Abstract
The apolipoprotein E (APOE) genotype is the major genetic risk factor for Alzheimer's disease (AD). We have access to cerebrospinal fluid (CSF) and plasma APOE protein levels from 641 individuals and genome-wide genotyped data from 570 of these samples. The aim of this study was to test whether CSF or plasma APOE levels could be a useful endophenotype for AD and to identify genetic variants associated with APOE levels. We found that CSF (P = 8.15 × 10−4) but not plasma (P = 0.071) APOE protein levels are significantly associated with CSF Aβ42 levels. We used Mendelian randomization and genetic variants as instrumental variables to confirm that the association of CSF APOE with CSF Aβ42 levels and clinical dementia rating (CDR) is not because of a reverse causation or confounding effect. In addition the association of CSF APOE with Aβ42 levels was independent of the APOE ɛ4 genotype, suggesting that APOE levels in CSF may be a useful endophenotype for AD. We performed a genome-wide association study to identify genetic variants associated with CSF APOE levels: the APOE ɛ4 genotype was the strongest single-genetic factor associated with CSF APOE protein levels (P = 6.9 × 10−13). In aggregate, the Illumina chip single nucleotide polymorphisms explain 72% of the variability in CSF APOE protein levels, whereas the APOE ɛ4 genotype alone explains 8% of the variability. No other genetic variant reached the genome-wide significance threshold, but nine additional variants exhibited a P-value <10−6. Pathway mining analysis indicated that these nine additional loci are involved in lipid metabolism (P = 4.49 × 10−9).
INTRODUCTION
The ɛ4 allele of the apolipoprotein E (APOE) gene is the most important genetic risk factor for late-onset Alzheimer's disease (AD). The two most common non-synonymous single nucleotide polymorphisms (SNPs) in APOE are rs7412 and rs429358, which define the APOE ε2 and ε4 alleles, respectively. APOE ε2 has a cysteine residue at codons 112 and 158, whereas APOE ε4 has an arginine residue at these codons. APOE ε3 has a cysteine at codon 112 and an arginine at codon 158. These amino acid substitutions have been hypothesized to change the structure and function of APOE in the periphery while little is known about how these amino acid changes may affect normal function of the APOE protein in the central nervous system. In humans, the most common allele is APOE ε3 [frequency in European-descendent healthy elderly individuals: 0.78 (1)]. APOE ε2 has been associated with a more than 50% reduction in the risk for AD, whereas APOE ε4 is associated with a 1.8–9.9-fold increase in the risk for AD (1). The APOE alleles combine to generate five possible genotypes listed in order of increasing risk for AD: ε2/2, ε2/3, ε3/3, ε2/4, ε3/4 and ε4/4 (2).
Several hypotheses for how APOE may affect the risk for the development of AD have been proposed, and most have focused on how APOE affects Aβ pathology. APOE ɛ4 carriers have more Aβ plaques (3) and more PIB retention (4) than APOE ɛ4 negative individuals. Similarly, mice expressing a human APOE ɛ4 allele on the background of an AD causing amyloid precursor protein mutation have earlier and more severe pathology than mice expressing APOE ɛ3 (5), while mice expressing the human APOE ɛ2 do not generate fibrillar amyloid deposits (6). In vitro studies indicate that APOE ɛ4 binds to Aβ with a higher affinity than APOE ɛ3 (7), and that APOE and Aβ may also compete for clearance through the same receptors (8). A recent study that used in vivo microdialysis in a mouse model of Aβ-amyloidosis expressing human APOE isoforms, demonstrated that the APOE isoforms differentially regulate the clearance of brain amyloid-β (9).
Most of the genome-wide genotyping chips do not include the SNPs that define the APOE isoforms. In the genome-wide association analyses (GWAS) for AD, the most significant SNP is rs2075650 (10–12), which is located within TOMM40, a gene close to APOE, shows a linkage disequilibrium with rs429358 (APOE ɛ4) of R2= 0.4 (D′ = 1). We have reported that the APOE genotype is strongly associated with cerebrospinal fluid (CSF) Aβ42 levels (13–15), supporting the results from animal studies that APOE influences risk for AD through an Aβ-dependent mechanism.
Given that the APOE ɛ4 genotype plays an important role in the neuropathology of AD as well as interest in identifying novel biomarkers and endophenotypes for AD, APOE protein levels have been examined as a potential biomarker for AD in several studies yielding mixed results. APOE protein levels have been associated with the APOE ɛ4 genotype in plasma and in brain (16–22). A recent study suggested that APOE plasma levels could be a good biomarker for AD, because APOE protein levels are associated with AD status, but most of this association was driven by age and APOE ɛ4 genotype (22). Additionally, several studies have failed to find an association between CSF APOE protein levels and risk for AD or APOE ɛ4 genotype, although these studies have been performed using relatively small sample sizes (18,20,23,24).
In this study, we used a large series of well-characterized individuals, in whom CSF and plasma APOE levels were measured in order to test whether APOE protein levels would be a useful endophenotype for AD. An endophenotype is a heritable quantitative trait that is correlated with disease but measurable in all individuals regardless of disease status. Ideally the endophenotype should have simpler genetic architecture so that SNPs in candidate genes may directly lead to changes in the endophenotype.
We also investigated genetic variants that may influence APOE protein levels in CSF and plasma. Identification of the genetic factors that are associated with APOE levels could potentially identify novel therapeutic targets or pathways that would enable a greater understanding of the role of APOE' in the pathogenic mechanisms of AD.
RESULTS
CSF and plasma APOE levels as a potential endophenotype for AD
No correlation between CSF and plasma APOE levels
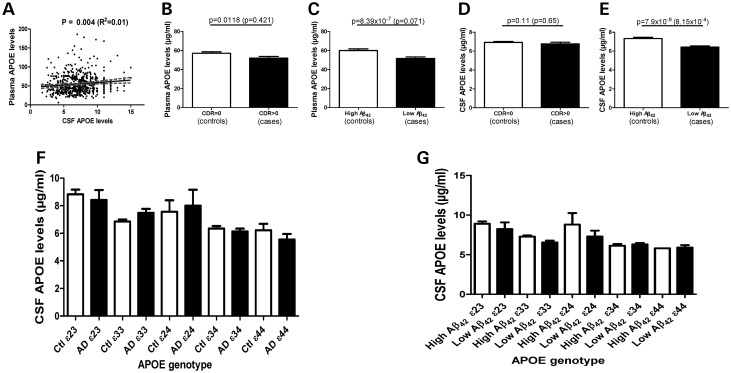
We first tested whether CSF and plasma APOE protein levels are correlated using data from 641 individuals in whom CSF and plasma APOE levels were measured in the same assay (Table 1). There was not a strong correlation between CSF and plasma APOE protein levels (R2= 0.01; P = 0.004, Fig. 1A), suggesting that these measures could individually have a different potential to be an endophenotype for AD.
Table 1.
Sample demographics
| Sample | Samples with biomarker level | Samples with GWAS | Age (years) | Male (%) | APOE ɛ4+ (%) |
|---|---|---|---|---|---|
| Knight-ADRC-CSF | |||||
| Controls | 239 | 197 | 71 ± 7 (53–91) | 35 | 32 |
| AD | 92 | 81 | 74 ± 7 (53–90) | 46 | 42 |
| ADNI-CSF | |||||
| Controls | 92 | 86 | 76 ± 5 (62–92) | 50 | 24 |
| MCI | 149 | 141 | 75 ± 6 (57–89) | 69 | 54 |
| AD | 69 | 65 | 75 ± 6 (56–88) | 56 | 61 |
| Knight-ADRC brain series | |||||
| Cases | 82 | 86 ± 7 (72–102) | 45 | 41 | |
| Controls | 39 | 85 ± 9 (64–107) | 41 | 23 | |
For cases the age represents age at the onset and for controls the age represents age at the last assessment.
Figure 1.
Correlation of CSF and plasma levels. CSF APOE levels are significantly lower in individuals with low CSF Aβ42 levels. CSF and plasma levels of APOE were measured on 641 samples from the Knight-ADRC and ADNI using the rules based medicine human discovery MAP panel version 1. (A) CSF and plasma APOE levels show very low correlation (R2= 0.01). Plasma (B) and CSF (D) APOE levels in clinically defined cases and controls. CSF Aβ42 was used to define individuals with likely Aβ brain deposition (cases or presymptomatic cases) and individuals without Aβ brain deposition (controls). Plasma (C) and CSF (E) APOE levels in individuals with low or high CSF Aβ42 levels. The P-value is for the association of plasma/CSF APOE levels without including APOE genotype as a covariate. The P-value in parenthesis was calculated including the APOE genotype as a covariate. (F) CSF APOE levels in cases and controls stratified by the APOE genotype. No significant differences were found between cases and controls in any strata. (G) CSF APOE levels in individuals with high and low CSF Aβ42 levels stratified by the APOE genotype.
