Abstract
A number of mouse models for spinal muscular atrophy (SMA) have been genetically engineered to recapitulate the severity of human SMA by using a targeted null mutation at the mouse Smn1 locus coupled with the transgenic addition of varying copy numbers of human SMN2 genes. Although this approach has been useful in modeling severe SMA and very mild SMA, a mouse model of the intermediate form of the disease would provide an additional research tool amenable for drug discovery. In addition, many of the previously engineered SMA strains are multi-allelic by design, containing a combination of transgenes and targeted mutations in the homozygous state, making further genetic manipulation difficult. A new genetic engineering approach was developed whereby variable numbers of SMN2 sequences were incorporated directly into the murine Smn1 locus. Using combinations of these alleles, we generated an allelic series of SMA mouse strains harboring no, one, two, three, four, five, six or eight copies of SMN2. We report here the characterization of SMA mutants in this series that displayed a range in disease severity from embryonic lethal to viable with mild neuromuscular deficits.
INTRODUCTION
Spinal muscular atrophy (SMA) is one of the leading causes of childhood mortality and the second most common autosomal recessive disorder affecting between 1:6000 and 1:10 000 live births (1). The molecular genetic defect in SMA gives rise to a range of clinical severity of disease observed in the human population. SMA is caused by mutations in the survival motor neuron gene (SMN1). In humans, an intrachromosomal duplication event gives rise to an additional copy of SMN, called SMN2, containing a single C-to-T transition in exon 7 distinguishing it from SMN1. This transition causes the aberrant splicing of 80–90% of SMN2 transcripts causing them to lack exon 7 (2). Because a small fraction of full-length (FL) SMN is generated from SMN2, the range in disease severity correlates with the number of copies of the SMN2 gene a patient possesses (3). Although SMN is a ubiquitously expressed protein that plays a role in RNA processing, metabolism and transport, deficiencies in SMN affect primarily the motor system in SMA. The most severe form of SMA manifests in death in infancy due to respiratory muscular weakness (4), whereas the least severe form of the disease occurs in adulthood and is associated with mild weakness of the proximal muscles (1). Because SMA is a disease of ‘low’ levels of SMN (as opposed to complete lack of SMN), it becomes a unique challenge to model in a mammalian system.
Mice harbor only a single Smn gene called Smn1, and targeted disruption of the murine Smn1 locus results in embryonic lethality (5) due to the lack of compensatory Smn. Examination of heterozygous null (Smn1+/−) animals revealed a very mild phenotype, with animals living a normal lifespan and exhibiting a low level of motor neuron degeneration (6). In an effort to circumvent the embryonic lethality and generate an animal with intermediate SMA phenotype, a human SMN2 transgene was added onto the Smn1−/− background (Smn1−/−;SMN2+/+), resulting in an animal that succumbs to disease in the first post-natal week (7). The further addition of a human SMN2 transgene lacking exon 7 onto this model (Smn−/−;SMN2+/+;SMNΔ7+/+) resulted in an animal with a mean lifespan of ∼14 days that exhibits progressive muscle weakness associated with neuromuscular junction (NMJ) denervation (8). The addition of the SMNΔ7 transgene extended the life of the severe model by ∼5–7 days, and thus this model became amenable for therapeutic screening and continues to be a mainstay model for drug development for SMA. Building on the knowledge garnered from these two early mouse models of SMA, other longer lived models have been developed that also combine targeted mutaganesis of the murine Smn1 locus with transgenic overexpression of human SMN2, or transgenic overexpression of genetically modified variants of SMN2 (9–10). In addition, a mouse model harboring a conditional knock out of Smn in motor neuron cell types was developed that displayed a reduced life expectancy along with functional motor deficits (11). Each of these modifications has resulted in an animal with a significantly longer lifespan than that of the SMNΔ7 model, though the conditional knock out model had an average lifespan of ∼30 days, significantly shorter than the other less severe models. In addition, common characteristics of long-lived SMA mouse models (those surviving >1 year) include necrotic tails and ears and seemingly mild NMJ pathology (9,12). Collectively, these models and others have provided invaluable insights into the pathogenesis of SMA, and have certainly provided the tools necessary for the development of a number of novel SMA-specific and non-SMA-specific therapeutics that have progressed to clinic, including but not limited to olesoxime (13), quinazoline (14) and antisense oligonucleotide therapy (15).
Using a targeted mutagenesis approach, the allelic series at the murine Smn1 locus was created with a 2-fold objective in mind: (i) to reduce the genetic complexity used in the current collection of mouse models by directly targeting a titration of SMN into a single locus; and (ii) through a combination of crosses within the series, to generate an intermediate mouse model of SMA exhibiting clinical features associated with disease pathology, thus providing clear outcome measures for therapeutic discovery. In the present work, we demonstrate the phenotypic analysis of viable combinations in the allelic series. Mutants in the allelic series exhibit diminished SMN expression coupled with mild neuromuscular deficits. In addition, the presentation of these new alleles adds to the collection of research tools available for SMA research.
RESULTS
Viability of mutant mice harboring the allelic series
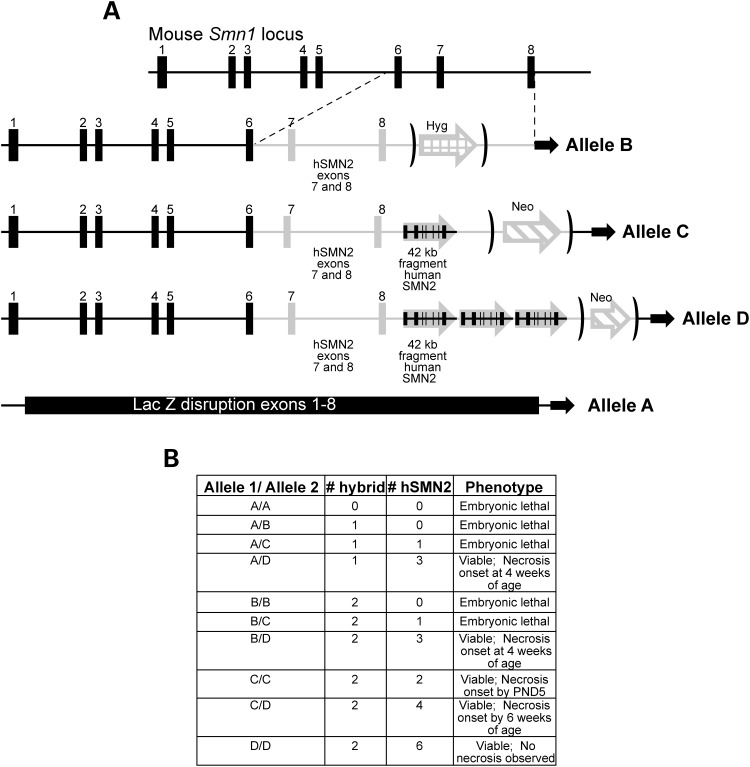
Targeted mouse lines for each allelic construct were obtained from Regeneron Pharmaceuticals (Tarrytown, NY, USA) and built using the VelociGene technology as described (16). As depicted in Figure 1, each construct in the allelic series contained zero to four copies of SMN, either through a hybrid (murine genomic Smn1 spanning exons 1 through 6 and human genomic SMN2 spanning exons 7 and 8) portion or combination of both hybrid and an FL 42 kb fragment of human SMN2. Animals harboring each allele were identified through PCR genotyping of tail DNA and selected for subsequent backcrossing to both C57BL/6J and FVB/NJ to create congenic lines (refer to Materials and Methods for specific stock numbers of congenic lines). Heterozygous animals in C57BL/6J congenic, FVB/NJ congenic and mixed background lines of the allelic series (designated Smn1A/+, Smn1B/+, Smn1C/+ and Smn1D/+) were intercrossed to assess the viability of homozygous mutants. As no significant differences were noted between the FVB/NJ and the C57BL/6J congenic lines, the majority of this paper will focus on the characterization of the allelic series on the mixed genetic background. Viability analysis of these initial heterozygous intercrosses of each of the alleles in the series revealed a sharp-line delineation between embryonic lethality and complete viability. As expected, no live homozygous mutants were generated from intercrosses of the Smn1A allele owing to its complete disruption of the Smn1 locus. The first viable mutants observed were homozygous for the Smn1C allele and harbored four relative copies of SMN (two hybrid and two hSMN2). As expected, animals homozygous for the Smn1D allele harboring eight copies of SMN (two hybrid and six hSMN2) were also viable. The design of the allelic series allows for the examination of a genetic titration of SMN levels in the murine system through the cross-pairing of different alleles creating compound heterozygotes at the Smn1 locus. Of the possible combinations of alleles, only those containing the Smn1D allele paired with any other allele demonstrated viablity as mutant compound heterozygotes (Fig. 1B). Each viable mutant contained not less than four copies of SMN either as a hybrid allele or as an FL SMN2. As a result of this analysis, further phenotypic characterization was performed on the Smn1C/C mutant, along with the Smn1D/A, Smn1D/B, Smn1D/C and Smn1D/D mutant combinations.
Figure 1.
Composition and phenotype of the new Smn allelic series. (A) The allelic series set is a combination of targeted mutations containing either (i) (bottom) complete disruption of the Smn1 locus via lacZ insertion (Smn1A allele); (ii) the insertion of a hybrid human genomic SMN2 exons 7 and 8 fused to murine exons 1–6 (Smn1B allele); (iii) a combination of the hybrid allele and one 42 kb fragment of genomic human SMN2 (Smn1C allele); and (iv) a combination of the hybrid allele and three copies of a 42 kb fragment of genomic human SMN2 (Smn1D allele). (B) The allelic series can be genetically manipulated to generate mutants containing zero to eight relative copies of SMN. Studies demonstrated animals with fewer than four relative SMN copies were embryonic lethal. Those with more than four copies were completely viable and presented with varying degrees of tail and ear necrosis inversely correlating with relative SMN copy number.
