Abstract
Smads are signal mediators for the members of the transforming growth factor-β (TGF-β) superfamily. Upon phosphorylation by the TGF-β receptors, Smad3 translocates into the nucleus, recruits transcriptional coactivators and corepressors, and regulates transcription of target genes. Here, we show that Smad3 activated by TGF-β is degraded by the ubiquitin–proteasome pathway. Smad3 interacts with a RING finger protein, ROC1, through its C-terminal MH2 domain in a ligand-dependent manner. An E3 ubiquitin ligase complex ROC1-SCFFbw1a consisting of ROC1, Skp1, Cullin1, and Fbw1a (also termed βTrCP1) induces ubiquitination of Smad3. Recruitment of a transcriptional coactivator, p300, to nuclear Smad3 facilitates the interaction with the E3 ligase complex and triggers the degradation process of Smad3. Smad3 bound to ROC1-SCFFbw1a is then exported from the nucleus to the cytoplasm for proteasomal degradation. TGF-β/Smad3 signaling is thus irreversibly terminated by the ubiquitin–proteasome pathway.
INTRODUCTION
Cytokines of the transforming growth factor-β (TGF-β) superfamily are multifunctional proteins that regulate growth, differentiation, apoptosis, and morphogenesis of various types of cells (Roberts and Sporn, 1990). TGF-β and related factors bind to two different types of serine/threonine kinase receptors, termed type I and type II. Type I receptor is activated by type II receptor upon ligand binding and mediates specific intracellular signals. Smads are the central signal mediators of the TGF-β superfamily (Heldin et al., 1997; Massagué and Wotton, 2000). Among the three different classes of Smads, receptor-regulated Smads (R-Smads) directly interact with type I receptors and become activated through phosphorylation of the C-terminal SSXS motif. R-Smads then form heteromeric complexes with common-partner Smads (Co-Smads) and translocate into the nucleus. Nuclear Smad complexes bind to transcriptional coactivators (p300 and CBP) or corepressors (e.g., TGIF, c-Ski, and SnoN) and regulate transcription of target genes (Attisano and Wrana, 2000; Miyazono, 2000). Inhibitory Smads (I-Smads) interfere with the activation of R-Smads by the serine/threonine kinase receptors and the formation of R-Smad/Co-Smad complexes. Smad2 and Smad3 are R-Smads activated by TGF-β and activin receptors, whereas Smad1, Smad5, and presumably Smad8 are activated by bone morphogenetic protein (BMP) receptors. Smad4 is the only Co-Smad in mammals, and Smad6 and Smad7 are I-Smads.
Ubiquitin-dependent protein degradation plays a key role in various biological processes, including signal transduction, cell cycle progression, and transcriptional regulation (Hershko and Ciechanover, 1998). Ubiquitination of proteins is induced by the action of an E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases. In the ubiquitin–proteasome pathway, E3 ligases play a crucial role in the recognition of target proteins and subsequent protein degradation by the 26S proteasomes. SCF complexes composed of Skp1, Cullins, and F-box proteins are a class of E3 ubiquitin ligases that participate in the degradation of a number of regulatory proteins. In the SCF complex, Cullin interacts with Skp1, and Skp1 in turn binds to an F-box protein. Recruitment of different F-box proteins into the SCF complexes may be important for the specific ubiquitination of certain target proteins (Laney and Hochstrasser, 1999).
A RING finger protein, ROC1 (also termed Rbx1 or Hrt1), has been identified as a Cullin-binding protein (Ohta et al., 1999; Seol et al., 1999; Tan et al., 1999). It has also been identified as a component of the von Hippel-Lindau tumor suppressor complex, which contains Cullin2, elongin B, and elongin C (Kamura et al., 1999). ROC1 binds to all mammalian Cullins and also acts as an adaptor to E2-conjugating enzymes (Skowyra et al., 1999). ROC1 thus plays a crucial role in the ubiquitination of target proteins as a fourth subunit of the SCF E3 ligase complex.
Smad2 activated by TGF-β has been shown to be degraded by the ubiquitin–proteasome pathway after translocation into the nucleus (Lo and Massagué, 1999). However, E3 ligases responsible for the ubiquitination of TGF-β–activated Smads and the mechanism for the ubiquitination of nuclear Smad2/3 have not been fully elucidated. Here we show that Smad3 activated by TGF-β interacts with the ROC1-SCFFbw1a complex containing an F-box protein Fbw1a (also termed βTrCP1) (Cenciarelli et al., 1999; Winston et al., 1999a), which triggers the ubiquitination and proteasomal degradation of Smad3. Interaction of p300 with nuclear Smad3 facilitates the Smad3-ubiquitination. We also show that Smad3 associated with the E3 ligase complex is translocated from the nucleus to the cytoplasm, where Smad3 is degraded by the 26S proteasome. The ROC1-SCFFbw1a complex is an E3 ligase for IκB and β-catenin, which participate in the NF-κB– and Wnt/Wingless-signaling pathways, respectively. Our present findings revealed that the ROC1-SCFFbw1a complex is also involved in the TGF-β/Smad3–signaling pathway.
MATERIALS AND METHODS
Transfection and Pulse–Chase Analysis
Transient transfection of DNA was performed using FuGENE6 (Roche Diagnostics, Indianapolis, IN). Transfected COS7 cells or human HaCaT cells were labeled for 10 min or 2 h at 37°C with 50 μCi/ml [35S]methionine and cysteine (Amersham Pharmacia Biotech, Buckinghamshire, UK) in methionine- and cysteine-free DMEM and chased in DMEM supplemented with fetal bovine serum for the time periods indicated as described previously (Miyazono et al., 1991; Lo and Massagué, 1999). Cells were then lysed and subjected to immunoprecipitation.
Immunoprecipitation and Immunoblotting
For inhibition of proteasomal degradation, cells were incubated with 50 μM MG132 (Peptide Institute, Osaka, Japan) for 2–4 h unless indicated otherwise and lysed with Nonidet P-40 lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40) containing 50 μM MG132. Immunoprecipitation and immunoblotting were performed as described by Kawabata et al. (1998). For immunoprecipitation of Smad3, antibody specific to Smad3 (Korchynskyi et al., 1999; gift of P. ten Dijke) or anti-Smad2 antibody, which cross-reacts with Smad3 (Transduction Laboratories, Lexington, KY) were used.
Yeast Two-Hybrid Assay
Yeast two-hybrid assay was performed as previously described (Akiyoshi et al., 1999). Briefly, EGY48, the host yeast strain, was transformed with a combination of the reporter (pSH18-34), a bait (pEG202 or ROC1 in pEG202), and a prey (pJG4-5 or Smad3C in pJG4-5). Yeast was selected on appropriate growth media, and then three independent colonies were assayed for β-galactosidase activity on 5-bromo-4-chloro-3-indolyl-β- D-galactoside.
