Abstract
Evaluation of docking results is one of the most important problems for virtual screening and in silico drug design. Modern approaches for the identification of active compounds in a large data set of docked molecules use energy scoring functions. One of the general and most significant limitations of these methods relates to inaccurate binding energy estimation, which results in false scoring of docked compounds. Automatic analysis of poses using self-organizing maps (AuPosSOM) represents an alternative approach for the evaluation of docking results based on the clustering of compounds by the similarity of their contacts with the receptor. A scoring function was developed for the identification of the active compounds in the AuPosSOM clustered dataset. In addition, the AuPosSOM efficiency for the clustering of compounds and the identification of key contacts considered as important for its activity, were also improved. Benchmark tests for several targets revealed that together with the developed scoring function, AuPosSOM represents a good alternative to the energy-based scoring functions for the evaluation of docking results.
Keywords: scoring, docking, virtual screening, CAR, AuPosSOM
Introduction
The modeling of protein ligand interactions by computational molecular docking is a widely used approach for virtual screening. One of the most challenging problems with this method lies in the identification of active compounds in large sets of docked molecules. Typically, ligand-binding affinity is estimated by scoring functions. Ideally, scoring functions should be able to select the correct pose for each molecule and arrange ligands in accordance with their affinity to the receptor. In practice however, estimation of binding energy is a very difficult task. Several studies have shown that although docking programs are usually able to provide poses with correct ligand conformation, the relevant estimation of binding affinity for the right pose remains a great challenge.1–6 Another problem posed by the scoring function approach, is the strong dependency of scoring efficiency on a particular data set.5,7 Some authors3 have suggested making trial runs for the target of interest using ligand sets with known activity in order to define the best suitable scoring functions. Various machine learning methods7–9 have been used for the construction of target dependent docking scoring functions. Consensus scoring methods, which offer promising prospects to solve the problem, take into account predictions from several scoring functions.10,11 However, if all scoring functions fail to estimate the binding affinity, the consensus will also yield poor results.12 Thus, the development of new approaches for a more efficient evaluation of docking results remains a pressing issue.
In the past decade, several methods based on binding mode contact analysis have been developed for the characterization of protein ligand interactions in docking experiments.13,14 This approach takes advantage of the fact that in the majority of cases, structurally similar ligands present high conservation of the binding modes to their receptor.15 The key idea of this method uses the similarity of binding modes to identify native like poses to select active compounds. The pharmacogram approach16 utilizes similarity to the pharmacophore grid to score docking poses and compounds. The grid is constructed using the best poses out of several best-docked compounds top-ranked by conventional scoring functions. The template-based method uses knowledge of the key receptor ligand interactions to focus docking searches on poses similar to the template.17,18 The template is generated from an experimental protein ligand complex structure. This method can be implemented as an additional term to the scoring function, and has been shown to improve results of docking evaluation. Analogously, the structure interaction fingerprint method13 uses ligands from cocrystal structures as a reference for the native contact set. Interactions found between a ligand and a receptor are presented as fingerprints which are constructed for all poses of all docked molecules and scored in accordance with the similarity to the reference as measured by the Tanimoto coefficient. The structure interaction fingerprint method has been shown to provide results superior to the conventional scoring function approach. However, the mandatory requirement of reference data for the structure interaction fingerprint method raises important limitations of this promising approach. The concept of contact fingerprint comparison was successfully applied in the maximum common binding mode approach for the selection of the native like poses of active molecules.14 Poses of docked ligands were scored according to the similarity of their binding modes and poses, the highest rank selected as native like conformations. The method performed better than individual scoring functions and did not require a reference structure.
Automatic analysis of poses using self-organizing maps (AuPosSOM)19 represents a new approach that uses the concept of contact fingerprint similarity for virtual screening. Kohonen’s self-organizing maps (SOM) method20 is applied for the unsupervised clustering of docked compounds,21 ligands and decoys then arranged in the hierarchal tree with respect to the similarity of binding modes. The problem of the correct pose selection is overcome by the use of statistical analysis of the contact information over all poses of the ligand. The AuPosSOM approach has been shown to provide good results for the tested data sets, clustering a significant number of active compounds separately from the decoys.19 However, preliminary knowledge of activity for at least a few active compounds is required for the selection of the correct cluster.