SMN expression levels in the allelic combinations
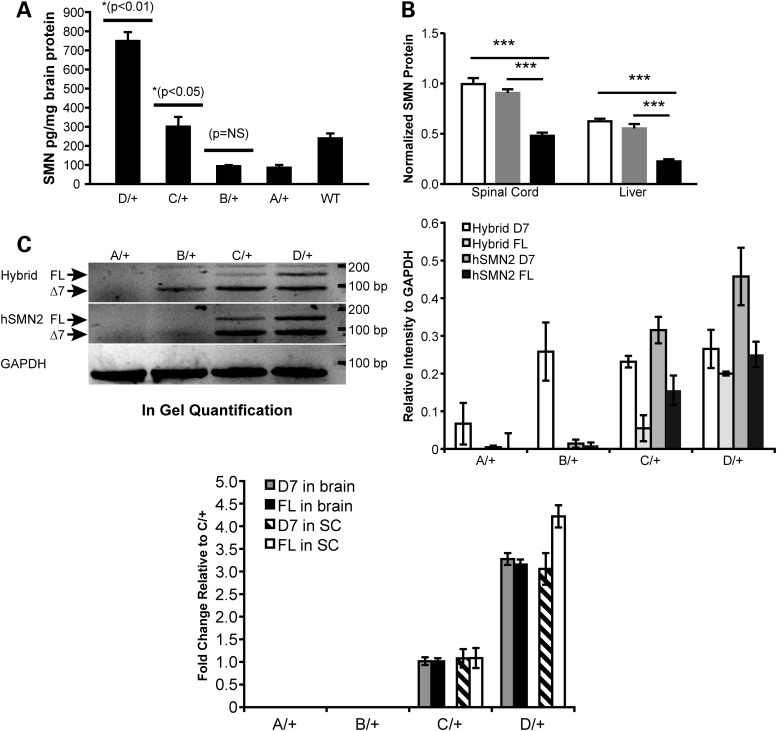
The severity of SMA in patients and of SMA-like phenotypes in mouse models is inversely correlated to the level of expression of SMN protein. The hybrid and human SMN2 genes knocked in the mouse Smn1 can be transcribed, a priori, into the FL and delta-7 (D7) mRNAs, of which the former only encodes a stable protein. To verify that the severity of phenotypes among the different alleles arises from the relative abundances of SMN, we undertook a comparison of the SMN protein levels across the allelic series. We had to consider two technical obstacles. The first one was the lethality of the homozygotes for the Smn1A and Smn1B alleles. The second was that the ELISA assay developed to measure SMN proteins utilizes a polyclonal antibody recognizing human SMN that also cross-reacts with murine SMN1, although with less efficiency. This raised the possibility that homozygous mutants with a lower level of total SMN mostly accounted for by the human genes would appear as producing more SMN than their wild-type (WT) counterpart. To circumvent these two difficulties, we measured SMN levels in heterozygotes for the different alleles, all being viable and expected to produce comparable level of endogenous mouse SMN. As shown in Figure 2A, animals heterozygous for the Smn1D allele express significantly more SMN protein (754.7 ± 40.9 pg/mg protein) compared with animals heterozygous for the Smn1C allele (t-test, P < 0.01). Animals heterozygous for the Smn1C allele (306.35 ± 10.3 pg/mg protein) expressed significantly more SMN than both Smn1A and Smn1B heterozygous animals (91.2 ± 8.5 and 99.5 ± 0.15 pg/mg protein, respectively) (t-test, P < 0.05). Animals harboring only endogenous murine Smn1 alleles expressed roughly twice the amount of SMN as animals heterozygous for the Smn1A (functional null) allele (245 ± 8.45 versus 91.2 ± 8.52 pg/mg protein). To further characterize the production of SMN protein by the C allele, total SMN protein expression in heterozygous, homozygous and WT C allele mice was performed by western blot with another antibody; this demonstrated a significant (P < 0.001) overall reduction of SMN in the spinal cord and the liver of homozygous mice compared with their heterozygote littermates (0.4807 ± 0.11 RU versus 0.9947 ± 0.22 RU and 0.2262 ± 0.08 RU versus 0.5541 ± 0.12 RU, normalized to WT spinal cord protein, respectively) and WT (Fig. 2B and image of the western blot in Supplementary Material, Fig. S2A).
Figure 2.
SMN expression in allelic series mutants. (A) Total SMN protein detected by ELISA in brain lysates (n = 2/genotype) of heterozygous Smn1D, Smn1C, Smn1B, Smn1A and Smn1+/+ animals demonstrated a titration correlating with total numbers of relative SMN copies per allele. Heterozygous Smn1C allele mice have significantly more SMN in the brain than both heterozygous Smn1B and heterozygous Smn1A (P < 0.05; t-test), and significantly less SMN in the brain than heterozygous Smn1D allele mice (P < 0.01; t-test). Smn1+/+ protein levels demonstrate a preferential detection of human SMN relative to murine SMN protein. (B) Western blot analysis of FL SMN in the spinal cord and liver extracts of homozygous Smn1C/Cmutants demonstrated reduced relative SMN total protein (mouse + human) compared with heterozygous and WT controls. SMN protein levels were significantly (P < 0.001) reduced in the spinal cord and liver of homozygous Smn1C/C mutants (black bars). No differences were observed between heterozygous (gray bars) and WT controls (open bars). Data are represented as the mean ± SEM (n = 12 animals/genotype mixed genders) normalized first to ATP5B protein, then to WT levels. (C) Semi-qPCR of hybrid and human SMN. FL-SMN and D7-SMN were visualized based on the difference of product size on ethidium bromide-stained agarose gel. Left panel: FL, 148 bp, delta7: 94 bp. Right panel: Quantification of different products normalized to Gapdh on three animals/genotype. (D) Real-time RT-PCR analysis of D7-SMN2 versus FL-SMN2 derived from human genomic portions across the allelic series in total RNA isolated from the brain and the spinal cord. Data are represented as fold change values normalized to Smn1C/+.
The comparable levels of SMN protein found in the Smn1A and Smn1B alleles suggested that the hybrid gene expresses very low levels of SMN. To address this, we turned toward the analysis of SMN mRNA, which provides a more precise distinction between the hybrid and human gene, as well as an assessment of the retention of exon 7. Semi-quantitative real-time PCR (qPCR) using mouse- and human-specific forward primers in exon 6 and a reverse primer in human exon 8 allowed us to separate the hybrid and human transcripts, as well as their respective D7 and FL variants. Figure 2C shows that the hybrid gene only produced D7 mRNA (Smn1B/+ upper gel). The predominance of exon 7 exclusion is likely the reason why the Smn1B allele phenotypically behaves like the knock-out allele (Smn1A). The human gene, however, present in the Smn1C and Smn1D alleles, efficiently produced FL RNAs, aside the more abundant D7 RNA. Our hybrid-specific assay also amplified a product of the size of the FL-SMN2 in the Smn1C and Smn1D alleles, but not in the Smn1B allele. Given that all three alleles share a single copy of the hybrid gene, and that this unexpected full length hybrid SMN2 product becomes more abundant as the FL-hSMN2 product increases in Smn1C/+ and Smn1D/+, we reasoned that that the mouse-specific exon 6 primer used to recognize the hybrid transcript also recognizes the human exon 6, albeit with a lower affinity. The limitation in our ability to distinguish between the hybrid and human SMN2 RNAs probably comes from the high homology (>80%) between the mouse Smn1 and human SMN2 sequences, especially in exon 6. Real-time PCR (Supplementary Material, Fig. S2B) proved more resistant to this cross-reactivity of the primers and confirmed that no human SMN2 was present in the Smn1A and Smn1B alleles, as well as that the amount of hybrid transcript was unchanged between the Smn1B, Smn1C and Smn1D alleles. This last observation confirms that the FL-SMN detected by our hybrid-specific RT-PCR (Fig. 2C) originates, in fact, from the human SMN2 gene.
We examined the relative abundance of FL-SMN and D7 SMN expressed from the human SMN2 genomic portions of Smn1C and Smn1D alleles by qPCR using previously described methods (17). As shown in Figure 2D, both FL-SMN and D7-SMN were proportionally increased in the Smn1D allele relative to Smn1C allele (D7 SMN brain: 3.21 ± 0.13 fold; spinal cord: 3.05 ± 0.34 fold; FL-SMN brain: 3.15 ± 0.11 fold; spinal cord 4.21 ± 0.24 fold). As expected, no human FL-SMN or D7-SMN was detected in the Smn1A and Smn1B alleles.
Together, these analyses show that Smn1B allele, which is embryonic lethal and contains only the hybrid portion of the alleles (Fig. 1), has truly a negligible impact on the phenotype, resulting from the loss-of-function of the murine Smn1 gene. Also, the mild SMA phenotype seen in Smn1C/C mutant mice is due to the preferential splicing of the D7-SMN gene product over the FL-SMN product. From a practical point of view, the D7 and FL transcripts can reliably be detected and compared in our allelic series, which makes it valuable to assess later the efficacy of therapeutic splicing modulators.
Body weight and necrosis in allelic series mutants
Cohorts of Smn1C/C mutant mice and littermate controls were generated by interbreeding mice heterozygous for the Smn1C allele. In addition, cohorts of mice representative of the Smn1D allelic series (Smn1D/A, Smn1D/B, Smn1D/C and Smn1D/D) were also generated for phenotypic analysis. Smn1C/C mutant mice along with Smn1D/A, Smn1D/B and Smn1D/C mutants presented with a progressive necrosis of peripheral body parts beginning with their tail and moving toward the hindlimbs and then to the pinnae of the ears (Supplementary Material, Fig. S1A). Interestingly, the genetic titration of SMN levels in each of these mutant animals corresponded to the onset of tail necrosis. In Smn1C/C mutants, tail swelling and shortening were evident as early as post-natal day (PND) 5 and progressed to complete loss of tail by 45 days of age. In comparison, Smn1D/A and Smn1D/B mutants exhibit necrotic tail onset at 4 weeks of age, whereas Smn1D/C mutants had a tail necrosis at 6 weeks, and no necrotic tails were observed in Smn1D/D mutants (depicted in Fig. 1B). Necrosis events in Smn1C/C mutants on a mixed genetic background (N2 on FVB) were also monitored at PsychoGenics, Inc. (PGI; Tarrytown, NY, USA). Similar to tail necrosis observations at The Jackson Laboratory (JAX; Bar Harbor, ME, USA), Smn1C/C mutants in this study first presented with tail necrosis at approximately PND12, and additionally the progression of necrosis to the toes and ears was also monitored at PGI (Supplementary Material, Fig. S1B).
To examine the lesions in the necrotic tails, Smn1C/C mutant mice and controls were sacrificed at 20, 30 and 40 days of age. Tails were cross-sectioned and stained with hematoxylin and eosin (H&E) for histological analysis. As depicted in Supplementary Material, Fig. S1C–F, tail necrosis in these animals was characterized by disorganization of muscle fiber bundles along with apparent loss of intact vasculature in the absence of an inflammatory lesion as the animals aged. Lateral vasculature seemed to be primarily affected in these mutants with a loss of integrity in the vessel wall which was most evident in the inner tunica intima of the small lateral vasculature (Supplementary Material, Fig. S1E, inset), whereas the ventral artery remained intact until the very latest stages of necrosis. To determine whether vasculature destruction precedes necrosis in mutant Smn1C/C mice, we used thermal imaging to gauge the integrity of the blood supply in the distal tail (where necrosis first appears). As shown in Supplementary Material, Fig. S1G and H, notable temperature differences were observed in pre-necrotic regions of the tail as early as 20 days of age. These data support the hypothesis that vascular destruction occurs before overt exterior necrosis.
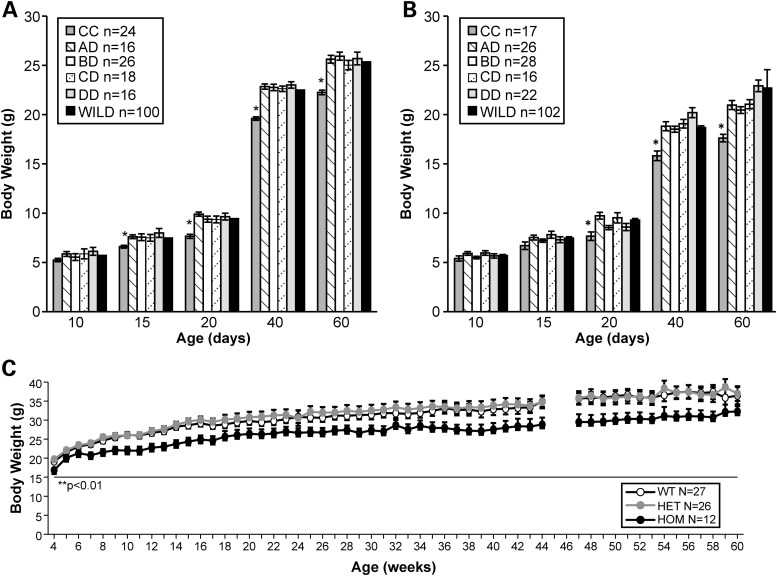
Body weight (BW) of both mutant and control littermates in all viable combinations of the allelic series was monitored through PND60. For these experiments and throughout this paper, PND0 is defined as the day the animals were born. As depicted in Figure 3A, by PND15, Smn1C/C males with an average BW of 6.59 ± 0.25 g were significantly smaller (P>0.05; t-test) than WT littermate controls (BW 7.48 ± 0.13 g), Smn1D/A (BW 7.56 ± 0.20 g), Smn1D/B (BW 7.56 ± 0.20 g), Smn1D/C (BW 7.49 ± 0.26 g) and Smn1D/D (BW 7.98 ± 0.40 g) mutants. Mutant Smn1C/C females with an average BW of 7.66 ± 0.43 g were also significantly smaller (P < 0.05; t-test) than WT littermate controls (BW 9.29 ± 0.15 g) as well as Smn1D/A (BW 9.73 ± 0.34 g), Smn1D/B (BW 8.53 ± 0.21 g), Smn1D/C (BW 9.53 ± 0.52 g) and Smn1D/D (BW 8.59 ± 0.36 g) mutants but at a slightly later time point (PND20) (Fig. 3B). BW analysis was also carried out separately at PGI on mixed genetic background Smn1C/C mutants with littermate controls. Smn1C/C animals were weighed from PND3 and every other day up to PND20. From birth until PND18, no genotype differences were noted in BW. Similar to data generated at JAX, at PND18, BW measures deviated where Smn1C/C mutant mice weighed significantly lighter than WT and heterozygote littermates (data not shown). As shown in Figure 3C, this analysis also demonstrated Smn1C/C mutants to weigh significantly less than littermate controls between 4 and 17 weeks of age (P < 0.0018) and this BW difference remains consistent through >1 year of age.