Binding of Smad3 to Biotinylated DNA
DNA-binding assay using biotinylated oligonucleotide was performed as described by Tada et al. (1999). Briefly, cells were treated or not with 3 ng/ml TGF-β in the presence or absence of MG132. Cell lysates were incubated with 30 pmol of biotinylated double-stranded 3xCAGA oligonucleotide and 12 μg poly dI-dC for 1 h. Proteins were precipitated with streptavidin-agarose for 30 min, washed, and detected by immunoblotting. For detection of ROC1 from nontransfected HaCaT cells, cell lysates from four 10-cm tissue culture dishes (Falcon, Becton Dickinson Labware, Franklin Lakes, NJ) were used, whereas those from a 10-cm tissue culture dish were used for detection of ROC1 from transfected COS7 cells.
Immunofluorescence Labeling
Immunohistochemical staining of Smad3C or full-length Smad3 in transfected COS7 cells was performed using anti-Myc, anti-FLAG, or anti-phospho-Smad3 antibodies followed by the incubation with fluorescein isothiocyanate-labeled goat anti-mouse immunoglobulin G as described by Ebisawa et al. (1999). Nuclei of the cells were stained by 4,6-diamidino-2-phenylindole. Intracellular localization was determined by confocal laser scanning microscopy.
RESULTS
Proteasomal Degradation of Activated Smad3
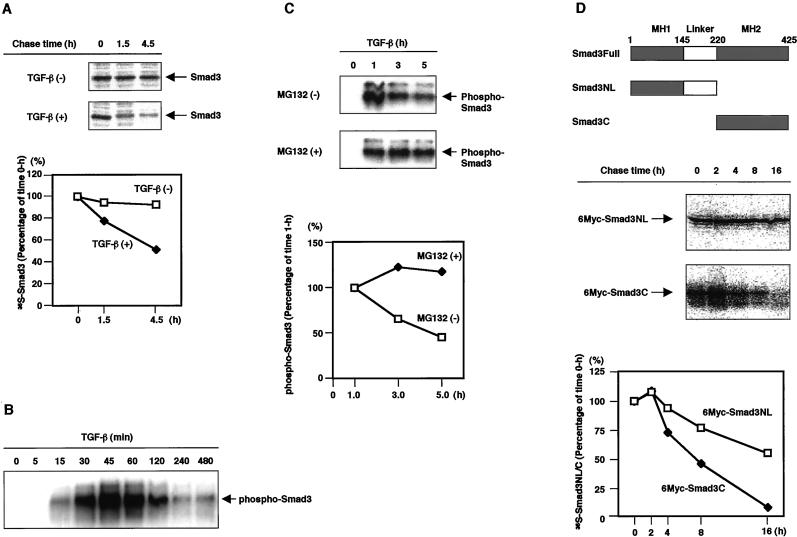
TGF-β potently inhibits the growth of HaCaT human keratinocyte cells and regulates the expression of various genes (Akiyoshi et al., 2001). To investigate the stability of the Smad3 protein, we first examined the turnover of endogenous Smad3 in HaCaT cells by pulse–chase analysis. Similar to Smad2 activated by TGF-β (Lo and Massagué, 1999), the half-life of Smad3 was shorter in the presence than in the absence of TGF-β (Figure 1A). Because the ligand stimulation was important for the rapid turnover of Smad3, we next investigated the turnover of Smad3 phosphorylated by TGF-β using an antibody specific to phospho-Smad3. Phosphorylated Smad3 was increased after TGF-β stimulation with a peak at 45 min and rapidly disappeared thereafter (Figure 1B). Similar to phosphorylated Smad2 (Lo and Massagué, 1999), dephosphorylation may not be the major reason for the rapid disappearance of phospho-Smad3, because a proteasome inhibitor MG132 almost completely abolished the decrease in the phospho-Smad3 protein (Figure 1C).
Figure 1.
TGF-β induces Smad3 turnover through proteasomal degradation. (A) Smad3 is more rapidly degraded in the presence than in the absence of TGF-β. HaCaT cells were metabolically labeled for 2 h, treated with TGF-β (5 ng/ml), and then chased for the time periods indicated. Cell lysates were immunoprecipitated by an anti-Smad3 antibody and analyzed by SDS-PAGE. The autoradiographic signals were quantified and the values were plotted relative to the 0-h values. (B) Rapid turnover of phosphorylated Smad3 upon TGF-β stimulation. HaCaT human keratinocytes were treated with TGF-β (5 ng/ml) for the indicated periods. Smad3 was then immunoprecipitated by an anti-Smad3 antibody (Transduction Laboratories), followed by immunoblotting using an antiphospho-Smad3 antibody (gift of P. ten Dijke). (C) Prevention of the rapid turnover of phosphorylated Smad3 by a proteasome inhibitor MG132. HaCaT cells were treated with TGF-β (5 ng/ml) in the presence or absence of 50 μM MG132. Phospho-Smad3 was detected as in B. The intensities of the immunobloted bands were quantified and the values were plotted relative to the 1-h values. (D) Comparison of the turnover of deletion mutants of Smad3. Structures of the Smad3 deletion mutants are shown (top). Full, NL, and C denote full-length, the MH1 domain and linker, and the MH2 domain, respectively. 6Myc-tagged Smad3NL or Smad3C in transfected COS7 cells was metabolically labeled for 10 min and then chased for the time periods indicated. Cell lysates were immunoprecipitated by a Myc antibody and analyzed by SDS-PAGE. The autoradiographic signals were quantified and plotted as in A.
Smurf1, a member of the HECT family of E3 ubiquitin ligases, ligand-independently induces the ubiquitination and degradation of BMP-specific Smad1 through binding to a PY motif in the linker region (Zhu et al., 1999). More recently, Smurf2, which is highly related to Smurf1, has been shown to interact with Smad2 through its PY motif upon TGF-β stimulation in vitro and leads to Smad2 degradation (Lin et al., 2000). We investigated whether the presence of the linker region is essential for the degradation of Smad3. The turnover of two deletion mutants of Smad3, Smad3NL containing the N-terminal MH1 domain and the linker region, and Smad3C containing the C-terminal MH2 domain, was compared in transfected COS7 cells by pulse–chase analysis. Interestingly, Smad3C lacking the linker region was less stable than Smad3NL (Figure 1D), and treatment with the proteasome inhibitor MG132 eliminated this degradation (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results). These findings suggest that, although Smurf-like molecules might be involved in the degradation of Smad3NL, other classes of E3 ligases may recognize the MH2 domain of Smad3 and regulate the ubiquitination and degradation of the TGF-β–activated Smad3.
