Abstract
The human X-linked macrosatellite DXZ4 is a large tandem repeat located at Xq23 that is packaged into heterochromatin on the male X chromosome and female active X chromosome and, in response to X chromosome, inactivation is organized into euchromatin bound by the insulator protein CCCTC-binding factor (CTCF) on the inactive X chromosome (Xi). The purpose served by this unusual epigenetic regulation is unclear, but suggests a Xi-specific gain of function for DXZ4. Other less extensive bands of euchromatin can be observed on the Xi, but the identity of the underlying DNA sequences is unknown. Here, we report the identification of two novel human X-linked tandem repeats, located 58 Mb proximal and 16 Mb distal to the macrosatellite DXZ4. Both tandem repeats are entirely contained within the transcriptional unit of novel spliced transcripts. Like DXZ4, the tandem repeats are packaged into Xi-specific CTCF-bound euchromatin. These sequences undergo frequent CTCF-dependent interactions with DXZ4 on the Xi, implicating DXZ4 as an epigenetically regulated Xi-specific structural element and providing the first putative functional attribute of a macrosatellite in the human genome.
INTRODUCTION
Estimates of repeat DNA content in the human genome range from half (1) to over two-thirds (2). These elements include the tandem repeats that consist of homologous DNA sequences that are organized into a head to tail arrangement. The size of the individual repeating unit can range from the simple microsatellite repeat of 1–6bp that are found scattered throughout our genome (3), to much larger repeat units of several kilobases (kb) that typically occupy a handful of chromosomal locations (4). The copy number of the individual repeat units is generally polymorphic from one individual to the next (5) accounting for why many of the smaller tandem repeats became the marker of choice for genetic studies (6), and explaining why some of the larger tandem repeats remain underrepresented or incomplete in the various builds of the human genome (7,8), a situation that is compounded by the inherent difficulty in the assembly of repetitive sequences (9).
Some of the largest tandem repeats in the human genome are the macrosatellites, also referred to as the megasatellites (4). The different macrosatellite repeats typically exist at one or two chromosomal locations such as the TAF11-like macrosatellite that is specific to chromosome 5p15.1 (10,11) or RS447 located at chromosomes 4p15 and 8pter (12). The macrosatellites are composed of individual repeat units of several kb in size and can consist of only a few to over one hundred individual repeat units. To date, only a few have been characterized in detail (4,10,11,13–16). What purpose these sequence elements have in our genome is unknown, but their contribution to disease susceptibility is clearly demonstrated by the link between contraction in the copy number of the 4q35 macrosatellite D4Z4 and onset of facioscapulohumeral muscular dystrophy (17).
Among the macrosatellites, one that is the subject of intense investigation in our laboratory is the X-linked repeat DXZ4. DXZ4 is composed of between 12 and 100 three-kb repeat units arranged in tandem at Xq23 (13,15). X-linkage means that in females, DXZ4 is exposed to the epigenetic process of X chromosome inactivation (XCI). XCI is the mammalian form of dosage compensation that serves to balance the levels of X-linked gene expression between the sexes (18). XCI occurs very early in development (19) and gene silencing is achieved at the chosen inactive X chromosome (Xi) through repackaging its chromatin into facultative heterochromatin (20). In response, however, DXZ4 adopts an unexpected chromatin organization that differs from that of the flanking chromosome. In males and on the female active X chromosome, DXZ4 is packaged into heterochromatin characterized by histone H3 trimethylated at lysine-9 (H3K9me3) (21–23) and DNA CpG hypermethylation (13,22). In contrast, DXZ4 on the Xi is packaged into euchromatin characterized by histone H3 dimethylated at lysine 4 (H3K4me2) (21,22,24) and CpG hypomethylation (13,22) and is bound by the zinc finger proteins CCCTC-binding factor (CTCF) (22,25) and Yin Yang-1 (YY1) (26). At metaphase, DXZ4 can readily be detected on the Xi as the most intense signal of H3K4me2 at Xq23 (24) at the distal edge of an extensive band of heterochromatin characterized by the histone variant macroH2A1 and proximal to a band of H3K9me3 (24,27). At interphase, H3K4me2 distribution clearly reveals DXZ4 as an intense focus or ‘dot’ (25) within the hypo-H3K4me2 territory of the Xi (21). Furthermore, the numerous bands of macroH2A1 and H3K9me3 that give the chromosome a striped pattern at metaphase by immunofluorescence (28) appear at interphase as two distinct chromosomal territories (27). Given that the multifunctional protein CTCF (29) plays a central role in the organization of chromosomal territories at interphase (30,31), it is conceivable that Xi-specific CTCF association with euchromatic DXZ4 at the boundary between two different heterochromatin territories could contribute to the organization of the Xi chromatin at interphase. However, this would likely require the presence of additional Xi-specific CTCF bound sequences elsewhere on the Xi. While bands of H3K4me2 have been reported at the distal edge of other macroH2A1 bands on the Xi (24), the sequence identity and binding of CTCF at these bands are unknown.