Figure 3.
Smn1C/C mutant exhibits the lowest BW in the allelic series. (A) The BW of mutants on an FVB/NJ congenic background in the allelic series along with WT littermate controls was monitored through 60 days of age at JAX. Smn1C/C mutant males weighed significantly less than WT controls, Smn1D/A, Smn1D/B, Smn1D/C and Smn1D/D mutants (P < 0.001) at each time point tested. (B) Female Smn1C/C mutants weighed significantly less than WT controls, Smn1D/A, Smn1D/B, Smn1D/C and Smn1D/D mutants by PND20 (P < 0.001). (C) Independent data collected at PGI on mixed genetic background Smn1C/C mutants also demonstrated significant BW (gender combined) differences compared with heterozygous and WT littermate controls at PND18 and PND20 (P < 0.0001). Animals were weighed at PND3 and then every other day from PND4 to PND20. (D) Animals were weighed once per week starting at week 4 through 61 weeks of age and were found to be significantly lighter than littermate controls (P < 0.0018). All animals were untreated. Data represent mean ± SEM.
Because of the degree of necrosis and BW differences observed in the Smn1C/C mutants in comparison with all other mutants in the series, we determined this to be the most severely affected viable mutant in the allelic series and thus further interrogated this mutant with respect to NMJ analysis and neuromuscular deficits. We did not observe an overall gender impact on the neuromuscular phenotype in the Smn1C/C mice. For the purposes of the following studies, males and females were grouped together.
NMJ and electrophysiological analysis in Smn1C/C mutants
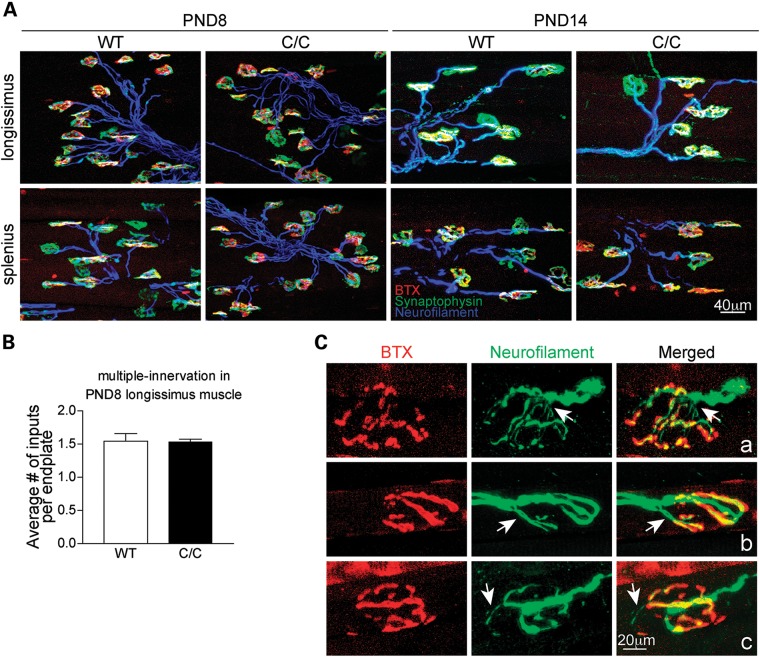
Previous work has demonstrated selective NMJ denervation in a group of clinically relevant muscles in the severe SMNΔ7 mouse model (18). To examine whether NMJ denervation occurs in milder SMA Smn1C/C mice, NMJ innervation patterns were examined with standard immunostaining in the splenius capitis and longissimus capitis muscles, the most severely affected muscles in SMNΔ7 mice. At PND8 and PND14, all NMJs were innervated and no nerve sprouting was observed (Fig. 4A). In addition, no difference in the number of nerve inputs per endplate was observed (Fig. 4B). Taken together, these findings are consistent with the observation in severe mouse models that synapse formation at the NMJ is not affected in SMA (18–20). To further investigate whether synapse maintenance is disrupted in Smn1C/C mice, NMJ morphology in the splenius, longissimus and semispinalis muscles was examined at PND90. We found that ∼10% of junctions in these muscles displayed abnormal NMJ morphology (Smn1+/+: 2.3 ± 0.3% versus Smn1C/C: 9.6 ± 0.6%, P < 0.05). As shown in Figure 4C, some junctions showed fragmented acetylcholine receptor (AChR) clusters, accompanied by abnormal thin nerve terminals as labeled with neurofilament antibodies (the arrow in Fig. 4Ca). Some junctions were innervated by multiple nerve terminals (the arrow in Fig. 4Cb). Furthermore, nerve terminal sprouting was also observed at some NMJs (the arrow in Fig. 4Cc). These results suggest that synaptic maintenance is mildly disrupted in some NMJs of the vulnerable muscles in Smn1C/C mice.
Figure 4.
NMJ formation is normal in Smn1C/C mutants but maintenance is modestly impaired in an age-related manner. (A) Confocal micrographs showing the morphology of NMJs in the longissimus capitis and splenius capitis muscles of WT and Smn1C/C mice at PND8 and PND14. Nerve terminals were labeled with anti-neurofilament (blue) and anti-synaptophysin (green) antibodies. AChRs were labeled with α-bungarotoxin BTX (red). (B) Quantification of the average number of nerve inputs per endplate in the longissimus muscle at PND8 (Smn1+/+: 1.5 ± 0.1 versus Smn1C/C: 1.5 ± 0.0). (C) Examples of confocal micrographs showing abnormal NMJ morphology of PND90 Smn1C/C mice. Error bars show SEM.
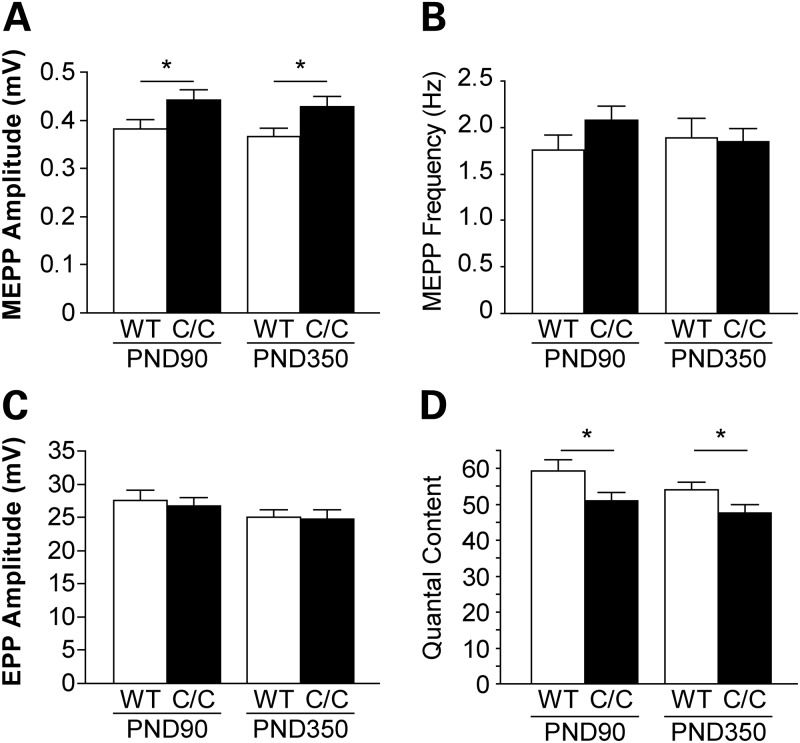
To examine neuromuscular transmission in Smn1C/C mice, we performed intracellular recording in the splenius capitis muscle of PND90 and PND350 mutant animals and WT controls. Muscle contraction was prevented by pre-incubation of µ-conotoxin, which blocks voltage-gated sodium channels in muscle (21). At PND90 and PND350, there was a slight increase in the spontaneous miniature endplate potential (MEPP) amplitude in Smn1C/C mice (Fig. 5A), which may be due to the higher input resistance of smaller muscle fibers in Smn1C/C mice (myofiber size : Smn1+/+: 1494.6 ± 18.7 µm2 versus Smn1C/C: 1176.0 ± 14.7 µm2, P < 0.05). There was no difference in the MEPP frequency (Fig. 5B) or the evoked endplate potential (EPP) amplitudes (Fig. 5C) in Smn1C/C compared with WT controls. At PND90, ∼8% (4 out of 51 junctions) of the junctions in the Smn1C/C splenius muscle were silent, where MEPPs could be recorded, but nerve stimulation did not elicit EPPs. At PND350, more silent junctions (15%, 9 out of 59 junctions) were observed in Smn1C/C splenius capitis muscles. In addition, in the remaining functional junctions that showed EPPs upon nerve stimulation, the quantal content, calculated by dividing the EPP amplitude over MEPP amplitude, was decreased by 13–14% in Smn1C/C mice at both ages (Fig. 5D). These observations suggest a reduced synaptic transmission efficacy in the vulnerable muscle of Smn1C/C mice.
Figure 5.
NMJs from Smn1C/C mutant display a reduction in synaptic efficacy. Intracellular recordings were performed in the splenius capitis muscle at PND90 and PND350. Quantification of synaptic potentials and efficacy: (A) MEPP amplitude (PND90: Smn1+/+ 0.38 ± 0.02 versus Smn1C/C 0.44 ± 0.02 mV, P < 0.05; PND350: Smn1+/+ 0.37 ± 0.02 versus Smn1C/C 0.43 ± 0.02 mV, P < 0.05). (B) MEPP frequency (PND90: Smn1+/+ 1.76 ± 0.17 versus Smn1C/C 2.08 ± 0.15 Hz, P = 0.15; PND350: Smn1+/+ 1.89 ± 0.22 versus Smn1C/C 1.85 ± 0.14 Hz, P = 0.88). (C) EPP amplitude (PND90: Smn1+/+ 27.50 ± 1.55 versus Smn1C/C 26.65 ± 1.27 mV, P = 0.67; PND350: Smn1+/+ 25.09 ± 1.05 versus Smn1C/C 24.72 ± 1.45 mV, P = 0.84). (D) Quantal content (PND90: Smn1+/+ 59.35 ± 3.06 versus Smn1C/C 50.96 ± 2.42, P < 0.05; PND350: Smn1+/+ 54.08 ± 1.98 versus Smn1C/C 47.59 ± 2.43, P < 0.05). There is an increase in the amplitude of MEPPs and a decrease in the quantal content in Smn1C/C NMJs. (n = 40–60 NMJs from four animals in each genotype). Error bars show SEM.
Motoneuron loss and central synapse loss in the spinal cord have been shown in a mouse model of severe SMA, SMNΔ7 (8,22–24). To examine whether such loss also occurs in Smn1C/C mice, we counted the number of motor axons in lumbar (L) 3 and L4 ventral roots. No significant difference was observed (Supplementary Material, Fig. S3A and B) in PND350 Smn1C/C mice comparing with WT. In addition, we performed axon counts in the motor branch of the femoral nerve of 7-month-old Smn1C/+ and Smn1C/C males. A slight decrease was observed (545 ± 2.17, 526 ± 4.76, t-test P = 0.0374) (Supplementary Material, Fig. S3D and E). Together with the absence of axon loss noted in the ventral roots, this is consistent with the dying back axonopathy of the motoneurons characteristic of SMA.