Physical Interaction of Smad3 with ROC1
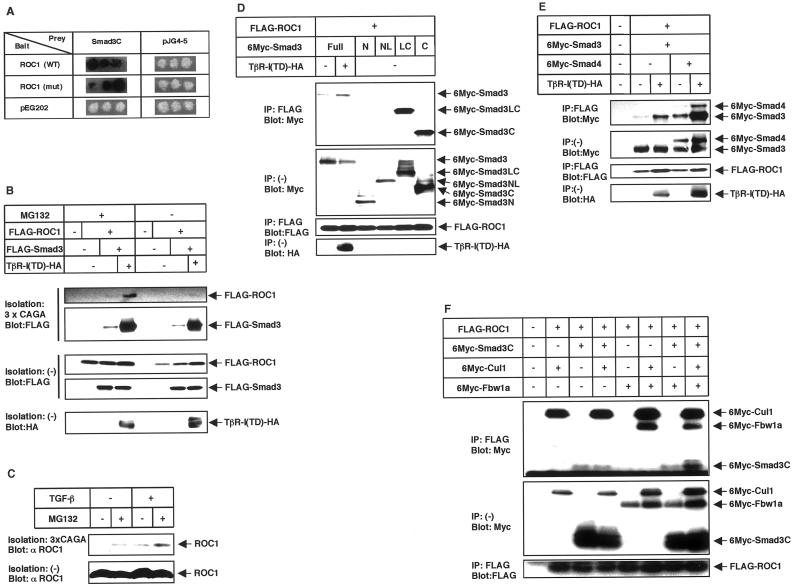
Of different E2-conjugating enzymes, UbcH5b and UbcH5c appear to participate in the ligand-dependent degradation of Smad2 (Lo and Massagué, 1999; Xu and Attisano, 2000) and Smad3 (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results). Because UbcH5 is involved in the ubiquitin ligase activity catalyzed by ROC1 (Iwai et al., 1999; Ohta et al., 1999), we studied the physical interaction of ROC1 with Smad3. We have found that ROC1 interacts with Smad3C in the yeast two-hybrid system (Figure 2A). A ROC1 mutant C75A/H77A, which has mutations in the RING finger domain and fails to recruit ROC1-associated ligase activity (Ohta et al., 1999), also interacted with Smad3C.
Figure 2.
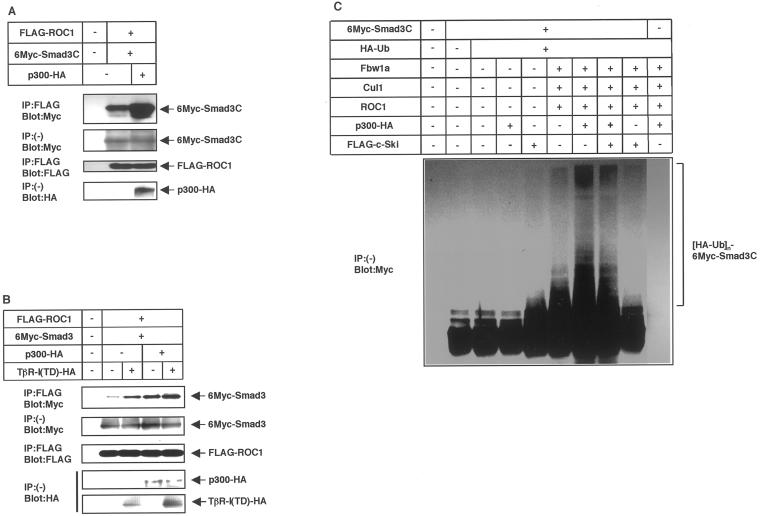
Physical interaction of ROC1 with Smad3. (A) Binding of Smad3C to ROC1 in the yeast two-hybrid system. Positive interactions yield a dark color on the indicator plates. Yeast with negative interaction remains white. Wild-type (WT) or a mutant (mut) form of ROC1 (C75A/H77A) were tested in the interaction assay. pEG202 and pJG4-5 are control vectors without inserts. (B) Interaction between Smad3 and ROC1 upon receptor stimulation in mammalian cells. COS7 cells were transfected with the indicated plasmids and lysed. Proteins were isolated by precipitation with a biotinylated oligonucleotide containing the Smad3-binding elements (3xCAGA), followed by immunoblotting (Blot) using FLAG antibody. The top two panels show the interaction. For the detection of ROC1, the film was exposed for a 15-times longer period than for that of Smad3. (C) Interaction of endogenous Smad3 with ROC1 in HaCaT human keratinocytes. HaCaT cells were treated with TGF-β (3 ng/ml), and ROC1 bound to Smad3 was detected by isolation of the biotinylated 3xCAGA DNA, followed by immunoblotting with a ROC1 antibody (gift of N. Shimbara). In C–F, the top panels show the interaction and the bottom panels show the expression of each protein. (D) Interaction of ROC1 with various Smad3 deletion constructs. In D–F, COS7 cells were transfected with the indicated plasmids and subjected to FLAG immunoprecipitation (IP) followed by Myc immunoblotting (Blot). Full, N, NL, LC, and C denote full-length, the MH1 domain, the MH1 domain and linker, the linker and MH2 domain, and the MH2 domain, respectively. (E) Smad4 does not affect the interaction of ROC1 with Smad3. Note that the expression level of TβR-I(TD) was higher in the presence than in the absence of Smad4 in this experiment. (F) Enhancement of the interaction between ROC1 and Smad3C in the presence of Cul1 and Fbw1a. Among the components of the ROC1-SCFFbw1a complex, Skp1 is abundantly expressed in COS7 cells. Therefore, it was not transfected in the present study.
The interaction between Smad3 and ROC1 was also examined in mammalian cells. When activated Smad3 was isolated using biotinylated DNA containing the Smad-binding elements (CAGA box; Dennler et al., 1998), ROC1 was coprecipitated with Smad3 upon stimulation by constitutively active TGF-β type I receptor [TβR-I(TD)] and the interaction between ROC1 and Smad3 was enhanced by the proteasome inhibitor MG132 (Figure 2B). Importantly, we also found that the interaction between ROC1 and Smad3 occurs under physiological conditions. ROC1 does not have an ability to bind to the CAGA DNA by itself (Figure 2B). However, when activated Smad3 was precipitated from nontransfected HaCaT cells by the biotinylated CAGA DNA, ROC1 was coprecipitated with Smad3 upon ligand stimulation (Figure 2C).
The interaction between Smad3 and ROC1 was also examined by immunoprecipitation followed by immunoblotting. Analysis using various Smad3 deletion mutants confirmed that Smad3 binds to ROC1 through the MH2 domain (Figure 2D). Because Smad3NL failed to interact with ROC1, and it was not ubiquitinated by ROC1-SCFFbw1a (see below; Figure 3E), we used Smad3C in some of the following experiments. Although Smad4 also interacts with Smad3 through the MH2 domain (Heldin et al., 1997), Smad4 did not interfere with the interaction of ROC1 with Smad3, and it was coimmunoprecipitated with Smad3 in the ROC1-immunoprecipitates (Figure 2E).