In this study, we developed a scoring function for the identification of the active compounds in the AuPosSOM clustered dataset without prior knowledge of the compounds’ activity. Additionally, we improved the efficiency of the clustering and key contact data analysis. Benchmark tests for several targets revealed that together with the newly developed scoring function, AuPosSOM represents a good alternative to energy-based scoring functions for the evaluation of docking results. Our method does not require reference data such as the crystal structure of the protein ligand complex.
Materials and methods
Datasets
Datasets for the benchmarking tests of AuPosSOM were obtained from the DUD 2.0 database (DUD – A Directory of Useful Decoys; Shoichet Laboratory, University of California, San Francisco, CA, USA).22 The DUD database contains ligands and decoys for 40 protein targets with the approximate active compounds:decoys ratio 1:36. Decoys are physicochemically similar but topologically dissimilar to the active ligands. Because of these properties, DUD is considered to be one of the most challenging datasets for virtual screening benchmarking tests currently available. Datasets for the following nine targets with different polar and geometric properties of the binding sites were selected for the evaluation of the AuPosSOM clustering efficiency: CDK2 (active compounds:decoys ratio 50:1779), COX1 (25:849), DHFR (201:3318), HIV protease (53:1885), HIV RT (40:1437), HSP90 (24:860), progesterone receptor (PR; 27:967), thrombin (65:2292), and trypsin (44:1544). Proteins with metal containing binding sites were not considered, as currently available docking programs do not efficiently treat interactions of ligands with metals. Corresponding protein structures for docking and ligands for definition of the docking search space were also retrieved from the DUD 2.0 database.
Docking
Protein molecules for docking were prepared using the Sybylx1.2 (SYBYL, Tripos International, St Louis, MO)23 and Chimera1.524 software programs. Mol2 files of the targets provided by DUD did not contain water molecules. Explicit hydrogens were added and AMBER-ff99SB25,26 charges were calculated for the receptor molecules. Ligand molecules provided by DUD were already assigned with the atom’s partial charges. This was calculated using the quantum mechanical approach for unbound ligands. Docking was performed by Surflex-Dock 2.0 from the Sybyl 1.223 package using mol2 files of targets and ligands. Protomol was generated with default parameters (threshold of 0.50 and bloat equal to 0). Each docking experiment was repeated 20 times yielding 20 docked poses. Ligand energy minimization prior to docking and all-atom-in-pocket minimization after docking was performed. Virtual screening with docking was performed on one Linux PC (quadricore Intel 2.66 GHz, 2 GB RAM). Four C Score23 scoring functions were utilized for the evaluation of the docking results using the conventional energy scoring approach: ChemScore, PMF, G-score and D-score.
Contact selection
Identification of the protein ligand contact set that represents the difference in the binding modes between decoys and active compounds is a key point for successful clustering. Five types of atom selections were tested:
-
Hydrogen bonds (HB).
Interactions were computed for all possible donor acceptor pairs. An interaction was considered as a HB when the D-H...A distance was 1.85 Å ± 0.65 Å and the D-H...A angle was 180° ± 80°.
-
Coulomb contacts.
Contacts were searched between polar atoms with partial charges of opposite signs and greater than the threshold values. Thresholds of 0.3, 0.5 and 0.7 e for the module of the partial charge were tested for thrombin and HIV protease. Oxygen atoms were included as negatively charged for all selections. A threshold value of 0.5 e was selected as the best one corresponding to the results of the clustering (Figure S1). Contacts were searched between the selected atoms at the distance less than the sum of Van der Waals radii plus constant. Four values of constant were tested: 0.0, 0.5, 1.0, 1.5 Å (Figure S2). The distance of 1 Å was selected as the best one. The best values of the thresholds for partial charges and distances were implemented for all the other targets.
-
Lipophilic contacts 1.
Contacts between protons with the module of partial charges less than 0.1 e and within the distance of the sum of Van der Waals radii plus 0.5 Å. For the partial charges used in this work the selection included mainly aliphatic protons.
-
Lipophilic contacts 2.
Contacts between the following atoms: carbons of CH2 and CH3 groups, chlorine, bromine, and iodine atoms (which are not ions). Contacts were searched between the selected atoms on the distance less than the sum of Van der Waals radii plus constant. Three thresholds were tested for the distance constants: 0, 0.5, and 1.0 Å (Figure S3). The threshold of 0.5 Å was selected as the best one.