CSF but not plasma APOE level is a potential endophenotype for AD
We first tested whether gender and age were associated with CSF and plasma APOE protein levels. CSF APOE protein levels were positively correlated with age (P = 9.89 × 10−6) and significantly higher in men compared with women (7.3 versus 6.5 µg/ml; P = 2.07 × 10−4). Age and gender explained 3.9% of the variability in CSF APOE levels. In contrast, gender but not age showed a significant association with plasma APOE protein levels with men having significantly lower levels of APOE than women (54.5 versus 56.3 µg/ml; P = 0.0083).
Finally, we tested whether CSF or plasma APOE protein levels were associated with case–control status for AD. Plasma APOE protein levels were significantly decreased in cases compared with controls (51.7 versus 57.1 µg/ml; P = 0.0118, Fig. 1B), but this association disappeared when the APOE genotype was included as a covariate (P = 0.421, see next section for full analysis for association with the APOE genotype). CSF APOE protein levels demonstrated a trend for association with case–control status (P = 0.11), but this association disappeared when the APOE genotype was included as a covariate (P = 0.65; Fig. 1D and E). Several studies have suggested that up to 30% of elderly non-demented control samples show some evidence of AD pathology at autopsy (25,26). Additionally, individuals with CSF Aβ42 levels less than 500 pg/ml in the Knight-ADRC-CSF cohort have evidence of Aβ deposition in the brain, as detected by PET-PIB (27). Similarly, CSF Aβ42 levels less than 192 pg/ml in the Alzheimer's Disease Neuroimaging Initiative (ADNI) series can be used as a proxy for brain Aβ deposition (28). Individuals with CSF Aβ42 levels below these thresholds could be classified as preclinical AD cases with the presumption that some evidence of fibrillar Aβ brain deposits would be detected (27,28). When these thresholds were used to classify samples into individuals with and without Aβ deposits in the brain we observed a different result. We found a very significant difference in plasma APOE protein levels between individuals with low and high CSF Aβ42 (low versus high = 59.99 versus 51.45 µg/ml; P = 8.39 × 10−7, including gender as covariate). However, when the APOE genotype was included as a covariate the difference in plasma APOE levels was no longer significant (P = 0.071, Fig. 1C), indicating that the association was driven by the APOE genotype. On the other hand, individuals with lower CSF Aβ42 levels that could also potentially have fibrillar Aβ deposits in the brain also had APOE levels that were significantly reduced (6.55 versus 7.38 µg/ml; P = 7.90 × 10−9, including age and gender as covariates; Table 2 and Fig. 2E), that remained significant even when the APOE genotype was included as a covariate (P = 8.15 × 10−4, Fig. 1E).
Table 2.
CSF APOE levels stratified by clinical case–control status or by CSF Aβ42 levels
| Controls ɛ23 | Cases ɛ23 | Controls ɛ33 | Cases ɛ33 | Controls ɛ24 | Cases ɛ24 | Controls ɛ34 | Cases ɛ34 | Controls ɛ44 | Cases ɛ44 | Controls | Cases | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | 8.9 ± 2.6 | 8.4 ± 2.3 | 6.9 ± 2.5 | 7.6 ± 2.5 | 7.5 ± 2.5 | 8.0 ± 2.0 | 6.4 ± 2.0 | 6.2 ± 2.0 | 6.2 ± 2.3 | 5.7 ± 1.8 | 6.9 ± 2.32 | 6.8 ± 2.3 |
| Range | 2.6–15.4 | 3.06–11 | 2.3–14.3 | 2.6–14.1 | 3.9–11.3 | 5.7–9.3 | 2.5–14 | 2.7–14.8 | 3.1–13 | 2.41–8.8 | 2.3–15.4 | 2.4 ± 14.8 |
| n | 49 | 11 | 233 | 62 | 9 | 3 | 123 | 86 | 25 | 22 | 439 | 184 |
| P-value | 0.608 | 0.128 | 0.721 | 0.562 | 0.376 | 0.167 (0.857) | ||||||
| High Aβ42 ɛ23 | Low Aβ42 ɛ23 | High Aβ42 ɛ33 | Low Aβ42 ɛ33 | High Aβ42 ɛ24 | Low Aβ42 ɛ24 | High Aβ42 ɛ34 | Low Aβ42 ɛ34 | High Aβ42 ɛ44 | Low Aβ42 ɛ44 | High Aβ42 | Low Aβ42 | |
| Mean ± SD | 9.0 ± 2.3 | 8.2 ± 3.08 | 7.3 ± 2.0 | 6.7 ± 2.58 | 8.1 ± 2.4 | 7.41 ± 2.3 | 6.2 ± 2.0 | 6.4 ± 2.0 | 5.8 | 4.9 ± 2.1 | 7.3 ± 2.2 | 6.5 ± 2.3 |
| Range | 4.0–15.4 | 2.6–14 | 2.7–13.0 | 2.3–14 | 3.9–11.0 | 3.9–11 | 3.0–14.8 | 2.6–14 | — | 2.4–13 | 2.7–15.4 | 2.3–14.3 |
| n | 47 | 13 | 174 | 121 | 4 | 8 | 59 | 150 | 1 | 46 | 286 | 338 |
| P-value | 0.078 | 0.003 | 0.133 | 0.354 | 0.834 | 7.90 × 10−9 (8.15 × 10−4) | ||||||
The table shows the mean, standard deviation and range for CSF APOE levels in µg/ml, as well as the number of samples in each stratum.
The P-value is calculated for normalized CSF APOE levels including age and gender as covariates. In the last analysis, which included all samples, the P-value in parenthesis included APOE genotype as a covariate as well.
Figure 2.
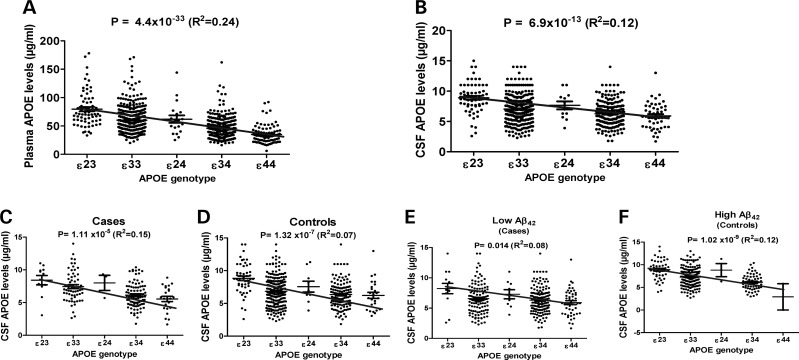
The APOE genotype influences plasma and CSF APOE levels. (A) The APOE genotype shows a strong association with plasma APOE levels and explains 24% of the variability in plasma APOE levels. Gender was included as a covariate in the analysis. (B) The APOE genotype shows a strong association with CSF APOE levels and explains 12% of the variability in CSF APOE levels. Age and gender were included in the analysis as covariates. (C) The APOE genotype is strongly associated with CSF APOE levels in clinically defined cases. (D) The APOE genotype is strongly associated with CSF APOE levels in clinically defined controls. (E) The APOE genotype is strongly associated with CSF APOE levels in individuals with low CSF Aβ42 levels. (F) The APOE genotype is strongly associated with CSF APOE levels in individuals with high CSF Aβ42 levels.
To confirm that the difference in CSF APOE protein levels in individuals with and without potential fibrillar Aβ brain deposits (measured as CSF Aβ42 as a proxy) is not driven by the APOE genotype, we stratified the samples by the APOE genotype (Fig. 1G). This analysis is less statistically powerful because each of the within genotype comparison groups is much smaller in size. Despite the decrease in statistical power, we found that CSF APOE levels are significantly lower in APOE 33 individuals with low CSF Aβ42 levels compared with APOE 33s with high Aβ42 levels (P = 0.003; Table 2). A similar trend was found in APOE 23 and 24 individuals, but not in 34 and 44 individuals. It is important to note that these individuals (with APOE 34 and 44 genotypes) already have lower CSF APOE levels. No difference was found in the plasma APOE levels in any APOE genotype group (Table 2).
APOE genotype and APOE levels influence Aβ42levels in CSF/plasma
CSF Aβ42 and APOE levels are strongly correlated
CSF Aβ42 has emerged as one of the most promising biomarkers for AD (25–27). Since CSF Aβ42 can be used as a proxy for PIB retention (24,26) and is correlated with plaque number in the brain (29), we investigated whether CSF or plasma APOE protein levels are associated with CSF Aβ42 levels.