RESULTS
Identification of large novel tandem repeats at 56 and 130 Mb on the X chromosome that map to bands of H3K4me2
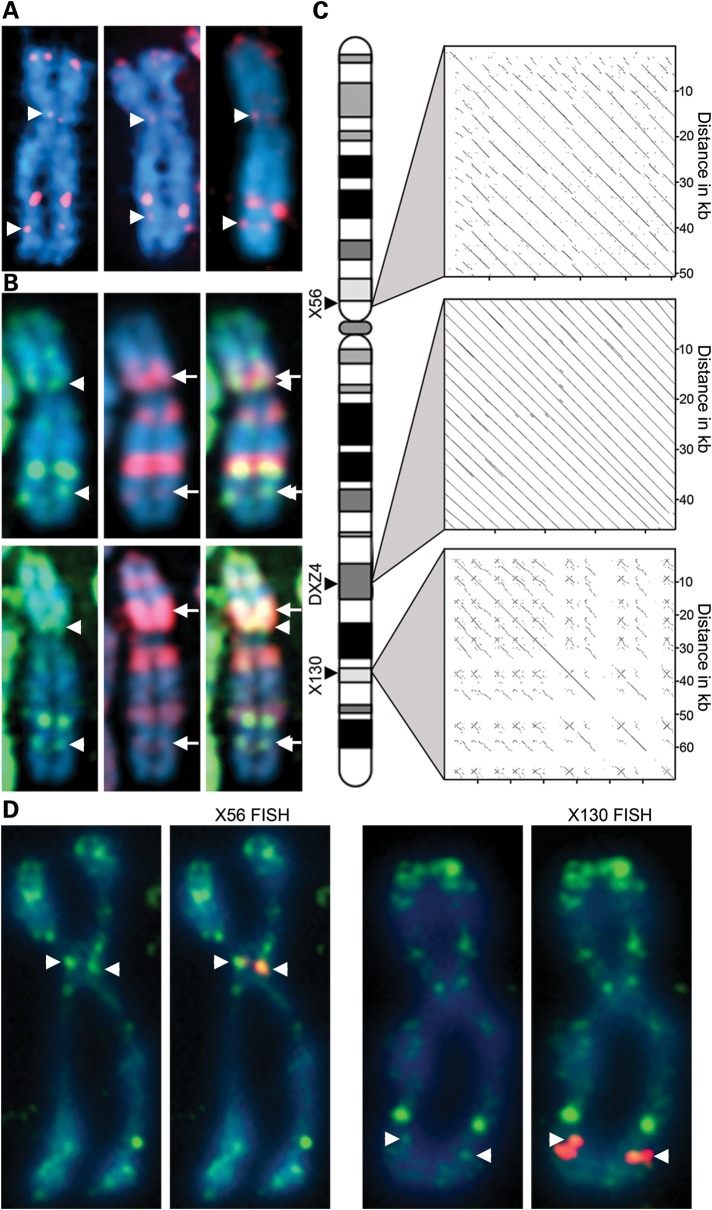
H3K4me2 at DXZ4 was originally identified through the observation of an intense signal of H3K4me2 on the long arm of the X chromosome at the distal edge of a major chromatin band characterized by the histone variant macroH2A1 (24). Prominent H3K4me2 signals at the tip of Xp are expected as this region corresponds to the pseudoautosomal region of the X that is shared with the Y chromosome (32). It is not dosage compensated resulting in gene expression from both the Xa and Xi (33) accounting for the euchromatin signal. Additional weaker bands of H3K4me2 are observed on the metaphase Xi that are conserved in primates (34); the most consistent signals were located at Xp11.21 and Xq26.2 (Fig. 1A). Like DXZ4, these signals reside at the distal edges of macroH2A1 bands relative to the Xp telomere (Fig. 1B). Given that DXZ4 is a macrosatellite, we sought to determine whether other tandem repeat DNA could underlie the H3K4me2 signals at Xp11.21 and Xq26.2. Using the UCSC genome browser (http://genome.ucsc.edu), we extracted large DNA fragments from each chromosomal band and aligned them pairwise against a repeat-masked version of the same sequence. At both locations, DNA sequences displaying extensive tandem repeat organization were identified at 56 Mb (X56) and 130 Mb (X130) (Fig. 1C). To confirm that these sequences corresponded to the locations of the H3K4me2 signals, we used a bacterial artificial chromosome (BAC) clone encompassing each tandem repeat to perform fluorescence in situ hybridization (FISH) on anti-H3K4me2 immunostained metaphase chromosomes. Both probes were inseparable from the H3K4me2 signals (Fig. 1D).
Figure 1.
Mapping of two novel tandem repeats at X56 and X130 to H3K4me2 bands on the inactive X chromosome (Xi). (A) Metaphase Xi showing H3K4me2 (red) merged with DAPI (blue). Signals at X56/X130 indicated by white arrowheads. (B) As above showing H3K4me2 (green) and macroH2A1 (red). White arrows indicate X56/130 proximal macroH2A1 bands. (C) Relative chromosomal location of X56/DXZ4/X130 and pairwise alignments of the corresponding DNA sequences (X56 = 56 781 370–56 833 369; DXZ4 = 114 959 731–115 005 842; X130 = 130 859 561–130 929 560; Hg19 coordinates). Pairwise alignments generated using YASS (57) using default alignment settings. (D) Metaphase Xi H3K4me2 distribution (green) merged with X56/X130 DNA FISH (red). Images in (A) were captured using a Zeiss Axiovert 200M, whereas the images in (D) were captured using a DeltaVision pDV with deconvolution, accounting for the sharper signals.
DXZ4 is composed of between 12 and 100 uninterrupted 3.0 kb repeat units that share 99% DNA sequence identity between adjacent monomers. Repeat units are 62% GC and <5% is repetitive, consisting of three polymorphic microsatellite repeats (13,15). In contrast, both X56 and X130 are less well organized. X56 spans 52.0 kb, is 54% GC and is composed of an imperfect ∼5.4 kb repeat unit that is arranged in tandem ∼10 times, with between 68 and 100% sequence identity between adjacent repeats. Approximately 15% of the tandem array is repeat masked with 9.6% corresponding to interspersed repeats (7.3% LINE) and 5.5% simple and low complexity repeats. X130 spans 70.0 kb, is 41% GC and is the least well-conserved tandem repeat of the three. No obvious consensus repeat unit is readily defined, with stretches of ∼2.0 kb sharing 68–85% nucleotide identity throughout the interval. X130 has higher repeat content (46.2%) with most of that accounted for by interspersed repeats (20.4% SINE and 16.7% LINE). The reduced homogeneity at the X56 and X130 tandem repeats relative to DXZ4 is reflected in the more dispersed nature of the pair-wise alignments shown in Figure 1C.
Universal expression of spliced transcripts spanning the X56 and X130 tandem repeats from the Xa
DXZ4 is transcribed from a promoter element contained within each tandem repeat unit (22). DXZ4 transcript can be detected in both sexes and in all human tissues examined (15). In females, most transcript originates from the Xa, a feature that is conserved in macaque (34). Close examination of the genomic locus for X56 and X130 reveals several GenBank messenger RNA (mRNA) for each that is annotated as Reference Sequence Gene LOC550643 at X56 and LOC286467 for X130. At X56, the primary transcript spans ∼90 kb and three exons are spliced together into a 0.7 kb mRNA (Fig. 2A, top map). The X56 mRNA contains a short open reading frame (ORF) that encodes a hypothetical 71 amino acid peptide. At X130, the primary transcript spans 130 kb and 13 exons are spliced into a 2.8 kb mRNA (Fig. 2A, bottom map). As with X56, the X130 mRNA contains a short ORF that is predicted to encode a hypothetical peptide of 181 amino acids. Interestingly, both mRNA initiate transcription on one side of the tandem repeat (proximal for X56 and distal for X130) and proceed across the full repeat before terminating on the opposite side. To confirm expression of X56 and X130, PCR was performed on complementary DNA (cDNA) synthesized from total RNA isolated from 20 different human tissues. Like DXZ4 (15), both X56 and X130 spliced mRNA were universally expressed, albeit with low levels in adult liver (Fig. 2B). Comparable levels of transcript could be detected in male and female samples (Fig. 2C). In order to determine if X56 and X130 were also expressed from the Xi, RNA FISH was performed in combination with a probe to the X inactive specific transcript (XIST) in order to define the territory of the Xi (35). RNA FISH signals for X56 and X130 were detected but were distant from and did not overlap the XIST territory, suggesting that transcription of both is likely restricted to the Xa (Fig. 2D). Examination of histone modifications using the UCSC genome browser (http://genome.ucsc.edu) (36) indicated a broad peak of histone H3 trimethylated at lysine 4 (H3K4me3) within the vicinity of X56 and X130 exon 1 (data not shown). H3K4me3 is associated with active promoters (37). We therefore cloned upstream of a promoterless luciferase reporter gene the DNA sequence immediately 5′ of exon 1 of X56 and X130. Lysates from cells transfected with either construct consistently gave >200-fold increase in luciferase activity relative to the promoterless luciferase construct confirming the location of the promoter for X56 and X130 (Fig. 2E).