-
All atom contacts.
Contacts between atoms of all types at the distance of the sum of Van der Waals radii.
Contacts were selected in such a way that one atom belonged to the protein and another one to the ligand. Contact search was accomplished for all datasets. Atom selection and search for the contacts between ligands and receptors were performed using modified “findclash” and “findhbond” modules of Chimera 1.5.24
AuPosSOM clustering
AuPosSOM clustering was made for all nine datasets using all types of contact selection. Fingerprints were presented as one dimensional vectors and were generated as previously described.19 Clustering was made using the unsupervised learning SOM algorithm implemented in AuPosSOM for each type of contact separately. Optimized parameters for clustering were obtained from Bouvier et al.19 The 4×5 SOM matrix was utilized providing that the resulting tree of clusters of compounds could contain up to 20 leaves.
A filtering step was added in order to increase the efficiency of the SOM procedure and to facilitate calculations. The final clustering algorithm included three steps: (1) first round of clustering, (2) contact filtering, and (3) second round of clustering. As the SOM algorithm uses random value for the generation of the initial map, calculations were repeated ten times to obtain representative results. Two filters were then introduced. The first filter removed contacts that were very weakly populated and could be considered as noise. This facilitates calculation and simplifies manual contact analysis of the clustering results without influencing the SOM efficiency. A contact was considered as weak if its average population over all the leaves of the tree was less than 0.02. This population is caused by non systematic contacts of ligands with receptors and can be treated as noise. The second filter was constructed to remove contacts with large average population in all leaves. This procedure simplifies the contact matrix and increases the efficiency of the SOM clustering. Mean values for the contact population were calculated for each leaf of the tree and the average value of the obtained mean values was calculated. A contact was considered as equally populated and removed from the contact matrix if the following condition was true for all leaves of the tree:
| (1) |
where Icontact mean leaf is the contact population’s mean value for the leaf, Itree is the contact population value averaged over mean values for all leaves, and α = 0 and β = 3.2 are empirically optimized constants.
AuPosSOM scoring function
An empirical scoring function was developed for the identification of the leaves containing active compounds. In accordance with the contact activity relationship (CAR) concept,19 it was proposed that active compounds should form more selective and highly populated contacts than decoys, as their affinity to the target is higher. In order to calculate scores for the leaves, Icontact mean leaf values were calculated for each contact over each leaf. Each contact was weighted (Wcontact leaf) for each leaf separately with respect to the following rules:
-
If the contact is defined as an equally populated contact with respect to the condition described below (equation (1); α = 0.05, β = 3.5), the negative value is assigned to the weight of the contact:
(2) -
If the contact is not defined as an equally populated contact, the contact is described as selective and the positive value is assigned to the weight:
(3) -
After the weight has been assigned Icontact mean leaf is compared to the maximum mean contact population of the contacts in the leaves. If Icontact mean leaf is higher than 90% of the maximum population, the contact is described as a highly populated contact and the assigned weight is increased:
(4) The score for the leaf (Sleaf) was defined as:
(5)
where I is the number of the contact and N is the total number of contacts in the vector.
Data analysis
The efficiency of AuPosSOM was evaluated in two ways:
-
Receiver operating characteristic (ROC).
ROC plots were calculated for both the AuPosSOM and the conventional scoring functions and compared.
True positive rate (TPR) and false positive rate (FPR) of ROC curve is defined as follows:
. Thus, the good point on the ROC curve may correspond to a large number of decoys in the leaf together with active compounds if the pool of decoys is large. This is the case for the DUD database. To deal with this problem, the best leaf of the tree was defined and evaluated.
-
Percentage of active compounds in the leaf.
Two characteristics for the leaves were calculated to evaluate clustering efficiency: (i) the percentage of active compounds in the leaf selected from all active compounds in the dataset (active from all actives), and (ii) the percentage of active compounds in the leaf selected from all compounds in the leaf (active from all in the leaf). The best leaf of the AuPosSOM tree was defined as the leaf containing the maximum number of active compounds. Parameters of the best leaves were compared for all contact types and datasets.
Two dimensional heat maps were used for visualization of the clustered vectors. This representation of the clustered vector matrix provides clear information about contact population distribution and allows for easily defined key contacts for the binding of the compounds clustered in the same leaf.