Lastly, we assessed whether synaptic input onto the motoneuron cell bodies in the spinal cord was affected, as reported in other severe SMA models. We found no significant loss in VGLUT1-positive synapses in L1–2 motoneurons in PND350 Smn1C/C mice when compared with control (Supplementary Material, Fig. S4).
Neuromuscular assessment in C/C mutant mice
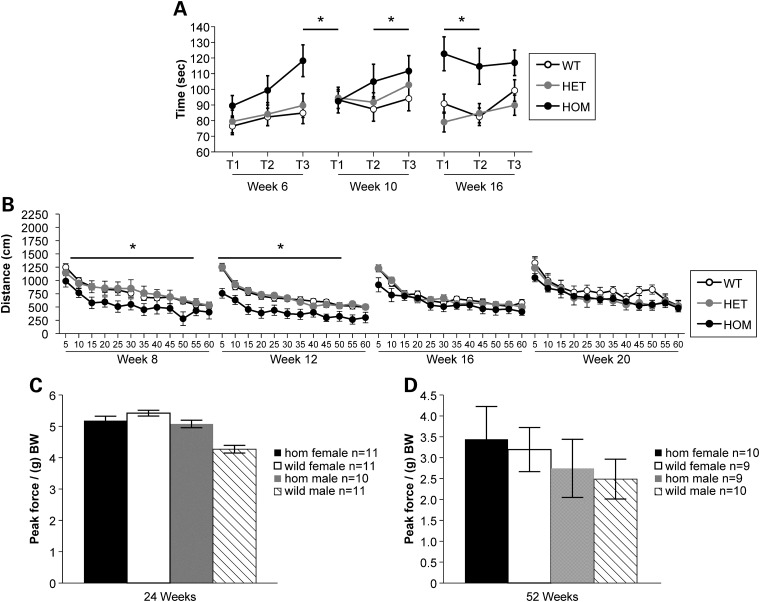
In order to assess any neuromuscular deficits present in the Smn1C/C mutant mice, cohorts of mutants and controls were generated and subjected to grip strength and rotarod analysis at two separate institutions: JAX and PGI. The assessments conducted were rotarod, open field (OF), von Frey and grip strength. Accelerated rotarod testing was performed at PGI, using mixed genetic background Smn1C/C mutants and littermate controls. The rotarod analysis was carried out at 6, 10 and 16 weeks of age. At each time point, n > 10 animals of mixed genders were tested and the data represented are of combined genders. As shown in Figure 6A, this repeated testing demonstrated that the homozygous Smn1C/C mutants consistently performed better (longer latency to fall) than WT (P < 0.0064) and heterozygote (P < 0.0074) littermates (trials combined) across all time points tested. At JAX, rotarod analysis was carried out at 1, 2, 3 and 4 months of age on cohorts of 10 female and 10 male Smn1C/C allele mixed genetic background mutants along with littermate controls. This analysis also revealed that Smn1C/C mutant animals performed significantly better at 1 month of age, indicated by a longer latency to fall (P < 0.005, Student's t-test) on the accelerating rotarod (average latency 55.93 ± 1.06 s) than WT (average latency 49.07 ± 1.78 s) (data not shown). In addition, homozygous mutants performed marginally better than WT littermate controls on the constant speed rotarod test at 1 month of age, with an average latency to fall of 53.96 ± 1.64 s compared with 48.26 ± 2.36 s for controls (P < 0.05) (data not shown). We continued testing in a longitudinal fashion on the same cohort of animals; no other significant differences were found.
Figure 6.
Behavioral testing demonstrates slight impairment of Smn1C/C mutant mice in OF behavior. (A) Rotarod testing at PGI demonstrated similar findings with mutants having a significantly longer latency to fall than both heterozygous and WT controls (P = 0.0074; P = 0.0064, respectively) on an accelerating rotarod. (B) Homozygous Smn1C/C mutants exhibit reduced total distance traveled at 8 and 12 weeks of age compared with littermate controls and improve to equal performance by 16 weeks of age on the OF test at PGI. (C and D) Neither homozygous Smn1C/C mutant males nor females exhibit diminished grip strength (as normalized to gram of BW) at either 24 weeks (C) or 12 months of age (D) at JAX. Data are represented as mean ± SD.
OF activity was also assessed in mixed genetic background Smn1C/C mutant mice and littermate controls at PGI. Animals were tested for 1h and data were recorded in 5 min time bins. At each time point, n > 10 animals of mixed genders were tested and the data represented are of combined genders. As shown in Figure 6B, at week 8 and week 12 the total distance traveled by homozygous mutants was significantly shorter than heterozygous (P = 0.0232 at week 8; P = 0.0093 at week 12) or WT (P = 0.0181 at week 8; P = 0.0093 at week 12) controls. By 16 weeks of age, homozygous mutants displayed no differences when compared with either heterozygous or WT controls, which could be linked to the completion of the major stage of paw necrosis which is observed in the Smn1C/C mutant mice.
Smn1C/C mutant mice were also assessed for forelimb grip strength, using the Digital Grip Strength Meter (Columbus Instruments, Columbus, OH, USA) at 24 and 52 weeks of age (Fig. 6C and D, respectively). No significant differences were observed between Smn1C/C mutants and WT controls at either 24 weeks of age or 52 weeks of age. Because BW can positively influence grip strength (25), peak force was normalized per gram of BW.
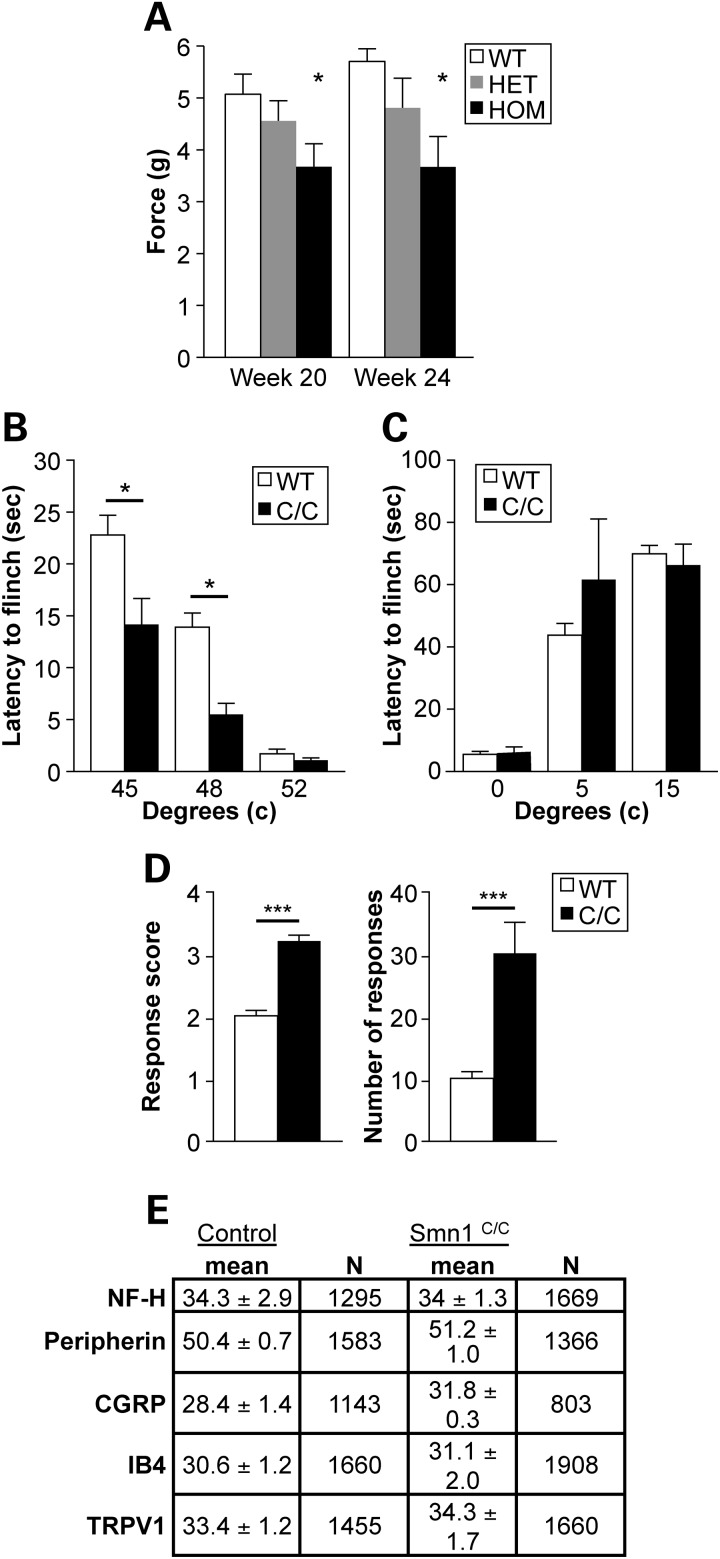
The von Frey filaments can be used to determine tactile sensitivity thresholds. Each nylon filament was designed to buckle to a U-shape at a specific force (g) when applied perpendicularly to a surface. Smn1C/C mutant animals and controls were subjected to plantar von Frey testing at PGI at 20 and 24 weeks of age. In each of these trials, paw withdrawal force was significantly lower in Smn1C/C mutant animals than in both heterozygous and WT controls (P = 0.0155), suggesting hyperalgesia in the Smn1C/C mutant line at this testing age (Fig. 7A). Von Frey testing was also carried out at the University of Southern California (USC) on 6–8-week-old Smn1C/C mutant mice and WT controls. Similar to the findings obtained by PGI, a statistically significant difference (P < 0.001) was observed in withdrawal thresholds (5.5 ± 0.1 and 6.3 ± 0.1 g) for Smn1C/C and WT mice, respectively (data not shown).
Figure 7.
Examination of heightened nociceptive responses in Smn1C/C mutant mice. (A) Homozygous Smn1C/C mutants exhibit increased nocioception at 20 and 24 weeks as assessed by von Frey at PGI. In each of these trials, paw withdrawal force was significantly lower in Smn1C/C mutant animals than both heterozygous and WT controls (P < 0.0155). (B) Smn1C/C mice exhibited an increase in sensitivity to moderately noxious heat with a significant decrease in hindpaw withdrawal latencies when the animals were placed on a metal plate heated to 45 (P < 0.01) and 48°C (P < 0.001). No differences were observed between Smn1C/C mutant animals and WT littermate controls in response to a high noxious stimulus of 52°C. (C) No differences were observed in behaviors of either genotype when exposed to noxious cold (5–15°C). (D) Smn1C/C mice exhibited robust sensitization in the evaporative cooling assay responding more vigorously with a score of 3.2 ± 0.1 compared with a score 2.0 ± 0.1 observed in WT mice, as well as in the number of responses in Smn1C/C and WT animals at 30.0 ± 5.0 and 10.0 ± 1.0 (P < 0.001), respectively. (E) Quantitation of specific markers in the somas of cutaneous nerve fibers in Smn1C/C and WT mice (n = 3/group) demonstrates no difference in the prevalence of any one fiber type. Data are the percentage of the PGP9.5 (pan neuronal marker) population of lumber dorsal root ganglion (DRG) neurons that were also immuno-reactive for a marker (NF-H, neurofilament marker of myelinated A-delta fibers; CGRP, calcitonin gene-related peptide-marker of peptidergic neurons; IB4, isolectin B4-marker of non-peptidergic neurons; TRPV1, transient receptor potential vanilloid 1 ion channel-expressed in nociceptors).
