Abstract
Facioscapulohumeral muscular dystrophy (FSHD), the most prevalent myopathy afflicting both children and adults, is predominantly associated with contractions in the 4q35-localized macrosatellite D4Z4 repeat array. Recent studies have proposed that FSHD pathology is caused by the misexpression of the DUX4 (double homeobox 4) gene resulting in production of a pathogenic protein, DUX4-FL, which has been detected in FSHD, but not in unaffected control myogenic cells and muscle tissue. Here, we report the analysis of DUX4 mRNA and protein expression in a much larger collection of myogenic cells and muscle biopsies derived from biceps and deltoid muscles of FSHD affected subjects and their unaffected first-degree relatives. We confirmed that stable DUX4-fl mRNA and protein were expressed in myogenic cells and muscle tissues derived from FSHD affected subjects, including several genetically diagnosed adult FSHD subjects yet to show clinical manifestations of the disease in the assayed muscles. In addition, we report DUX4-fl mRNA and protein expression in muscle biopsies and myogenic cells from genetically unaffected relatives of the FSHD subjects, although at a significantly lower frequency. These results establish that DUX4-fl expression per se is not sufficient for FSHD muscle pathology and indicate that quantitative modifiers of DUX4-fl expression and/or function and family genetic background are determinants of FSHD muscle disease progression.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant disease marked by progressive muscle atrophy in specific muscle groups (1,2). FSHD is one of the most prevalent myopathies, affecting ∼1 of every 7500–14 000 adults, and can afflict both children and adults (3). The most common form of FSHD, FSHD1 (MIM 158900), accounts for >95% of reported cases and results from a range of contractions within the chromosome 4q35 localized macrosatellite D4Z4 repeat array (4–6). At the 4q35 locus, normal individuals contain >10 D4Z4 repeats (and often >30) on both chromosomes whereas individuals with FSHD1 have between 1 and 10 repeats on one chromosome. The contraction likely causes changes in the epigenetic status of the chromatin leading to misexpression of a gene or genes (7–10). The far less common form, FSHD2 (MIM 158901), is unlinked genetically to 4q35 but presents with the same clinical symptoms as FSHD1 (11,12). Both forms of FSHD are exclusively linked to one of two types of the chromosome 4q subtelomeres (4qA), indicating that the lesion itself is not sufficient for pathology. Despite differences in genetic lesion, FSHD1 and FSHD2 may share a common pathogenic mechanism in which aberrant DNA hypomethylation within the 4q35 locus occurs and likely affects gene regulation in both types of FSHD (11). Overall, FSHD, by all indications, is an autosomal dominant gain-of-function disease with a strong epigenetic component.
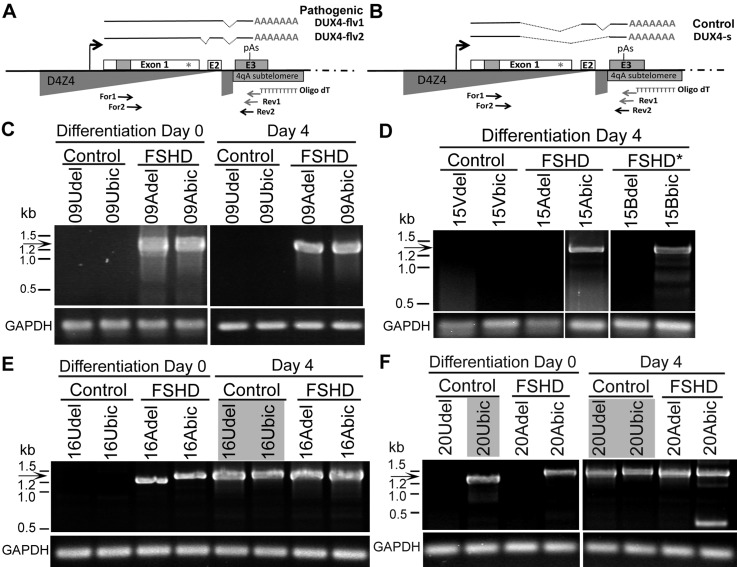
Each D4Z4 repeat unit within the 4q35 array contains a copy of the DUX4 gene (13). Recent studies have led to a new model for DUX4-mediated FSHD pathology (Fig. 1A and B) (14–17). In this model, it is only the DUX4-fl (full-length) mRNA alternative splice variant encoded by the distal-most 4q35 D4Z4 repeat that is stably expressed and pathogenic. The reduction in the number of D4Z4 repeat units below a threshold of 11 in the 4q35 array in FSHD1 results in epigenetic changes leading to alternative splicing of the DUX4 transcript to produce the DUX4-fl mRNA. Stabilization of this mRNA is due to a 4qA-specific polyadenylation signal (PAS) residing in a subtelomeric exon distal to the array. This exon becomes spliced into the DUX4-fl message (thereby explaining the requirement for a 4qA subtelomere to develop FSHD1 and FSHD2), and the DUX4-FL protein is produced from this stable, polyadenylated mRNA (14). Since the DUX4-FL protein can act as a transcription factor to induce ectopic expression in skeletal muscle of a large number of genes (16) and can be highly cytotoxic to somatic cells (18–21), its aberrant expression in skeletal muscle, even though restricted to a small percentage of myonuclei at any one time (15), may lead to progressive muscle cell death or dysfunction and ultimately to overt pathology. Nonetheless, expression of the DUX4-FL protein need not lead to pathology and likely has a non-pathogenic function in humans since it is normally expressed in the testis (15). An additional alternatively spliced short mRNA isoform, termed DUX4-s (short), which does not encode the transcriptional activation domain of DUX4-FL, is widely expressed in somatic cells; however, it is strictly the aberrant expression of the DUX4-fl isoform in myogenic cells that is linked to both FSHD1 and FSHD2 (Fig. 1A and B) (14–16).
Figure 1.
Polyadenylated DUX4-fl mRNA was expressed in cultures of myogenic cells derived from FSHD and control subjects. Schematic for DUX4 mRNA splicing and detection by RT-PCR based on the model (15) for (A) FSHD and (B) control cells. Locations of oligonucleotide primers are indicated with arrows. Nested RT-PCR analysis of polyadenylated DUX4 mRNA from CD56+ myogenic cells derived from biceps (bic) and deltoid (del) muscle biopsies for (C) cohort 09, (D) cohort 15, (E) cohort 16 and (F) cohort 20 following differentiation for 0 or 4 days, as indicated. All RT-PCRs were repeated at least three times, all products were sequenced and the presence of spliced DUX4-fl mRNA (arrow) was confirmed in cultures from FSHD subjects (09Adel, 9Abic, 15Abic, 15Bbic, 16Adel, 16Abic, 20Adel, 20Abic) and from control individuals not containing an FSHD D4Z4 contraction (16Udel, 16Ubic, 20Udel, 20Ubic). Gray boxes indicate DUX4-fl expression in unaffected cell cultures. RT–PCR for GAPDH mRNA expression controlled for integrity of the mRNA and first strand cDNA synthesis.
Previous studies found that unaffected healthy myogenic cells and tissues expressed DUX4-s but neither DUX4-fl mRNA nor protein was ever detected (15,22). We have now carried out a study of a larger number of unaffected and FSHD samples and we report, in contrast to the previous work, that DUX4-fl mRNA and protein are in fact expressed by myogenic cells and muscle tissue from certain healthy unaffected individuals as well as from genetically diagnosed FSHD1 individuals. However, consistent with its suggested role in FSHD pathogenesis, we found that DUX4-fl was significantly more likely to be expressed in FSHD1 than in unaffected cells and tissues. Our finding that DUX4-fl can be expressed by genetically unaffected nuclei suggests that DUX4-fl expression per se is not sufficient to induce FHSD pathology, but the higher expression of DUX4-fl in FSHD than unaffected nuclei suggests that quantitative modifiers of DUX4-fl expression and/or function, including family genetic background, are determinants of FSHD muscle disease progression.