Figure 2.
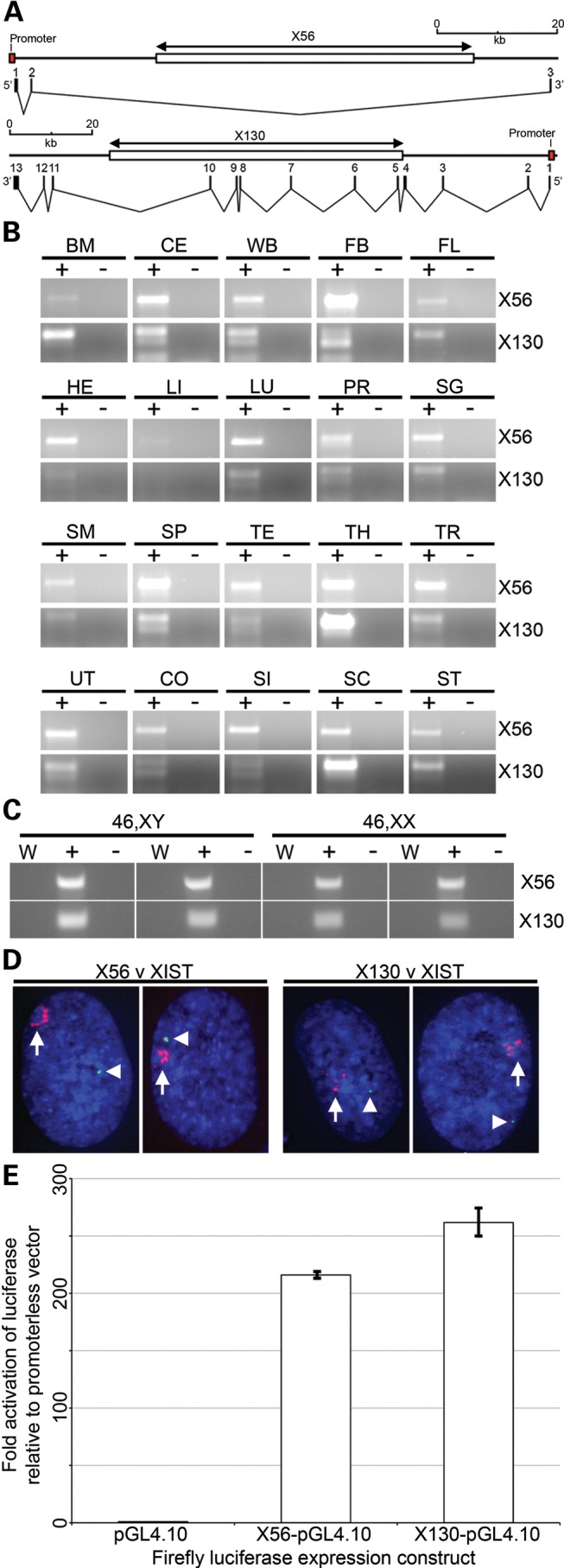
Genomic organization and expression of X56 and X130. (A) Schematic maps showing the location of the tandem array (large white box indicated above by the X56 and X130 double-headed arrow) and transcriptional units at each location. The location of the promoter is indicated by the labelled red box and exon structure for the X56 (3 exons) and X130 (13 exons) transcripts are indicated directly beneath each map. The scale is given above each schematic and regions represented are X chromosome coordinates 56 755 370–56 845 001 bp for X56 and 130 835 825–130 965 880 bp for X130 based on Hg19. (B) Expression of X56 (671 bp product, PCR between exons 1 and 3) and X130 (301 bp product, PCR between exons 1 and 3) in cDNA prepared from total RNA extracted from a broad range of human tissues. Samples include cDNA template prepared with reverse transcriptase (+) and without (−). Tissues are: BM, bone marrow; CE, cerebellum; WB, whole brain; FB, fetal brain; FL, fetal liver; HE, heart; LI, liver; LU, lung; PR, prostate; SG, salivary gland; SM, skeletal muscle; SP, spleen; TE, testis; TH, thymus; TR, trachea; UT, uterus; CO, colon; SI, small intestine; SC, spinal cord; ST, stomach. (C) Expression of X56 and X130 in male (1139 and 1140) and female (hTERT-RPE1 and IMR90) cDNA. Samples as above, also showing a no-template water control (W). (D) RNA FISH analysis of X56 and X130 expression (green signals indicated by white arrow heads) relative to XIST RNA (red signal indicated by white arrow) in hTERT-RPE1 cells merged with DAPI (Blue). Images represent deconvolved sections. (E) Bar-chart indicating luciferase activity in 293 cell extracts 72h after transfection with the promoterless luciferase vector (pGL4.10) or the same vector containing an 852 bp fragment from the putative X56 promoter or 760 bp fragment from the putative X130 promoter. Fold activation of luciferase activity is shown to the left relative to the promoterless vector. Data represent replicated experiments each performed in triplicate.