To further interrogate the nocioceptive phenotype in Smn1C/C mutant mice, a series of mechanical and thermal behavioral assays were carried out at USC using 6–8-week-old Smn1C/C mutants (with no evidence of paw necrosis) and age-matched littermates. We found that Smn1C/C mice exhibited an increase in sensitivity to moderately noxious heat with a significant decrease in hindpaw withdrawal latencies when the animals were placed on a metal plate heated to 45°C (P < 0.01) and 48°C (P < 0.001) (Fig. 7B). However, there were no differences in these animals' response to a high noxious stimulus of 52°C. Similarly, we found no differences in behaviors of either genotype when exposed to noxious cold (0–15°C; Fig. 7C). However, Smn1C/C mice exhibited robust sensitization in the evaporative cooling assay, a more moderate cold stimulus (26,27). Smn1C/C mice responded more vigorously in this assay with a response score of 3.2 ± 0.1 compared with a score of 2.0 ± 0.1 observed in WT mice, as well as in the number of responses in Smn1C/C and WT animals at 30.0 ± 5.0 and 10.0 ± 1.0 (P < 0.001), respectively (Fig. 7D). The increased sensitivity of Smn1C/C mutants is unlikely due to a change at the afferent level of the sensory nervous system itself. First, we did not notice any neuropathy in peripheral sensory nerves such as the femoral nerve (Supplementary Material, Fig. S3D and E). Second, we interrogated Smn1C/C mice with markers for specific nociceptor subtypes including: neurofilament marker of myelinated A-delta fibers (NF-H), peripherin, calcitonin gene-regulated peptide (CGRP), isolectin B4 (IB4) and transient receptor potential vanilloid 1 (TRPV1) ion channel, and found no differences between Smn1C/C mutants and controls (Fig. 7E).
Cardiac function in Smn1C/C mutants
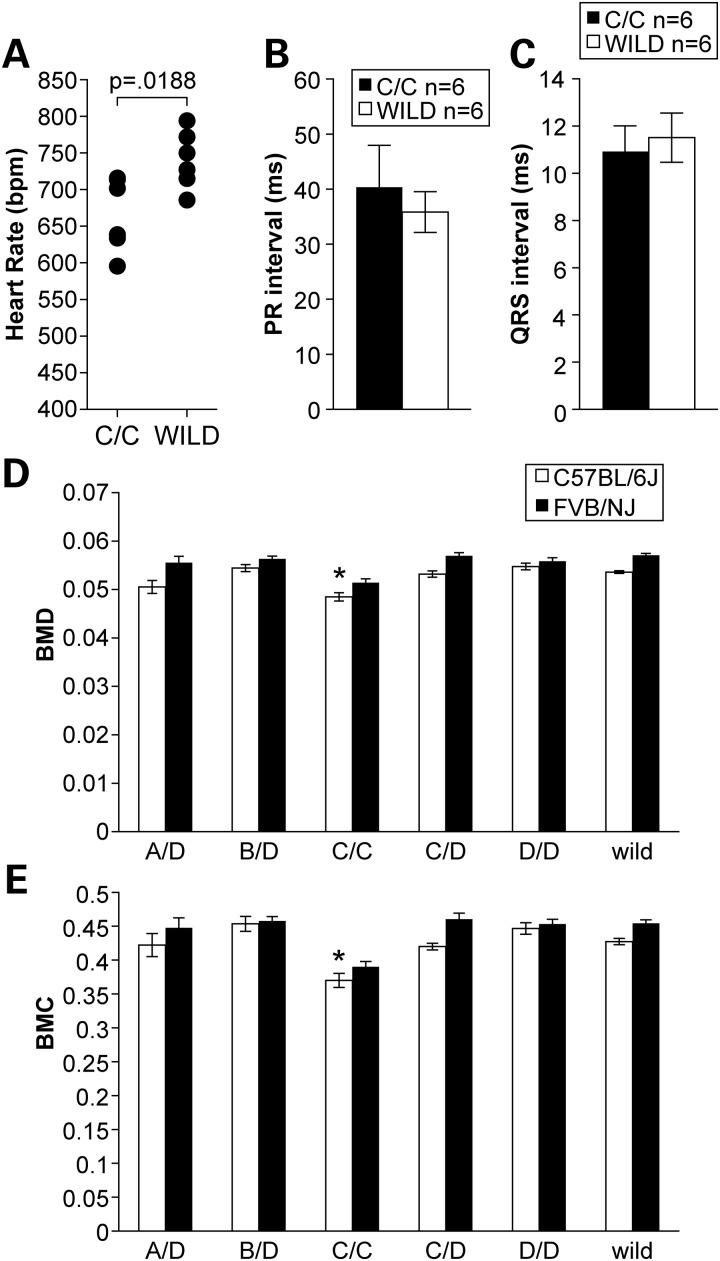
Previous work has demonstrated that the severe SMNΔ7 mouse model of SMA displays cardiac arrhythmia and functional deficits (28). Such deficits are hypothesized to be caused by decreased sympathetic innervation leading to an autonomic imbalance. To determine whether Smn1C/C mutants displayed similar cardiac defects found in the SMNΔ7 model, we performed conscious electrocardiography (ECG) and echocardiography on 9–month-old Smn1C/C mutants and littermate controls. Similar to previous findings in the severe SMNΔ7 mouse model, Smn1C/C mutants exhibited a significantly lower (P < 0.05) average heart rate (666 ± 50 b.p.m.) than littermate controls (740 ± 39 b.p.m.) (Fig 8A). Heart block is considered a common cause of bradyarrhythmia and can be detected by elongation of the PR interval. However, in contrast to the bradyarrhythmic phenotype displayed by the SMNΔ7 model, homozygous Smn1C/C mutants did not have a significantly longer PR interval, though the PR intervals in Smn1C/C mutants did have a tendency to be longer than WT controls (Fig. 8B). In addition, homozygous Smn1C/C mutants also did not exhibit a longer QRS interval, another characteristic that is known to be associated with bradyarrhythmia (Fig. 8C). Although the Smn1C/C mutants do appear to have a significantly lower heart rate compared with controls, this phenotype does not seem to manifest in overt bradyarrythmia. Further echocardiography tests were performed on Smn1C/C mutant mice and controls to measure overall heart function. Smn1C/C mutant mice were not statistically different from WT control littermates with respect to overall blood flow velocities and chamber sizes (data not shown).
Figure 8.
Cardiovascular assessment and body composition analysis of Smn1C/C mutants. (A) Conscious ECG recordings of 9-month-old Smn1C/C mutants (n = 6) and WT littermate controls (n = 6) of mixed genders revealed a significant decrease in heart rate (P < 0.05). (B–C). No increase in PR interval duration or subsequent QRS interval elongation was detected. (D) Total body DEXA analysis revealed that Smn1C/C mutants in the allelic series demonstrate significantly diminished bone mineral density. (E) Bone mineral content (*P < 0.05, t-test), on both the FVB/NJ and C57BL/6J genetic backgrounds when compared with WT controls as well as other mutants in the allelic series.
Body composition of mutants in the allelic series
Owing to their diminished BW, we sought out to determine whether or not mutants in the allelic series had a change in their overall body composition including fat mass, lean mass and bone mineral density and bone mineral content by dual-energy X-ray absorbance (DEXA) analysis. At 6 months of age, cohorts of 10 mutants from each line in the viable series (Smn1C/C, Smn1D/A, Smn1D/B, Smn1D/C and Smn1D/D) on both congenic FVB/NJ and C57BL/6J congenic backgrounds along with littermate controls were subjected to full-body DEXA analysis. As shown in Figure 8D and E, Smn1C/C mutants on both the congenic C57BL/6J genetic background and the FVB/NJ genetic background exhibit significantly (P < 0.05) lower bone mineral density and bone mineral content than both other mutants in the allelic series as well as WT controls. Data are represented as mean ± SEM of pooled genders.
DISCUSSION
In this study, we presented extensive characterization of a novel series of mouse models created by genetically engineering varying copies of human SMN2 into the murine Smn1 locus. We performed crosses within the allelic series that would generate mutants with no, one, two, three, four, five, six or eight relative copies of SMN through a combination of a hybrid (genomic mouse exons 1–6; genomic human exon 7–8) and FL human SMN2 genomic DNA segment. Surprisingly, mutants generated in this series were either embryonic lethal (three or less relative SMN copies), or completely viable (four or more relative SMN copies) with a mild neuromuscular phenotype. Of the viable mutants in the series, the Smn1C/C mouse with two copies of the hybrid allele and two copies of the FL SMN2 allele demonstrated the most severe phenotype characterized by reduced BW, peripheral necrosis, mild progressive NMJ abnormalities and electrophysiological defects, diminished OF activity, decreased bone mineral density and evidence of cardiac abnormalities. Interestingly, the Smn1D/A mutant in the allelic series with an equivalent relative copy number of SMN reflected in a different portion of hybrid and human SMN2 (1:3, respectively) copy numbers displayed a much milder phenotype. We reason that this is due to the reduced amount of FL-SMN produced by the hybrid portion of this construct.
Tail and ear necrosis is a predominant phenotype observed in the allelic series of SMA mouse models. Necrosis is a perplexing feature that has been observed in multiple mild mouse models of SMA (13,29,30). Our observations now make clear that there is an inverse correlation between necrosis severity and SMN protein levels as demonstrated by the segmented delay in the onset of tail necrosis with proportional increases in SMN levels. These observations conform well to the established theory that increases in SMA disease severity are associated with decreased SMN levels (3,31). Histologically, the deterioration of the tail vasculature walls is evident, and we believe that this destruction precedes necrosis as thermal imaging of distal tail of Smn1C/C mutants demonstrates diminished blood flow prior to the exterior necrosis. Although necrosis is not a predominant clinical finding in patients with SMA, there have been rare documented cases of patients exhibiting peripheral necrosis (32). Moreover, it has also been demonstrated that treating severe SMA model mice with compounds that upregulate SMN expression allows the animals to live long enough to develop tail and ear necrosis (33–36). Taken together, these data suggest that although necrosis is not a common symptom seen in the human SMA patient population, its severity in the mouse model inversely correlates with SMN copy number, and that severity can be modulated with compounds that upregulate SMN expression and thus this phenotype is a true barometer in the animal model reflective of overall SMN expression. Because the allelic series contains a titrated gene dose of SMN, these animal models can be useful to ascertain the effectiveness of therapeutics with respect to the number of gene targets available.
Although the NMJ innervation patterns in vulnerable muscles of the Smn1C/C mutant mice are normal during early development in comparison with the severe SMNΔ7 model, some abnormalities could be detected in ∼10% of the junctions examined characterized by fragmented AChR clusters and abnormally thin nerve terminals in older mutants. Electrophysiological analysis of vulnerable muscles revealed an age-related increase in non-functional junctions and a decrease in quantal content. In addition, we found that axon number was slightly but significantly decreased in the motor branch of the femoral nerve, consistent with the ‘dying back’ neuropathy associated with mild SMA (37). Despite these mild NMJ perturbances, Smn1C/C mice functioned well on multiple tests of neuromuscular deficit including rotarod, grip strength and OF, demonstrating only significant deficits prior to 16 weeks of age in the OF. We hypothesized that this obvious improvement in OF activity could be due to the mutant animals achieving a steady-state level of necrosis in their paws and subsequent reduction of pain levels promoting an increase in movement. Smn1C/C mutant mice did appear to have a heightened nocioceptive response as assessed by the von Frey test. Smn1C/C mice exhibited increased sensitivity to both thermal and mechanical stimuli. Smn1C/C mice were robustly more sensitive to both heat and cold in what are considered innocuous to moderately noxious stimuli, but do not show these heightened sensations at highly noxious temperatures. Additionally, we found no difference in the prevalence of the expression of specific markers in the somas of cutaneous nerve fibers, and we demonstrated that there was no axon loss in a peripheral sensory nerve (sensory branch of the femoral nerve) of the Smn1C/C mutants. Taken together, these data do not suggest that Smn1C/C mice are purely hyperalgesic (increased sensitivity to noxious stimuli), but are allodynic (increased sensitivity to normally non-painful stimuli), similar to several clinically relevant chronic pain models (38), and this allodynia phenotype likely originates in the central nervous system rather than in the periphery. However, it remains to be determined whether this phenotype is due to changes at the level of the afferent neuron itself, or as a consequence of the necrosis associated with the SMN disease. This observation is important when using behavioral testing as an outcome measure in mild SMA models exhibiting necrosis, as pain associated with necrosis may confound interpretation of results. Regardless, these results suggested that in addition to motor deficits, SMN deficiencies in the Smn1C/C mice may also lend this strain as a potential model of chronic pain.