We found a very significant positive correlation between plasma APOE protein levels and CSF Aβ42 levels (P = 2.63 × 10−7), but this association was driven by the APOE genotype. When the APOE genotype was included as a covariate, the P-value for the association of plasma APOE protein levels with CSF Aβ42 levels was 0.13. In contrast, CSF APOE protein levels demonstrated a much stronger association with CSF Aβ42 (P = 3.11 × 10−14; Supplementary Material, Fig. S1), which was significant even when the APOE genotype was included as a covariate (P = 2.84 × 10−8). To confirm that CSF APOE protein levels and Aβ42 are correlated independently of the APOE genotype, we stratified the samples by the APOE genotype and reanalyzed the data (Supplementary Material, Fig. S1). Within APOE 33 individuals CSF APOE protein levels and Aβ42 showed a strong correlation (P = 3.28 × 10−5). A nominally significant association was found in the 23 and 24 strata (P = 0.03, and 0.04, respectively). Previously, we reported that the APOE genotype is strongly associated with CSF Aβ42 (13–15). A model including age, gender and APOE genotype explains 14% of the variability in CSF Aβ42. A model including also CSF APOE protein levels, explains 18.5% of the variability in CSF Aβ42 measurements, indicating that APOE protein levels can explain additional variability in CSF Aβ42 levels independent of the APOE genotype.
Mendelian randomization
We use Mendelian randomization to try to determine whether CSF APOE levels are actually in the causal pathway for AD. Since changes in Aβ42 levels have been causally linked to disease in Mendelian forms of AD, we first tested whether APOE levels are causally linked to CSF Aβ42 levels. Mendelian randomization refers to the random assortment of alleles inherited by offspring from their parents at conception (30,31). This random assortment of inherited alleles has been likened to a randomized clinical trial, in which the research subjects are randomly allocated to different genotypes rather than to medical interventions (30). According to Mendelian randomization, genetic variants associated with CSF APOE levels are randomly transmitted to the offspring and largely free from reverse causation and confounding. Therefore, we used the SNPs most significantly associated with CSF APOE levels (see section Genome-wide association analysis) and a genetic risk score (see section Materials and Methods) as instrumental variables. Age, gender, series and clinical dementia rating (CDR) were included as covariates or endogenous predictors (Fig. 3). The F-statistics value indicates that the APOE genotype, rs2075650, the genetic risk score were appropriate instrumental variables (F = 20.41, 12.69 and 16.68 in the first-stage regression, respectively). The strong positive association between CSF APOE and Aβ42 levels in the general linear model analyses (β = 0.33, R2= 0.096, P= 3.11 × 10−14) was confirmed using 2SLS (β = 1.82, R2= 0.073, P= 5.00 × 10−9) with the APOE genotype as the instrumental variable. Similar results were obtained with limited information maximum likelihood (LIML) methods (β = 0.99, R2= 0.11, P= 2.63 × 10−11). Similar results and conclusions were obtained when using rs2075650 or the genetic risk score as instrumental variables (P= 4.58 × 10−4 and 2.85 × 10−7, respectively).
Figure 3.
Mendelian randomization: graphical illustration of the structural model.
Finally, we analyzed whether APOE levels are in the casual pathway of AD by using CDR or clinical status at lumbar puncture (LP) as the dependent variable. A significant association was found for CSF APOE levels and CDR when using the APOE genotype (P= 3.06 × 10−5), rs2075650 (P= 0.006), or the genetic risk score (P= 0.0284) as instrumental variables. Significant associations were found when using case–control status as the dependent variable. Initially, we used CSF Aβ42 levels as a proxy for Aβ42 deposition (preclinical and clinical AD). Several studies have reported a high correlation between low CSF Aβ42 and PET-PIB positive signals (27,28), and non-demented individuals with low CSF Aβ42 are also more likely to convert to AD cases (27,28). Therefore we repeated the Mendelian Randomization analysis using CDR at last assessment instead of CDR at LP. In these analyses, we found a more significant association between CSF APOE levels and last CDR than when using CDR at LP. A significant association was found for CSF APOE levels and CDR when using the APOE genotype (P= 2.06 × 10−6), rs2075650 (P= 0.0013) or the genetic risk score (P= 0.0063) as instrumental variables. To take into account the potential pleiotropic effect of APOE genotypes, we created a genetic score without the SNP in APOE. In this case, the association of CSF APOE genotype and CDR was weaker than previous analysis, but still significant (P= 0.0361).
The results of the instrumental variable analysis provide support for a non-confounded and unbiased association between CSF APOE levels and CSF Aβ42 levels, CDR and case–control status. This approach is analogous to methods used in randomized clinical trials to infer an unbiased effect on treatment. Together, these results clearly suggest that CSF APOE protein levels are a potential endophenotype for AD and are much more closely linked to AD pathology than plasma APOE protein levels.
Genetic variants associated with CSF APOE protein levels
APOE genotype influences CSF and plasma APOE levels but not APOE mRNA levels
To determine whether the APOE genotype is associated with CSF and plasma APOE protein levels, we used an additive model, coding APOE genotype into five levels according to genetic risk (APOE ɛ22 = 0, ɛ23 = 1, ɛ33 = 2, ɛ24 = 3, ɛ34 = 4, ɛ44 = 5). The APOE genotype showed a very strong association with plasma (P = 4.4 × 10−33; Fig. 2A) and CSF (P = 6.9 × 10−13; Fig. 2B) APOE protein levels. Individuals with APOE genotypes associated with a higher risk for disease (34 and 44) exhibit a dose-dependent lowering of CSF and plasma APOE protein levels. This association was independent of the case–control status, and the presence of Aβ deposition in the brain (Fig. 2). A model for CSF APOE protein levels including age (P = 3.51 × 10−5), CSF Aβ42 (P = 2.65 × 10−6) and APOE genotype (P = 6.9 × 10−13), explained 12% of the variability in CSF APOE protein levels. The APOE genotype alone accounts for 8.2% of the variability in CSF APOE protein levels. The fact that all of the variables in the model were significantly associated with CSF APOE protein levels indicates that age, Aβ brain pathology and APOE genotype may influence CSF APOE levels independently. Independent of the status for Aβ brain pathology as defined by CSF Aβ42 levels, we found the APOE genotype to be associated with CSF APOE protein levels (Fig. 2E and F). To determine whether the association between APOE genotype and CSF APOE protein levels is driven by isoform differences in APOE transcription, we measured APOE mRNA levels in parietal lobe tissue from 121 individuals. Age, gender, postmortem interval and CDR were included as covariates. No association between APOE genotype and APOE mRNA levels were observed (R2= 0.05; P = 0.28). We also stratified our samples by case–control status, and found no association between APOE genotype and APOE mRNA levels in the parietal cortex from either cases or controls (Supplementary Material, Fig. S2). A model for plasma APOE protein levels including gender, CSF Aβ42 and APOE genotype (P = 4.4 × 10−33) explained 24% of the variability in plasma APOE protein levels. The APOE genotype alone explains 17.7% of the variability in plasma APOE levels.
Fine mapping APOE gene region
Our previous results clearly indicated that the APOE genotype is significantly associated (passing genome-wide correction) with CSF and plasma APOE protein levels. However, age, CSF Aβ42 levels and APOE genotype only explain 12% of the variability in CSF APOE protein levels; suggesting that other factors, including cis-acting genetic variants may also influence APOE protein levels. Recent studies have suggested that other variants in the APOE-TOMM40 region may also be associated with risk or age of AD onset, although these latter findings are controversial (32,33).
We extracted genotype data for the TOMM40-APOE genomic region from the imputed GWAS data and tested for an association using three different models: the first model included only age, and gender as covariates, a second model included the APOE genotype as a covariate, and the final model analyzed only individuals who are APOE ɛ33. It is important to note that in the first model, the individual SNPs that define either the presence of the APOE4 allele or the presence of the APOE2 allele (rs429358 and rs7412) are not present because they are not in the Illumina chip and they did not pass the imputation quality control (QC). For the first model, the top SNPs rs769449, rs2075650 showed a very significant association (P = 9.58 × 10−7 and 1.67 × 10−6), but less significant than the full APOE genotype (P = 6.9 × 10−13). When the APOE genotype was included as a covariate these SNPs no longer show a significant association with CSF APOE levels (P>0.3). In addition, neither these SNPs nor any other SNPs in this region demonstrated an association with CSF APOE protein levels in APOE ɛ33 subjects (data not shown). These results suggest that the APOE genotype accounts for the majority of the genetic association with CSF APOE protein levels in this gene region. We also analyzed whether the TOMM40 poly-T repeat (rs10524523) showed association with CSF APOE protein levels. We found no association of rs10524523 with CSF APOE protein levels when age, gender and APOE genotype were included as covariates in the full data set (P = 0.85) or in the APOE33 carriers (P = 0.75).