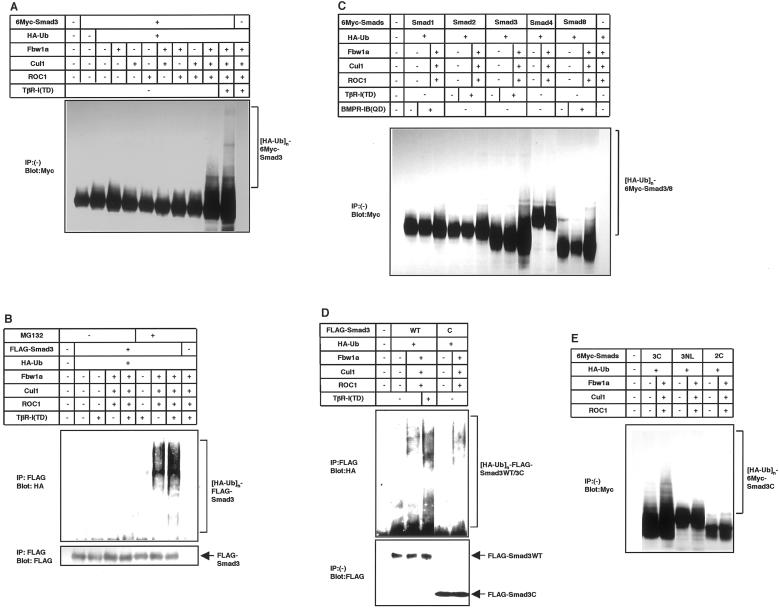
Figure 3.
Ubiquitination of Smad3 by the ROC1-SCFFbw1a complex in vivo. (A and B) Ligand-dependent ubiquitination of Smad3 is induced by the ROC1-SCFFbw1a complex. COS7 cells were transfected with the indicated plasmids. For the detection of ubiquitinated proteins, cells were treated with 50 μM MG132 for 4 h before cell lysis unless indicated. Lysates from cells were directly subjected to anti-Myc immunoblotting (A) or subjected to anti-FLAG immunoprecipitation followed by anti-HA immunoblotting (B). Expression levels of Smad3 are shown in the bottom (B). Polyubiquitination species of Smad3 are indicated ([HA-Ub]n-6Myc or FLAG-Smad3). (C) Smad3 and Smad8 are preferentially degraded by the ROC1-SCFFbw1a complex. COS7 cells were transfected with the indicated plasmids, and ubiquitination was determined as in A. A short version of Smad8 lacking a part of linker region was used in the present study. (D and E) Ubiquitination occurs on the MH2 domain of Smad3. In D, COS7 cells were transfected with the indicated plasmids and treated with MG132, and ubiquitination was detected as in B. Expression levels of wild-type (WT) Smad3 and Smad3C are shown at the bottom. In E, ubiquitination of Smad3C was compared with that of Smad3NL and Smad2C containing the MH2 domain of Smad2 in transfected COS7 cells as in A.
ROC1 is a component of the ROC1-SCFFbw1a complex. We found that Cul1 and Fbw1a, but not Skp1, weakly interact with Smad3C in transfected COS7 cells (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results). Moreover, we observed that, among several F-box proteins, only Fbw1a efficiently induced ubiquitination of Smad3 (see below; Figure 4D). We therefore cotransfected Cul1 and Fbw1a together with ROC1 and examined the interaction of ROC1 with Smad3C. Interestingly, coimmunoprecipitation of Smad3C with ROC1 was dramatically enhanced in the presence of Cul1 and Fbw1a (Figure 2F), suggesting that the other components of the ROC1-SCFFbw1a complex may facilitate the interaction of ROC1 with Smad3.
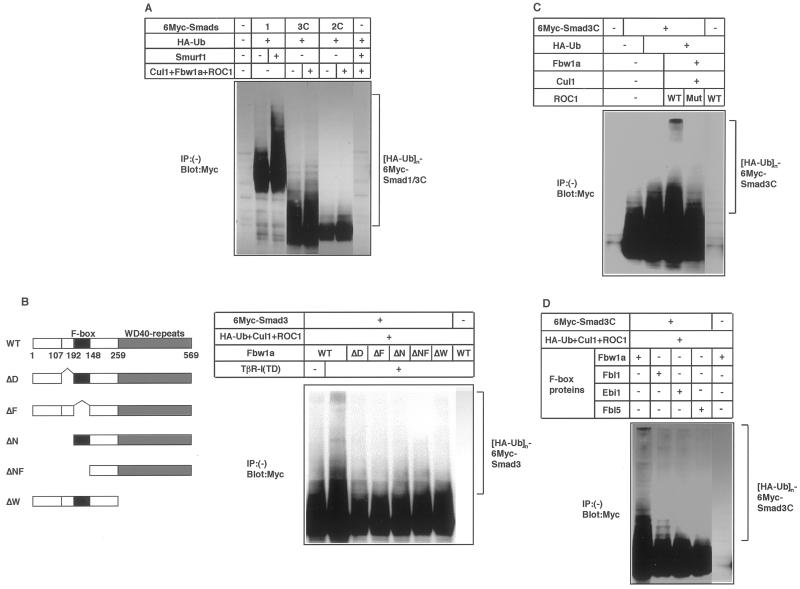
Figure 4.
Ubiquitination of Smad3 requires integrity of the ROC1-SCFFbw1a complex. (A) Comparison of the Smad1 ubiquitination by Smurf1 and Smad3 ubiquitination by ROC1-SCFFbw1a. 293T cells were transfected with the indicated plasmids and cell lysates were subjected to anti-Myc immunoblotting. (B) Ligand-dependent ubiquitination of Smad3 was examined using various Fbw1a mutants. Structures of the Fbw1a mutants are shown (left). Ubiquitination of Smad3C was determined in COS7 cells as in A. WT, D, F, N, NF, and W denote wild-type, dimerization domain, F-box, N-terminal domain, N-terminal domain and F-box, and WD40 repeats, respectively. (C) Ubiquitination of Smad3C by the wild-type (WT) or C75A/H77A mutant (mut) ROC1 was examined in COS7 cells. (D) Ubiquitination of Smad3C by various F-box proteins in the presence of ROC1 and Cul1 in COS7 cells. Fbw1a and hEbi1 are the F-box proteins with WD40 repeats, whereas Fbl1/Skp2 and Fbl5 have leucine-rich repeats.