Results
Comparison of the AuPosSOM scoring and conventional scoring functions efficiency
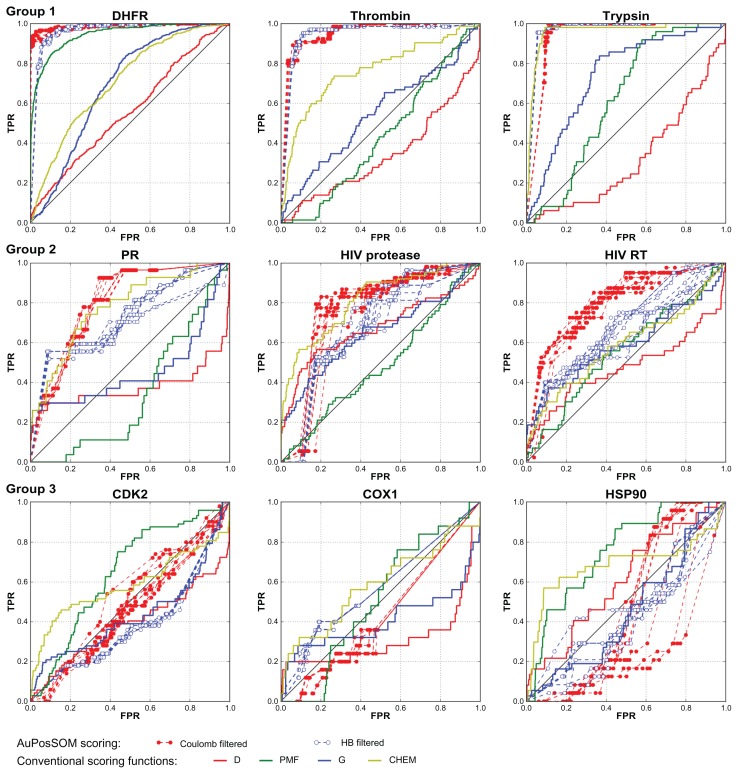
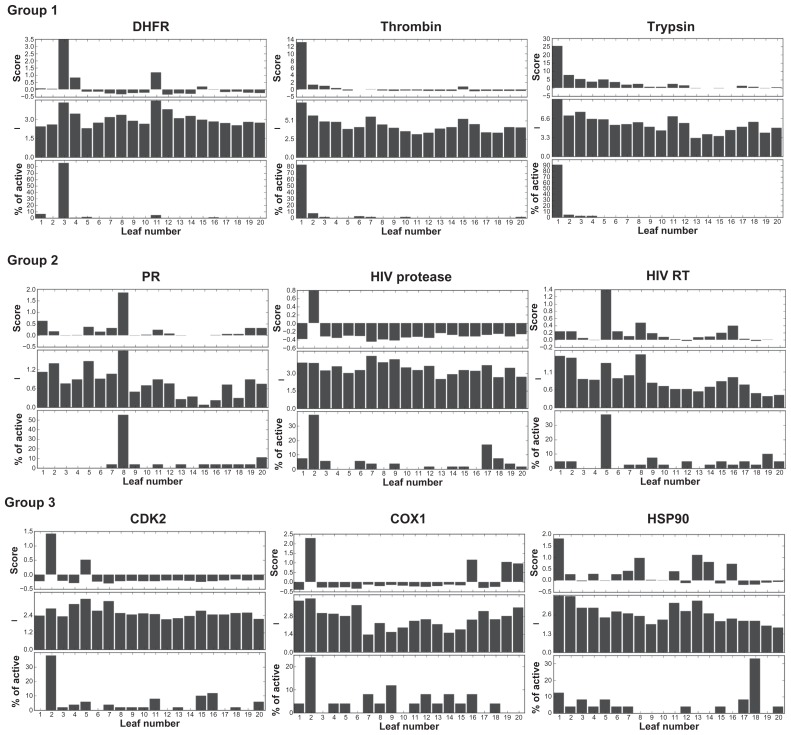
Results of the docking for nine DUD datasets were evaluated using AuPosSOM clustering followed by scoring with the scoring function developed in this study. Comparison of ROC curves for the AuPosSOM and four conventional scoring functions revealed that for eight out of nine targets, the results yielded by AuPosSOM are as good as (or better than) the results obtained using the energy scoring approach (Figure 1 and Table 1). Tested datasets can be divided into three groups based on clustering efficiency and ROC curves for scoring: DHFR, thrombin and trypsin (group 1) for which AuPosSOM was very efficient; progesterone receptor, HIV RT and HIV protease (group 2) for which AuPosSOM provided reasonable results; and HSP90, CDK2 and COX1 (group 3) for which AuPosSOM scoring did not identify a significant number of active compounds with clustering efficiency less than that for group 2.