RESULTS
To generate well-controlled clinical materials for analyses of FSHD pathogenesis, open biopsies of deltoid and biceps muscles were recovered from multiple cohorts of FSHD donors and their first-degree relatives (23). For 15 cohorts, samples were obtained from at least one affected FSHD individual with a shortened D4Z4 array and clinically verified muscle weakness and at least one control first-degree relative with an uncontracted D4Z4 repeat array (Table 1). Six additional cohorts (05, 15, 27, 28, 29, 30) contained two related adult subjects (age 49–70 years), both with a diagnostic FSHD1 deletion at chromosome 4q35, yet with only one subject showing muscle weakness, while the other had yet to exhibit weakness in either their deltoid or biceps. For this analysis, these non-manifesting, but genetically FSHD1, subjects were treated as FSHD affected (Tables 1 and 2 and Supplementary Material, Table S1). Each donor provided two biopsies: one from the biceps, a muscle often more severely affected early on in FSHD, and one from the deltoid, a muscle often exhibiting less pathology in FSHD. Myogenic cells were derived from biopsies by cell culture and FACS was used to isolate low passage CD56+ primary myogenic cells for culture (23,24). For this study, we have investigated expression of the FSHD candidate gene DUX4 in muscle biopsies and cultures of myogenic cells from these cohorts.
Table 1.
Clinical characteristics of FSHD subjects and unaffected donors
| Donora | Familial relations | Gender | Age at enrollment (years) | EcoRI/BlnI sizesb (kb) | Deltoid strengthc | Biceps strengthc |
|---|---|---|---|---|---|---|
| 03A | Proband | F | 40 | 20 kb (4qA), 57kb (4qB) | 5/5 | 4+/4+ |
| 03U | Sister of 03A | F | 42 | 80kb (4qB), >112kb (4qA) | 5/5 | 5/5 |
| 05A | Proband | F | 55 | 25 kb (4qA), 67kb | 5/5 | 5/5 |
| 05B | Son of 5A | M | 19 | 25 kb (4qA), 94kb | 4/4+ | 4/4 |
| 05C | Brother of 5A | M | 49 | 25 kb (4qA), 67 kb | 5/5 | 5/5 |
| 07A | Proband | F | 18 | 29 kb (4qA), 53 kb (4qA) | 4+/4+ | 5/5 |
| 07U | Mother of 07A | F | 49 | 34 kb (4qB), 53 kb (4qA), >112 kb (4qB)e | 5/5 | 5/5 |
| 09A | Proband | F | 31 | 25 kb (4qA), >112 kb | 5/5 | 4+/4+ |
| 09U | Mother of 09A | F | 57 | 47 kb (4qB), >112 kb (4qA) | 5/5 | 5/5 |
| 10A | Proband | F | 48 | 24 kb (4qA), 46 kb | 5/5 | 5/5 |
| 12A | Daughter of 12B | F | 22 | 18 kb (4qA), 63 kb (4qA) | 4+/4+ | 4+/4+ |
| 12Bd | Proband | M | 49 | 18 kb (4qA), >112 kb | 4+/4+ | 4+/4+ |
| 12U | Daughter of 12B | F | 24 | >112 kb (4qA), >112 kb (4qA) | 5/5 | 5/5 |
| 12Vd | Sister of 12B | F | 45 | >112 kb, >112 kb | 5/5 | 5/5 |
| 13A | Father of 13B | M | autopsy | 14 kb (4qA)f | nd | nd |
| 13B | Proband | F | 42 | 16 kb (4qA), >112 kb | 5/5 | 4+/4 |
| 13U | Mother of 13B | F | 63 | 57 kb (4qB), >112 kb | 5/5 | 5/5 |
| 14A | Proband | M | 50 | 19 kb (4qA), >60 kb | 5/5 | 3+/4 |
| 14B | Brother of 14A | M | 53 | 19 kb (4qA), >60 kb (4qA) | 4+/4+ | 4+/4+ |
| 14V | Sister of 14A | F | 49 | 60 kb (4qA), 112 kb (4qB) | 5/5 | 5/5 |
| 14W | Brother of 14A | M | 47 | 72 kb (4qB)f | 5/5 | 5/5 |
| 15A | Proband | M | 66 | 28 kb (4qA), >112 kb (4qB) | 5/5 | 4+/5 |
| 15B | Brother of 15A | M | 69 | 28 kb (4qA), >112 kb (4qB) | 5/5 | 5/5 |
| 15V | Sister of 15A | F | 60 | >107 kb (4qB), >145 kb (4qB) | 5/5 | 5/5 |
| 16A | Proband | F | 56 | 20 kb, 97 kb | 5−/5− | 4−/4+ |
| 16U | Sister of 16A | F | 60 | 56 kb (4qB), 93 kb (4qA), 97 kb (4qA)e | 5/5 | 5/5 |
| 17A | Proband | M | 23 | 19 kb (4qA), 87 kb (4qA) | 4+/5 | 3/4 |
| 17U | Brother of 17A | M | 21 | 97 kb (4qB), >112 kb (4qA) | 5/5 | 5/5 |
| 17V | Father of 17A | M | 50 | 90 kb (4qA), >112 kb (4qB) | 5/5 | 5/5 |
| 18A | Proband | F | 36 | 21 kb (4qA), >112 kb (4qB) | 5/5 | 4+/4+ |
| 18U | Brother of 18A | M | 37 | 57 kb (4qB), >112 kb (4qB) | 5/5 | 5/5 |
| 19A | Proband | M | 65 | 22 kb (4qA), >112kb (4qA) | 4+/4+ | 4+/4+ |
| 19U | Daughter of 19A | F | 41 | 79kb (4qA), >112kb (4qA) | 5/5 | 5/5 |
| 20A | Proband | M | 28 | 20 kb (4qA), 39kb (4qA), 48kb (4qB)e | 5/5 | 5/5 |
| 20U | Mother of 20A | F | 48 | 39kb (4qA), 65kb (4qB) | 5/5 | 5/5 |
| 21A | Proband | F | 82 | 26 kb (4qA), >112kb (4qA) | 4+/4+ | 4+/4+ |
| 21B | Daughter of 21A | F | 59 | 26 kb (4qA), 40kb (4qA) | 5/5 | 4+/4+ |
| 21U | Daughter of 21A | F | 48 | 63kb (4qB), >112 (4qA) | 5/5 | 5/5 |
| 22A | Proband | F | 71 | 27 kb (4qA), >112kb (4qA) | 5/2 | 4/4 |
| 22U | Daughter of 22A | F | 43 | 60kb (4qB), >112kb (4qA) | 5/5 | 5/5 |
| 23A | Proband | M | 27 | 18 kb (4qA), 45kb (4qA) | 5/5 | 2+/2+ |
| 23U | Father of 23A | M | 59 | 45kb (4qA), >112kb (4qB) | 5/5 | 5/5 |
| 27A | Proband | F | 39 | 12 kb (4qA), 47kb (4qA), 52kb (4qB)e | 5/5 | 4/4 |
| 27B | Mother of 27A | F | 59 | 12 kb (4qA), 47kb (4qA), 69kb (4qB) | 5/5 | 5/5 |
| 28A | Proband | M | 44 | 29 kb (4qA), 75kb (4qB) | 5/5 | 5/5 |
| 28B | Father of 28A | M | 68 | 29 kb (4qA)f | 5/5 | 5/5 |
| 29A | Proband | M | 39 | 30 kb (4qA)f | 5/5 | 5/5 |
| 29B | Mother of 29A | F | 70 | 30 kb (4qA)f | 5/5 | 5/5 |
| 30A | Proband | M | 57 | 30 kb (4qA), >112kb (4qB) | 5/5 | 5/5 |
| 30B | Sister of 30A | F | 59 | 32 kb (4qA), 89kb (4qB) | 5/5 | 5/5 |
aDonors are designated by cohort (family) number (01, 03, etc.) followed by A, B or C for the FSHD subjects or U, V or W for the unaffected first degree relative(s).
bFSHD1 was confirmed by presence of a shortened 4qA type D4Z4 repeat array identified by an EcoRI/BlnI restriction fragment of <35 kb (Supplementary Material). Shortened repeat arrays are shown in bold.
cMuscle strength is presented using a modified MRC scale where 5 is full strength and side of biopsy (right/left) is underlined.
dSubjects 12B and 12V were originally named 11A and 11U for internal analyses, but were renamed for publication to reflect familial relationship.
eSubjects with three BlnI resistant chromosomes (designated as chromosome 4q-type array) showed only one BlnI-sensitive chromosome (designated as chromosome 10-type array) resulting in an apparent chromosome 4q:10q array ratio of 3:1.
fSubjects with only one BlnI resistant array (designated as chromosome 4q-type array) showed three BlnI-sensitive arrays (designated as chromosome 10-type array) resulting in an apparent chromosome 4q:10q array ratio of 1:3.
Table 2.