ROC1-SCFFbw1a Induces Ubiquitination of Smad3
Ubiquitination of Smad3 by ROC1-SCFFbw1a was investigated in vivo. Smad3 was transfected into COS7 cells together with ROC1, Cul1, Fbw1a, and hemagglutinin (HA)-tagged ubiquitin. Although ROC1 alone did not efficiently induce the Smad3 ubiquitination (Figure 3A), addition of Cul1 and Fbw1a dramatically enhanced the ubiquitination, which was facilitated by TβR-I(TD). Ubiquitination of Smad3 by ROC1-SCFFbw1a was observed in the presence, but very weakly in the absence, of proteasome inhibitors MG132 (Figure 3B) or lactacystin (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results), indicating the proteasomal degradation of ubiquitinated Smad3. Ubiquitination of Smad3 was detected even in the absence of TβR-I(TD) (Figure 3, A and B), probably because Smad3 transfected into mammalian cells spontaneously translocates into the nucleus (Zhang et al., 1997; Xu et al., 2000; see also Figure 6B). The ubiquitinated species obtained by ubiquitin immunoblotting appeared to differ from those obtained by Smad3 immunoblotting (Figure 3, A and B), because the HA antibody directed to HA-ubiquitin may have preferentially reacted with polyubiquitinated Smad3.
Figure 6.
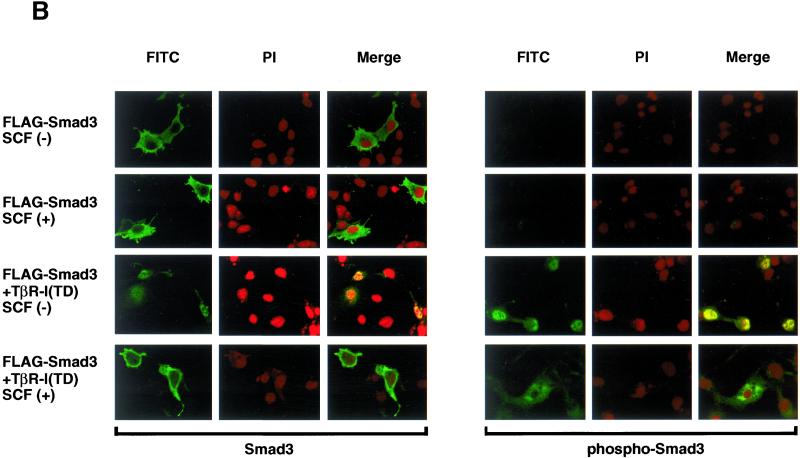
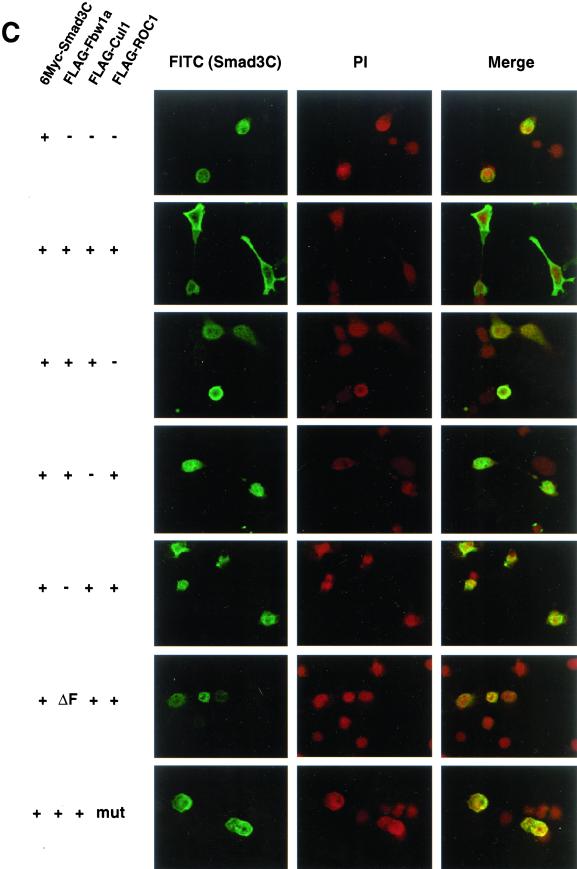
Smad3 is exported from the nucleus by the ROC1-SCFFbw1a complex. (A) Localization of 6Myc-Smad3C was examined in the absence (top) or presence of the ROC1-SCFFbw1a complex (middle) or both ROC1-SCFFbw1a and p300 (bottom). Anti-Myc staining for 6Myc-Smad3C (green) and nuclear staining by 4,6-diamidino-2-phenylindole (PI; red) were performed in transfected COS7 cells. (B) Subcellular localization of full-length FLAG-Smad3 stimulated or not with TβR-I(TD) was examined in the absence or presence of the ROC1-SCFFbw1a complex (SCF). Smad3 was detected by anti-FLAG (left; green) or anti-phospho-Smad3 staining (right; green) in transfected COS7 cells. Nuclear staining (PI; red) was performed as in A. (C) Localization of 6Myc-Smad3C was determined in the absence and presence of various components of ROC1-SCFFbw1a (top five panels) and in the presence of the Fbw1a mutant (ΔF) or the ROC1 mutant C75A/H77A (mut) (bottom two panels). 6Myc-Smad3C (green) and the nucleus (red) were stained as shown in A.
We next examined which R-Smads are ubiquitinated by ROC1-SCFFbw1a. Of different R-Smads, Smad3 and Smad8 were preferentially ubiquitinated by ROC1-SCFFbw1a (Figure 3C). In contrast, Smad2 and Smad4 were only minimally ubiquitinated by ROC1-SCFFbw1a (Figure 3E). In agreement with the finding that the MH2 domain of Smad3 is responsible for the interaction with ROC1, ubiquitination of the MH2 domain of Smad3 (Smad3C) was induced by ROC1-SCFFbw1a in a manner similar to that of the full-length Smad3 (Figure 3D). In contrast, the MH1 and linker region of Smad3 (Smad3NL) was not ubiquitinated by ROC1-SCFFbw1a (Figure 3E).
The degree of ubiquitination of Smad1 by Smurf1 and that of Smad3 by ROC1-SCFFbw1a were compared in transfected COS7 cells or 293T cells. Ubiquitination of Smad1 by Smurf1 was only very weakly observed in COS7 cells (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results), whereas it was detected in 293T cells, which occurred to an extent similar to that of the Smad3C-ubiquitination induced by ROC1-SCFFbw1a (Figure 4A).