Figure 1.
ROC curves for AuPosSOM and conventional scoring functions for nine DUD datasets divided in three groups with respect to scoring efficiency.
Note: AuPosSOM ROC curves are presented for ten independent runs of clustering with filtering for HB and Coulomb contact selections.
Table 1.
Enrichment factors for AuPosSOM HB contact selection and four scoring functions
| AuPosSOM HB | D | PMF | G | CHEMscore | |
|---|---|---|---|---|---|
| DHFR | 7.32 | 1.61 | 7.36 | 1.19 | 2.46 |
| Thrombin | 9.46 | 0.00 | 0.00 | 2.90 | 5.43 |
| Trypsin | 9.02 | 0.50 | 0.67 | 2.19 | 7.73 |
| PR | 6.00 | 3.00 | 0.00 | 3.40 | 4.25 |
| HIV protease | 3.30 | 2.44 | 1.96 | 3.20 | 5.95 |
| HIVRT | 4.30 | 2.24 | 1.96 | 3.35 | 3.07 |
| CDK2 | 2.80 | 1.08 | 1.32 | 1.91 | 3.23 |
| COX1 | 1.86 | 1.64 | 0.00 | 2.30 | 2.63 |
| HSP90 | 1.06 | 2.09 | 3.66 | 0.79 | 2.50 |
Notes: Number of scored compounds with the highest scores for the calculation of the enrichments factors was defined as number of compounds in the best leaf of the tree (leaf with the highest score). AuPosSOM scoring data for the selected datasets is presented on Figure 6.
Abbreviations: AuPosSOM, Automatic analysis of poses using self-organizing maps; HB, hydrogen bonds.
AuPosSOM ROC curves for the group 1 datasets demonstrated a high level of active compound identification. The results for DHFR and thrombin datasets are better than those of the best energy scoring (PMF function for DHFR and ChemScore for thrombin). Only ChemScore is comparable to AuPosSOM scoring for the trypsin dataset. The group 2 datasets were challenging for both clustering and energy-scoring approaches. For the progesterone receptor dataset AuPosSOM scoring ROC curves are better than those of three scoring functions. HB contact selection and filtered coulomb contacts provided clustering scores that were as good as the best scoring functions for HIV protease. Energy scoring did not yield meaningful results for the HIV RT dataset. AuPosSOM scoring for coulomb contact selection was more efficient than energy function scoring. Both AuPosSOM and energy scoring approaches did not provide ROC plots that were significantly different from the random one for CDK2 and COX1 datasets. For the HSP90 dataset, AuPosSOM scoring was worse than three energy scoring functions, while energy scoring was not very efficient.
Clustering efficiency
Coulomb and HB contact selections were the most efficient and provided the highest level of clustering of active compounds (Table 2). More than 80% of the active compounds were clustered in one leaf for the best contact selection for the targets from group 1. The best result was obtained for the DHFR coulomb contact dataset, with almost 90% of active compounds clustered in one leaf and only 5% of decoys from all compounds in the leaf. Clustering for progesterone receptor, HIV RT and HIV protease provided worse enrichment for the active compounds. The percentage of the active compounds in the best leaf from all compounds in the leaf was relatively low, between 14.5% to 18.5% for the best contact set. At the same time, more than 47% of the active compounds were clustered together for the best contact sets. HB selection gave the best results for progesterone receptor and the coulomb contact set was the best for HIV RT and HIV protease. HSP90, CDK2 and COX1 datasets appeared to be the most difficult to evaluate. A high number of decoys were clustered in the best leaves for CDK2 and COX1 targets. The evaluation of the HSP90 dataset was better for energy scoring functions compared to the AuPosSOM scoring, clustering yielding reasonable results for this target. The absence of populated contacts made it possible to cluster 55.2% of the active compounds in one leaf for the filtered matrix of coulomb contact selection. They formed 9% of the compounds of the cluster.
Table 2.