Summary of DUX4-fl expression assays
| Control samples | Shortest 4qAa (kb) | RT-PCR | RT-PCR | DUX4 ICC | RT-PCR |
|---|---|---|---|---|---|
| Day 0 | Day 4 | Cell culture | Biopsy | ||
| 03Udel | >112 | − | − | + | Nd |
| 03Ubic | >112 | − | − | − | − |
| 07Udel | 53 | − | + | + | − |
| 07Ubic | 53 | − | + | − | − |
| 09Udel | >112 | − | − | − | − |
| 09Ubic | >112 | − | − | + | − |
| 12Udel | >112 | + | + | + | − |
| 12Ubic | >112 | − | − | − | − |
| 15Vdel | NAb | − | − | − | − |
| 15Vbic | NAb | − | − | − | − |
| 16Udel | 93 | − | + | − | − |
| 16Ubic | 93 | − | + | + | − |
| 17Udel | >112 | nd | nd | nd | − |
| 17Ubic | >112 | nd | nd | nd | − |
| 17Vdel | 90 | − | + | + | − |
| 17Vbic | 90 | − | − | − | + |
| 18Udelb | NAb | − | − | − | − |
| 18Ubicb | NAb | − | − | − | − |
| 20Udel | 39 | − | + | − | + |
| 20Ubic | 39 | + | + | − | + |
| FSHD Samples | 4qA (kb) EcoRI/BlnI | RT−PCR | RT−PCR | DUX4 ICC | RT−PCR |
| Day 0 | Day 4 | Cell Culture | Biopsy | ||
| 03Adel | 20 | − | + | − | nd |
| 03Abic | 20 | + | + | + | + |
| 05Adel | 25 | nd | nd | nd | + |
| 05Abic | 25 | nd | nd | nd | + |
| 05Bdel | 25 | + | + | nd | + |
| 05Cdel | 25 | nd | nd | nd | + |
| 05Cbic | 25 | − | + | − | + |
| 07Adel | 29 | + | − | + | − |
| 07Abic | 29 | − | + | + | − |
| 09Adel | 25 | + | + | + | + |
| 09Abic | 25 | + | + | + | + |
| 12Adel | 18 | + | + | + | + |
| 12Abic | 18 | − | + | + | + |
| 15Adel | 28 | − | + | − | + |
| 15Abic | 28 | + | − | + | + |
| 15Bdel | 28 | − | − | + | + |
| 15Bbic | 28 | + | + | + | + |
| 16Adel | 20 | + | + | + | + |
| 16Abic | 20 | + | + | + | + |
| 17Adel | 19 | + | + | + | + |
| 17Abic | 19 | − | + | + | + |
| 18Adel | 21 | + | + | + | + |
| 18Abic | 21 | − | + | + | + |
| 20Adel | 20 | + | + | + | + |
| 20Abic | 20 | + | + | + | + |
| 27Adel | 12 | − | − | − | − |
| 27Abic | 12 | − | + | − | − |
| 27Bdel | 12 | + | − | − | − |
| 27Bbic | 12 | − | − | − | − |
| 28Adel | 29 | + | + | + | − |
| 28Abic | 29 | − | + | + | + |
| 28Bdel | 29 | − | + | + | nd |
| 28Bbic | 29 | − | + | + | + |
| 29Adel | 30 | − | + | + | + |
| 29Abic | 30 | + | + | + | + |
| 29Bdel | 30 | + | + | + | + |
| 29Bbic | 30 | + | + | + | + |
| 30Adel | 30 | − | + | nd | nd |
| 30Abic | 30 | − | + | nd | nd |
| 30Bdel | 32 | + | + | nd | nd |
| 30Bbic | 32 | + | + | nd | nd |
+, DUX4-fl positive in at least one experiment; −, DUX4-fl negative in all repetitions; nd = not determined.
aShortest fragment from EcoRI/BlnI digestion.
bNot applicable (NA); no 4qA alleles detected
Reverse transcription polymerase chain reaction (RT-PCR) and sequencing of reaction products was used to analyze DUX4 mRNA expression in cultured myogenic cells derived from nine complete family cohorts (03, 07, 09, 12, 15, 16, 17, 18, 20), including cells derived from both biceps and deltoid of FSHD subjects and their unaffected first-degree relatives. The six cohorts containing at least two genetically FSHD1 subjects were similarly assayed, in all totaling 38 genetically FSHD and 18 unaffected control cell strains. Because DUX4-fl is up-regulated during myogenesis in FSHD-derived cells (15), we used RT-PCR to identify DUX4 mRNAs in cultures both prior to differentiation (Differentiation Day 0) and after 4 days of myogenic differentiation (Differentiation Day 4) (Fig. 1 and Table 2). In every case, the complete sequencing of the PCR reaction product confirmed that 4q35-derived DUX4-fl had been detected. For certain cohorts, such as 03 and 09, our results were consistent with the previous studies (14,15,22) in that we detected DUX4-fl mRNA only in FSHD cells and never in unaffected control cells (Fig. 1C). Overall, 35 of 38 FSHD affected cell cultures from the 20 subjects tested expressed DUX4-fl mRNA, consistent with previous studies in which most, but not all, FSHD cell cultures were reported to express DUX4-fl mRNA (14,15).
In contrast to the previous studies, however, we also detected DUX4-fl mRNA in eight cultures from unaffected subjects in five (07, 12, 16, 17, 20) of the nine cohorts that we examined. For example, in cohorts 07, 16 and 20, cells derived from both muscle biopsies of unaffected subjects (07Udel, 07Ubic, 16Udel, 16Ubic, 20Ude and 20Ubic) expressed DUX4-fl mRNA. Consistent with DUX4-fl mRNA being up-regulated during myogenesis, DUX4-fl mRNA was detected in 8 of the 18 differentiated control cultures and only in 2 cases (12Udel and 20Ubic) was DUX4-fl mRNA also detected in undifferentiated cultures. Significantly, DUX4-fl was never detected in cells derived from control subjects 15V and 18U who had only 4qB subtelomeric alleles that lack exon 3 and the PAS required to produce a stable DUX4-fl mRNA (14), further supporting the specificity of the RT-PCR for 4qA DUX4-fl mRNA transcripts and the lack of cross contamination in our samples.
It is notable that each RT-PCR repetition did not necessarily reproduce DUX4 detection (Supplementary Material, Table S1), which is consistent with the finding that DUX4 mRNAs, when expressed, are in very low abundance (15). Thus, as is the case for the digital PCR method of quantifying low abundance templates (25), individual reactions from the same cDNA could be either positive or negative for DUX4-fl. Overall, myogenic cell cultures were designated as positive if DUX4-fl mRNA was detected in any of the three or more repetitions and negative if DUX4-fl mRNA was never detected (Table 2). By this measure, we found polyadenylated DUX4-fl mRNA in 35 of 38 FSHD-derived myogenic cell strains, 8 of 18 control myogenic cell strains and never in the DUX4 non-permissive control myogenic cells from subjects (15V and 18U). Together, these results establish that DUX4-fl mRNA expression is not restricted to FSHD-derived myogenic cells.
The identities of the control unaffected cells used for these studies were reconfirmed by microsatellite analysis and the absence of FSHD1 lesions was confirmed by Southern blotting analysis. In all cases, the myogenic cells were confirmed to be genotypic matches to the original blood sample from each donor subject and/or the biopsies from each donor subject. In addition, RT-PCR products in every PCR were sequenced to confirm the identity of the product as 4qA-derived DUX4-fl by six distinguishing polymorphisms between 4qA and 10qA (See Materials and Methods) (26) and, when possible, matched to the particular subject based on additional 4qA sequence polymorphisms. Therefore, these findings confirm our conclusion that DUX4-fl mRNA expression is not exclusive to FSHD-derived myogenic cells.