Polyubiquitination of Smad3 was investigated using Fbw1a and ROC1 mutants. Ubiquitination was observed when wild-type Fbw1a, but not Fbw1a mutants (Hattori et al., 1999; Kitagawa et al., 1999; Suzuki et al., 2000), were used (Figure 4B). The ROC1 mutant C75A/H77A, which binds to Smad3C (Figure 2A) but lacks the ROC1-associated ligase activity (Ohta et al., 1999), did not induce the ubiquitination of Smad3C either (Figure 4C). These findings suggest that integrity of the ROC1-SCFFbw1a complex is essential for the Smad3 ubiquitination. We examined whether other E3 ligases induce the ubiquitination of Smad3. Fbw1a and hEbi1 are F-box proteins containing WD40 repeats, whereas Fbl1 (also termed Skp2) and Fbl5 have leucine-rich repeats (Cenciarelli et al., 1999; Dong et al., 1999; Winston et al., 1999a). Of these F-box proteins, only Fbw1a efficiently induced the ubiquitination of Smad3C (Figure 4D).
p300 Facilitates the Ubiquitination of Smad3
Ubiquitination of Smad3 appears to be initiated after the nuclear translocation of Smad3. Because Smad oligomers in the nucleus interact with a transcriptional coactivator p300 through the MH2 domain (Feng et al., 1998; Janknecht et al., 1998; Nishihara et al., 1998), we investigated whether the recruitment of p300 regulates the ubiquitination of Smad3. In transfected COS7 cells, p300 enhanced the interaction between ROC1 and Smad3C or full-length Smad3 (Figure 5, A and B). Moreover, p300 dramatically enhanced the ubiquitination of Smad3C (Figure 5C) and Smad3 (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results). In contrast, coexpression of Smad4 did not significantly enhance the ubiquitination of Smad3 (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results).
Figure 5.
Interaction with ROC1-SCFFbw1a and ubiquitin-dependent degradation of Smad3 are enhanced by p300. (A and B) Physical interaction of Smad3C (A) or the full-length Smad3 (B) with ROC1 is enhanced by p300 in COS7 cells. Interaction was examined by FLAG immunoprecipitation (IP) followed by Myc immunoblotting (Blot). The top panel shows the interaction and the bottom panels the expression of each protein as indicated. (C) Ubiquitination of Smad3C was induced by p300 but not by c-Ski. Lysates from transfected COS7 cells were directly subjected to anti-Myc immunoblotting. Polyubiquitination species of Smad3C are indicated ([HA-Ub]n-6Myc-Smad3C).
c-Ski interacts with Smad3 and represses the transcriptional responses through recruitment of histone deacetylases (Akiyoshi et al., 1999). c-Ski binds to the MH2 domain of Smad3 and competes with p300 for the Smad3 binding. In contrast to p300, c-Ski did not facilitate the ubiquitination of Smad3C (Figure 5C); moreover, it repressed the Smad3 ubiquitination accelerated by p300. Thus, the interaction of Smad3 with p300, but not with c-Ski, triggers the ubiquitination of Smad3.
Smad3 Is Exported to the Cytoplasm by the ROC1-SCFFbw1a Complex
Although nuclear translocation and recruitment of p300 may trigger the ubiquitination of Smad3, it is unknown where the Smad3 bound to ROC1-SCFFbw1a is ultimately located for degradation. We have transfected Smad3 with or without the ROC1-SCFFbw1a complex into COS7 cells and determined its subcellular localization. R-Smads lacking the MH1 and linker regions are mainly located in the nucleus and induce constitutive activation of target genes (Baker and Harland, 1996; Zhang et al., 1997; Xu et al., 2000). Smad3C is predominantly located in the nucleus, although weak staining of the cytoplasm could also be observed (Figure 6A). However, in the presence of ROC1-SCFFbw1a, Smad3C was mainly observed in the cytoplasm. Interestingly, the cytoplasmic translocation of Smad3C was further facilitated in the presence of p300.
Full-length Smad3 was detected both in the cytoplasm (Figure 6B) and in the nucleus (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results) as reported previously (Zhang et al., 1997), and nuclear translocation of Smad3 was induced by TβR-I(TD) (Figure 6B). Phosphorylated Smad3 was not observed without ligand stimulation and detected in the nucleus only after stimulation by TβR-I(TD). Similar to Smad3C, full-length Smad3 was, both in the absence and presence of TβR-I(TD), predominantly detected in the cytoplasm when cotransfected with ROC1-SCFFbw1a.
Cytoplasmic transportation of nuclear Smad3 is dependent on the integrity of the ROC1-SCFFbw1a complex, because the absence of ROC1, Cul1, or Fbw1a failed to induce the cytoplasmic relocation of Smad3C (Figure 6C). Moreover, in the presence of mutant forms of Fbw1a (ÆF) or ROC1 (C75A/H77A), Smad3C was mainly stained in the nucleus. Thus, binding of ROC1-SCFFbw1a to Smad3 may trigger the cytoplasmic relocation and ultimate degradation of Smad3 by the 26S proteasomes.
DISCUSSION
It has been reported that TGF-β signaling is irreversibly terminated by ubiquitin-dependent degradation of the activated Smad2 (Lo and Massagué, 1999). Here we showed that Smad3 is also degraded in a ligand-dependent manner. E2-conjugating enzymes including UbcH5b/c have been suggested to be involved in the degradation of Smad2 (Lo and Massagué, 1999; Xu and Attisano, 2000). We demonstrated that the E3 ligase complex ROC1-SCFFbw1a interacts with activated Smad3 through its MH2 domain and induces the ubiquitination and proteasomal degradation of Smad3 (Figure 7).
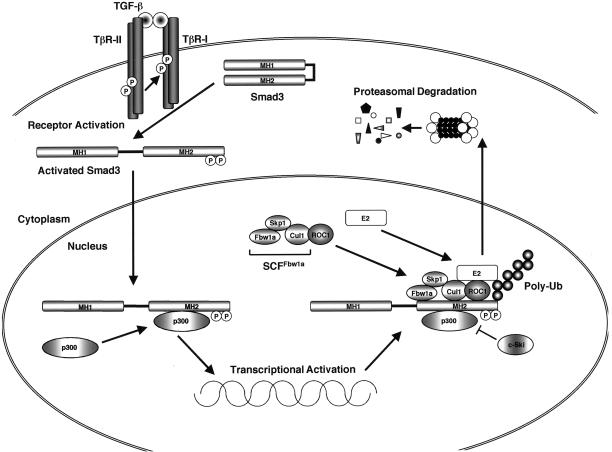
Figure 7.
Interaction of nuclear Smad3 with ROC1-SCFFbw1a and export to the cytoplasm. This is a schematic representation of a model for TGF-β-dependent Smad3 degradation by ROC1-SCFFbw1a based on the results described in this paper. Upon activation by TGF-β receptors, Smad3 forms a complex with Smad4 and translocates into the nucleus. Transcription of target genes is facilitated by the association of the transcriptional coactivator p300 with Smad3. The ROC1-SCFFbw1a complex binds to the MH2 domain of Smad3, which is enhanced by p300. ROC1 may be the primary binding component for Smad3, and Cul1 and Fbw1a may also facilitate the interaction of ROC1 with Smad3. E2 ubiquitin-conjugating enzymes function together with ROC1-SCFFbw1a to link ubiquitin to Smad3. Nuclear Smad3 is then exported to the cytoplasm and is ultimately degraded by the 26S proteasomes.