Genome-wide association analysis
Next, to test whether there is evidence for trans-acting SNPs that may influence CSF or plasma APOE protein levels, we performed genome-wide association analyses using data from 570 individuals from the Knight-ADRC and ADNI cohorts
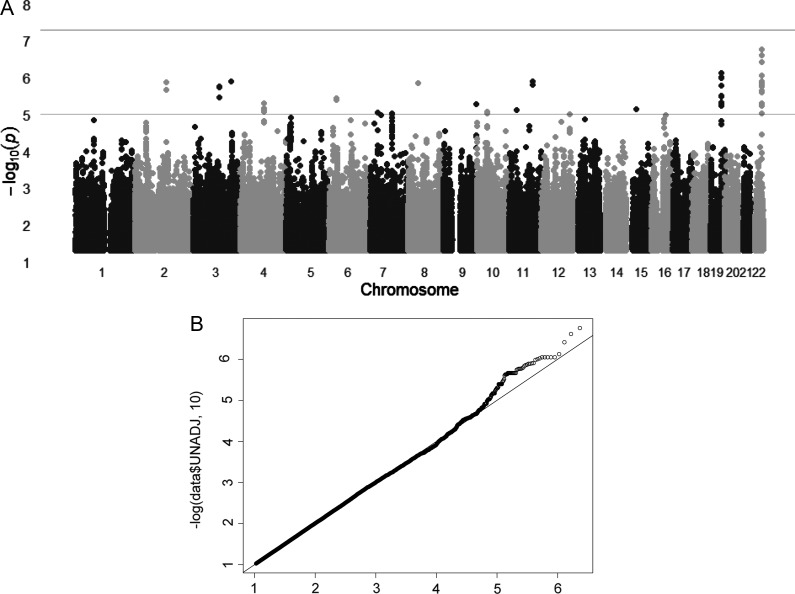
For CSF APOE protein levels, none of the tested SNPs, even in the APOE-TOMM40 region, showed genome-wide significant association (Fig. 4A). The SNPs that tag APOE ɛ4 (e.g. rs769449) showed a P-value of 9.58 × 10−7. However, as explained above, we have shown that the APOE genotype, determined by two SNPs, is genome-wide significant in this data set (P = 6.9 × 10−13, Fig. 2B). SNPs in five different chromosomal regions showed a more significant or similar association than the SNPs on the GWAS chip that tag APOE ɛ4. Another four chromosomal regions were suggestive of an association (P< 1 × 10−6) with CSF APOE protein levels (Table 3). The fact that there are nine loci showing a similar P-value to rs2075650 (APOE), which we know is tagging a real signal, and that two of those signals are located in genes with similar function (PNPLA5 and PLA2G4E are both phospholipases) may indicate that the association of some of these SNPs are not a type I error.
Figure 4.
The Manhattan plot and the Q–Q plot for the GWAS for CSF APOE levels. (A) Within each chromosome, shown on the x-axis, the results are plotted left to right from the p-terminal end. Horizontal dashed lines indicate P-value thresholds of 1 × 10–5 and 5 × 10–8 (genome-wide significance). (B) Quantile–quantile (Q–Q) plot. No evidence of systematic inflation of P-values was found (λ = 1.007). The plots compare additive model statistics to those expected under the null distribution using fixed-effects for all analyzed imputed SNPs passing quality control criteria in the studies.
Table 3.
GWAS top hits for CSF APOE levels
| CHR | SNP | P-value | MAF | Gene | Location | Imputed/genotyped |
|---|---|---|---|---|---|---|
| 22 | rs138097 | 1.68E − 07 | 0.29 | SULT4A1 | Intron | Imp. |
| 22 | rs138062 | 2.34E − 07 | 0.29 | SULT4A1 | Intron | Imp. |
| 22 | rs138079 | 8.60E − 07 | 0.15 | SULT4A1 | Intron | Imp. |
| 22 | rs138087 | 8.60E − 07 | 0.15 | SULT4A1 | Intron | Imp. |
| 22 | rs80628 | 8.60E − 07 | 0.15 | SULT4A1 | Intron | Imp. |
| 22 | rs80637 | 8.60E − 07 | 0.15 | SULT4A1 | Intron | Imp. |
| 22 | rs470088 | 8.60E − 07 | 0.15 | SULT4A1 | Intron | Imp. |
| 22 | rs138064 | 1.25E − 06 | 0.14 | SULT4A1 | Intron | Imp. |
| 22 | rs138069 | 1.25E − 06 | 0.15 | SULT4A1 | Intron | Imp. |
| 22 | rs129655 | 1.38E − 06 | 0.15 | 5’ PNPLA5 | Imp. | |
| 22 | rs470093 | 1.64E − 06 | 0.15 | PNPLA5 | utr-3 | 1.67E − 06 |
| 22 | rs138084 | 1.70E − 06 | 0.15 | SULT4A1 | Intron | Imp. |
| 22 | rs5764317 | 2.22E − 06 | 0.14 | 5’ SULT4A1 | Imp. | |
| 22 | rs138053 | 2.22E − 06 | 0.14 | 5’ SULT4A1 | Imp. | |
| 22 | rs138086 | 2.28E − 06 | 0.15 | SULT4A1 | Intron | Imp. |
| 22 | rs80627 | 2.28E − 06 | 0.27 | SULT4A1 | Intron | Imp. |
| 22 | rs138127 | 2.39E − 06 | 0.15 | 5’ PNPLA5 | Imp. | |
| 22 | rs138105 | 4.90E − 06 | 0.27 | SULT4A1 | Intron | Imp. |
| 22 | rs138125 | 5.72E − 06 | 0.16 | 5’ PNPLA5 | Imp. | |
| 22 | rs138119 | 9.05E − 06 | 0.16 | 5’ PNPLA5 | Imp. | |
| 19 | rs769449 | 9.58E − 07 | 0.19 | APOE | Intron | Imp. |
| 19 | rs2075650 | 1.67E − 06 | 0.21 | TOMM40 | Intron | 1.71E − 06 |
| 19 | rs34342646 | 3.02E − 06 | 0.19 | PVRL2 | Intron | Imp. |
| 19 | rs12972970 | 3.24E − 06 | 0.19 | PVRL2 | Intron | Imp. |
| 19 | rs71352238 | 4.82E − 06 | 0.20 | 5’ TOMM40 | Imp. | |
| 19 | rs12972156 | 4.95E − 06 | 0.19 | PVRL2 | Intron | Imp. |
| 19 | rs34404554 | 5.44E − 06 | 0.21 | TOMM40 | Intron | Imp. |
| 19 | rs11556505 | 5.72E − 06 | 0.21 | TOMM40 | Synonymous | Imp. |
| 3 | rs6441306 | 1.22E − 06 | 0.44 | 5’ IFT80 | Imp. | |
| 11 | rs1793382 | 1.28E − 06 | 0.38 | Gene desert | 1.06E − 06 | |
| 11 | rs1690600 | 1.62E − 06 | 0.38 | Gene desert | Imp. | |
| 2 | rs12470837 | 1.37E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs12469076 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs9711604 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs12473669 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs10928155 | 2.20E − 06 | 0.17 | 3’ MGC50273 | 2.23E − 06 | |
| 2 | 2:132588372 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | 2:132586866 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs4558626 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs2314388 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs6709732 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs6740860 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 2 | rs6709893 | 2.20E − 06 | 0.17 | 3’ MGC50273 | Imp. | |
| 4 | rs283411 | 5.07E − 06 | 0.07 | ADH1C | Intron | Imp. |
| 4 | rs283404 | 6.86E − 06 | 0.07 | 3’ ADH1C | Imp. | |
| 4 | rs166892 | 6.86E − 06 | 0.07 | 3’ ADH1C | Imp. | |
| 4 | rs1229852 | 6.86E − 06 | 0.07 | 3’ ADH1C | Imp. | |
| 4 | rs283422 | 6.86E − 06 | 0.07 | 3’ ADH1C | Imp. | |
| 4 | rs283421 | 6.86E − 06 | 0.07 | 3’ ADH1C | Imp. | |
| 4 | rs283420 | 6.86E − 06 | 0.07 | 3’ ADH1C | Imp. | |
| 4 | rs72681907 | 7.04E − 06 | 0.07 | 3’ ADH1C | Imp. | |
| 4 | rs283418 | 7.42E − 06 | 0.07 | 3’ ADH1C | Imp. | |
| 4 | rs284780 | 8.36E − 06 | 0.08 | 3’ ADH1C | Imp. | |
| 15 | rs12232304 | 7.50E − 06 | 0.07 | PLA2G4E | Intron | 4.39E − 06 |
| 10 | rs947690 | 8.62E − 06 | 0.25 | 5’ RET | Imp. | |
| 10 | rs3004258 | 8.94E − 06 | 0.25 | 5’ RET | Imp. | |
| 10 | rs1547930 | 9.43E − 06 | 0.25 | 5’ RET | Imp. | |
| 7 | rs73310519 | 8.86E − 06 | 0.10 | PDE1C | Intron | Imp. |
| 7 | rs9942607 | 9.39E − 06 | 0.48 | 3’ FZD1 | Imp. | |
| 7 | rs10252817 | 9.39E − 06 | 0.48 | 3’ FZD1 | 9.42E − 06 | |
| 7 | rs9942608 | 9.39E − 06 | 0.48 | 3’ FZD1 | Imp. | |
| 12 | rs2101238 | 9.56E − 06 | 0.21 | NOS1 | Intron | Imp. |
The table shows the chromosome (chr.) and rs. number, the P-value, minor allele frequency (MAF), gene, SNP location respect to the gene and whether the SNPs was imputed or directly genotyped for the most significant hits in the CSF APOE GWAS analysis. If the SNP was directly genotyped, the P-value for the direct genotype is shown.