Chromatin characterization of X56 and X130 reveal Xi-specific CTCF bound euchromatin
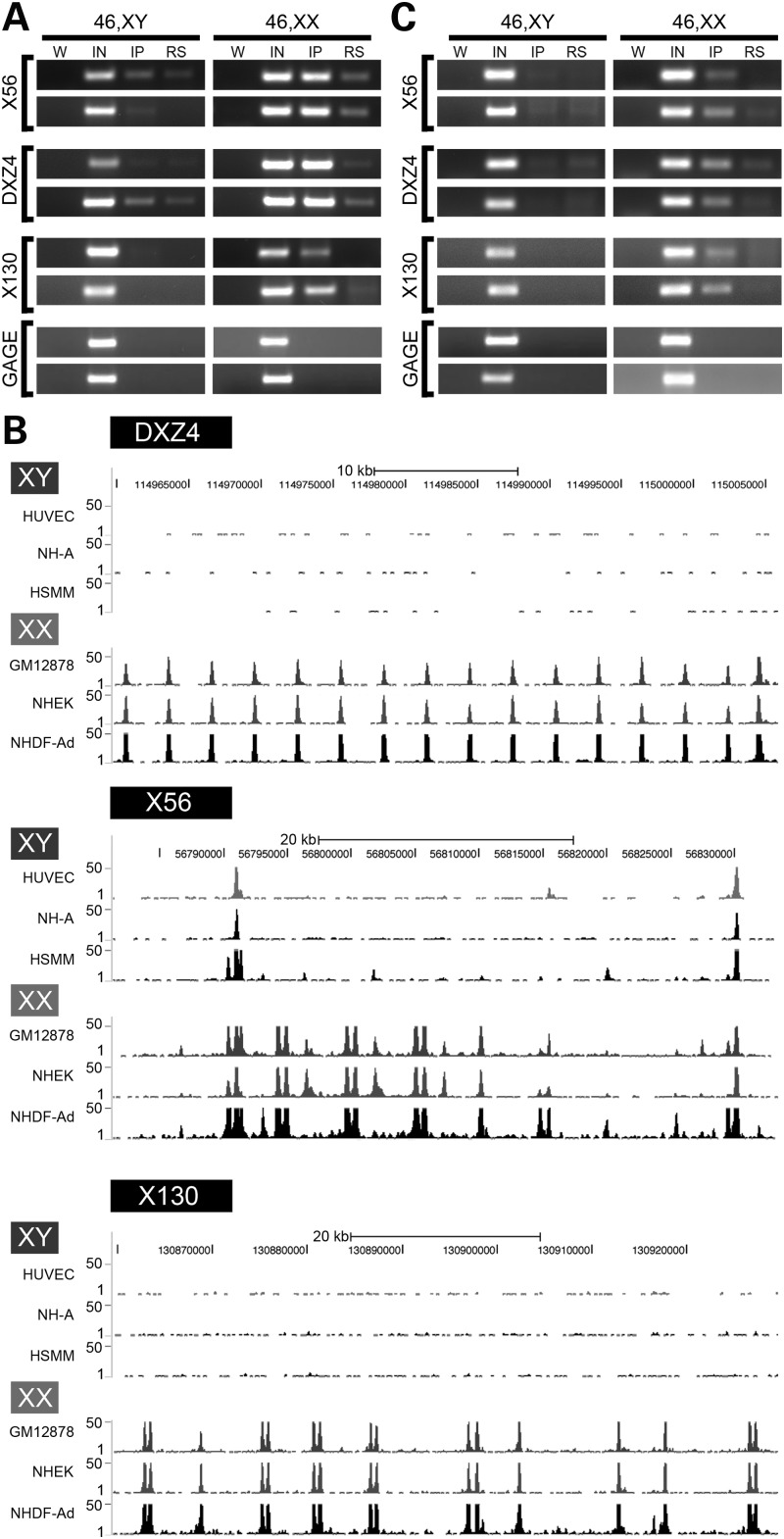
H3K4me2 is a characteristic of DXZ4 chromatin on the Xi (22). To determine whether H3K4me2 at X56 and X130 was restricted to the Xi in the same manner as DXZ4, we performed chromatin immunoprecipitation (ChIP) on several independent normal diploid male and female cell lines. Like that of DXZ4, the presence of H3K4me2 was female specific (Fig. 3A). Our interpretation of these data is that the tandem repeats at X56 and X130 show signatures of euchromatin on the Xi only. Given the similarity of genomic and chromatin organization of X56 and X130 to that of DXZ4, we looked for evidence of CTCF association with both tandem repeats. Initially, we took a bioinformatic approach and examined the publicly available ENCODE data for CTCF ChIP combined with high-throughput sequencing. Like DXZ4, both X56 and X130 show female-specific CTCF association (Fig. 3B). CTCF ChIP assayed by PCR confirmed female specificity (Fig. 3C).
Figure 3.
Chromatin characterization of the X56 and X130 intervals. (A) H3K4me2 ChIP assessed by PCR to X56, DXZ4 and X130 for two independent males and females. W, water; IN, input; IP, anti-H3K4me2/CTCF; RS, rabbit serum. Beneath these three are ChIP PCR for the GAGE locus as a negative control for H3K4me2 and CTCF. The sequence of oligonucleotides used for PCR is listed in Supplementary Material, Table S1. (B) CTCF ChIP Seq profiles covering intervals of DXZ4, X56 and X130 for three independent male and three independent female samples. Images were extracted from the UCSC Genome Browser (http://genome.ucsc.edu) that display data from the Encode histone modifications by Broad Institute ChIP Seq (36). Regions displayed correspond to DXZ4 (114 959 731–115 005 842), X56 (56 781 370–56 833 369) and X130 (130 859 561–130 929 560). Coordinates are based on human genome build Hg19. The peaks shown represent signal enrichment calculated from the density of sequence tags overlapping a 25 bp window. For each region (DXZ4, X56, and X130), the males (XY) are shown at the top, followed by the females (XX). Scale and chromosome coordinates are given above each profile. (C) CTCF ChIP assessed by PCR as for part (A).
Xi-specific long-range intrachromosomal interactions between DXZ4 and the 56 Mb and 130 MB tandem repeats
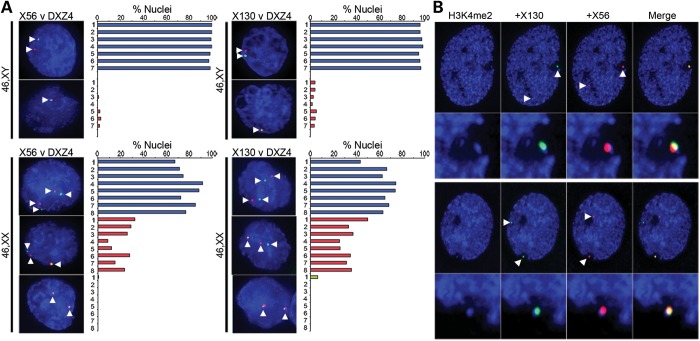
CTCF is implicated in many functions, including mediating long-range chromatin interactions (38). Given the Xi-specific association of CTCF with DXZ4, X56 and X130, we hypothesized that the three sequences interact specifically on the Xi. To test this hypothesis, we first performed FISH with DXZ4 and X56 or X130 in seven independent male and eight female cell lines. DXZ4 signal rarely overlapped with X56 or X130 in males but overlapped in a significant number of female cells at one set of alleles (P = 0.0001 for DXZ4-X56; P < 0.0001 for DXZ4-X130 using the two-sample t-test) (Fig. 4A). To confirm that the pair of overlapping signals corresponded to cis interactions on the Xi, we performed FISH with X56 and X130 combined with H3K4me2 in a female cell line. In 100% of cells (n = 102), overlap between X56 and X130 occurred within the hypo-H3K4me2 territory of the Xi at the DXZ4 ‘dot’ (25) (Fig. 4B). Furthermore, these data indicate that the DXZ4 H3K4me2 ‘dot’ actually corresponds to various combinations of the H3K4me2 signals originating from DXZ4, X56 and X130, as well as potentially other, as yet to be defined, Xi H3K4me2 signals (24).
Figure 4.