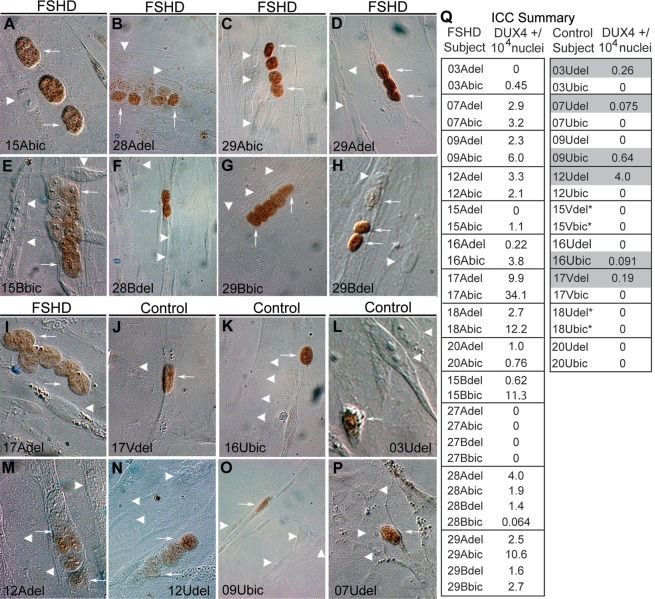
Consistent with the mRNA expression results, we found that the DUX4-FL protein is also expressed in myogenic cells from both unaffected and FSHD subjects. Using immunocytochemistry with mAb P4H2 (15,27) to analyze DUX4-FL expression in cultures after 4–6 days of differentiation (Fig. 2 and Table 2), we found DUX4-FL positive nuclei in 6 (03Udel, 07Udel, 09Ubic, 12Udel, 16Ubic and 17Vdel) of the 18 unaffected cultures that we examined, which were from 6 different genetically unaffected individuals. We also detected DUX4-FL nuclei in 26 of 33 FSHD cultures, which were from 14 of the 17 FSHD subjects tested. We conclude that DUX4-FL protein expression is not exclusive to FSHD-derived myogenic cells, supporting the conclusion of our DUX4 RNA expression analysis.
Figure 2.
DUX4-FL protein was detected in FSHD and control myogenic cells. Differentiated CD56+ myogenic cells derived from FSHD subjects (A–H, I and M), or control subjects (J–L and N–P) were immunostained for DUX4-FL (brown). DUX4-FL positive nuclei (white arrows) and DUX4-FL negative nuclei (white arrowheads) were observed. (Q) Summary of DUX4-FL immunostaining; gray shading indicates unaffected controls expressing DUX4-FL; asterisk indicates negative control cells with two non-permissive B haplotype subtelomeres.
On average, we found that DUX4-FL+ nuclei, though always rare, were more frequent in FSHD than unaffected cell cultures (Fig. 2Q). In the 26 FSHD cultures with positive staining, we found that 0.047% ± 0.026 (ave ± SE) or ∼1 in 2000 nuclei were DUX4-FL+, with a high of 1 in ∼300 for 17Abic. This very low frequency of DUX4-FL+ nuclei was similar to that seen in the previous study in which expression was found only in FSHD cultures (15). Here, however, we also found DUX4-FL expression in unaffected cultures, though in the 6 of 14 chromosome 4qA-containing cultures with positive staining, only 0.009 ± 0.007% or 1 in ∼11 000 of the nuclei were DUX4-FL+, with a high of 1 in 2500 for 12Udel. We confirmed specificity of DUX4-FL immunostaining in several ways. First, we used transfection assays to confirm that mAb P4H2 reacted with DUX4-FL but not DUX4-s. Second, we found that DUX4-FL protein was never detected in cells from two control individuals (15V and 18U) who had two 4qB alleles. Third, in repeated assays of differentiated 07Ubic cultures in which we examined more than one million nuclei, we never detected a DUX4-FL-positive nucleus. Finally, we also did not detect DUX4-FL in several additional unaffected cell strains despite analyzing more than one million nuclei in aggregate. Thus, the detection of DUX4-FL in myogenic cultures was highly specific and supports the conclusion that despite overall low numbers of expressing nuclei, DUX4-FL protein expression is not an unregulated event. In addition, this confirmed that DUX4-FL protein expression per se, as with the DUX4-fl mRNA expression, does not perfectly correlate with FSHD.
Overall, FSHD cell cultures had a significantly higher frequency of DUX4-FL positive nuclei than unaffected cell cultures (Fig. 3; p= 0.001, likelihood-ratio test for Poisson regression). Furthermore, the frequency of DUX4-FL positive nuclei was higher in FSHD than in unaffected cultures in six of the seven cohorts in which we examined both FSHD and unaffected cells, with cohort 12 being the exception. The difference in frequencies between muscle types was not significant (p= 0.4), although the difference in frequencies between FSHD and unaffected cell cultures was somewhat larger in biceps than in deltoid (p= 0.03 for interaction term). In summary, for myogenic cells, we found DUX4-fl mRNA and/or protein to be expressed in unaffected control cells from seven of nine cohorts, with cells from the two control donors (15V and 18U) with the non-permissive 4qB/B subtelomeres consistently being the sole exceptions. In addition, DUX4-fl mRNA and/or protein was detected in myogenic cells from all 20 FSHD subjects analyzed. For each assay that we used (RT-PCR and immunostaining), we detected DUX4-fl at a significantly higher frequency in FSHD than in unaffected cells.
Figure 3.
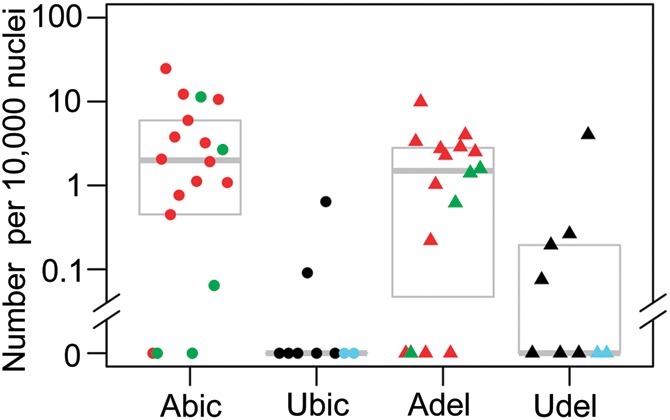
The total numbers of DUX4-FL expressing nuclei are significantly higher in myogenic cultures from affected (A) versus unaffected (U) subjects. Counts of DUX4-FL positive nuclei, determined by ICC, per 10 000 nuclei are shown. From left to right, columns show FSHD biceps (A.bic, n= 18, including 5 asymptomatic in green), unaffected biceps (U.bic, n= 9, including 2 4qB/4qB in blue), FSHD deltoid (A.del, n= 16, including 4 asymptomatic in green), and unaffected deltoid samples (U.del, n= 9 including 2 4qB/4qB in blue). Gray boxes show range from 25th percentile to 75th percentile, with the median shown as a bold grey line. Within each column, samples are ordered by increasing length of shortest 4qA EcoRI/BlnI fragment, with 4qB/4qB haplotypes at far right (ties broken arbitrarily). The difference between affected and unaffected samples is highly significant (p = 0.001; likelihood-ratio test), the difference between muscle types is non-significant (p = 0.4), and the interaction between muscle type and disease status is mildly significant (p = 0.03). (All P-values exclude the 4qB/4qB samples, which lack DUX4-FL permissive alleles.)
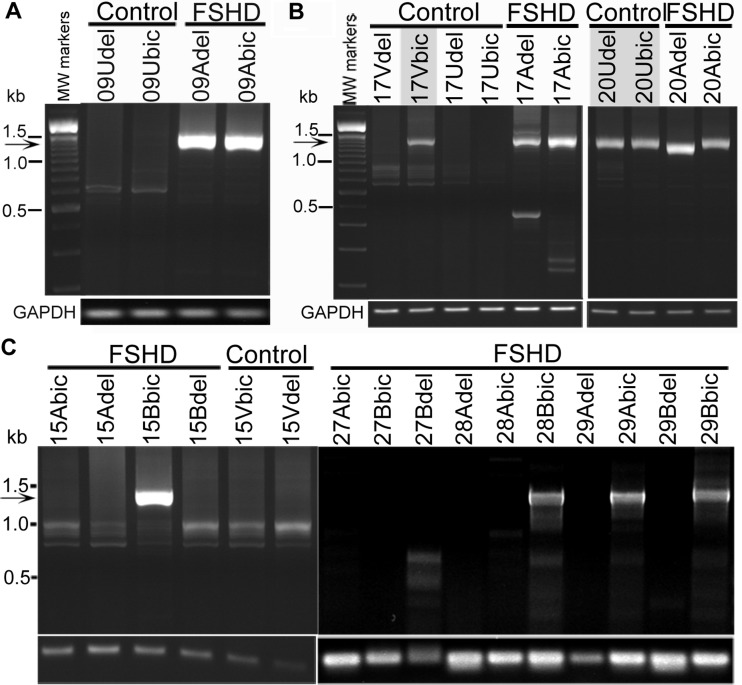
We next analyzed DUX4-fl mRNA expression directly in muscle biopsies, including those from which cultured cells were derived. These studies also detected DUX4-fl expression in biceps and deltoid muscles of both unaffected control subjects and FSHD subjects (Figs 4 and 5, Supplementary Material, Fig. S1; summarized in Supplementary Material, Table S1). For unaffected subjects, we analyzed 26 biopsies from 14 individuals (in 12 cohorts) that had the permissive 4qA subtelomere, and we found DUX4-fl mRNA expression in three (∼12%) of the biopsies assayed (17Vbic, 20Udel and 20Ubic), which were from two unaffected individuals in two cohorts (Fig. 4). Furthermore, an additional eight biopsies from four unaffected subjects lacking any detectable permissive 4qA subtelomeres (13U, 14W, 15V and 18U) were consistently negative for DUX4-fl mRNA. By comparison, for FSHD subjects, we analyzed 59 biopsies from 32 individuals (21 cohorts) and found DUX4-fl mRNA expression in 46 (∼78%) of the assayed biopsies, whereas no DUX4-fl mRNA was detected in 13 of the biopsies. These FSHD biopsies without detectable DUX4-fl mRNA included the biceps biopsy from subject 13A and 27A, deltoid biopsy from subject 28A and both the deltoid and biceps biopsies from subjects 7A, 14A, 14B, 27B and 30B (Supplementary Material, Table S1). We conclude that DUX4-fl mRNA expression is not exclusive to FSHD muscle; however, the percentage of muscle biopsies that expressed detectable DUX4-fl mRNA was significantly greater for FSHD than unaffected biopsies (p < 0.0005 for both biceps and deltoid, Fisher's exact test).