To confirm whether other SNPs contribute to the phenotypic variance in CSF APOE protein levels, we used genomic-partitioning analysis (34) to estimate the proportion of phenotypic variance explained by genome-wide SNPs. Our analyses indicate that 49.7% of the total variability of CSF APOE protein levels can be explained by genetic factors. Our previous analyses indicate that CSF APOE protein levels are strongly associated with age and gender. When CSF APOE protein levels were corrected by age, gender and principal component factors, the genome-wide SNPs explain 72% of the remaining variability in CSF APOE levels. When SNPs located in the APOE region with a P < 1 × 10−5 were removed, the remaining genome-wide SNPs explain 64% of the variability in APOE protein levels. Similarly, when the APOE genotype was included as a covariate the rest of the SNPs explain 63% of the variability in CSF APOE levels. This indicates that the APOE locus explains between 8 and 9% of the CSF APOE levels. This value is very close to the amount of variability explained by the APOE genotype calculated above (8.2%, see section APOE genotype influences CSF and plasma APOE levels but not APOE mRNA levels). In addition, our analysis indicates that the SNPs with a P-value < 1 × 10−6 explain an additional 10.52% of the variability in CSF APOE levels, which is higher than the APOE genotype (8.2%). Finally, we performed a pathway analysis of the top hits (Table 3) discarding the SNPs located in the APOE gene region. We used the SNP function portal to perform gene set enrichment analysis (GSEA). Interestingly, the most significant GSEA node was ‘cellular lipid metabolism’ (P = 4.49 × 10−9; Supplementary Material, Table S1). These results strongly suggest that some of these SNPs may constitute a real signal for CSF APOE levels. However, our study is underpowered to identify such variants at genome-wide significant levels.
For plasma APOE protein levels only rs2075650, passed the genome-wide significant threshold (P = 1.91 × 10−17, Supplementary Material, Fig. S3). As in the case of CSF APOE levels, the full APOE genotype showed a much more significant association with a P-value of 4.4 × 10−33. None of the SNPs with a P<1 × 10−4 was in common between the GWAS for plasma and CSF APOE protein levels, suggesting that APOE production and regulation in plasma and CSF may be different and that genetic variants has different effect in this different tissues.
DISCUSSION
To our knowledge, this is the largest study to examine CSF APOE protein levels in a human population and to determine what role APOE genotype has on influencing these levels. In total, we had access to CSF and plasma APOE protein levels from 641 individuals and genome-wide genotype data from 570 of these samples.
There are several important points that should be highlighted regarding the potential of CSF APOE protein levels as a relevant endophenotype for AD. First there is a very low correlation between plasma and CSF protein APOE levels (Fig. 1A) confirming the previous assumption that APOE synthesized in the periphery does not cross the blood–brain barrier to any large degree (35). There is great interest in identifying novel biomarkers for AD, and a lot of effort has been expended on the identification of plasma biomarkers because plasma is easier to obtain than CSF (14,22). Previous studies suggest that plasma APOE may be a potential biomarker for AD (22). In those studies, plasma APOE protein levels were found to be lower in AD cases compared with controls, but most of this association was driven by the APOE genotype. Additionally, a weak association between plasma APOE protein levels and amyloid in the brain as determined by a positive PIB signal (P = 0.016) was observed. In our study, we found similar results; the plasma APOE levels tend to be lower in individuals with AD (P = 0.018, and P = 0.421, without and with the APOE genotype as a covariate, Fig. 1B) and in individuals with fibrillar Aβ deposition (P = 8.39 × 10−7, and P = 0.071, without and with the APOE genotype as a covariate, Fig. 1C). Plasma APOE levels also showed a trend to be correlated with CSF Aβ42 levels (P = 2.63 × 10−7, and 0.13, without and with the APOE genotype as a covariate). However, we found that the CSF APOE levels showed a much stronger association with fibrillar Aβ brain deposition (P = 7.9 × 10−9, and P = 8.15 × 10−4, without and with the APOE genotype as a covariate, Fig. 1E) and more significant correlation with CSF Aβ42 levels (P = 3.11 × 10−14 and P = 2.84 × 10−8, without and with the APOE genotype as a covariate), than plasma APOE levels, even when the APOE genotype was included as a covariate. These results suggest that CSF APOE protein levels are more powerful than plasma levels as a potential endophenotype for AD.
CSF Aβ42 levels are a well-established biomarker for AD and several reports have demonstrated a reduction in CSF Aβ42 levels are significantly correlated to AD. Moreover, the changes in CSF Aβ42 levels, can be used to predict disease progression, progression rate and are also correlated with the presence of amyloid in the brain as determined by PIB binding (27,36–38). The identification of factors modifying CSF Aβ42 levels could facilitate a greater understanding of the pathogenic mechanisms involved in AD and potentially identify new therapeutic targets/pathways. We, and others have reported that the APOE genotype is the main genetic factor associated with CSF Aβ42 levels (13,15). In this study, we demonstrate that APOE levels are associated with CSF Aβ42 levels independently of the APOE genotype and when both the genotype and protein levels are included in the same model, the additional variability in the CSF Aβ42 levels is explained. In our analysis an additional 4% of the variability in CSF Aβ42 levels is explained by the APOE protein levels. We also used Mendelian randomization to analyze whether CSF APOE levels are actually in the causal pathway for AD. We used the most significant SNPs for CSF APOE levels and a genetic score as instrumental variables. Examination of F-statistics from the first-stage regressions to evaluate the strength of the instruments [all F-statistics were greater than 10 (range 10.37–18.39)] indicated sufficient strength to ensure the validity of instrumental variable methods in these data. Although these results suggest that CSF APOE levels are part of the causal pathway for AD, our analyses do not preclude potential that APOE ɛ2,3,4 genotype exhibits pleiotropic effects on both CSF APOE levels and AD status. To avoid this problem, we created a genetic score without the SNPs in the APOE region to use it as instrumental variable. In this analysis, we also found a significant association between CSF APOE levels and CDR. The results of the instrumental variable analysis provide a non-confounded and unbiased association between CSF APOE levels and CSF Aβ42 levels, CDR and case–control status. In all the trials, we found a strong association and correlation of CSF APOE with CSF Aβ42 levels confirming that this association is not a reverse causation or confounding effect, supporting the hypothesis that APOE protein levels affect the risk for AD by affecting Aβ pathology. These results suggest that CSF APOE protein levels could be a useful endophenotype for AD, and that by identifying the genetic factors that modify APOE protein levels, it may be possible to influence, indirectly, the Aβ42 pathology and the risk for AD.
Although CSF and plasma APOE levels show a low correlation (R2= 0.01), in both cases the APOE genotype is strongly associated with APOE protein levels (Fig. 1), and this association is stronger in the plasma. In fact a large portion of the variability in plasma APOE is explained by the APOE genotype (R2= 0.177). The association of the APOE genotype with APOE protein levels may be driven by the fact the APOE isoforms show different affinities for the LDL receptor. There are several reports supporting this hypothesis. It has been reported that APOE4 and APOE3 readily bind to the low-density lipoprotein receptor, whereas APOE2 binds poorly to the LDL receptor (39–41). This hypothesis also would explain why we found a strong association of the APOE genotype with protein levels but not with gene expression. In addition, since APOE receptors are highly expressed in the brain and the liver, this could explain why the APOE genotype is strongly associated with CSF and plasma levels, but not with gene expression.
This is the first time that the APOE genotype has been reported to be associated with CSF APOE protein levels (P = 6.9 × 10−13; Fig. 1). There are several studies in which the association of the APOE genotype with APOE plasma levels has been consistently reported (16–21). In contrast, previous studies found no association between APOE genotype and CSF APOE levels including a report from our group (14,16,18,19). One of the reasons for this lack of association may be the smaller sample size used in those studies. Another possible explanation is that the earlier studies used different antibodies to quantify the APOE protein levels. We had access to APOE protein levels measured in the same individuals by ELISA [reported by Wahrle et al. (24)] and by the rules based medicine (RBM) assay used in this study (n = 22). We found a very low correlation between the APOE protein levels measured using these different assays (R2= 0.09). To determine which method more accurately measures the APOE protein levels, we used mass-spectrometry to quantify APOE in the same samples. We found a strong correlation between the mass-spectrometry and RBM data (R2= 0.79; n = 27), but not with the mass-spectrometry and the ELISA data (R2= 0.22; n = 55). These results indicate that the RBM assay more accurately reflects the APOE protein levels and explains why we did not find the association of the APOE genotype with protein levels in our previous study. It also highlights the importance of validating the assays using independent methods.
