemics, with severe neurological manifestations and fatal pulmonary complications. If HEV-71 is identified, preventive measures must be taken to stop transmission. Viral surveillance is therefore important.
Although serious outbreaks of HFMD have occurred in many Asian countries, there are few reports from India. This could be due to many reasons. Primarily, HFMD cases tend to be benign; hence, patients may not seek clinical care. Second, physicians may be unaware of the clinical features of HFMD. Third, as treatment is supportive, the expenses incurred for laboratory diagnosis can often be prohibitive. Most patients consult private practitioners at private clinics, with poor documentation of cases. Last but not the least, most laboratories do not offer diagnostic tests for HFMD.
In 1997, in Sarawak (Malaysia), 29 previously healthy children aged <6 years (median, 1.5 years) died of rapidly progressive cardiorespiratory failure during an outbreak of hand, foot, and mouth disease, caused primarily by HEV-71. The unique features of the outbreak were the rapid onset and progression of cardiac and pulmonary failure in previously healthy children and no clinical features were identified that could reliably predict the severe course of the disease resulting in death.[3]
In 2000, HEV-71 caused the largest HFMD epidemic recorded to date in Singapore, an epidemic that involved mainly young children <4 years of age. Children older than 10 years of age were also affected, and four deaths (two HFMD and two non-HFMD cases) were documented, which were associated with HEV-71 and extremes of the clinical spectrum of HEV-71, including non-specific febrile illness, myocarditis, and encephalitis.[2]
There is much concern with respect to HEV-71-related HFMD, due to its neuropathogenicity. It is believed that unusual clinical complications, including interstitial pneumonitis and associated deaths, can be attributed to a repertoire of viral genetic variations affecting the virulence and tropism of the etiological agents. Hence, molecular diagnosis of isolates, as in this study, will not only help to know the epidemiological trends of the disease, but will be beneficial to the development of vaccines and therapeutic agents in managing neurological complications.
Viral culture is the gold standard for laboratory diagnosis of EV infections.[14] HEV-71 and CV-A16, the important etiological agents of HFMD, usually produce CPE in RD, and Vero cell lines.[1] We have used a nested PCR to detect the enterovirus genome sequences. Isolates have been identified as CV-A16, using PCR-based analysis. The inability of the sequencing primers to amplify most, but not all isolates, probably reflects the genetic diversity of the isolates. These results underscore the need for development of improved primers to detect genetically diverse isolates.
Enterovirus (EV) isolation can be attempted from a wide range of samples, including rectal and throat swabs and swabs from vesicles and ulcers. During HFMD outbreaks, isolation rates are best in swabs from the throat and vesicles (if present). In the absence of vesicle swabs, throat and rectal swabs give optimal isolation rates. Most of our isolates were from vesicle swabs [Table 1]. Caution must be exercised with rectal and throat swab isolates as they may represent asymptomatic carriage. Limited sampling is advocated in developing countries with limited resources. Even if samples from multiple sites are used, they can be investigated in a stepwise manner, with the most useful sample being tested first.[15]
An HEV-71-related HFMD outbreak has been documented in Calicut between October 2003 and February 2004. Of the 81 suspected pediatric cases, a specific neutralization assay on 19 showed a significant rise in the EV71 antibody titer. Reports of an HFMD outbreak from Nagpur between September 2005 and April 2006 were based on a clinical diagnosis made on four children with laboratory diagnosis of CV-A16 by RT-PCR only in one patient.[11,12]
CV-A16 causes large outbreaks, interspersed with periods of quiescence.[16] The period of quiescence will depend on the build-up of the cohort of non-immune individuals. The outbreak in Singapore was largely contained by instituting measures like closure of child-care centers, repeated public health education through the mass media on observance of good personal hygiene, and keeping children away from crowds.[2]
CONCLUSION