Figure 4.
Polyadenylated DUX4-fl mRNA was expressed in muscle biopsies from control subjects and FSHD subjects. Polyadenylated and spliced DUX4-fl mRNA (arrow) was identified by nested RT-PCR and confirmed by sequencing in muscle biopsies. (A) Cohort 09 shows DUX4-fl exclusively in FSHD subjects. (B) Cohorts 17 and 20 show DUX4-fl mRNA expression in control biopsies from unaffected subjects without the FSHD-linked D4Z4 deletion. (C) Cohorts 15, 28 and 29 show DUX4-fl mRNA expression in biopsies from FSHD subjects exhibiting no apparent weakness in the biopsied muscle (15Bbic, 28Bbic, 29Bbic). RT-PCRs for GAPDH mRNA controlled for mRNA integrity and first strand cDNA synthesis.
Figure 5.
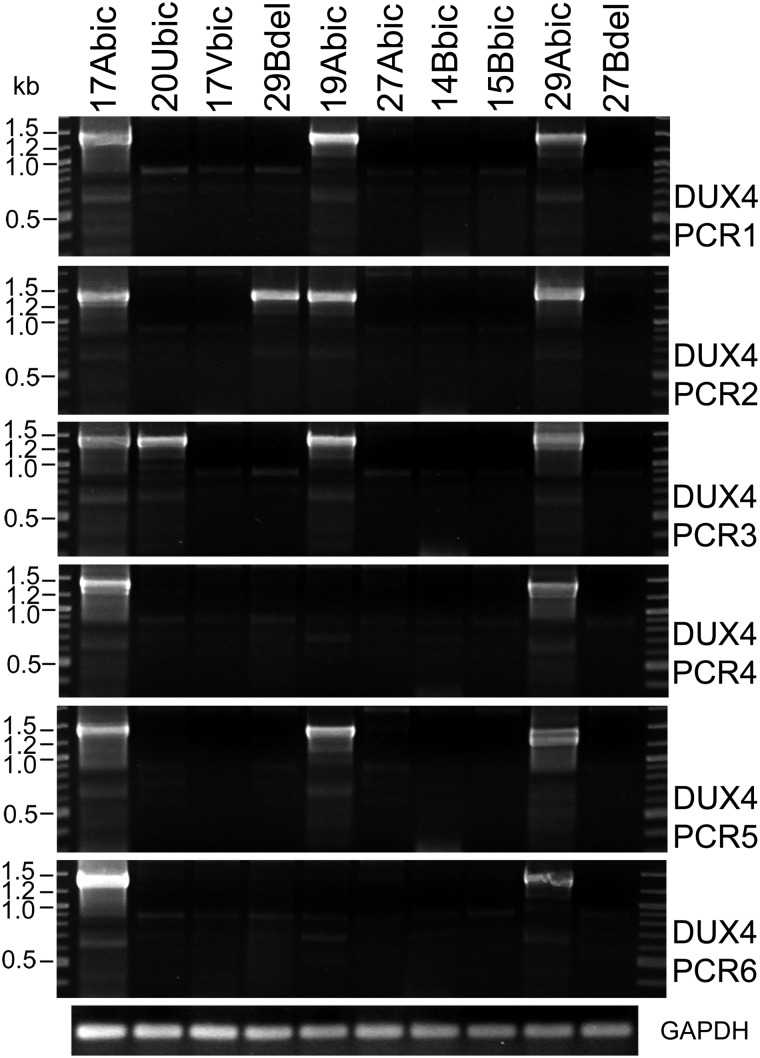
Digital RT-PCR shows variation in DUX4-fl mRNA detection due to low levels of transcript in muscle biopsy. Blind RT-PCRs were performed in six replicates using mRNA derived from muscle biopsies from the indicated subjects. GAPDH RT-PCRs controlled for mRNA integrity and cDNA synthesis.
To assess potential differences in the absolute levels of DUX4-fl transcript present in any one biopsy sample and to identify potential quantitative differences in DUX4-fl mRNA between FSHD affected and control biopsies, we performed digital PCR on blinded samples (Fig. 5 and Supplementary Material, Table S1). As with the cell culture RT-PCR data, the levels of DUX4-fl mRNA in biopsy samples were extremely low resulting as expected in sometimes positive and sometimes negative results for certain samples (20Ubic, 29Bdel and 19Abic). In contrast, other samples were always positive (17Abic and 29Abic) or always negative (27Abic, 14Bbic and 27Bdel). Overall, these experimental PCR replicates provide a relative quantitative analysis of DUX4-fl mRNA in which we find that DUX4-fl mRNA is, usually but not always, more abundant in FSHD biopsies compared with unaffected biopsies.
DUX4-fl expression in biopsies, as with the myogenic cell culture data, was not unique to FSHD samples (summarized in Table 3). Our finding of DUX4-fl mRNA in multiple unaffected muscle biopsies suggests that the mechanisms underlying expression of DUX4-fl in unaffected myogenic cell cultures were not simply a result of changes that arose as a consequence of culturing. Therefore, we conclude that DUX4-fl mRNA and/or protein expression is not exclusive to FSHD muscle and myogenic cells. Nonetheless, a consistent finding from each of our assays was that DUX4-fl was significantly more likely to be detected in FSHD than unaffected cells and tissues.
Table 3.
Summary of DUX4-fl expression
| Source | Subjects | Muscle origin | DUX4-fl (+) | DUX4-fl mRNA (+) | DUX4-FL protein (+) | DUX4-fl mRNA and protein (+) |
|---|---|---|---|---|---|---|
| Cell cultures | FSHD | biceps | 19/20 | 18/19 | 15/18 | 14/17 |
| FSHD | deltoid | 18/19 | 17/19 | 12/16 | 11/16 | |
| Controla | biceps | 4/9 | 3/9 | 2/9 | 1/9 | |
| Controla | deltoid | 6/9 | 5/9 | 4/9 | 3/9 | |
| 4qB only | biceps | 0/2 | 0/2 | 0/2 | 0/2 | |
| 4qB only | deltoid | 0/2 | 0/2 | 0/2 | 0/2 | |
| Biopsies | FSHD | biceps | 23/30 | 23/30 | nd | nd |
| FSHD | deltoid | 23/29 | 23/29 | nd | nd | |
| Controla | biceps | 2/17 | 2/17 | nd | nd | |
| Controla | deltoid | 1/17 | 1/17 | nd | nd | |
| 4qB only | biceps | 0/4 | 0/4 | nd | nd | |
| 4qB only | deltoid | 0/4 | 0/4 | nd | nd | |
| Total DUX4-fl status | FSHD | 30/32 | 30/32 | 14/17 | 14/17 | |
| Controla | 7/18 | 5/18 | 6/9 | 4/9 | ||
| 4qB only | 0/4 | 0/4 | 0/2 | 0/2 |
DUX4-fl (+) indicates DUX4-fl was detected by either RT-PCR or ICC (or both).
DUX4-fl mRNA indicates DUX4-fl mRNA was detected by RT-PCR from either cell culture or biopsy mRNA (or both).