From a genetic point of view, the most interesting finding was that the APOE genotype shows a genome-wide significant association with CSF APOE protein levels (P = 6.9 × 10−13; Fig. 1), but single-SNP analysis using SNPs on the GWAS chip detected a much weaker association in this region (P = 9.58 × 10−7; Fig. 3 and Table 3). This result indicates that the two SNPs that encode the APOE genotype capture more genetic information than the single SNPs. Similar effects could be happening in other regions of the genome and could explain part of the missing heritability in AD. Finally, in our genome-wide analysis, we found several other SNPs (Table 3) that could represent real signals, but did not reach genome-wide significance. Our genome partitioning analysis indicates that a large proportion (72%) of the variability in CSF APOE levels is explained by SNPs other than the APOE genotype. Specifically the SNPs (nine different loci) that showed a P-value < 1 × 10−6 explain 10% of the variability of CSF APOE. In addition our pathway analysis indicates that these loci are involved in lipid metabolism (Supplementary Material, Table S1). Specifically PNPLA5, ADH1C and NOS1 have been shown to be involved in lipid metabolism regulation (42) or genetic variants in these genes are associated with the risk for cardiovascular disease (43) or stroke (44). All of these results strongly suggest that some of these signals are real. However, it will be necessary to perform genetic analyses in a larger series of samples with CSF APOE levels and GWAS data, or alternatively to perform in vitro experiments to determine whether the candidate genes identified in this study alter APOE levels.
In summary, we demonstrate that CSF APOE protein levels, but not plasma APOE levels, are associated with lower CSF Aβ42 levels, independent of the APOE genotype, suggesting that CSF APOE levels may be a useful endophenotype for studies of AD. Furthermore, our genetic studies show that APOE genotype explains only a small proportion of the genetic variance in CSF levels and identifies some promising candidate genes that may also influence CSF APOE levels.
MATERIALS AND METHODS
Subjects
Association with CSF and plasma APOE levels was tested in a series of 331 samples from the Knight-ADRC and 310 samples from ADNI (see ADNI materials and methods). APOE protein levels were determined using Rules Based Medicine (Human Discovery MAP, v1.0). For the Knight-ADRC samples CSF and plasma were collected as described previously (27,36,45). Cases received a diagnosis of dementia of the Alzheimer's type, using criteria equivalent to the National Institute of Neurological and Communication Disorders and Stroke-Alzheimer's Disease and Related Disorders Association for probable AD (46,47). Controls received the same assessment as the cases but were non-demented. All individuals were of European descent and written consent was obtained from all participants.
Expression studies were carried out using cDNA obtained from tissue samples that represented the parietal lobe from 82 AD cases and 39 cognitively normal individuals (CDR = 0) obtained through the Knight-ADRC Neuropathology Core. A summary of the demographics of all subjects is shown in Table 1.
Genotyping
Rs7412 and rs429358 which define the APOE ε2/ε3/ε4 isoforms were genotyped using Taqman genotyping technology, as previously described (13,33,48–52). The poly-T repeat located in intron 6 of TOMM40 was genotyped as previously described (33). In each plate, we also included positive controls representative of each genotype and several samples were included on different plates and on different days to confirm genotype. The genotyping success rate was >98% with a reproducibility rate that was greater than 99%.
For 278 Knight-ADRC samples, we had access to genome-wide genotype data. These samples were genotyped with the Illumina 610 or the Omniexpress chip. The ADNI samples (n = 292) were genotyped with the Illumina 610 chip. Prior to association analysis, all samples and genotypes underwent stringent QC. Genotype data were cleaned by applying a minimum call rate for SNPs (98%) and individuals (98%) and minimum minor allele frequencies (MAF) (0.02). SNPs not in the Hardy–Weinberg equilibrium (P< 1 × 10−6) were excluded. For the Knight-ADRC samples, which were genotyped on two chips, genotype and sample quality thresholds were applied within the subset of the individuals genotyped on each chip.
We tested for unanticipated duplicates and cryptic relatedness using pairwise genome-wide estimates of proportion identity-by-descent. When a pair of identical samples or a pair of samples with cryptic relatedness was identified, the sample from the Knight-ADRC or samples with a higher number of SNPs passing QC were prioritized. Eigenstrat (53) was used to calculate principal component factors for each sample and confirm the ethnicity of the samples.
Imputation
The 1000 genome data (June 2011 release) and the Beagle software were used to impute up to 6 million SNPs. SNPs with a Beagle R2 of 0.3 or lower, an MAF lower than 0.05, out of the Hardy–Weinberg equilibrium (P< 1 × 10−6), a call rate lower than 95% or a Gprobs score lower than 0.90 were removed. A total of 5 815 690 SNPs passed the QC process.
Gene expression
Quantification of gene expression was done by real-time PCR as previously described (13).
Total RNA was extracted from a tissue sample representing the parietal lobe of 82 AD cases and 39 non-demented individuals, using the RNeasy mini kit (Qiagen) following the manufacturer's protocol. cDNA was prepared from total RNA, using the High-Capacity cDNA Archive kit (ABI). Gene expression was analyzed by real-time PCR, using an ABI-7500 real-time PCR system. TaqMan assays were used to quantify TOMM40 [Hs01587378_mh, exon boundary 4–5 (NM-001128916.1 )] and APOE [Hs00171168_m1, exon boundary 3–4 (NM_000041.2)] mRNA levels. Primers and TaqMan probe for the reference gene, GAPDH, were designed over exon–exon boundaries, using Primer Express software, Version 3 (ABI) (sequences available on request). Cyclophilin A (ABI: 4326316E) was also used as the reference gene. Each real-time PCR run included within-plate duplicates and each experiment was performed, at least twice for each sample. Real-time data were analyzed using the comparative Ct method. The Ct values for each sample were normalized with the Ct value for the housekeeping genes, GADPH and cyclophilin, and were corrected for the PCR efficiency of each assay (54), although the efficiency of all reactions was close to 100%. Only samples with a standard error of <0.15% were analyzed. We calculated the mean between the gene expression normalized with GADPH and cyclophilin. TOMM40 expression normalized by GADPH and cyclophilin showed a correlation coefficient of 0.77 (P = 4.20 × 10−22), whereas APOE expression normalized by GADPH and cyclophilin showed a correlation coefficient of 0.62 (P = 2.71 × 10−12).
Statistical analyses
Association tests between APOE genotype and CSF and plasma APOE protein levels and APOE mRNA expression were performed using SAS v9.2 and PLINK. For the statistical analyses, the APOE genotype was re-coded as follows ε2/2= 0, ε2/3= 1, ε3/3= 2, ε2/4= 3, ε3/4= 4 and ε4/4= 5.
Association with gene expression
APOE mRNA expression values were log transformed to approximate a normal distribution. Analysis of the covariance (ANCOVA) was used to test for association between APOE genotype and gene expression levels. Stepwise regression analysis was used to determine the significant covariates (age, gender, postmortem interval, APOE genotype and CDR).
Association with CSF and plasma APOE levels
Association with CSF and plasma APOE levels was performed as previously reported (13,15,33,50,51,55). Briefly, plasma and CSF APOE protein levels were transformed to approximate a normal distribution. Stepwise regression analysis was used to determine the significant covariates for each tissue and series. Age and gender showed a significant association with CSF APOE protein values and were included in the model. Gender but not age showed a significant association with plasma APOE protein values and was included in the model. The residuals for each series resulting after correction by the covariates were then analyzed together. No significant differences in the residuals from the different series were found, indicating that the differences in the CSF APOE protein levels due to the use of different platforms were corrected by using the residuals. The normalized CSF and plasma APOE values were tested for association using an additive model in PLINK, including age, gender and principal component factors as covariates. The genomic inflation factor for the GWAS was 1, indicating that there is no inflation of the P-values due to population stratification or a non-normal distribution of the trait.
Mendelian randomization
Mendelian randomization is a form of instrumental variable analysis in which genotypes, which can be assumed to be independent of environmental exposures, are used as the instrumental variables. The current system is an ideal case for the application of Mendelian randomization as APOE e2,3,4 genotype is strongly and reliably associated with CSF APOE levels, but completely independent of factors that may confound the relationship between CSF APOE levels and both CSF Aβ levels and clinical status.
We used instrumental variable methods to obtain estimates of the causal (non-confounded and unbiased) association between CSF APOE levels and CSF Aβ42 levels, CDR and case–control status. We initially used CDR at lumbar puncture for the analysis. As CSF Aβ42 levels were used as a proxy for Aβ deposition, and presyntomatic individuals, we also used CDR at the last assessment. For the ADNI samples, the CDR at the last assessment was taken 36 months after the LP. The average follow-up time for the Knight-ADRC samples was 3.2 years (13).