Parameters of the best leaves of the trees for all datasets
| Target | HB | Coulomb contacts | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||||
| Not filtered | Filtered | Not filtered | Filtered | |||||||||||||
|
|
|
|
|
|||||||||||||
| Active from all active, % | Active from all in the leaf, % | Active from all active, % | Active from all in the leaf, % | Active from all active, % | Active from all in the leaf, % | Active from all active, % | Active from all in the leaf, % | |||||||||
| DHFR | 85.1 | ±0.22 | 43.01 | ±0.86 | 77.82 | ±0.22 | 55.54 | ±0.30 | 90 | ±0.10 | 89.82 | ±1.12 | 89.36 | ±0.29 | 95.04 | ±0.53 |
| Thrombin | 81.52 | ±0.21 | 26.2 | ±0.25 | 78.03 | ±0.36 | 28.89 | ±0.39 | 83.64 | ±0.51 | 26.65 | ±0.62 | 80 | ±0.21 | 36.78 | ±1.61 |
| Trypsin | 89.56 | ±0.34 | 41.85 | ±0.12 | 93.33 | ±0.00 | 44.83 | ±0.27 | 67.33 | ±0.39 | 25.99 | ±0.16 | 68.89 | ±0.00 | 27.88 | ±0.21 |
| PR | 51.43 | ±0.77 | 14.54 | ±1.04 | 52.86 | ±0.62 | 14.48 | ±0.60 | 28.57 | ±0.00 | 10.26 | ±0.87 | 31.79 | ±2.75 | 10.22 | ±1.58 |
| HIV protease | 39.44 | ±1.77 | 9.63 | ±1.01 | 50 | ±0.83 | 19.14 | ±0.54 | 28.15 | ±4.24 | 9.79 | ±1.94 | 67.96 | ±1.56 | 18.53 | ±2.25 |
| HIVRT | 34.39 | ±2.98 | 10.8 | ±1.59 | 38.05 | ±1.62 | 10.14 | ±0.57 | 37.56 | ±4.52 | 14.57 | ±1.02 | 47.07 | ±2.90 | 15.46 | ±1.16 |
| CDK2 | 32.2 | ±3.94 | 6.55 | ±0.80 | 54.6 | ±2.87 | 5.2 | ±0.18 | 18 | ±4.82 | 8.18 | ±4.61 | 20.6 | ±2.54 | 2.71 | ±0.28 |
| COX1 | 17.69 | ±3.53 | 4.18 | ±0.71 | 50 | ±0.00 | 2.45 | ±0.00 | 27.31 | ±4.02 | 11.91 | ±2.83 | 61.54 | ±0.00 | 3.51 | ±0.04 |
| HSP90 | 22.8 | ±3.15 | 7.37 | ±2.39 | 26 | ±2.29 | 8.73 | ±3.13 | 24 | ±3.65 | 12.34 | ±2.25 | 55.2 | ±3.21 | 9.05 | ±0.83 |
| Lipophilic contacts 1 | Lipophilic contacts 2 | |||||||||||||||
| DHFR | 17.33 | ±1.62 | 23.56 | ±6.29 | 15,35 | ±0.89 | 10.81 | ±4.65 | 16.49 | ±2.40 | 11.6 | ±3.98 | 26.53 | ±1.01 | 15.47 | ±0.77 |
| Thrombin | 40.15 | ±1.77 | 13.51 | ±0.83 | 44.85 | ±0.35 | 13.5 | ±0.92 | 51.21 | ±0.40 | 13.05 | ±0.24 | 80.3 | ±0.00 | 6.56 | ±0.00 |
| Trypsin | 68.22 | ±1.66 | 30,71 | ±0.63 | 67.33 | ±1.80 | 32.03 | ±0.44 | 54.44 | ±2.56 | 21.85 | ±0.25 | 80 | ±0.00 | 5.86 | ±0.00 |