DUX4-FL protein indicates DUX4-FL protein was detected by ICC in cell culture.
nd, not determined.
aFour subjects or eight biopsies with no 4qA alleles detected.
DISCUSSION
Here, we used a new large collection of biopsies and myogenic cells from cohorts of FSHD subjects and their unaffected relatives to independently investigate the DUX4-FL expression model of FSHD pathogenesis (14,15). Aberrant DUX4-FL expression in skeletal muscle is expected to induce ectopic expression of multiple genes and, if expression is high enough, to be potentially cytotoxic (16,18–21). In skeletal muscle, DUX4-FL was previously detected only in FSHD cells and tissues (14,15,22). Therefore, it was surprising that we found DUX4-fl mRNA and protein expression in muscle and myogenic cells from unaffected donors, all of which had only genetically unaffected sized 4q35 alleles and full muscle strength. Our extensive set of control experiments and tests for specificity showed that we indeed detected DUX4-FL in some unaffected myogenic cells and muscle tissues. Furthermore, upon performing a statistical analysis, the previous report that failed to detect DUX4-fl mRNA in unaffected myoblasts or myotubes using a smaller sample set (n = 4) of 4qA-containing controls (15), was consistent with DUX4-fl being detectable in up to 50% of controls (Wilson's 95% confidence interval). Although the probability of detecting DUX4-fl mRNA in a sample depends both on the sensitivity of the assay, cell culture conditions and on the number of replicate RT-PCR runs, if we assume the same sensitivity and culture conditions then the previously published results from Snider et al. are consistent with our detection of DUX4-fl mRNA in 2 out of 16 control myoblast cultures and 8 out of 16 control differentiated cultures (Table 2), from 5 of 8 unaffected subjects (p = 0.61 and p = 0.05, respectively, by Monte Carlo simulations, assuming 4 replicates per sample) (15). Similarly, the previously reported biopsy data that did not detect DUX4-fl in 9 unaffected subjects is consistent with DUX4-fl being detectable in up to 30% of controls (Wilson's 95% confidence interval), and is not significantly different from our detection of DUX4-fl mRNA expression in 3 out of 34 control biopsies (from 2 out of 18 unaffected individuals, of which only 3 lacked the permissive 4qA allele; p = 0.36 assuming four replicates per sample). Even though the seemingly disparate results of these two studies are statistically consistent with each other, our use of a larger number of samples, resulting in positive RT-PCR results, produced a significantly different interpretation: DUX4-fl mRNA and protein expression is not exclusive to FSHD. We did, however, find that DUX4-fl was significantly more likely to be expressed in FSHD than unaffected muscle cells and tissues.
Our familial cohort study also indicates that DUX4-fl mRNA and protein expression in cells from genetically FSHD1 subjects does not necessarily correlate with decreased muscle strength. Individuals in six of the cohorts (05C, 15B, 27B, 28B, 29B and 30B) showed no apparent muscle weakness in their biceps or deltoid muscles at the time the biopsies were taken (Table 1), yet five of these six subjects (05C, 15B, 28B, 29B, 30B) showed relatively robust DUX4-fl expression (Figs 2 and 3C, Table 2), and overall the DUX4-fl expression for these six did not differ significantly from that of the clinically affected FSHD subjects (p> 0.02 in all assays; likelihood-ratio test; Table 2, Supplementary Material, Table S1). For example, the age of subject 15B was 69 years with no clinical weakness perceptible to subject or investigator at the time of biopsy but was found during the study to have a 28 kb FSHD 4qA allele (Table 1). The relatively high frequency of DUX4-FL positive nuclei in subject 15B (1 in ∼1500 nuclei compared with the 1 in ∼9000 nuclei found in myogenic cells from his 66-year-old FSHD manifesting brother or the average 1 in ∼2000 nuclei for FSHD as a group) and expression in the 15B biceps biopsy (Fig. 4) was not associated with a more severe disease in the subject. This suggests that DUX4-fl mRNA and protein expression in a particular muscle and at a particular time during the course of the disease does not correlate well with FSHD clinical manifestation of the disease. Whether a longitudinal series of biopsies taken over the multi-year course of the disease would show a tighter correlation could not be answered with our set of single time-point biopsies.
Together, our findings unexpectedly revealed that polyadenylated DUX4-fl mRNA and DUX4-FL protein were expressed in myogenic cells and muscle biopsies from both unaffected controls and FSHD subjects, though at a greater frequency in FSHD. Our findings provide evidence for a modified model of FSHD pathogenesis in which detectable DUX4-fl expression alone is not sufficient for FSHD pathology. Revealing that expression of DUX4-fl mRNA and protein does not require a D4Z4 deletion, nor does it necessarily lead to FSHD, requires a new view of FSHD pathogenesis. Integrating these new findings with the existing model, we propose that DUX4-fl mediated pathology is regulated by modifiers acting at the level of DUX4-fl mRNA expression, splicing, and/or protein function.
Nonetheless, the evidence that DUX4 expression plays a necessary role in FSHD muscle weakness and pathology is still compelling considering the exclusive linkage of FSHD with permissive 4qA subtelomeres (28) that are uniquely capable of producing stable polyadenylated DUX4-fl mRNA (14). A recent study has also suggested that some 4qB subtelomeres may be permissive for FSHD progression, but it was not determined if DUX4-FL might have been stably expressed within the context of these particular 4qB alleles (29). DUX4-fl modifiers could perhaps include epigenetic regulators such as DNA methylation, environmental signals, post-translational modifications, and/or proteins (including the dominant negative DUX4-s) that regulate DUX4-FL expression levels or affect its function. In particular, modifiers might act to increase or decrease the frequency or level of DUX4-fl expression and/or DUX4-FL function throughout the lifetime and at different sites within muscles. In this model, if DUX4-FL levels increased beyond a tolerated threshold then overt pathology would result.
Supporting this DUX4-fl modifier model, we found that DUX4-fl was expressed by more FSHD than unaffected nuclei in myogenic cell cultures (p = 0.001; likelihood-ratio test), and was detected by RT-PCR with greater frequency in FSHD than control samples in differentiated myogenic cell cultures as well as in myoblasts and biopsies (p = 0.0005, p = 0.002 and p = 0.000001, respectively; likelihood-ratio test), suggesting that FSHD muscle is more prone to DUX4-fl expression than unaffected muscle. For each assay, the effect of muscle type (biceps, deltoid) on DUX4-fl detection was non-significant (p > 0.1), as was interaction with disease status aside from a mildly significant interaction in myogenic cell cultures (p = 0.03; likelihood ratio test), where the difference between DUX4-fl detection in FSHD and control samples was slightly larger in biceps than in deltoid. In addition, our use of first-degree relatives as unaffected controls raises the intriguing possibility that modifiers linked genetically to family background may influence DUX4-fl expression and FSHD pathology. Consistent with this idea, some FSHD families in our studies and others' have unaffected members with shortened D4Z4 arrays in combination with permissive 4qA alleles (29–31).
Our study thus shows that expression of DUX4-fl mRNA or protein, though likely necessary for FSHD pathogenesis, is clearly not a sufficient condition. These results thus raise the possibility that quantitative modifiers of DUX4-fl expression and/or function, and family genetic background are determinants of FSHD muscle disease progression. Such modifiers of DUX4-fl in FSHD, once identified, would represent new therapeutic targets to influence DUX4-fl expression and block FSHD disease progression.
MATERIALS AND METHODS
Human subjects biopsy and primary cell culture
This study was approved by The Johns Hopkins School of Medicine Institutional Review Board. Open muscle biopsy was performed on both the biceps and deltoid muscles of the FSHD affected and unaffected donors yielding ∼1 g tissue per biopsy. Each donor with a clinical diagnosis of FSHD1 was confirmed by the University of Iowa Diagnostic Laboratories to have a contracted D4Z4 array on a 4qA allele by pulse-field gel electrophoresis (PFGE) and Southern blotting (32). Donors with FSHD1 were identified based on the combined presence of (i) clinical symptoms, (ii) shortened 4q35 D4Z4 arrays and (iii) a 4qA subtelomeric allele. All such confirmed FSHD1 individuals in our study had EcoRI/BlnI alleles of <32 kb corresponding to ∼8.5 or fewer repeats and thus below the ≤10 repeat threshold for a pathogenic allele. One 48-year-old subject with no clinical symptoms (20U) had an ∼39 kb/4 qA allele (corresponding to ∼10.7 repeats) that her son (20A) inherited along with a shorter ∼20 kb/4 qA allele from the father. It is possible that the ∼39 kb allele could be proved pathogenic (despite the current absence of clinical pathology in 20U), but such an outcome cannot change the central conclusion of this study. Extended film exposures of the PFGE analysis followed by extensive analysis of the segregation of alleles confirmed that none of the unaffected subjects showed somatic mosaicism. In addition, the diagnostic PFGE was repeated for subjects 03U, 07U, 17V and 20U using blood and 03U and 07U using myoblasts confirming EcoRI/BlnI fragment sizes and absence of mosaicism. Certain subjects displayed a 3:1 or 1:3 ratio of chromosome 4q-type to chromosome10q-type D4Z4 arrays (based on sensitivity to BlnI digestion, Table 1); however, because the total number of 4q and 10q alleles always equaled four, we found no indication of somatic mosaicism (26,32).