ROC1 binds to all isoforms of Cullins (Kamura et al., 1999; Ohta et al., 1999; Seol et al., 1999; Tan et al., 1999), and by recruiting E2-conjugating enzymes, it plays a critical role in the ubiquitination of target proteins. We demonstrated that ROC1 interacts with Smad3 activated by TGF-β in nontransfected HaCaT keratinocytes (Figure 2C). Moreover, our results revealed that the other components of the ROC1-SCFFbw1a complex facilitate the interaction with Smad3.
SCFFbw1a is an E3 ligase for IκB and β-catenin, which are involved in the NF-κB- and Wnt/Wingless-signaling pathways, respectively (Yaron et al., 1998; Spencer et al., 1999; Winston et al., 1999b). Fbw1a recognizes the phosphorylated DSGψXS motif (ψ is a hydrophobic residue) in IκB and β-catenin through the C-terminal WD40 repeats (Hattori et al., 1999; Kitagawa et al., 1999; Suzuki et al., 2000). However, the DSGψXS motif is not present in Smad3. Moreover, phosphorylation of Smad2 and Smad3 may not be essential for the ligand-dependent degradation (Lo and Massagué, 1999; Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results). Thus, the mode of interaction of Smad3 with ROC1-SCFFbw1a appears to be different from that of IκB and β-catenin. Interestingly, it has recently been reported that the NF-κB p105 precursor is ubiquitinated by the SCFFbw1a complex through a recognition motif distinct from that of IκB and β-catenin (Orian et al., 2000).
Smad2 and Smad3 are structurally highly similar to each other and activated by the TGF-β/activin pathway. In contrast to Smad3, however, Smad2 was not polyubiquitinated by the ROC1-SCFFbw1a complex (Figure 3, C and E). Interestingly, Lin et al. (2000) reported that Smurf2 degrades activated Smad2, but not Smad3, suggesting that they may be degraded by distinct E3 ligases. With regard to Smad3, only Fbw1a induced the ubiquitination of Smad3 among several F-box proteins examined in the present study (Figure 4D). Because there are more than 30 F-box proteins in mammals (Cenciarelli et al., 1999; Regan-Reimann et al., 1999; Winston et al., 1999a), however, it remains to be determined whether other F-box proteins may also induce the Smad3 ubiquitination. Notably, Smad8, an R-Smad in the BMP pathway, was also efficiently ubiquitinated by ROC1-SCFFbw1a (Figure 3C), although Smad8 is structurally less related to Smad3 than Smad2. Smad1, which is structurally related to Smad8 and activated by the BMP pathway, is degraded by Smurf1 via interaction through the PY motif in the linker region (Zhu et al., 1999). However, Smad8 lacks the PY motif in the linker region. Thus, similar to Smads 2 and 3, Smads 1 and 8 may be ubiquitinated through mechanisms different from each other.
In the nucleus, the Smad oligomers recruit the transcriptional coactivators p300/CBP (Feng et al., 1998; Janknecht et al., 1998; Nishihara et al., 1998). p300 and CBP are structurally related proteins with histone acetyltransferase activity. Through acetylation of core histones and possibly other proteins, p300 and CBP loosen the chromatin structure and accelerate transcription of target genes (Travers, 1999). Interestingly, the presence of p300 dramatically enhanced the ubiquitination of Smad3 by the ROC1-SCFFbw1a complex (Figure 5). Although p300 induces protein acetylation, Smad3 is not acetylated by p300 under physiological conditions (Fukuchi, Imamura, Chiba, Ebisawa, Kawabata, Tanaka, and Miyazono, unpublished results). The mechanism for the enhancement of Smad3 ubiquitination by p300 remains to be elucidated. Our results thus indicate that p300 accelerates the transcriptional response induced by Smad3, but at the same time, it may also initiate the degradation process of Smad3 and limit the signaling activity of Smad3.
Smad2 and Smad3 interact with transcriptional corepressors, including c-Ski, SnoN and TGIF (Massagué and Wotton, 2000). c-Ski competes with p300 for the interaction with Smad3, recruits histone deacetylases to the Smad complexes, and represses the transcription of target genes induced by TGF-β (Akiyoshi et al., 1999). SnoN is structurally related to c-Ski and also recruits histone deacetylases (Nomura et al., 1999). Interestingly, upon TGF-β stimulation, SnoN and c-Ski are degraded by proteasomes, although the half-life of c-Ski is longer than that of SnoN (Stroschein et al., 1999; Sun et al., 1999). However, Smad3 is not degraded together with SnoN or c-Ski under these conditions. In agreement with these findings, recruitment of c-Ski did not affect the ubiquitination of Smad3 (Figure 5C). Thus, the Smad3 ubiquitination may be initiated when the transcriptional activation occurs by the recruitment of p300 to the Smad complexes; it remains to be elucidated whether interaction with transcriptional corepressors affects the ultimate fate of nuclear Smad3.
Nuclear translocation of Smad2 triggers its degradation by the ubiquitin–proteasome pathway (Lo and Massagué, 1999); however, it has not been determined where in the cell Smad2 is degraded. We have shown in the present study that nuclear Smad3 is located in the cytoplasm after binding to the ROC1-SCFFbw1a complex (Figure 6, A and B). Function of ROC1-SCFFbw1a may be essential for the nuclear export of Smad3, because in the absence of the components of ROC1-SCFFbw1a, Smad3C remains in the nucleus (Figure 6C). Notably, the ROC1 mutant C75A/H77A failed to induce the nuclear export of Smad3C, suggesting the importance of ubiquitin ligase activity in this process. A nuclear localization signal has been identified in the MH1 domain of R-Smads (Xiao et al., 2000); however, this motif may not be involved in the nuclear export of Smad3 by ROC1-SCFFbw1a, because the nuclear export was observed in both full-length Smad3 and Smad3C lacking the MH1 and linker regions.
p27, a cyclin-dependent kinase inhibitor, is exported from the nucleus to the cytoplasm for proteasomal degradation. A 38-kDa protein JAB1 has been shown to induce the nuclear export of p27, resulting in the acceleration of its proteasomal degradation (Tomoda et al., 1999). The p53 tumor-suppressor protein is also exported to the cytoplasm by MDM2, for which polyubiquitination or other activities associated with the MDM2 RING-finger domain may be required (Boyd et al., 2000; Geyer et al., 2000). It should be determined in the future whether ROC1-SCFFbw1a by itself can induce the nuclear export of Smad3 or whether other proteins may participate together with ROC1-SCFFbw1a in the nuclear export of Smad3.