| PR | 24.64 | ±3.44 | 11.88 | ±3.73 | 32.86 | ±1.72 | 5.42 | ±2.53 | 26.07 | ±1.99 | 6.99 | ±2.03 | 48.93 | ±2.05 | 4.46 | ±0.43 |
| HIV protease | 25.56 | ±2.63 | 8.78 | ±1.07 | 19.63 | ±1.69 | 6 | ±0.58 | 41.85 | ±3.71 | 11.91 | ±1.06 | 29.63 | ±1.44 | 3.24 | ±0.36 |
| HIVRT | 27.32 | ±2.13 | 6.68 | ±1.92 | 38.29 | ±3.79 | 5.58 | ±0.41 | 26.34 | ±5.54 | 8.77 | ±1.48 | 35.37 | ±2.50 | 4.46 | ±0.44 |
| CDK2 | 29 | ±2.24 | 8.87 | ±2.71 | 29.8 | ±3.63 | 4.5 | ±0.61 | 17.2 | ±3.49 | 6.17 | ±1.18 | 36 | ±0.00 | 2.11 | ±0.08 |
| COX1 | 21.92 | ±1.76 | 6.17 | ±1.45 | 30.38 | ±6.07 | 6.13 | ±2.09 | 20 | ±1.54 | 5.88 | ±0.62 | 24.62 | ±1.88 | 4.71 | ±0.80 |
| HSP90 | 24.8 | ±3.56 | 7.24 | ±1.56 | 20 | ±2.53 | 12.82 | ±4.48 | 20.8 | ±1.66 | 6.77 | ±4.29 | 21.2 | ±2.15 | 5.05 | ±2.39 |
| All atom contacts | ||||||||||||||||
| DHFR | 72.92 | ±1.37 | 35.08 | ±1.29 | 72.48 | ±0.38 | 44.32 | ±0.85 | ||||||||
| Thrombin | 11.97 | ±1.31 | 7.86 | ±2.50 | 73.33 | ±1.46 | 5.71 | ±0.44 | ||||||||
| Trypsin | 44 | ±4.18 | 34.63 | ±4.45 | 33.78 | ±3.10 | 15.45 | ±4.67 | ||||||||
| PR | 36.07 | ±2.72 | 19.72 | ±2.11 | 31.43 | ±0.81 | 3.26 | ±0.19 | ||||||||
| HIV protease | 21.48 | ±2.95 | 6.79 | ±1.17 | 22.78 | ±1.90 | 7.45 | ±0.75 | ||||||||
| HIVRT | 36.34 | ±4.29 | 10.31 | ±0.99 | 53.66 | ±0.00 | 4.69 | ±0.04 | ||||||||
| CDK2 | 20 | ±2.37 | 10.77 | ±2.48 | 33.2 | ±2.23 | 4.18 | ±0.43 | ||||||||
| COX1 | 29.62 | ±2.46 | 10.57 | ±1.94 | 50.38 | ±2.07 | 5.79 | ±0.36 | ||||||||
| HSP90 | 20 | ±1.79 | 5.71 | ±2.59 | 24.4 | ±0.77 | 5.58 | ±5.43 | ||||||||
Notes: Average values over ten SOM runs are calculated and RMSD values are pointed out. Data for the clustering before and after application of filters is presented. The best results are pointed out in bold for each target.
Abbreviation: HB, hydrogen bonds; SOM, self-organizing maps; RMSD, root mean square deviation.
Clustering can be characterized by the ROC plots created using information about distances between the leaves in the tree to add points in the ROC space. Distance information points out the similarity of the clusters, the best leaf of the tree used as the starting point. These ROC plots provide results superior to the ones derived from AuPosSOM scoring (Figure S5).