Biopsies were designated with the number of the cohort (e.g. 01), whether FHSD-affected (A for the first affected donor in the cohort, B for the second, etc.) or unaffected control (U for the first control donor in the cohort, V for the second, etc.), and the muscle (bic = biceps or del = deltoid) from which the biopsy was obtained; e.g. ‘03Abic’ biopsy and derived cells originated from the biceps of the first FSHD-affected donor in cohort 03. Primary muscle cell strains were established from portions of muscle biopsies, enriched for CD56 positive myogenic cells and propagated and differentiated as described (24).
For RNA studies, cells were propagated by daily feeding with HMP medium [Ham's F-10 medium supplemented with 20% characterized FBS (Hyclone), 1.2 mm CaCl2 (Sigma-Aldrich), 0.5% chick embryo extract and 1% antibiotics/antimycotics (Cellgro)] or LHCN medium [4:1 DMEM:Medium 199 supplemented with 15% characterized FBS (Hyclone), 0.02 m HEPES (Sigma-Aldrich), 0.03 µg/ml ZnSO4 (Sigma), 1.4 µg/ml Vitamin B12 (Sigma-Aldrich), 0.055 µg/ml dexamethasone (Sigma-Aldrich), 1% antibiotics/antimycotics (Cellgro), 2.5 ng/ml hepatocyte growth factor (Chemicon International) and 10 ng/ml basic fibroblast growth factor (Millipore)]. When >90% confluent, plates were rinsed with PBS (Cellgro) and cells were switched to differentiation medium [4:1 DMEM:Medium 199 supplemented with 2% horse serum (Hyclone), 2 mm l-glutamine (Gibco), 1% antibiotics/antimycotics (Cellgro), 10 mm HEPES (Gibco) and 1 mm sodium pyruvate (Gibco)] for up to 7 days. Cultures were rinsed two times with PBS and cells were removed with cell lifters (Costar), collected by centrifugation, snap frozen in liquid nitrogen, and stored at −80°C. Cells for immunocytochemistry were propagated on gelatin-coated four-well chamber slides (Thermo) by daily feeding with HMP medium until >90% confluent and then switched to differentiation medium as described above.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated from cultured cells using an RNeasy Mini Kit (Qiagen) and DNase I-treated on the column using the RNase-Free DNase Set (Qiagen) before eluting with 30 µl of RNase-free water. First-strand cDNA was synthesized on 1–1.5 µg total RNA using Superscript III Reverse Transcriptase (Invitrogen) and an oligo (dT)16 DNA primer at 55°C for 1 h. Primary PCR was performed with Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Scientific) and 8% Dimethyl Sulfoxide using 10% of the first strand reaction with the following cycling conditions: 98°C 2 min followed by 25 cycles of 98°C for 15 s, 62°C for 20 s, 72°C for 1 min. To ensure specificity, nested PCR using 8–10% of the primary PCR was used with the following cycling conditions: 98°C 2 min and 30 cycles of 98°C for 15 s, 62°C for 15 s, 72°C for 50 s. The oligonucleotide primers for nested RT-PCRs [previously documented (15)] were 14A and 183 for primary PCR and 15A and 184 for the nested round of PCR. All RT-PCR products were gel-purified, cloned using the pGEM-T Easy Vector System (Promega) and sequenced in both directions. Sequences confirmed the identity of PCR products as properly spliced DUX4-fl thus eliminating the possibility of PCR artifacts. Six chromosome 4qA specific changes compared with chromosome 10qA DUX4 sequence AL732375 were used to verify all transcripts as being derived from chromosome 4q as follows: relative to bp1 being A of the ATG; bp1209 CTCCGC(ch4) versus 6 nt deletion (ch10); bp1421 T(ch4) versus C(ch10); bp 1447 C(ch4) versus A(ch10); bp1536 C(ch4) versus G(ch10); bp1579 T(ch4) versus G(ch10); bp1601 G(ch4) versus C(ch10). Available SNPs were able to additionally confirm the subject origin of the RT-PCR products. Individual polymorphic changes in DUX4-fl transcript are available upon request. In all cases, RT-PCR products were dependent upon the presence of the reverse transcriptase in the first strand synthesis controlling against genomic DNA or plasmid contamination.
Total RNA was isolated from frozen muscle biopsies using Trizol (Invitrogen) and BCP (Molecular Research Center) after grinding in dry ice with a mortar and pestle. Total RNA (5 µg) was subjected to two DNase I (Ambion) treatments for 15 min each at 37°C in the presence of RNaseOUT (Invitrogen), purified with the RNeasy Kit (Qiagen) and RNA was eluted with 60 ul EB buffer and the volume was reduced with speed vacuum. cDNA was synthesized using the RevertAid first strand cDNA synthesis kit (Fermentas) using oligo(dT)18 primers in 20 µl reactions and diluted by adding 45 µl of nuclease-free water. DUX4-fl transcripts were detected by nested PCR approach as described above which ensures specificity of the reaction for the 4q35-derived DUX4 transcripts. For the extended cycle RT-PCRs, the primary PCR was 25 cycles followed by 30 or 40 cycles of nested PCR. All PCR products were sequenced and specificity of reaction products was confirmed as above. In addition, RT-PCR from muscle biopsies was performed independently, including from RNA preparation through PCR, in two different laboratories, one at BBRI and one at Children's Hospital, each producing similar results.
Immunocytochemistry
Cells were propagated on gelatin-coated 4-well chamber slides (Thermo) until >90% confluent and then switched to differentiation medium for 4–6 days. Cells were rinsed two times with PBS, fixed with 2% formaldehyde for 7–10 min at room temperature (RT), washed three times with PBS, permeabilized with 1% Triton-X100 (Sigma) in PBS for 15 min, and incubated in blocking solution [2% horse serum (Gibco), 2% goat serum (Gibco), and 2% BSA (EMD) in PBS + 0.1% Triton-X100] for 30–60 min at RT. Cells were incubated overnight at 4°C with a 1:50 dilution in blocking solution of the P4H2 mouse mAb (15,27). A horseradish peroxidase-linked secondary antibody system and diaminobenzidine substrate (Vectastain ABC Elite, Vector Laboratories) was used for detection essentially according to manufacturer's directions. Cells were subsequently co-stained for desmin (clone D33, Dako; 1:100 or Clone DE-U-10, Sigma; 1:100) with Alexa fluor-conjugated secondary antibodies (Molecular Probes; anti-rabbit IgG-546 or 594 and anti-mouse IgG-488) diluted 1:300 in blocking solution. Nuclei were stained with bisbenzimide and were scanned manually for DUX4-FL-positive cells using differential interference contrast microscopy (DIC) or bright-field optics and imaged using a Leica DMR microscope with a DC300F camera and DM-IL software and/or a Nikon E800 system with Spot camera and software version 4.6 (Diagnostic Instruments, Inc). The number of nuclei per well was approximated for each cell strain by counting 10 random fields of known area at 10× and extrapolating to the total area of the well. A total between 60 000 and 150 000 nuclei were screened for each cell culture.
Statistical Analysis
The per-replicate detection probability of DUX4-fl mRNA by RT-PCR was fit with a mixed-effect binomial logistic regression model (33) using the R package lme4 (34), with fixed effects for disease status and muscle type (including an interaction term), and random effects for cohort, individual and biopsy (or cell culture). The random effects account for correlations in DUX4-fl detection between family members, between biceps and deltoid from a single individual, and between multiple RT-PCR replicates for a single biopsy (or cell culture), respectively. An analogous mixed-effect model was fit to the ICC count data using lme4, but with Poisson rather than binomial regression (33) with an offset term to account for variation in total number of nuclei counted, and with an additional replicate-level random effect to account for overdispersion between replicate counts from the same cell-culture. Significance of fixed effects was assessed using likelihood-ratio tests, and reported P-values for FSHD versus unaffected comparisons exclude the unaffected individuals with no 4qA alleles having been detected.