With respect to the improved performance of Smn1C/C mutant animals on the rotarod apparatus, studies have demonstrated that rotarod performance is sensitive to BW, with mice of smaller strains having longer latencies to fall than mice of larger strains and females having a longer latency than males (39). We have determined that Smn1C/C mutant mice are significantly smaller than littermate controls by PND15 (Fig. 3A–C), and in addition Smn1C/C mutant mice exhibited very mild (if any) NMJ pathology, and modest reductions in muscle quantal content, which would likely not contribute to a coordination deficit.
Interestingly, Smn1C/C mutant mice presented with significantly lower heart rates compared with WT controls. Although this phenotype did not seem to present as overt bradyarrythmia as has been described for the severe SMNΔ7 (28), it is a clinically relevant phenotype as recent studies have brought to light the presence symptomatic bradycardia in longer lived patients with severe SMA (40). This clinical presentation is certainly one that will need to be addressed as new therapeutics further extends patient survival. Taken together, the presence of vascular necrosis and cardiac anomalies in the Smn1C/C mutant mice pointed toward autonomic nervous system (ANS) involvement in SMA pathology. Whether or not primary ANS involvement is an integral part of the phenotype of SMA remains unclear. Smn1C/C mutant mice may provide a useful tool to investigate this question.
Smn1C/C mutant mice also presented with differences in both bone mineral density and bone mineral content. This finding is significant in that it has been previously reported that pathological fractures were common in children with neuromuscular disorders (41). Furthermore, analysis of patients with varying neuromuscular disorders revealed that those with SMA exhibited the lowest bone mineral densities as a result of osteopenia (41). For these reasons, the Smn1C/C mutant animal may be useful in the study of lower bone mineral density associated with SMA pathology.
The hope for the allelic series generated by targeted disruption of the murine Smn1 locus with varying amounts of SMN was to provide a series of research models that could recapitulate the entire spectrum of disease severity seen in SMA. Unfortunately, only the two extremes of this spectrum were generated with embryonic lethality and complete viability being represented. Despite this, we now have a collection of alleles to add to our ‘toolbox’ with a clear definable amount of SMN that exhibit splicing defects, leading to the reduction in FL-SMN that is directly responsible for a distinct phenotype associated with each level of expression. These alleles afford the opportunity to test therapeutic intervention when multiple drug targets are available, or just a single target is available. Also the presence of multiple phenotypes with relevance to SMA like SMN depletion, DEXA deficits and mild neuromuscular abnormalities allows for biomarker and endpoint exploration in this model. The clear phenotypes present in the viable mutants of the allelic series provide a barometer for SMN expression and can certainly lend additive insights into the knowledge gained from current mild models of SMA. Lastly, because these alleles were engineered at a single locus, genetic manipulation using the allelic series becomes simple and straight forward, allowing for the easy incorporation of these alleles into other SMA mouse models of interest.
MATERIALS AND METHODS
Gene targeting
Four gene targeting events at the murine Smn1 locus were engineered by Regeneron Pharmaceuticals, using the VelociGene technology on 129S6/SvEvTac- and C57BL/6Tac-derived F1H4 ES cells as described (16). These targeting events included: (i) a complete disruption of the Smn1 locus by replacement of exons 1 through 8, with lacZ and an FRT site remaining from the deletion of a selection marker. This allele is referred to as Smn1tm2Mrph (VelociGene Allele Identification Number VG5064; MGI ID 3794065) and is commonly known, and referred to in this paper, as the ‘Smn1A’ allele. (ii) A 2.2 kb segment of mouse genome containing exons 7 and 8 of the mouse Smn1 (survival motor neuron 1) gene was replaced with a 1.3 kb fragment of human genomic DNA containing exons 7 and 8 of the human SMN2 (survival of motor neuron 2, centromeric) gene. A selection cassette located downstream from the human SMN2 polyadenylation signal was removed by FLPe expression in ES cells leaving an FRT site at the downstream junction between human and mouse DNA. This allele is referred to as Smn1tm4(SMN2)Mrph (VG5113; MGI ID 3794200) and is commonly known, and referred to in this paper, as the ‘Smn1B’ allele. (iii) Disruption of the murine Smn1 locus containing two tandem Smn1/SMN2 genes. The first is a hybrid gene in which a 2.2 kb segment of mouse genome containing exons 7 and 8 of the mouse Smn1 gene was replaced with a 1.3 kb fragment of human genomic DNA containing exons 7 and 8 of the human SMN2 gene. The second is a full 42 kb copy of the human SMN2 gene. A selection cassette located downstream from the human SMN2 polyadenylation signal was removed by FLPe expression in ES cells leaving an FRT site at the downstream junction between human and mouse DNA. This allele is referred to as Smn1tm5(Smn1/SMN2)Mrph (VG5114; MGI ID 3794202) and is commonly known, and referred to in this paper, as the ‘Smn1C’ allele. (iv) Disruption of the murine Smn1 locus containing four tandem SMN genes. The first is a hybrid gene in which a 2.2 kb segment of mouse genome containing exons 7 and 8 of the mouse Smn1 gene was replaced with a 1.3 kb fragment of human genomic DNA containing exons 7 and 8 of the human SMN2 gene. The next three are identical FL, 42 kb copies of the human SMN2 gene. A selection cassette located downstream from the final human SMN2 polyadenylation signal was removed by FLPe expression in ES cells leaving an FRT site at the downstream junction between human and mouse DNA. This allele is referred to as Smn1tm6(SMN2)Mrph (VG5115; MGI ID 3846546) and is commonly known, and referred to in this paper, as the ‘Smn1D’ allele. Each of these alleles is depicted schematically in Figure 1. In addition, these alleles are available as the following stock numbers from JAX: Smn1A allele (007955 FVB.Cg-Smn1tm2Mrph/J, 007963 B6.Cg-Smn1tm2Mrph/J); Smn1B allele (008713 FVB.129(B6)-Smn1tm4(SMN2)Mrph/J, 008453 B6.129-Smn1tm4(SMN2)Mrph/J); Smn1C allele (008714 B6.129-Smn1tm5(Smn1/SMN2)Mrph/J, 008604 FVB.129(B6)-Smn1tm5(Smn1/SMN2)Mrph/J); Smn1D allele (009378 B6.129-Smn1tm6(SMN2)Mrph/J, 009381 FVB.Cg-Smn1tm6(SMN2)Mrph/J).
Genotyping and animal husbandry: JAX
For each construct, appropriately targeted ES cells were injected into eight-cell embryos to generate F0 mice, using the VelocImouse technology at Regeneron Pharmaceuticals as described (42). The resulting B6;129 mixed genetic background founder lines were sent to JAX and screened for the presence of the respective mutations using the following primer sets: (i) A allele: mSmn1A F 5′-GAACTAGAAGACAGGTGGAG-3′ and mSmn1A R 5′-GTCTGTCCTAGCTTCCTCACTG-3′; multiplexed with mSmn1 F 5′-TGGGAGTCCATCCATCCTAAGTC and mSmn1R 5′-GCTAAGAAAATGACAATTGCACATTTG-3′; (ii) B allele: mSmn1B F 5′-GGATAACCTAGGCATACTGCACTGT-3′ and mSmn1B R 5′-TCTGTGGACACCAGTTAAACTTGACT-3′ multiplexed with mSmn1(9111) F 5′-GTCCCTGGTCGACAAGAACAG-3′ and mSmn1(9112) R 5′-ACGCTCTGCTGCTGACTTAGG-3′; (iii) C allele: mSmn1C F 5′-TACCCAGATGCAGTGCTCTTGTAG-3′ and mSmn1C R 5′-CCTTATGGCATAGACACCAACTTCT-3′ multiplexed with previously listed mSmn1(9111) and mSmn1(9112); (iv) D allele: mSmn1D F 5′-GATGGCTTTCTGATGCTACAACAC-3′ and mSmn1D R 5′-CCAATATTCACAGAGGCCAACTG-3′ multiplexed with previously listed mSmn1(9111) and mSmn1(9112). At JAX, all animals were housed under specific pathogen-free conditions in rooms with a 12 h light/12 h dark cycle at temperature of 18–23°C and 40–60% humidity. Food and water were provided ad libitum for the duration of the study. All experiments were conducted in accordance with the protocols described by the National Institutes of Health's Guide for the Care and Use of Animals and were approved by JAX's, PGI's and USC's respective institutional animal care and use committees. The scope of the work at JAX included breeding and maintenance and general phenotypic assessment (BW/survival) of the independent allelic lines, gene expression, protein expression assessed by ELISA, intercrossing between allelic lines; behavioral and neuromuscular testing including grip strength and rotarod; and other testing including DEXA and conscious ECG. NMJ analysis and muscle electrophysiology was carried out at USC along with mechano- and thermo-sensation testing. Gene expression, protein expression and neuromuscular assessment (including rotarod, OF and von Frey testing) were performed at PGI.
Animal husbandry: PGI
Smn1C/C mice (JAX stock number 8604 strain name FVB.129(B6)-Smn1tm5(Smn1/SMN2)Mrph/J-N2 generation) were obtained from JAX and were bred and maintained at PGI. All mice were housed on OptiMICE® racks in an enriched environment. A 12–12 light/dark cycle and a room temperature of 20–23°C were maintained, with relative humidity maintained ∼50%. Food and water were provided ad libitum for the duration of the study. Mice were tail-snipped and toe-tattooed at PND3, where PND0 refers to the date of birth. Genotyping was performed by JAX or by a third party. At weaning age, animals were subcutaneously injected with microchips (Datamars, TIP8010) for identification purposes and weaned with 4–5 mice per cage with mixed genotypes.
Animal husbandry: USC
All experimental procedures were carried out in compliance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. The protocols were approved by the Institutional Animal Care and Use Committee of the University of Southern California (Protocol 11136).
SMN gene and protein expression: PGI
The spinal cord and the liver from 13-week-old Smn1C/C mice were dissected and flash-frozen with isopentane on dry ice. Total RNA extraction was performed using RNeasy 96 Universal Tissue Kit® (Qiagen, Valencia, CA, USA), following manufacturer's protocol. After elution, RNA was quantified using Quan-iT RiboGreen RNA Kit® (Life Technologies, Carlsbad, CA, USA). One microgram of total RNA was reverse-transcribed into cDNA. The reactions were allowed to proceed at room temperature for 10 min, 55°C for 30 min, then inactivated at 85°C for 5 min in GeneAmp PCR Systems 9700 Thermal Cycler (Applied Biosystems, Foster City, CA, USA). cDNA samples were diluted 10-fold with RNase-Free water for qPCR assays.
Five microliters of the diluted cDNA were amplified using 2× FastStart Universal Probe Master Rox, Universal Probe Library Probe (Roche Applied Science, Indianapolis, IN, USA), and 200 mm gene specific primers—HPLC-purified (Sigma-Aldrich, St Louis, MO, USA) in 25 µl reaction volume. The reactions were run on the ABI 7900HT Sequence Detection System (Applied Biosystems). qCPR conditions were 95°C for 10 min for the activation of FastStart Taq DNA Polymerase followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The Universal ProbeLibrary Assay Design Centre (Roche Diagnostics) was used to design PCR primers directed against the human SMN2 gene and a housekeeping gene, ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide (mAtp5b) for normalizing SMN2 expression level. The assays comprised sequence-specific primers (Sigma-Aldrich) which were used in conjunction with a 6-FAM fluorescent-labeled LNA probe (Universal ProbeLibrary, Roche Diagnostics). Primers were as follows: hSMN2 5′-GGGTTTGCTATGGCGATG and 3′-TCATCCCAAATGTCAGAATCAT (Universal ProbeLibrary # 15) and mAtp5b 5′-GGCACAATGCAGGAAAGG and 3′-TCAGCAGGCACATAGATAGCC (Universal ProbeLibrary # 77).