We used as instrument the APOE genotype or rs2075650, which were the SNPs explaining the largest proportion of CSF APOE in our sample (see GWAS analysis and results). We also used a genetic score as instrument. We constructed a ‘CSF APOE genetic score’ combining the CSF APOE raising alleles at each of top typed seven loci for our GWAS analysis (Table 3). We also created a genetic score without the SNP in the APOE gene signal to eliminate potential pleiotropic effects of the APOE genotype with a risk for AD and CSF APOE levels. The genetic risk score was defined as before (56). Age, gender, series and CDR were included in the model as covariates. We used two-stage least squares to fit the instrumental variable models in the main analyses, but checked results with limited information maximum likelihood and the generalized method of moments (57). We examined F-statistics from the first-stage regressions to assess the strength of the instruments. The F-statistic in a simple linear regression model is derived from the proportion of the variation explained by the genetic variant in the phenotype given the sample size. Values greater than 10 are often taken to indicate sufficient strength to ensure the validity of instrumental variable methods (58). This approach is analogous to methods used in randomized clinical trials to infer an unbiased on-treatment effect. All analyses were performed with SAS statistical software, version 9.1 (SAS Institute, Cary, USA).
Bioinformatics analyses
We used Pupasuite (59), the SNP Function Portal (http://brainarray.mbni.med.umich.edu/Brainarray/Database/SearchSNP/) and the SNP and CNV Annotation Database (http://www.scandb.org) to perform the SNP annotation and to identify the putative functional SNPs. We also used the SNP function portal to perform the Gene-ontology pathway analysis.
Genome partitioning: We used the algorithm GCTA (genome-wide complex trait analysis) to estimate the proportion of phenotypic variance explained by genome-wide SNPs (34).
ADNI materials and methods
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.ucla.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The principal investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California—San Francisco. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the USA and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55–90, to participate in the research, approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early AD to be followed for 2 years.
CSF and plasma APOE levels from the ADNI series were also measured using the rules based medicine panel. Clinical and biochemical data were obtained for 292 samples from the ADNI website. For up-to-date information, see www.adni-info.org.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. The authors have read the journal's policy and have the following conflicts: K.B. and E.H.P. are employed by Pfizer. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
FUNDING
This work was supported by Pfizer and grants from the National Institutes of Health (P30-NS069329-01, R01-AG035083, P50 AG05681) and the Alzheimer's Association (MNIRG-11-205368). Some of the samples used in this study were genotyped by the ADGC and GERAD. ADGC is supported by grants from the NIH (#U01AG032984) and GERAD from the Wellcome Trust (GR082604MA) and the Medical Research Council (G0300429).
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec, Inc.; Bristol-Myers Squibb Company; Eisai, Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer, Inc.; Servier; Synarc, Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the Rev August 16 2011 University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514 and the Dana Foundation.
Supplementary Material
REFERENCES
- 1.Bertram L., McQueen M., Mullin K., Blacker D., Tanzi R. (Accessed 1/26/2012) The AlzGene Database. Alzheimer Research Forum. Available at: http://www.alzgene.org . [Google Scholar]
- 2.Kauwe J.S., Wang J., Mayo K., Morris J.C., Fagan A.M., Holtzman D.M., Goate A.M. Alzheimer's disease risk variants show association with cerebrospinal fluid amyloid beta. Neurogenetics. 2009;10:13–17. doi: 10.1007/s10048-008-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmechel D.E., Saunders A.M., Strittmatter W.J., Crain B.J., Hulette C.M., Joo S.H., Pericak-Vance M.A., Goldgaber D., Roses A.D. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90:9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris J.C., Roe C.M., Xiong C., Fagan A.M., Goate A.M., Holtzman D.M., Mintun M.A. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann. Neurol. 2010;67:122–131. doi: 10.1002/ana.21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fryer J.D., Demattos R.B., McCormick L.M., O'Dell M.A., Spinner M.L., Bales K.R., Paul S.M., Sullivan P.M., Parsadanian M., Bu G., et al. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J. Biol. Chem. 2005;280:25754–25759. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- 6.Fryer J.D., Taylor J.W., DeMattos R.B., Bales K.R., Paul S.M., Parsadanian M., Holtzman D.M. Apolipoprotein E markedly facilitates age-dependent cerebral amyloid angiopathy and spontaneous hemorrhage in amyloid precursor protein transgenic mice. J. Neurosci. 2003;23:7889–7896. doi: 10.1523/JNEUROSCI.23-21-07889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strittmatter W.J., Weisgraber K.H., Huang D.Y., Dong L.M., Salvesen G.S., Pericak-Vance M., Schmechel D., Saunders A.M., Goldgaber D., Roses A.D. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kounnas M.Z., Moir R.D., Rebeck G.W., Bush A.I., Argraves W.S., Tanzi R.E., Hyman B.T., Strickland D.K. LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell. 1995;82:331–340. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- 9.Castellano J.M., Kim J., Stewart F.R., Jiang H., DeMattos R.B., Patterson B.W., Fagan A.M., Morris J.C., Mawuenyega K.G., Cruchaga C., et al. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci. Transl. Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina V., et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cruchaga C., Kauwe J.S., Mayo K., Spiegel N., Bertelsen S., Nowotny P., Shah A.R., Abraham R., Hollingworth P., Harold D., et al. SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer's disease. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001101. pii: e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauwe J.S., Jacquart S., Chakraverty S., Wang J., Mayo K., Fagan A.M., Holtzman D.M., Morris J.C., Goate A.M. Extreme cerebrospinal fluid amyloid beta levels identify family with late-onset Alzheimer's disease presenilin 1 mutation. Ann. Neurol. 2007;61:446–453. doi: 10.1002/ana.21099. [DOI] [PubMed] [Google Scholar]
- 15.Kauwe J.S., Cruchaga C., Mayo K., Fenoglio C., Bertelsen S., Nowotny P., Galimberti D., Scarpini E., Morris J.C., Fagan A.M., et al. Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proc. Natl. Acad. Sci. USA. 2008;105:8050–8054. doi: 10.1073/pnas.0801227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eto M., Watanabe K., Iwashima Y., Morikawa A., Oshima E., Sekiguchi M., Ishii K. Apolipoprotein E phenotypes and plasma lipids in young and middle-aged subjects. Tohoku. J. Exp. Med. 1986;148:25–34. doi: 10.1620/tjem.148.25. [DOI] [PubMed] [Google Scholar]
- 17.Gregg R.E., Zech L.A., Schaefer E.J., Stark D., Wilson D., Brewer H.B., Jr. Abnormal in vivo metabolism of apolipoprotein E4 in humans. J. Clin. Invest. 1986;78:815–821. doi: 10.1172/JCI112645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terrisse L., Poirier J., Bertrand P., Merched A., Visvikis S., Siest G., Milne R., Rassart E. Increased levels of apolipoprotein D in cerebrospinal fluid and hippocampus of Alzheimer's patients. J. Neurochem. 1998;71:1643–1650. doi: 10.1046/j.1471-4159.1998.71041643.x. [DOI] [PubMed] [Google Scholar]
- 19.Danik M., Champagne D., Petit-Turcotte C., Beffert U., Poirier J. Brain lipoprotein metabolism and its relation to neurodegenerative disease. Crit. Rev. Neurobiol. 1999;13:357–407. doi: 10.1615/critrevneurobiol.v13.i4.20. [DOI] [PubMed] [Google Scholar]
- 20.Fukumoto H., Ingelsson M., Garevik N., Wahlund L.O., Nukina N., Yaguchi Y., Shibata M., Hyman B.T., Rebeck G.W., Irizarry M.C. APOE epsilon 3/ epsilon 4 heterozygotes have an elevated proportion of apolipoprotein E4 in cerebrospinal fluid relative to plasma, independent of Alzheimer's disease diagnosis. Exp. Neurol. 2003;183:249–253. doi: 10.1016/s0014-4886(03)00088-8. [DOI] [PubMed] [Google Scholar]
- 21.Riddell D.R., Zhou H., Atchison K., Warwick H.K., Atkinson P.J., Jefferson J., Xu L., Aschmies S., Kirksey Y., Hu Y., et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J. Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta V.B., Laws S.M., Villemagne V.L., Ames D., Bush A.I., Ellis K.A., Lui J.K., Masters C., Rowe C.C., Szoeke C., et al. Plasma apolipoprotein E and Alzheimer disease risk: the AIBL study of aging. Neurology. 2011;76:1091–1098. doi: 10.1212/WNL.0b013e318211c352. [DOI] [PubMed] [Google Scholar]