Consistency with results from previous studies was assessed with Monte Carlo bootstrap simulations (35) n = 1000 runs), resampling first biopsies (or cell cultures) then RT-PCR replicates, both with replacement, from the empirical distribution for unaffected individuals (Supplementary Material, Table S1, again excluding those with no 4qA alleles detected). Data for biceps and deltoid were combined for these simulations, and reported results for biopsies were based on the 25+30 cycle nested RT-PCR data.
Microsatellite genotyping
Genomic DNA was isolated from cell lines or muscle biopsies using the Wizard SV Genomic DNA Purification System kit (Promega) according to the manufacturer's protocol. A set of 12 microsatellite markers (D1S2842, D2S206, D3S1565, D4S424, D5S641, D6S460, D9S171, D10S547, D11S987, D15S117, D16S503 and D20S889) from the ABI panel was used and the PCR products were analyzed on an ABI Prism 3730 DNA Analyzer (Perkin Elmer). The genotypes were determined using GeneMapper software (Applied Biosystems) and compared with the reference DNA isolated from leukocytes previously obtained from the same subjects.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grant no. 5U54HD060848 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development that supports the Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center for FSHD Research directed by C.P.E.; by grant no. MDA216652 from MDA to C.P.E.; by grant no. AR055877 from NIAMS/NIH and the FSH Society Cape Cod Walk and Roll research grant no. FSHS-22011-06 to P.L.J.; and by grant no. HL064641 from NHLBI/NIH, no. AR060328 from NIAMS/NIH and the Muscular Dystrophy Association to J.B.M.; and by grants from the Thoracic Foundation (Boston, MA) to S.H. and P.L.J.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank all participating subjects and their families for making this study possible, Mr Daniel Perez and the FSH Society for family outreach, Drs Doris Leung and Genila Bibat for assistance with muscle biopsy, Kendal Hanger for technical support, and Linda Geng and Dr Stephen J. Tapscott for their generous gift of the P4H2 monoclonal antibody.
Conflict of Interest statement. The authors declare no conflicts of interest.
REFERENCES
- 1.Padberg G.W. Facioscapulohumeral Disease [thesis] Leiden, the Netherlands: Leiden University; 1982. [Google Scholar]
- 2.Tawil R. Facioscapulohumeral muscular dystrophy. Neurotherapeutics. 2008;5:601–606. doi: 10.1016/j.nurt.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orphanet. Prevalence of rare diseass: bibliographic data in Orphanet Report Series, Rare Diseases collection. 2011. Inserm. http://www.orpha.net/orphacom/cahiers/docs/GB/Prevalence_of_rare_diseases_by_alphabetical_list.pdf .
- 4.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 5.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 6.Cabianca D.S., Gabellini D. The cell biology of disease: FSHD: copy number variations on the theme of muscular dystrophy. J. Cell Biol. 2010;191:1049–1060. doi: 10.1083/jcb.201007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Greef J.C., Wohlgemuth M., Chan O.A., Hansson K.B., Smeets D., Frants R.R., Weemaes C.M., Padberg G.W., van der Maarel S.M. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69:1018–1026. doi: 10.1212/01.wnl.0000271391.44352.fe. [DOI] [PubMed] [Google Scholar]
- 8.Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A., et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ottaviani A., Rival-Gervier S., Boussouar A., Foerster A.M., Rondier D., Sacconi S., Desnuelle C., Gilson E., Magdinier F. The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet. 2009;5:e1000394. doi: 10.1371/journal.pgen.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neguembor M.V., Gabellini D. In junk we trust: repetitive DNA, epigenetics and facioscapulohumeral muscular dystrophy. Epigenomics. 2010;2:271–287. doi: 10.2217/epi.10.8. [DOI] [PubMed] [Google Scholar]
- 11.de Greef J.C., Lemmers R.J., van Engelen B.G., Sacconi S., Venance S.L., Frants R.R., Tawil R., van der Maarel S.M. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 2009;30:1449–1459. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 12.de Greef J.C., Lemmers R.J., Camano P., Day J.W., Sacconi S., Dunand M., van Engelen B.G., Kiuru-Enari S., Padberg G.W., Rosa A.L., et al. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology. 2010;75:1548–1554. doi: 10.1212/WNL.0b013e3181f96175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 14.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Maarel S.M., Tawil R., Tapscott S.J. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol. Med. 2011;17:252–258. doi: 10.1016/j.molmed.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O., et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Wuebbles R.D., Long S.W., Hanel M.L., Jones P.L. Testing the effects of FSHD candidate gene expression in vertebrate muscle development. Int. J. Clin. Exp. Pathol. 2010;3:386–400. [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2011;69:540–552. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F., et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsumagari K., Chang S.C., Lacey M., Baribault C., Chittur S.V., Sowden J., Tawil R., Crawford G.E., Ehrlich M. Gene expression during normal and FSHD myogenesis. BMC Med. Genom. 2011;4:67. doi: 10.1186/1755-8794-4-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homma S., Chen J.C., Rahimov F., Beermann M.L., Hanger K., Bibat G.M., Wagner K.R., Kunkel L.M., Emerson C.P., Jr., Miller J.B. A unique library of myogenic cells from facioscapulohumeral muscular dystrophy subjects and unaffected relatives: family, disease and cell function. Eur. J. Hum. Genet. 2012;20:404–410. doi: 10.1038/ejhg.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadler G., Chen J.C.J., Wagner K.R., Ducellier J.R., Shay J.W., Emerson C.P., Wright W.E. Establishment of clonal myogenic cell lines from severely affected dystrophic muscles—CDK4 maintains the myogenic population. Skel. Musc. 2011;1:12. doi: 10.1186/2044-5040-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sykes P.J., Neoh S.H., Brisco M.J., Hughes E., Condon J., Morley A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- 26.Lemmers R.J., van der Vliet P.J., van der Gaag K.J., Zuniga S., Frants R.R., de Knijff P., van der Maarel S.M. Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete interchromosomal sequence transfers in human evolution. Am. J. Hum. Genet. 2010;86:364–377. doi: 10.1016/j.ajhg.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geng L.N., Tyler A.E., Tapscott S.J. Immunodetection of human double homeobox 4. Hybridoma (Larchmt) 2011;30:125–130. doi: 10.1089/hyb.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmers R.J., de Kievit P., Sandkuijl L., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 29.Scionti I., Fabbri G., Fiorillo C., Ricci G., Greco F., D'Amico R., Termanini A., Vercelli L., Tomelleri G., Cao M., et al. Facioscapulohumeral muscular dystrophy: new insights from compound heterozygotes and implication for prenatal genetic counselling. J. Med. Genet. 2012;49:171–178. doi: 10.1136/jmedgenet-2011-100454. [DOI] [PubMed] [Google Scholar]
- 30.Piko H., Molnar M.J., Herczegfalvi A., Mayer P., Karcagi V. [Role of associated alleles and hypomethylation status in the clinical expression of facioscapulohumeral muscular dystrophy] Orv. Hetil. 2011;152:1576–1585. doi: 10.1556/OH.2011.29179. [DOI] [PubMed] [Google Scholar]
- 31.Wohlgemuth M., Lemmers R.J., van der Kooi E.L., van der Wielen M.J., van Overveld P.G., Dauwerse H., Bakker E., Frants R.R., Padberg G.W., van der Maarel S.M. Possible phenotypic dosage effect in patients compound heterozygous for FSHD-sized 4q35 alleles. Neurology. 2003;61:909–913. doi: 10.1212/wnl.61.7.909. [DOI] [PubMed] [Google Scholar]
- 32.Lemmers R.J., van der Wielen M.J., Bakker E., Padberg G.W., Frants R.R., van der Maarel S.M. Somatic mosaicism in FSHD often goes undetected. Ann. Neurol. 2004;55:845–850. doi: 10.1002/ana.20106. [DOI] [PubMed] [Google Scholar]
- 33.Gelman A., Hill J. Data Analysis Using Regression and Multilevel/Hierarchical Models. Cambridge, UK: Cambridge University Press; 2007. [Google Scholar]
- 34.Baayen R.H., Davidson D.J., Bates D.M. Mixed-effects modeling with crossed random effects. J. Mem. Lang. 2008;56:390–412. [Google Scholar]
- 35.Efron B. Bootstrap methods: another look at the jackknife. Ann. Stat. 1979;7:1–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.