Interactions between DXZ4-X56 and DXZ4-X130. (A) FISH analysis of DXZ4 (green) and X56 or X130 (red) in male (46,XY) or female (46,XX) nuclei counterstained with DAPI. Signals are indicated by white arrowheads. The percentage of nuclei with FISH signals comparable with the examples given are indicated graphically to the right of the image. Independent male (1–7) and female (1–8) cell lines and number of nuclei scored are indicated in Supplementary Material, Table S2. (B) Two representative examples of H3K4me2 (blue) combined with DNA FISH to X130 (green) and X56 (red). Beneath each example is a blow-up of the Xi territory.
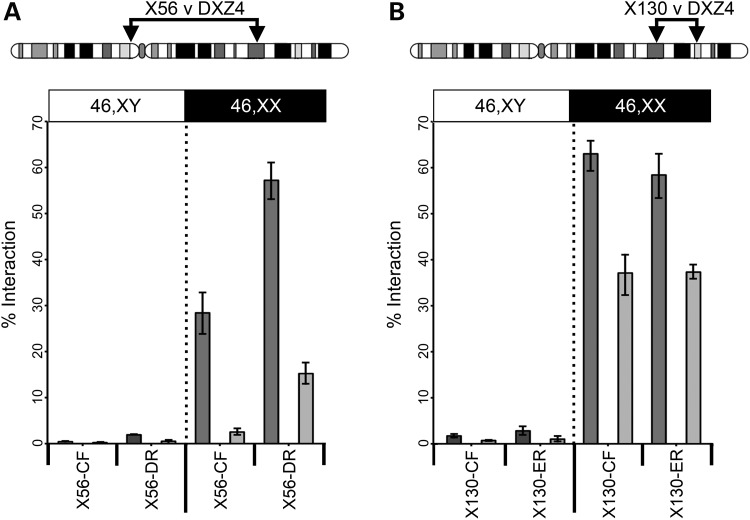
Other large tandem repeat DNA exists on the human X chromosome (39). The GAGE locus, located at Xp11.23 (40), rarely interacted with DXZ4; male and female samples did not differ significantly (P = 0.4077; Table 1), indicating that the interactions we observed among DXZ4, X56 and X130 were not a general feature of tandem repeats on the Xi. Furthermore, the frequency of observed interaction between DXZ4 and X130 was not related to their close physical proximity, because FISH with a BAC probe at a comparable distance proximal to DXZ4 rarely colocalized with DXZ4 and revealed no significant difference between the sexes (P = 0.3863; Table 1). To confirm chromatin interactions between DXZ4 and X56/X130, we used the procedure of chromosome conformation capture (3C). Oligonucleotide primers were designed to HindIII fragments of DXZ4, X56 and X130 that are bound by CTCF (Supplementary Material, Fig. S1). PCR products were only observed for female samples (Supplementary Material, Fig. S2) that, upon cloning and sequencing, were confirmed to be DXZ4-X56 or DXZ4-X130 hybrids. Quantification confirmed frequent female-specific interactions between CTCF-bound sequences at X56/X130 and DXZ4 (Fig. 5).
Table 1.
Frequency of interaction between DXZ4 and the GAGE locus (located at 49.3Mb according to Hg19) or BAC clone RP11-402K9 (located at 99.3Mb according to Hg19), in various male and female cells
| Cell line | Sex | Probe used with DXZ4 | Non-interacting (%) | Single-interacting (%) | Double-interacting (%) | n |
|---|---|---|---|---|---|---|
| hTERT-BJ1 | M | GAGE | 100.00 | 0.00 | — | 66 |
| GM07030 | M | GAGE | 93.80 | 6.20 | — | 127 |
| GM06982 | M | GAGE | 97.73 | 2.27 | — | 132 |
| IMR90 | F | GAGE | 100.00 | 0.00 | 0.00 | 40 |
| GM12802 | F | GAGE | 89.60 | 10.40 | 0.00 | 125 |
| GM21770 | F | GAGE | 91.82 | 8.18 | 0.00 | 110 |
| GM07030 | M | RP11-402K9 | 90.40 | 9.60 | — | 121 |
| GM06982 | M | RP11-402K9 | 93.50 | 6.50 | — | 124 |
| GM07030 | M | RP11-402K9 | 96.70 | 3.30 | — | 125 |
| GM012802 | F | RP11-402K9 | 92.50 | 7.50 | 0.00 | 120 |
| hTERT-RPE1 | F | RP11-402K9 | 89.20 | 10.80 | 0.00 | 65 |
Figure 5.
Interaction frequencies between DXZ4-X56 and DXZ4-X130. Chromosome conformation capture analysis for (A) X56 v DXZ4 and (B) X130 v DXZ4. Above each image is an ideogram of the X chromosome showing the location of the two regions being tested for interaction by 3C. Beneath the ideogram is a graph showing a significant difference in the percentage of interaction relative to BAC controls between DXZ4 and two different regions of X56 (CF and DR) and X130 (CF and ER) in two independent male (left side) and female (right side) samples (P = 0.0335 X56-CF; P = 0.0048 X56-DR; P = <0.0001 X130-CF and X130-ER. P-values calculated using the two-sample t-test). Oligonucleotides used for PCR are listed in Supplementary Material, Table S1. Detailed restriction maps for the regions tested are shown in Supplementary Material, Figure S1.
Intrachromosomal interactions between DXZ4 and X130 are mediated by CTCF
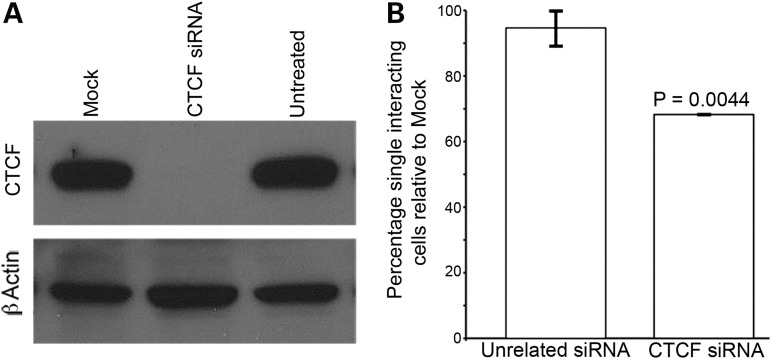
Given the importance of CTCF in mediating higher-order chromatin organization in the genome (31), we sought to determine whether CTCF mediates the Xi-specific interactions we observe by reducing the levels of the CTCF protein using small-interfering RNA (siRNA; Fig. 6A). Relative to a mock transfection, interaction frequencies between DXZ4 and X130 were significantly lower (P = 0.0044) but not when the procedure was performed with an unrelated siRNA (P = 0.6149) (Fig. 6B). Others have reported a reduction in the levels of H3K4me2 at CTCF-binding sites when CTCF protein levels are reduced by siRNA (41). We found that CTCF RNAi resulted in a significant reduction in the number of nuclei showing overlap between Xi DXZ4 and the H3K4me2 signal (P = 0.0136, Supplementary Material, Fig. S3), consistent with a role for CTCF in maintaining local euchromatin (38).