Total RNA from the whole brain of C57BL6 mice was reverse-transcribed (as described above). cDNAs from multiple reverse transcription reactions were pooled together and used to create qPCR standard curves for the SMN2 and Atp5b genes and were also served as calibrator to normalized plate-to-plate variations (same dilution factors are use to dilute calibrator and cDNA samples). PCR efficiencies were calculated using the equation of PCR efficiency = 10(1/−slope). Each sample cDNA (diluted 1:10) was assayed in triplicates and the Ct values were averaged. Values greater than 0.5 times the standard deviations were discarded. Relative quantity of the PCR product (relative to the calibrator) was calculated as follows: relative quantity of hSMN2 = PCR efficiencyCt calibrator − Ct sample; relative quantity of mAtp5b = PCR efficiencyCt calibrator − Ct sample. Relative expression of hSMN2 was normalized against the mAtp5b transcript level. Finally, hSMN2 mRNA expression was then normalized against the hSMN2 transcript level in the spinal cord of heterozygous mice.
The western tissues were homogenized in an ice-cold modified RIPA buffer. After centrifugation, the protein lysates were isolated. The protein content was determined using Bio-Rad's DC Assay Kit® (Bio-Rad, Hercules, CA, USA). Ten micrograms of protein lysates were denatured for 5 min at 95°C in the presence of Laemmli buffer/2-mercaptoethanol followed by separation using 4–20% SDS–PAGE Criterion Gels (Bio-Rad). After electrophoresis, proteins were transferred from gel to Hybond-LFP PVDF membranes (GE Healthcare Bioscience, Piscataway, NJ, USA) by electroblotting. The protein-PVDF membranes were stained with Ponceau S solution to confirm protein presence on the PVDF membrane; protein-PVDF membranes were then briefly rinsed with water to removed stain. Non-specific binding of primary and secondary antibodies were blocked by incubating the membrane with 5% w/v dried milk in 1× TBST for 1h at room temperature. After a brief rinse in 1× TBST, the blots were probed with primary antibody for SMN (Cat. No. 6100647; clone 8/SMN) (BD Transduction Labs, Sparks, MD, USA) or mATP5B (1:10 000) (Abcam, Cambridge, MA, USA), prepared in 1% w/v dried milk in 1× TBST for 1h at room temperature. Blot membranes were washed 3 times for 15 min with 1× TBST and probed with secondary antibodies, anti-rabbit IgG conjugated to HRP (1:1000) for SMN or anti-mouse IgG conjugated to HRP (1:1000) for mAtp5b, prepared in 1% w/v dried milk in 1× TBST for 1h at room temperature. Blot membranes were then washed three times for 5 min with 1× TBST. Bound antibodies were detected by using ECL Plus Western Detection Kit (GE Healthcare Bioscience) and by scanning blots in Typhoon 9410 scanner (GE Healthcare Bioscience).
The scanned images from the Typhoon were analyzed with the ImageQuantTL software version 7.0 (GE Healthcare Bioscience) using the rolling method. Normalized quantity of SMN protein was calculated as follows: corrected quantity of SMN protein/corrected quantity of mAtp5b. Normalized level of target gene was then normalized to that of smn_WT. Data were expressed as mean ± SEM and were analyzed by analysis of variance (ANOVA) followed by post hoc comparisons when appropriate. An effect was considered significant if P < 0.05. Values which fell above or below 2 standard deviations from the mean were considered statistical outliers and removed from the analysis.
Assessment of delta 7 SMN2 (D7-SMN) and FL SMN2 (FL-SMN) transcripts: JAX
Total RNA was extracted from the brain and the spinal cord of heterozygous Smn1A, Smn1B, Smn1C and Smn1D mice using Trizol (Invitrogen, Grand Island, NY, USA) according to the manufacturer's directions. Two and a half micrograms of RNA was reverse-transcribed using SuperScript (II) First Strand RT-PCR Kit (Invitrogen). To detect both the FL-SMN and the D7-SMN produced from the hybrid portion of the allelic series constructs a SYBR green (Invitrogen) qPCR assay using a forward primer in mouse exon 6: SmnEx6F 5′-GTCTGGATGACACTGATGCCC-3′ and a reverse primer in human exon 8: SMN2Ex8R 5′-CAATGAACAGCCATGTCCAC-3′ was utilized. The human SMN2 genomic portion of the allelic series was detected using the same reverse primer in exon 8 paired with a human-specific forward primer in exon 6 SMN2ex6F 5′-GGCTATCATACTGGCTATTATATGG-3′. All transcripts were normalized to Gapdh, using the primer pairs: Gapdh F: 5′-ACCCAGAAGACTGTGGATGG-3′ and Gapdh R: 5′-GGATGCAGGGATGATGTTCT-3′. We used previously described primers (17) in a Sybr green qPCR assay to distinguish the FL-SMN and the D7-SMN produced from the human genomic SMN2 portions of the allelic series. All reactions (hybrid-specific versus human-specific and FL-SMN2 versus D7-SMN2) were run on an ABI 7500 (Applied Biosystems). For in-gel quantification, PCR using the aforementioned primer sets were run for 25 cycles and reaction products were visualized on a standard 4% agarose gel and relative amounts of each product were quantified with ImageJ.
ELISA determination of SMN protein expression
Additionally, brain tissues were extracted from 6–8-week-old heterozygous mutant animals of the indicated genotypes and analyzed for total SMN by enzyme-linked immunosorbent assay (ELISA) (SMN ELISA Kit, Enzo Life Sciences; Farmingdale, NY, USA) according to the manufacturer's protocol. Values were normalized per milligram of total protein.
Histology
Tail biopsies
Mice at the indicated time points were euthanized and entire tail samples were excised. Tails were fixed in Bouin's, sectioned and stained with H&E. Representative sections from each mouse at each time point were examined by a board-certified veterinary pathologist.
Cutaneous nerve fiber staining
Three Smn1C/C mice and three WT littermates, all aged 6–8 weeks old, were anesthetized and L3-5 DRGs and hindpaw glabrous skin were carefully dissected and post-fixed for 1 h at 4°C in the same fixative. DRG sections were double-stained with 1:500 rabbit anti-human protein gene product (PGP) 9.5 (A01398; Genscript, Piscataway, NJ, USA), or 1:500 guinea pig anti-human PGP 9.5 (AB5898; Millipore, Temecula, CA, USA), and one of the following primary antibodies: 1:500 guinea pig anti calcitonin gene-related peptide (CGRP; T-5027; Peninsula, San Carlos, CA, USA), 1:300 rabbit anti-NF-H (sc-22909; Santa Cruz Biotechnology, Santa Cruz, CA, USA), 1:500 rabbit anti-peripherin (AB1530; Millipore), 1:500 rabbit anti-TRPV1 (RA14113; Neuromics, Edina, MN, USA). To detect IB4 binding, we included 1:500 Griffonia simplicifolia isolectin GS-IB4-Alexa 568 (I-21412; Invitrogen) during secondary antibody incubations. Digital images were acquired on a Zeiss (Jena, Germany) Axo Imager M2 with the Apotome attachment. Numbers of cells with positive staining were counted using the ImageJ software and percentages of overlap between different makers were calculated. Differences between genotypes were tested for significance using a two-sample independent t-test or one-way ANOVA, as appropriate. All data are listed means ± SEM.
Axon counts
To perform the axon counts, the femoral nerve and ventral root from Smn1C/C and Smn1C/+ (n = 4/genotype) were excised, fixed and cross-sectioned at 500 nm followed by staining with Toluidine Blue. Axons were counted and measured using the Analyze Particle Plug-in of ImageJ.
Immunohistochemistry of NMJs
Mice were anaesthetized by intraperitoneal injections of ketamine/xylazine (100 mg/kg ketamine/10 mg/kg xylazine) and transcardially perfused with Ringer's solution followed by 4% paraformaldehyde. Muscles were dissected and labeled with anti-neurofilament (1:2000; Chemicon) and anti-synaptophysin (1:200, Invitrogen) antibodies for nerve terminals and Alexa Fluor 594-conjugated α-bungarotoxin (Invitrogen) for AChRs. Fluorescently labeled NMJs were observed with epifluorescence or confocal microscopy. For imaging of NMJs, Z-stack images of immunostained whole-mount muscles were obtained at sequential focal planes 1 μm apart using the Zeiss LSM 510 META confocal microscope. Illustrated images are flattened projections of Z-stack images.
Electrophysiology of NMJs
Conventional intracellular recordings were performed as described previously (22). In brief, muscles were dissected in normal Ringer's solution. Muscle contraction was blocked by pre-incubating the muscle in 2–3 μm μ-conotoxin for 30 min. EPPs were elicited by 1 Hz train through a suction electrode and recorded via a glass pipette filled with 3 m KCl. Data were acquired and analyzed using the pClamp8 software and the Minianalysis software. The mean quantal content was calculated by a direct method (18). Student's t-test was used for comparison of means between SMA and WT controls.
Assessment of neuromuscular deficit at JAX
Animals of the indicated genotypes were assessed for neuromuscular deficit using gait, grip strength and rotarod analysis.
Grip strength
Grip strength was assessed using the Chatillon-Ametek Digital Force Gauge, DFIS 2 (Columbus Instruments) to determine the strength exerted by the forelimbs of an animal in response to a constant downward force. The grip-strength meter was positioned vertically, with the triangular metal transducer situated 40 cm above a foam platform. The mouse was raised toward the triangular transducer and it instinctively grasped the bar. Once an appropriate grip was assumed, the animal was pulled straight down from the bar. Peak force was measured in kilograms for three consecutive trials and an average was calculated for each animal.
Rotarod
Rotarod testing was carried out using the Economex Accelerating Rota-Rod (Columbus Instruments). Mice were placed on a rod of 3.5 cm in diameter which is situated 40 cm above a foam platform. Each mouse was isolated from the next with a 30 cm wide and 60 cm high gray PVC wall. The mice were acclimated to the instrument by being placed on the stationary rod for three consecutive trials, with a 1min cutoff. Each mouse was then re-placed onto the rod at a constant speed of 4.0 r.p.m. for three consecutive trials, again, each limited to 1 min. Finally, to measure balance performance, the mice were re-positioned on the rod with a constant speed of 4.0 r.p.m. while an acceleration of 0.1 r.p.m./s was applied over a 5min period, for three consecutive trials. Latency to fall was recorded for each trial.
Assessment of neuromuscular deficit: PGI
Animals of each genotype were assessed for motor function deficit using rotarod, OF and von Frey tests. Testing was conducted between 8:30 AM and 2:00 PM. Acclimation time prior testing was 1 h.
Rotarod
Animals of the indicated genotypes were placed on an accelerating rod (Rotamex, OH, USA). On testing days of week 6, 10 and 16, animals were allowed one training session (3–5 min), followed by three test runs. Animals were allowed 1 h of rest time between each test or training run. The latency to fall off the rod was recorded.
Automated OF
The OF chambers were acrylic square chambers (27 × 27 cm2; Med Associates, Inc., St Albans, VT, USA) surrounded by infrared photobeams. The activity was recorded in 5 min time bins by an automated system that counts successive photobeam breaks. Animals were tested for 1 h sessions at weeks 8, 12, 16 and 20.
Mechanical plantar test (von Frey method)
The mechanical stimuli (von Frey filaments) were part of a calibrated set of Touch Test® sensory evaluator pens of increasing bending strength. The kit consisted of 20 Semmes-Weinstein Monofilaments that range from 1.65 to 6.65 in evaluator size, which translates to 0.008–300 mN bending force. Animals were placed into the vF apparatus, one at a time. Three consecutive filament tip applications were applied. Once animal felt pain, it withdrew its paw from the filament tip. Animals were tested at weeks 20 and 24.