HFMD is a relatively unknown disease in India. As the clinical manifestations may be atypical and varied, clinical diagnosis may not suffice. Laboratory confirmation with molecular analysis is important as genetic recombination may produce strains with high pathogenic potential. Molecular typing of enteroviruses, unlike serological typing, is less cumbersome, important for epidemiological reasons and helps to document serotype-specific clinical features. Early detection and confirmation helps patient management, reduces hospitalization, prevents spread to susceptibles, excludes other infectious causes and eliminates unnecessary antibiotic usuage. Routine surveillance will provide additional information on the epidemiology of HFMD in India.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Russo DH, Luchs A, Machado BC, Carmona Rde C, Timenetsky Mdo C. Echovirus 4 associated to hand, foot and mouth disease. Rev Inst Med Trop Sao Paulo. 2006;48:197–9. doi: 10.1590/s0036-46652006000400004. [DOI] [PubMed] [Google Scholar]
- 2.Chan KP, Goh KT, Chong CY, Teo ES, Lau G, Ling AE. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg Infect Dis. 2003;9:78–85. doi: 10.3201/eid1301.020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan LG, Parashar UD, Lye MS, Ong FG, Zaki SR, Alexander JP, et al. Deaths of children during an outbreak of hand, foot, and mouth disease in Sarawak, Malaysia: Clinical and pathological characteristics of the disease. Clin Infect Dis. 2000;31:678–83. doi: 10.1086/314032. [DOI] [PubMed] [Google Scholar]
- 4.Wang JR, Tuan YC, Tsai HP, Yan JJ, Liu CC, Su IJ. Change of major genotype of enterovirus 71 in outbreaks of hand-foot-and-mouth disease in Taiwan between 1998 and 2000. J Clin Microbiol. 2002;40:10–5. doi: 10.1128/JCM.40.1.10-15.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang LY, Lin TY, Hsu KH, Huang YC, Lin KL, Hsueh C, et al. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet. 1999;354:1682–6. doi: 10.1016/S0140-6736(99)04434-7. [DOI] [PubMed] [Google Scholar]
- 6.Singh S, Poh CL, Chow VT. Complete sequence analyses of enterovirus 71 strains from fatal and non-fatal cases of the hand, foot and mouth disease outbreak in Singapore (2000) Microbiol Immunol. 2002;46:801–8. doi: 10.1111/j.1348-0421.2002.tb02767.x. [DOI] [PubMed] [Google Scholar]
- 7.Mori J, Clewly JP. Polymerase chain reaction and sequencing for typing rhinovirus RNA. J Med Virol. 1994;44:323–9. doi: 10.1002/jmv.1890440403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oberste MS, Maher K, Kilpatrick DR, Flemister MR, Brown BA, Pallansch MA. Typing of Human Enterovirus by partial sequencing of VP1. J Clin Microbiol. 1999;37:1288–93. doi: 10.1128/jcm.37.5.1288-1293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen SC, Chang HL, Yan TR, Cheng YT, Chen KT. An eight-year study of epidemiologic features of enterovirus 71 infection in Taiwan. Am J Trop Med Hyg. 2007;77:188–91. [PubMed] [Google Scholar]
- 10.Ang LW, Koh BK, Chan KP, Chua LT, James L, Goh KT. Epidemiology and control of hand, foot and mouth disease in Singapore, 2001-2007. Ann Acad Med Singapore. 2009;38:106–12. [PubMed] [Google Scholar]
- 11.Sasidharan CK, Sugathan P, Agarwal R, Khare S, Lal S, Jayaram Paniker CK. Hand-foot-and-mouth disease in Calicut. Indian J Pediatr. 2005;72:17–21. doi: 10.1007/BF02760573. [DOI] [PubMed] [Google Scholar]
- 12.Saoji VA. Hand, foot and mouth disease in Nagpur. Indian J Pediatr. 2008;74:133–5. doi: 10.4103/0378-6323.39697. [DOI] [PubMed] [Google Scholar]
- 13.Manayani DJ, Cherian T, Murali N, Finny GJ, Green J, Brown D, et al. Evaluation of a one-tube RT-PCR system for detection of enteroviruses. J Clin Virol. 2002;24:25–30. doi: 10.1016/s1386-6532(01)00234-7. [DOI] [PubMed] [Google Scholar]
- 14.Nijhuis M, van Maarseveen N, Schuurman R, Verkuijlen S, de Vos M, Hendriksen K, et al. Rapid and sensitive routine detection of all members of the genus enterovirus in different clinical specimens by real-time PCR. J Clin Microbiol. 2002;40:3666–70. doi: 10.1128/JCM.40.10.3666-3670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ooi MH, Solomon T, Podin Y, Mohan A, Akin W, Yusuf MA, et al. Evaluation of different clinical sample types in diagnosis of human enterovirus 71-associated hand-foot-and-mouth disease. J Clin Microbiol. 2007;45:1858–66. doi: 10.1128/JCM.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosoya M, Kawasaki Y, Sato M, Honzumi K, Hayashi A, Hiroshima T, et al. Genetic diversity of Coxsackievirus A16 associated with hand, foot and mouth disease epidemics in Japan from 1983 to 2003. J Clin Microbiol. 2007;45:112–20. doi: 10.1128/JCM.00718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