- 23.Bekris L.M., Millard S.P., Galloway N.M., Vuletic S., Albers J.J., Li G., Galasko D.R., DeCarli C., Farlow M.R., Clark C.M., et al. Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J. Alzheimers Dis. 2008;13:255–266. doi: 10.3233/jad-2008-13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahrle S.E., Shah A.R., Fagan A.M., Smemo S., Kauwe J.S., Grupe A., Hinrichs A., Mayo K., Jiang H., Thal L.J., et al. Apolipoprotein E levels in cerebrospinal fluid and the effects of ABCA1 polymorphisms. Mol. Neurodegener. 2007;2:7. doi: 10.1186/1750-1326-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider J.A., Aggarwal N.T., Barnes L., Boyle P., Bennett D.A. The neuropathology of older persons with and without dementia from community versus clinic cohorts. J. Alzheimers Dis. 2009;18:691–701. doi: 10.3233/JAD-2009-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price J.L., Morris J.C. Tangles and plaques in nondemented aging and “preclinical” Alzheimer's disease. Ann. Neurol. 1999;45:358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 27.Fagan A.M., Mintun M.A., Mach R.H., Lee S.Y., Dence C.S., Shah A.R., LaRossa G.N., Spinner M.L., Klunk W.E., Mathis C.A., et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann. Neurol. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 28.Jagust W.J., Landau S.M., Shaw L.M., Trojanowski J.Q., Koeppe R.A., Reiman E.M., Foster N.L., Petersen R.C., Weiner M.W., Price J.C., et al. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tapiola T., Alafuzoff I., Herukka S.K., Parkkinen L., Hartikainen P., Soininen H., Pirttila T. Cerebrospinal fluid {beta}-amyloid 42 and tau proteins as biomarkers of Alzheimer-type pathologic changes in the brain. Arch. Neurol. 2009;66:382–389. doi: 10.1001/archneurol.2008.596. [DOI] [PubMed] [Google Scholar]
- 30.Nitsch D., Molokhia M., Smeeth L., DeStavola B.L., Whittaker J.C., Leon D.A. Limits to causal inference based on Mendelian randomization: a comparison with randomized controlled trials. Am. J. Epidemiol. 2006;163:397–403. doi: 10.1093/aje/kwj062. [DOI] [PubMed] [Google Scholar]
- 31.Didelez V., Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat. Methods Med. Res. 2007;16:309–330. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 32.Roses A.D., Lutz M.W., Amrine-Madsen H., Saunders A.M., Crenshaw D.G., Sundseth S.S., Huentelman M.J., Welsh-Bohmer K.A., Reiman E.M. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2010;10:375–384. doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cruchaga C., Nowotny P., Kauwe J.S., Ridge P.G., Mayo K., Bertelsen S., Hinrichs A., Fagan A.M., Holtzman D.M., Morris J.C., et al. Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch. Neurol. 2011;68:1013–1019. doi: 10.1001/archneurol.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J., Manolio T.A., Pasquale L.R., Boerwinkle E., Caporaso N., Cunningham J.M., de Andrade M., Feenstra B., Feingold E., Hayes M.G., et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linton M.F., Gish R., Hubl S.T., Butler E., Esquivel C., Bry W.I., Boyles J.K., Wardell M.R., Young S.G. Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J. Clin. Invest. 1991;88:270–281. doi: 10.1172/JCI115288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fagan A.M., Roe C.M., Xiong C., Mintun M.A., Morris J.C., Holtzman D.M. Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch. Neurol. 2007;64:343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- 37.Fagan A.M., Head D., Shah A.R., Marcus D., Mintun M., Morris J.C., Holtzman D.M. Decreased cerebrospinal fluid Abeta(42) correlates with brain atrophy in cognitively normal elderly. Ann. Neurol. 2009;65:176–183. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snider B.J., Fagan A.M., Roe C., Shah A.R., Grant E.A., Xiong C., Morris J.C., Holtzman D.M. Cerebrospinal fluid biomarkers and rate of cognitive decline in very mild dementia of the Alzheimer type. Arch. Neurol. 2009;66:638–645. doi: 10.1001/archneurol.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahley R.W., Huang Y., Rall S.C., Jr. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J. Lipid. Res. 1999;40:1933–1949. [PubMed] [Google Scholar]
- 40.Mamotte C.D., Sturm M., Foo J.I., van Bockxmeer F.M., Taylor R.R. Comparison of the LDL-receptor binding of VLDL and LDL from apoE4 and apoE3 homozygotes. Am. J. Physiol. 1999;276:E553–E557. doi: 10.1152/ajpendo.1999.276.3.E553. [DOI] [PubMed] [Google Scholar]
- 41.Altenburg M., Arbones-Mainar J., Johnson L., Wilder J., Maeda N. Human LDL receptor enhances sequestration of ApoE4 and VLDL remnants on the surface of hepatocytes but not their internalization in mice. Arterioscler. Thromb. Vasc. Biol. 2008;28:1104–1110. doi: 10.1161/ATVBAHA.108.164863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baulande S., Langlois C. Proteins sharing PNPLA domain, a new family of enzymes regulating lipid metabolism. Med. Sci. 2010;26:177–184. doi: 10.1051/medsci/2010262177. [DOI] [PubMed] [Google Scholar]
- 43.Drogan D., Sheldrick A.J., Schutze M., Knuppel S., Andersohn F., di Giuseppe R., Herrmann B., Willich S.N., Garbe E., Bergmann M.M., et al. Alcohol consumption, genetic variants in alcohol deydrogenases, and risk of cardiovascular diseases: a prospective study and meta-analysis. PLoS ONE. 2012;7:e32176. doi: 10.1371/journal.pone.0032176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manso H., Krug T., Sobral J., Albergaria I., Gaspar G., Ferro J.M., Oliveira S.A., Vicente A.M. Variants within the nitric oxide synthase 1 gene are associated with stroke susceptibility. Atherosclerosis. 2012;220:443–448. doi: 10.1016/j.atherosclerosis.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 45.Shaw L.M., Vanderstichele H., Knapik-Czajka M., Clark C.M., Aisen P.S., Petersen R.C., Blennow K., Soares H., Simon A., Lewczuk P., et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann. Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berg L., McKeel D.W., Jr, Miller J.P., Storandt M., Rubin E.H., Morris J.C., Baty J., Coats M., Norton J., Goate A.M., et al. Clinicopathologic studies in cognitively healthy aging and Alzheimer's disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch. Neurol. 1998;55:326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 47.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 48.Cruchaga C., Fernandez-Seara M.A., Seijo-Martinez M., Samaranch L., Lorenzo E., Hinrichs A., Irigoyen J., Maestro C., Prieto E., Marti-Climent J.M., et al. Cortical atrophy and language network reorganization associated with a novel progranulin mutation. Cereb. Cortex. 2009;19:1751–1760. doi: 10.1093/cercor/bhn202. [DOI] [PubMed] [Google Scholar]
- 49.Koch W., Ehrenhaft A., Griesser K., Pfeufer A., Muller J., Schomig A., Kastrati A. TaqMan systems for genotyping of disease-related polymorphisms present in the gene encoding apolipoprotein E. Clin. Chem. Lab. Med. 2002;40:1123–1131. doi: 10.1515/CCLM.2002.197. [DOI] [PubMed] [Google Scholar]
- 50.Kauwe J.S., Cruchaga C., Bertelsen S., Mayo K., Latu W., Nowotny P., Hinrichs A.L., Fagan A.M., Holtzman D.M., Goate A.M. Validating predicted biological effects of Alzheimer's disease associated SNPs using CSF biomarker levels. J. Alzheimers Dis. 2010;21:833–842. doi: 10.3233/JAD-2010-091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cruchaga C., Graff C., Chiang H.H., Wang J., Hinrichs A.L., Spiegel N., Bertelsen S., Mayo K., Norton J.B., Morris J.C., et al. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch. Neurol. 2011;68:581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruchaga C., Haller G., Chakraverty S., Mayo K., Vallania F.L., Mitra R.D., Faber K., Williamson J., Bird T., Diaz-Arrastia R., et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer's disease families. PLoS One. 2012;7:e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 54.Muller P.Y., Janovjak H., Miserez A.R., Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. 1376, 1378–1379. [PubMed] [Google Scholar]
- 55.Kauwe J.S., Cruchaga C., Karch C.M., Sadler B., Lee M., Mayo K., Latu W., Su'a M., Fagan A.M., Holtzman D.M., et al. Fine mapping of genetic variants in BIN1, CLU, CR1 and PICALM for association with cerebrospinal fluid biomarkers for Alzheimer's disease. PLoS ONE. 2011;6:e15918. doi: 10.1371/journal.pone.0015918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Voight B.F., Peloso G.M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M.K., Hindy G., Holm H., Ding E.L., Johnson T., et al. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet. 2012 doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawlor D.A., Harbord R.M., Sterne J.A., Timpson N., Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 58.Timpson N.J., Lawlor D.A., Harbord R.M., Gaunt T.R., Day I.N., Palmer L.J., Hattersley A.T., Ebrahim S., Lowe G.D., Rumley A., et al. C-reactive protein and its role in metabolic syndrome: Mendelian randomisation study. Lancet. 2005;366:1954–1959. doi: 10.1016/S0140-6736(05)67786-0. [DOI] [PubMed] [Google Scholar]
- 59.Conde L., Vaquerizas J.M., Dopazo H., Arbiza L., Reumers J., Rousseau F., Schymkowitz J., Dopazo J. PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic. Acids Res. 2006;34:W621–W625. doi: 10.1093/nar/gkl071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.