Figure 6.
Intrachromosomal interactions are dependent on CTCF. (A) CTCF and b-actin western-blot analysis of whole-cell extracts from mock, untreated and CTCF siRNA-treated hTERT-RPE1 cells. (B) Change in the percentage of overlapping DXZ4 and X130 FISH signals in hTERT-RPE1cells treated with CTCF siRNA or an with unrelated siRNA relative to mock-treated cells. Data represent two independent experiments scoring 53 + 67 mock-treated nuclei, 56 + 61 CTCF-treated nuclei and 67 + 54 nuclei treated with unrelated siRNA. P-value calculated using the two samples t-test.
DISCUSSION
We report the identification and characterization of the DNA sequence (X56 and X130) underlying two H3K4me2 signals that reside at the distal edge of macroH2A1 bands on the Xi. Like DXZ4 (13), X56 and X130 correspond to large tandem repeat DNA that each are bound by CTCF specifically on the Xi. Both X56 and X130 are transcribed, but unlike DXZ4 (22) both transcripts are spliced and transcription initiates from a defined promoter outside of the tandem repeat that drives transcription across the repeat before terminating at a sequence on the opposite side. Similar to DXZ4 (15), transcription of X56 and X130 appears to be a feature of the Xa allele. It is possible that transcription of X56 and X130 is directly linked to CTCF association. Perhaps the process of transcription across the tandem repeat prevents CTCF binding, whereas the lack of transcription from the Xi permits association, acting as the switch between the epigenetic states of the Xa and Xi alleles. Given the short ORF for both and the fact that both are only predicted proteins it is conceivable that both are long non-coding RNAs (42), a possibility that warrants further investigation.
Intriguingly, DXZ4 on the Xi makes frequent contact with X56 and X130, suggesting that similar to its role in mediating long-range inter- and intra-chromosomal interactions throughout the genome (31), CTCF could provide a structural role for the Xi mediated through DXZ4. CTCF RNAi significantly reduced interactions between DXZ4 and X130 to a degree comparable with that described by others investigating long-range CTCF-mediated chromatin interactions (31). However, not all interactions were lost. One possible explanation could be that as tandem repeats, DXZ4 and X130 possess numerous CTCF-binding sites. Even a substantial reduction in protein levels might still result in enough residual CTCF to maintain interactions. Alternatively, more than just CTCF might be involved in establishing and maintaining interactions with DXZ4. Other factors are known to mediate chromatin looping such as SATB1 (43), or the cohesin complex that colocalizes with many CTCF sites throughout the genome (44–46). Given that cohesin has been reported at DXZ4 (47), we favor this as a possible explanation for the residual interactions seen in the CTCF RNAi-treated cells.
At interphase, the spatially distinct bands of macroH2A1 and H3K9me3 that are observed at metaphase (24,27,28) appear to cluster together into a bipartite structure (27,28), with one territory defined by macroH2A1 and the other by heterochromatin protein 1 (HP1) (25) that recognizes and binds to H3K9me3 (48–50). Previously, we proposed that this organization for the Xi was achieved through folding of the chromosome to bring like-chromatin types together (27). In light of the data we present here, we refine this model in the schematic image shown in Figure 7. In this model, we propose that euchromatic forms of DXZ4, X56 and X130 permit binding of CTCF to the Xi but not Xa alleles. Long-range interactions between the CTCF bound tandem repeats assist in segregating the HP1 and mH2A1 chromatin and maintaining the bipartite organization, potentially accounting for the alternate three-dimensional organization of the Xi relative to the Xa (51). A recent report investigating the spatial organization of the genome (30) revealed that CTCF is a frequent feature at the boundary of topological domains. Examination of the authors’ publically available data set for DXZ4, X56 and X130 confirm that each of these DNA sequences reside at boundary elements (Supplementary Material, Fig. S4), supporting our model.
Figure 7.
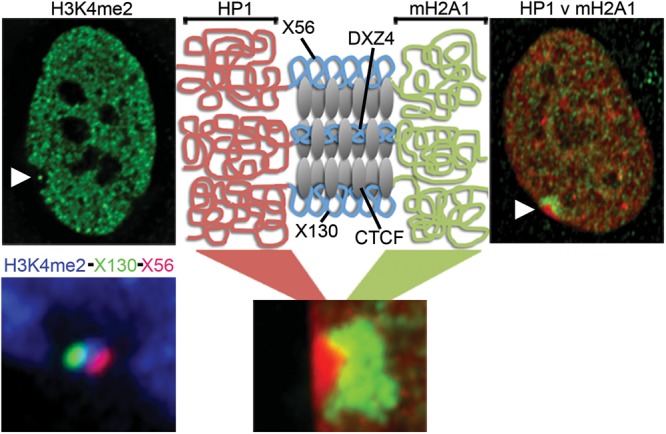
Model to explain the structural role of Xi-specific CTCF bound DXZ4, X56 and X130. Far top left image is a female nucleus showing the distribution of H3K4me2 (green) and the hole with a ‘dot’ at the Xi (white arrow head). Bottom left image is a close up example of the Xi-hole with a ‘dot’ in female cells showing the distribution of H3K4me2 (blue) combined with DNA FISH signals for X56 (red) and X130 (green). Far top right image shows the nuclear distribution of mH2A1 (green) merged with HP1g (red) in a female nucleus with the location of the Xi indicated by the white arrow-head. A close up example of HP1g (red) and mH2A1 (red) at the Xi is shown at the bottom in the middle. Directly above this is a schematic representation of chromatin at and flanking DXZ4, X56 and X130 showing HP1, mH2A1 or CTCF bound chromatin (red, green and blue squiggly lines, respectively).
Numerous novel large tandem repeats have been described in the human genome (4,10,11,13–17), but the purpose these sequences serve in our genome remains unknown despite a clear link to disease susceptibility (52). Collectively, the data we present here support a role for the macrosatellite DXZ4 as a Xi-specific structural element. Although the use of siRNA has yielded insight into the relationship between CTCF and DXZ4, the global impact of CTCF siRNA necessitates more focused analysis intended to decipher the function of DXZ4. With the advent of innovative genome-engineering technologies (53,54), the logical next step is to manipulate the array directly to probe DXZ4 function and shed light on this enigmatic sequence.