Our present study demonstrates the ultimate destination of Smad3 in the TGF-β–signaling pathway. Upon activation by the TGF-β receptors, Smad3 forms a heteromeric complex with Smad4 and translocates into the nucleus, where it regulates transcription of target genes. Smad3 recruits transcriptional coactivators p300 and CBP, which facilitate the transcriptional activation of target genes, but also triggers the interaction of Smad3 with the ROC1-SCFFbw1a complex. Smad3 is then exported to the cytoplasm and is degraded by the 26S proteasomes; TGF-β/Smad3 signaling is thus irreversibly terminated by the ubiquitin–proteasome pathway. Perturbation of the TGF-β/Smad–signaling pathway results in the cellular resistance to the growth inhibitory activity of TGF-β and progression of tumors. It will be important to determine how the degradation of Smad3 by ROC1-SCFFbw1a is regulated under physiological and pathological conditions.
ACKNOWLEDGMENTS
We thank P. ten Dijke and N. Shimbara for antibodies to Smad3 and ROC1, respectively, and L. Tsuda, K. Iwai, and M. Pagan for the cDNAs for F-box proteins. We are grateful to Y. Inada, A. Hanyu, A. Nishitoh-Sakai, and Y. Yuuki for technical help. This study was supported by Grants-in-Aid for Scientific Research and Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sport, Science, and Technology of Japan. T.I. is supported by Uehara Memorial Foundation and Public Trust Haraguchi Memorial Cancer Research Fund.
REFERENCES
- Akiyoshi S, Inoue H, Hanai J-i, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with Smads. J Biol Chem. 1999;274:35269–35278. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- Akiyoshi S, Ishii M, Miyazono K, Aburatani H, Kawabata M. Targets of transcriptional regulation of transforming growth factor-β: expression profile analysis using oligonucleotide arrays. Jpn J Cancer Res. 2001;92:257–268. doi: 10.1111/j.1349-7006.2001.tb01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- Baker JC, Harland RM. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Tsuda L, Zavitz KH, Lin M, Li S, Carthew RW, Zipursky SL. ebi regulates epidermal growth factor receptor signaling pathways in Drosophila. Genes Dev. 1999;13:954–965. doi: 10.1101/gad.13.8.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisawa T, Tada K, Kitajima I, Tojo K, Sampath TK, Kawabata M, Miyazono K, Imamura T. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J Cell Sci. 1999;112:3519–3527. doi: 10.1242/jcs.112.20.3519. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Zhang Y, Wu RY, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- Hattori K, Hatakeyama S, Shirane M, Matsumoto M, Nakayama K. Molecular dissection of the interactions among IκBα, FWD1, and Skp1 required for ubiquitin-mediated proteolysis of IκBα. J Biol Chem. 1999;274:29641–29647. doi: 10.1074/jbc.274.42.29641. [DOI] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, ten Dijke P. TGF-β signaling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW, Klausner RD, Pause A. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R, Wells NJ, Hunter T. TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Elledge SJ, Conaway RC, Harper JW, Conaway JW. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynskyi O, Landstrom M, Stoika R, Funa K, Heldin C-H, ten Dijke P, Souchelnytskyi S. Expression of Smad proteins in human colorectal cancer. Int J Cancer. 1999;82:197–202. doi: 10.1002/(sici)1097-0215(19990719)82:2<197::aid-ijc8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng X-H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- Lo RS, Massagué J. Ubiquitin-dependent degradation of TGF-β-activated Smad2. Nat Cell Biol. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K. Positive and negative regulation of TGF-β signaling. J Cell Sci. 2000;113:1101–1109. doi: 10.1242/jcs.113.7.1101. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Olofsson A, Colosetti P, Heldin C-H. A role of the latent TGF-β1-binding protein in the assembly and secretion of TGF-β1. EMBO J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara A, Hanai J-i, Okamoto N, Yanagisawa J, Kato S, Miyazono K, Kawabata M. Role of p300, a transcriptional coactivator, in signaling of TGF-β. Genes Cells. 1998;3:613–623. doi: 10.1046/j.1365-2443.1998.00217.x. [DOI] [PubMed] [Google Scholar]
- Nomura T, Khan MM, Kaul SC, Dong HD, Wadhwa R, Colmenares C, Kohno I, Ishii S. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Orian A, Gonen H, Bercovich B, Fajerman I, Eytan E, Israel A, Mercurio F, Iwai K, Schwartz AL, Ciechanover A. SCFβ-TrCP ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J. 2000;19:2580–2591. doi: 10.1093/emboj/19.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan-Reimann JD, Duong QV, Jackson PK. Identification of novel F-box proteins in Xenopus laevis. Curr Biol. 1999;9:R762–R763. doi: 10.1016/S0960-9822(00)80006-8. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Sporn MB. The transforming growth factor-βs. In: Sporn MB, Roberts AB, editors. Peptide Growth Factors and Their Receptors, part I. Heidelberg, Germany: Springer-Verlag; 1990. pp. 419–472. [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ, Shevchenko A, Deshaies RJ. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor-β signaling. Proc Natl Acad Sci USA. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Chiba T, Suzuki T, Fujita T, Ikenoue T, Omata M, Furuichi K, Shikama H, Tanaka K. Homodimer of two F-box proteins βTrCP1 or βTrCP2 binds to IκBα for signal-dependent ubiquitination. J Biol Chem. 2000;275:2877–2884. doi: 10.1074/jbc.275.4.2877. [DOI] [PubMed] [Google Scholar]
- Tada K, Inoue H, Ebisawa T, Makuuchi M, Kawabata M, Imamura T, Miyazono K. Region between α-helices 3 and 4 of the Mad homology 2 domain of Smad4: Functional roles in oligomer formation and transcriptional activation. Genes Cells. 1999;4:731–741. doi: 10.1046/j.1365-2443.1999.00293.x. [DOI] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- Travers A. How to put a HAT on the histones. Curr Biol. 1999;9:R23–R25. doi: 10.1016/s0960-9822(99)80037-2. [DOI] [PubMed] [Google Scholar]
- Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. A family of mammalian F-box proteins. Curr Biol. 1999a;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999b;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Liu X, Henis YI, Lodish HF. A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc Natl Acad Sci USA. 2000;97:7853–7858. doi: 10.1073/pnas.97.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Attisano L. Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor β signaling by targeting Smads to the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 2000;97:4820–4825. doi: 10.1073/pnas.97.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Chen Y-G, Massagué J. The nuclear import function of Smad2 is masked by SARA and unmasked by TGFβ-dependent phosphorylation. Nat Cell Biol. 2000;2:559–562. doi: 10.1038/35019649. [DOI] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC4 as a central mediator of Smad function. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]