Assessment of neuromuscular deficit: USC
von Frey testing
Adult mice from both genotypes (6–8 weeks old) were habituated in a plastic chamber on a mesh floor and then an electronic von Frey filament (IITC Life Sciences, CA, USA) was applied to the hindpaw to measure the withdrawal force. Stimuli were repeated five times with a 5 min rest period between each paw, and a withdrawal caused by the stimulation was considered a positive response.
Hot and cold plate assay
Mice were first pre-habituated (at room temperature) to the apparatus, which consisted of a Plexiglas chamber containing one metal platform, which was thermostatically controlled (43). The temperature of the surface was set to temperatures 45, 48 or 52°C for heat and 5, 10, and 15°C for cold and the latency to first response of hindpaw lifting, flinching, shaking or licking was recorded with a cutoff of 60 s for heat and 120 s for cold to prevent injury.
Acetone-evoked evaporative cooling assay
The evaporative cooling assay was performed as described (26). Briefly, mice were habituated in an elevated chamber with a mesh floor for 15min, and then a droplet of acetone was applied to one hindpaw. Mice were stimulated five times with an inter-stimulation period of 4min per mouse, alternating paws between stimulations, and responses were video-recorded for later quantification by an observer blind to the experimental conditions. Behaviors were scored according to the following scale: 0, no response; 1, brief lift, sniff, flick or startle; 2, jumping, paw shaking; 3, multiple lifts, paw lick; 4, prolonged paw lifting, licking, shaking or jumping; 5, paw guarding. The number of behaviors was also recorded over the 1 min test period. Statistical significance for all behavioral assays were assessed using either the paired or unpaired Student's t-test or one-way ANOVA, as appropriate (26,44).
Conscious ECG
Cardiac electrical activity (ECG) was recorded from conscious mice gently restrained in a Murine ECG Restrainer (QRS Phenotyping, Inc., Calgary, Alberta, Canada) using gel-foot pads to collect the signal. The signal acquisition was collected using PowerLab (AD Instruments, Golden, CO, USA). After the acclimation period, the ECG signal was recorded for up to 5 min or until a steady baseline signal was recorded for a 20–30 s interval. A signal average was also produced using 15 s of consecutive ECG tracings at two different time intervals during the lowest anesthetic concentration. The signal average (SA-ECG) was used to determine the heart rate (b.p.m.), P-duration (ms), T-height (mV), RR interval (ms), QT duration (ms), PR duration (ms), QRS height (mV), QRS duration (ms) and P- height (mV).
DEXA and thermal imaging
Body composition of mice of the indicated genotypes was determined by DEXA using the LUNAR PIXImus mouse densitometer. Mice were anesthetized with 2% tribromoethanol prior to scanning. After scanning, mice were allowed to recover from anesthesia with an external source of heat and then returned to their home cage. For infra-red imaging studies, Thermal Signature Analysis (TSA) using ImagIR (Seahorse Bioscience North Billerica, MA, USA) was utilized. Both superficial and core temperatures were recorded on Smn1C/C mutant and control mice simultaneously over a 10 min period. For imaging, animals were induced with 5.0% isoflurane in oxygen delivered at 0.8 l/min and placed on a heated platform set to 35°C. Mice were maintained at 1.5% isoflurane for the remainder of the experiment.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a grant from the Spinal Muscular Atrophy Foundation to [C.M.L. (JAX); C.P.K. (USC)]. This work is also supported by National Institutes of Health grants NS054069 and NS071364 (to D.D.M.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Karen Chen, Kathleen McCarthy and Sergey Paushkin from the SMA Foundation for their thoughtful review of this manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet. 1978;15:409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 4.Dubowitz V. Very severe spinal muscular atrophy (SMA type 0): an expanding clinical phenotype. Eur. J. Paediatr. Neurol. 1999;3:49–51. doi: 10.1053/ejpn.1999.0181. [DOI] [PubMed] [Google Scholar]
- 5.Schrank B., Gotz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jablonka S., Schrank B., Kralewski M., Rossoll W., Sendtner M. Reduced survival motor neuron (Smn) gene dose in mice leads to motor neuron degeneration: an animal model for spinal muscular atrophy type III. Hum. Mol. Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 7.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 8.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 10.Tsai M.S., Chiu Y.T., Wang S.H., Hsieh-Li H.M., Li H. Abolishing Trp53-dependent apoptosis does not benefit spinal muscular atrophy model mice. Eur. J. Hum. Genet. 2006;14:372–375. doi: 10.1038/sj.ejhg.5201556. [DOI] [PubMed] [Google Scholar]
- 11.Donnelly E.M., Quach E.T., Hillery T.M., Heeke B.L., Snyder B.R., Handy C.R., O'Connor D.M., Boulis N.M., Federici T. Characterization of a murine model of SMA. Neurobiol. Dis. 2012;45:992–998. doi: 10.1016/j.nbd.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 12.Monani U.R., Pastore M.T., Gavrilina T.O., Jablonka S., Le T.T., Andreassi C., DiCocco J.M., Lorson C., Androphy E.J., Sendtner M., et al. A transgene carrying an A2G missense mutation in the SMN gene modulates phenotypic severity in mice with severe (type I) spinal muscular atrophy. J. Cell Biol. 2003;160:41–52. doi: 10.1083/jcb.200208079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin L.J. Olesoxime, a cholesterol-like neuroprotectant for the potential treatment of amyotrophic lateral sclerosis. IDrugs. 2010;13:568–580. [PMC free article] [PubMed] [Google Scholar]
- 14.Butchbach M.E., Singh J., Thorsteinsdottir M., Saieva L., Slominski E., Thurmond J., Andresson T., Zhang J., Edwards J.D., Simard L.R., et al. Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum. Mol. Genet. 2010;19:454–467. doi: 10.1093/hmg/ddp510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G., et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valenzuela D.M., Murphy A.J., Frendewey D., Gale N.W., Economides A.N., Auerbach W., Poueymirou W.T., Adams N.C., Rojas J., Yasenchak J., et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 17.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes. Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling K.K., Gibbs R.M., Feng Z., Ko C.P. Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2012;21:185–195. doi: 10.1093/hmg/ddr453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGovern V.L., Gavrilina T.O., Beattie C.E., Burghes A.H. Embryonic motor axon development in the severe SMA mouse. Hum. Mol. Genet. 2008;17:2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray L.M., Lee S., Baumer D., Parson S.H., Talbot K., Gillingwater T.H. Pre-symptomatic development of lower motor neuron connectivity in a mouse model of severe spinal muscular atrophy. Hum. Mol. Genet. 2010;19:420–433. doi: 10.1093/hmg/ddp506. [DOI] [PubMed] [Google Scholar]
- 21.Cruz L.J., Gray W.R., Olivera B.M., Zeikus R.D., Kerr L., Yoshikami D., Moczydlowski E. Conus geographus toxins that discriminate between neuronal and muscle sodium channels. J. Biol. Chem. 1985;260:9280–9288. [PubMed] [Google Scholar]
- 22.Ling K.K., Lin M.Y., Zingg B., Feng Z., Ko C.P. Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS One. 2010;5:e15457. doi: 10.1371/journal.pone.0015457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mentis G.Z., Blivis D., Liu W., Drobac E., Crowder M.E., Kong L., Alvarez F.J., Sumner C.J., O'Donovan M.J. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69:453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park G.H., Maeno-Hikichi Y., Awano T., Landmesser L.T., Monani U.R. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J. Neurosci. 2010;30:12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks S.P., Dunnett S.B. Tests to assess motor phenotype in mice: a user's guide. Nat. Rev. Neurosci. 2009;10:519–529. doi: 10.1038/nrn2652. [DOI] [PubMed] [Google Scholar]
- 26.Knowlton W.M., Daniels R.L., Palkar R., McCoy D.D., McKemy D.D. Pharmacological blockade of TRPM8 ion channels alters cold and cold pain responses in mice. PLoS One. 2011;6:e25894. doi: 10.1371/journal.pone.0025894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bautista D.M., Siemens J., Glazer J.M., Tsuruda P.R., Basbaum A.I., Stucky C.L., Jordt S.E., Julius D. The menthol receptor TRPM8 is the principal detector of environmental cold. Nature. 2007;448:204–208. doi: 10.1038/nature05910. [DOI] [PubMed] [Google Scholar]
- 28.Heier C.R., Satta R., Lutz C., DiDonato C.J. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum. Mol. Genet. 2010;19:3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh C.T., Chang C.F., Lin E.Y., Tsai T.H., Chiang Y.H., Ju D.T. Spontaneous spinal epidural hematomas of cervical spine: report of 4 cases and literature review. Am. J. Emergency Med. 2006;24:736–740. doi: 10.1016/j.ajem.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Gavrilina T.O., McGovern V.L., Workman E., Crawford T.O., Gogliotti R.G., DiDonato C.J., Monani U.R., Morris G.E., Burghes A.H. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum. Mol. Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 32.Araujo Ade Q., Araujo M., Swoboda K.J. Vascular perfusion abnormalities in infants with spinal muscular atrophy. J. Pediatr. 2009;155:292–294. doi: 10.1016/j.jpeds.2009.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narver H.L., Kong L., Burnett B.G., Choe D.W., Bosch-Marce M., Taye A.A., Eckhaus M.A., Sumner C.J. Sustained improvement of spinal muscular atrophy mice treated with trichostatin A plus nutrition. Ann. Neurol. 2008;64:465–470. doi: 10.1002/ana.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer K., Marquis J., Trub J., Nlend Nlend R., Verp S., Ruepp M.D., Imboden H., Barde I., Trono D., Schumperli D. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum. Mol. Genet. 2009;18:546–555. doi: 10.1093/hmg/ddn382. [DOI] [PubMed] [Google Scholar]
- 35.Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H., et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Passini M.A., Bu J., Roskelley E.M., Richards A.M., Sardi S.P., O'Riordan C.R., Klinger K.W., Shihabuddin L.S., Cheng S.H. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito Y., Shibata N., Saito K., Kobayashi M., Osawa M. New insights into the pathogenesis of spinal muscular atrophy. Brain Dev. 2011;33:321–331. doi: 10.1016/j.braindev.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Scholz J., Woolf C.J. Can we conquer pain? Nat. Neurosci. 2002;5(5 Suppl.):1062–1067. doi: 10.1038/nn942. [DOI] [PubMed] [Google Scholar]
- 39.McFadyen M.P., Kusek G., Bolivar V.J., Flaherty L. Differences among eight inbred strains of mice in motor ability and motor learning on a rotorod. Genes Brain Behav. 2003;2:214–219. doi: 10.1034/j.1601-183x.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- 40.Bach J.R. Medical considerations of long-term survival of Werdnig-Hoffmann disease. Am. J. Phys. Med. Rehabil. 2007;86:349–355. doi: 10.1097/PHM.0b013e31804b1d66. [DOI] [PubMed] [Google Scholar]
- 41.Khatri I.A., Chaudhry U.S., Seikaly M.G., Browne R.H., Iannaccone S.T. Low bone mineral density in spinal muscular atrophy. J. Clin. Neuromuscul. Dis. 2008;10:11–17. doi: 10.1097/CND.0b013e318183e0fa. [DOI] [PubMed] [Google Scholar]
- 42.Poueymirou W.T., Auerbach W., Frendewey D., Hickey J.F., Escaravage J.M., Esau L., Dore A.T., Stevens S., Adams N.C., Dominguez M.G., et al. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat. Biotechnol. 2007;25:91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- 43.Daniels M.J., Varghese T. Dynamic frame selection for in vivo ultrasound temperature estimation during radiofrequency ablation. Phys. Med. Biol. 2010;55:4735–4753. doi: 10.1088/0031-9155/55/16/008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knowlton W.M., Bifolck-Fisher A., Bautista D.M., McKemy D.D. TRPM8, but not TRPA1, is required for neural and behavioral responses to acute noxious cold temperatures and cold-mimetics in vivo. Pain. 2010;150:340–350. doi: 10.1016/j.pain.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.