MATERIALS AND METHODS
Cells
Telomerase immortalized cell lines hTERT-RPE1 (C4000-1 46,XX retinal pigment epithelia) and hTERT-BJ1 (C4001-1 46,XY foreskin fibroblast) were originally obtained from Clontech. Both are now available from the American Type Culture Collection (ATCC). The male hepatocellular carcinoma cell line HepG2 (HB-8065) and the female embryonic kidney cell line 293 (CRL-1573) were obtained from ATCC as were the fetal lung fibroblast primary cells IMR-90 (CCL-186, 46,XX) and WI-38 (CCL-75, 46,XX) and the male primary fibroblast cells CCD-1139Sk (CRL-2708, 46,XY) and CCD-1140Sk (CRL-2714, 46,XY). B-Lymphocyte cell lines were obtained from the Coriell Institute for Medical Research (www.coriell.org/). Normal male cell lines include: GM06982, GM07033, GM07026, GM07030 and GM08729. Normal female cell lines include: GM21770, GM07059, GM12802, GM08728 and GM07011. All cells were maintained as recommended by the suppliers.
Metaphase chromosome preparation, immunofluorescence and FISH
Cytospun metaphase chromosomes were prepared and indirect immunofluorescence performed essentially as described (24). Indirect immunofluorescence combined with direct-labeled FISH was performed as described previously (26). BACs that were used for generating FISH probes include: RP11-818I17 (X56 tandem repeat), RP11-754H22 (X130 tandem repeat), 2272M5 (DXZ4 repeat), RP11-281B18 (GAGE locus) and RP11-402K9. BACs were obtained from Research Genetics (Life Technologies Corp.). Interphase interactions defined by FISH (Figure 4) were scored as positive if the center of the two FISH signals were within 270 nm. Statistical significance was calculated using the two-sample t-test with equal variance using GraphPad software (www.graphpad.com). Antibodies used for indirect immunofluorescence include rabbit anti-H3K4me2 (07-030; Millipore) and rabbit anti-macroH2A1 (55). Alexa-Fluor® conjugated secondary antibodies were obtained from Life Technologies Corporation. DNA was counterstained using ProLong® Gold antifade reagent supplemented with DAPI (Life Technologies Corp.). Images were collected either using a Zeiss Axiovert 200 m fitted with an AxioCam MRm and were managed using AxioVision 4.4 software (Carl Zeiss microimaging), or on a DeltaVision pDV. Delta Vision images were deconvolved with softWoRx 3.7.0 (Applied Precision) and compiled with Adobe Photoshop CS2 (Adobe Systems).
Chromatin immunoprecipitation (ChIP)
ChIP was performed essentially as described (26), with the exception that B-lymphoblast cells were fixed for 5min in a final formaldehyde concentration of 0.75% instead of 1.0% for 10min. Oligonucleotides used to assess ChIP by PCR are given in Supplementary Material, Table S1, and were obtained from Eurofins MWG Operon. Antibodies used for ChIP include anti-CTCF (07-729; Millipore) and anti-H3K4me2 (07-030; Millipore). PCR was performed using either OneTaq® 2x Master Mix (NEB) or HotStar Taq (Qiagen) according to the manufacturers’ recommendations.
Chromosome conformation capture (3C)
3C was performed essentially as described (56). BAC clones RP11-818I17 (X56 tandem repeat), RP11-754H22 (X130 tandem repeat) and 2272M5 (DXZ4 repeat) were used to generate appropriate 3C controls. Quantitation of PCR products was performed using a Chemidoc XRS and Quantity One-4.6.5 1-D Analysis Software (Bio-Rad). Statistical significance was calculated using the two-sample t-test with equal variance using GraphPad software (www.graphpad.com). 3C PCR products were verified as X56-DXZ4 or X130-DXZ4 hybrids by TA cloning into pDrive (Qiagen) followed by DNA sequence analysis.
RNA interference (RNAi)
RNAi was performed on hTERT-RPE1 cells using DharmaFECTTM 4 and ON-TARGETplus SMARTpool to human CTCF (L-020165-00-0005), a non-targeting control (D-001210-01), an unrelated siRNA to YY1 (L-011796-00) and mock transfected where no siRNA was added. Transfections were performed according to the manufacturers’ recommendations (Dharmacon, Thermo Scientific). A total of 5 × 104 or 6 × 104 cells (seeded into microtiter plates or onto cover slips in microtiter plates for FISH) were transfected with the above siRNAs three times over 72h. Cells on cover slips were processed for FISH as per usual (described above) or collected for RNA and protein isolation. Levels of CTCF RNA were consistently reduced by >82% as assessed qRT-PCR using iQ™ SYBR Green Supermix on a CFX96 (Bio-Rad) using QuantiTect SYBR Green PCR primer sets to CTCF (QT00045437) and GAPDH (QT01192646)(Qiagen). Whole cell extracts were prepared from cell pellets by resuspension in lysis buffer for 15min on ice [50 mm Tris (8.0), 150 mm NaCl, 1% NP-40 and 1.5 mm EDTA supplemented with protease inhibitors] before removing insoluble material by centrifugation at >20 000g at 4°C for 15min. The supernatant was transferred to a fresh tube before adding 1 volume of Laemmli loading buffer, heating at 90°C for 4min and separating on a 5–20% Tris-glycine SDS polyacrylamide gel. Protein was transferred to PVDF membrane by western blotting and specified proteins detected using standard techniques. Detection of CTCF was achieved using a goat polyclonal anti-CTCF antibody (G-8)(sc-271474, Santa Cruz Biotechnology) or beta-actin using a rabbit polyclonal anti-beta-actin (G046, Applied Biological Materials Inc.).
Reverse transcription PCR and RNA FISH
RNA FISH was performed on hTERT-RPE1 cells grown directly on slides essentially as described (34). Spectrum Red and Spectrum Green direct-labeled probes were prepared by nick translation according to the manufacturers’ instructions (Abbott Molecular). Probes consisted of a BAC clone to X56 (RP11-818I17, covering 56 737 116–56 945 365 of the X chromosome, Hg19) and X130 (RP11-754H22, covering 130 809 347–130 961 832 of the X chromosome, Hg19). All of the X56 transcript is contained within RP11-818I17, and all but exon-1 of the X130 transcript is contained within RP11-754H22. Total RNA was isolated from cells using the NucleoSpin RNA II kit (Machery-Nagel). First-strand cDNA was prepared from 2μg of total RNA with random hexamers with and without M-MuLV reverse transcriptase (RT) according to the manufacturer's instructions (NEB). cDNAs prepared with and without RT were used as templates for PCR with either OneTaq® master mix (NEB) or HotStar Taq (Qiagen) with the primers listed in Supplementary Material, Table S1. Human tissue total RNA was obtained from Clontech (636643). Residual genomic DNA was removed by pre-treating the RNA with DNaseI (Invitrogen) for 20min at room temperature, before heat inactivating the DNaseI at 70°C in the presence of 2.5 mm EDTA for 15min. First-strand cDNA was prepared and assessed by PCR as described above.
Promoter luciferase assay
DNA fragments initiating in and extending upstream of X56 and X130 exon-1 were generated by PCR with Platinum®Taq (Life Technologies Corp.) and cloned into pDrive (Qiagen). Inserts were verified by DNA sequencing before subcloning into the NheI and HindIII sites of pGL4.10[luc2] (Promega). The promoterless pGL4.10[luc2] firefly luciferase reporter construct and pGL4.10[luc2] containing putative X56 an X130 promoter sequences were cotransfected in triplicate on two separate occasions with the Renilla-luciferase expression vector pGL4.74[hRluc/TK] (Promega) into 293 cells by means of Lipofectamine 2000 (Life Technologies Corp.). Cells were assayed for luciferase activity on a Glomax-20/20 Luminometer (Promega) 72h after transfection with the dual-luciferase reporter assay system, according to the manufacturer's recommendations (Promega).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (GM073120 to B.P.C.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. and J. Cochran for assistance with statistical analysis, R. Rizkallah for technical suggestions and J. H. Dennis for critical discussion. We are indebted to A. Thistle for critically evaluating the manuscript.
Conflict of Interest statement. None declared.
References
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409: 860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 4.Warburton P.E., Hasson D., Guillem F., Lescale C., Jin X., Abrusan G. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics. 2008;9:533. doi: 10.1186/1471-2164-9-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hannan A.J. Tandem repeat polymorphisms: modulators of disease susceptibility and candidates for ‘missing heritability. Trends Genet. 2010;26:59–65. doi: 10.1016/j.tig.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Bruford M.W., Wayne R.K. Microsatellites and their application to population genetic studies. Curr. Opin. Genet. Dev. 1993;3:939–943. doi: 10.1016/0959-437x(93)90017-j. [DOI] [PubMed] [Google Scholar]
- 7.Eichler E.E., Clark R.A., She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet. 2004;5:345–354. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- 8.Rudd M.K., Willard H.F. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Treangen T.J., Salzberg S.L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 2012;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruce H.A., Sachs N., Rudnicki D.D., Lin S.G., Willour V.L., Cowell J.K., Conroy J., McQuaid D.E., Rossi M., Gaile D.P., et al. Long tandem repeats as a form of genomic copy number variation: structure and length polymorphism of a chromosome 5p repeat in control and schizophrenia populations. Psychiatr. Genet. 2009;19:64–71. doi: 10.1097/YPG.0b013e3283207ff6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tremblay D.C., Alexander G., Jr, Moseley S., Chadwick B.P. Expression, tandem repeat copy number variation and stability of four macrosatellite arrays in the human genome. BMC Genomics. 2010;11:632. doi: 10.1186/1471-2164-11-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gondo Y., Okada T., Matsuyama N., Saitoh Y., Yanagisawa Y., Ikeda J.E. Human megasatellite DNA RS447: copy-number polymorphisms and interspecies conservation. Genomics. 1998;54:39–49. doi: 10.1006/geno.1998.5545. [DOI] [PubMed] [Google Scholar]
- 13.Giacalone J., Friedes J., Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet. 1992;1:137–143. doi: 10.1038/ng0592-137. [DOI] [PubMed] [Google Scholar]
- 14.Kogi M., Fukushige S., Lefevre C., Hadano S., Ikeda J.E. A novel tandem repeat sequence located on human chromosome 4p: isolation and characterization. Genomics. 1997;42:278–283. doi: 10.1006/geno.1997.4746. [DOI] [PubMed] [Google Scholar]
- 15.Tremblay D.C., Moseley S., Chadwick B.P. Variation in array size, monomer composition and expression of the macrosatellite DXZ4. PLoS ONE. 2011;6:e18969. doi: 10.1371/journal.pone.0018969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 17.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 18.Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 19.Lee J.T. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 2011;12:815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 20.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 21.Boggs B.A., Cheung P., Heard E., Spector D.L., Chinault A.C., Allis C.D. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 22.Chadwick B.P. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 2008;18:1259–1269. doi: 10.1101/gr.075713.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters A.H., Mermoud J.E., O'Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 24.Chadwick B.P., Willard H.F. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J. Cell Biol. 2002;157:1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chadwick B.P., Willard H.F. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum. Mol. Genet. 2003;12:2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- 26.Moseley S.C., Rizkallah R., Tremblay D.C., Anderson B.R., Hurt M.M., Chadwick B.P. YY1 associates with the macrosatellite DXZ4 on the inactive X chromosome and binds with CTCF to a hypomethylated form in some male carcinomas. Nucleic Acids Res. 2012;40:1596–1608. doi: 10.1093/nar/gkr964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chadwick B.P., Willard H.F. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl Acad. Sci. USA. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chadwick B.P. Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis. Chromosoma. 2007;116:147–157. doi: 10.1007/s00412-006-0085-1. [DOI] [PubMed] [Google Scholar]
- 29.Filippova G.N. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 30.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., Lee C.W., Ye C., Ping J.L., Mulawadi F., et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graves J.A., Wakefield M.J., Toder R. The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum. Mol. Genet. 1998;7:1991–1996. doi: 10.1093/hmg/7.13.1991. [DOI] [PubMed] [Google Scholar]
- 33.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 34.McLaughlin C.R., Chadwick B.P. Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol. 2011;12:R37. doi: 10.1186/gb-2011-12-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clemson C.M., McNeil J.A., Willard H.F., Lawrence J.B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:1–17. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Ohlsson R., Bartkuhn M., Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119:351–360. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross M.T., Grafham D.V., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gjerstorff M.F., Ditzel H.J. An overview of the GAGE cancer/testis antigen family with the inclusion of newly identified members. Tissue Antigens. 2008;71:187–192. doi: 10.1111/j.1399-0039.2007.00997.x. [DOI] [PubMed] [Google Scholar]
- 41.Hou C., Dale R., Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl Acad. Sci. USA. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Cai S., Lee C.C., Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 44.Rubio E.D., Reiss D.J., Welcsh P.L., Disteche C.M., Filippova G.N., Baliga N.S., Aebersold R., Ranish J.A., Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stedman W., Kang H., Lin S., Kissil J.L., Bartolomei M.S., Lieberman P.M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 47.Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A., et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 49.Jacobs S.A., Taverna S.D., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis D., Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 51.Eils R., Dietzel S., Bertin E., Schrock E., Speicher M.R., Ried T., Robert-Nicoud M., Cremer C., Cremer T. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Maarel S.M., Tawil R., Tapscott S.J. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol. Med. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bogdanove A.J., Voytas D.F. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 54.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chadwick B.P., Willard H.F. Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet. 2001;10:1101–1113. doi: 10.1093/hmg/10.10.1101. [DOI] [PubMed] [Google Scholar]
- 56.Miele A., Gheldof N., Tabuchi T.M., Dostie J., Dekker J. Mapping chromatin interactions by chromosome conformation capture. Curr. Protoc. Mol. Biol. 2006 doi: 10.1002/0471142727.mb2111s74. Chapter 21, Unit 21 11. [DOI] [PubMed] [Google Scholar]
- 57.Noe L., Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 2005;33:W540–W543. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.