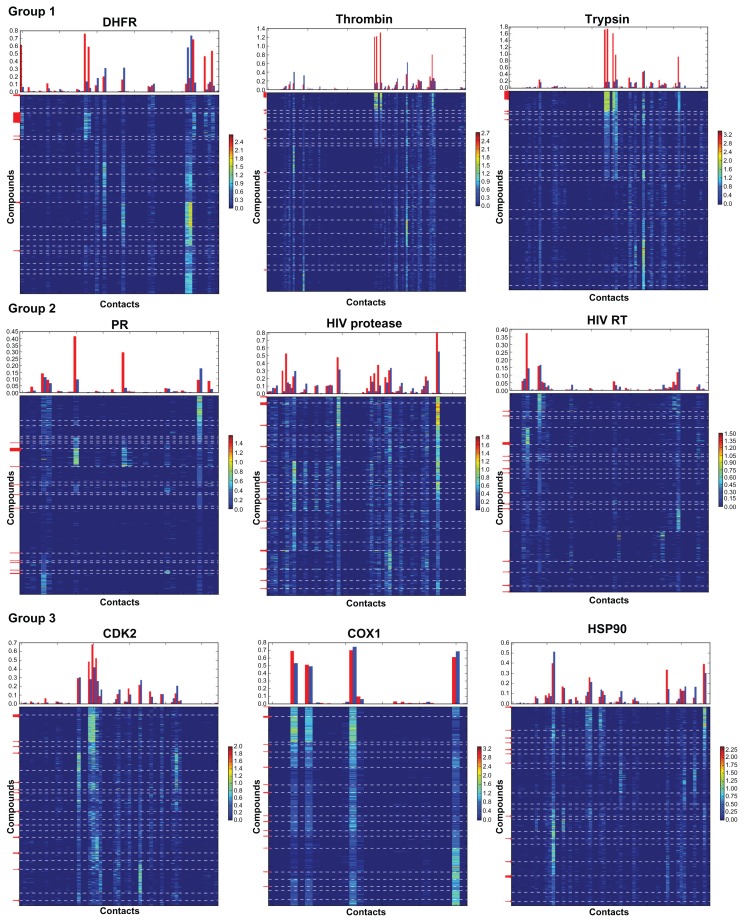
The observed set of contacts represents the properties of the protein binding site and ligands. Thus, the probability of obtaining a good clustering for a particular contact selection correlates with the properties of the receptor binding site. For example, COX1 bears a hydrophobic active site buried inside the protein globe. Only four atoms participate in the formation of HB with the compounds from the DUD dataset (Figure 2). These contacts are present in all leaves, providing a highly similar geometry of the polar fragment organization in the docked conformations for both active compounds and decoys, and a low chance for good clustering. The hydrophobic contact set (Figure S4) represents a significantly more heterogeneous contact population distribution, implying that hydrophobic contacts should play a key role in the activity, although there is still not enough difference in contact vectors for successful identification of the active compounds. The progesterone receptor’s binding site also bears a small number of HB (6 atoms form HB contacts with the average population over all active compounds higher than 0.05 (Figure 2)), while the dispersion of populations for the contacts between the leaves is much higher than that for the COX1 HB contact set. 53% of the active compounds form two very specific contacts, which makes their clustering in one leaf possible. Trypsin contains a polar active site27–29 indicating that the analysis of the HB and coulomb contacts should give good results. Indeed, five atoms form very intense and specific HB with the active compounds (the average population over all active compounds higher than 0.8 (Figure 2)). Notably, more than 93% of the active compounds were clustered in one leaf.
Figure 2.
Contact fingerprints for HB contacts for nine tested datasets.
Notes: Top panels represent an average population for the contacts over all active compounds (red bars) and all decoys (blue bars). Bottom panels represent heat maps for 2D fingerprints: rows correspond to the contact vectors of the compounds, columns correspond to the contacts with the atoms of the receptor, populations of the contacts are pointed out by different colors, and legends on the right sides of the plots represent the calibration of the colors for the contact population. Compounds are clustered with respect to the SOM tree for the HB contact dataset. Borders of the leaves are pointed out by white dashed lines. Red arrows on the left sides of the plots point out positions of the vectors for active compounds. Data without filtering is presented.
The number of decoys clustered with the active compounds is relatively high, exceeding 50% for even the best targets (except DHFR). This problem can be attributed to the similarity of the contacts between the active compounds and some decoys, for example as is the case for the thrombin and trypsin HB contact sets. Another reason for this observation is the size of the SOM matrix that allows for the maximum of 20 clusters. This may not be enough for large datasets; although it was demonstrated that increasing the matrix size does not lead to better clustering.16,18 This problem may be fixed by constructing the subtree for the best leaf.
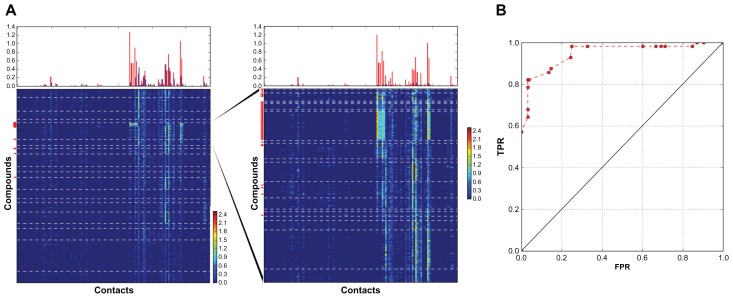
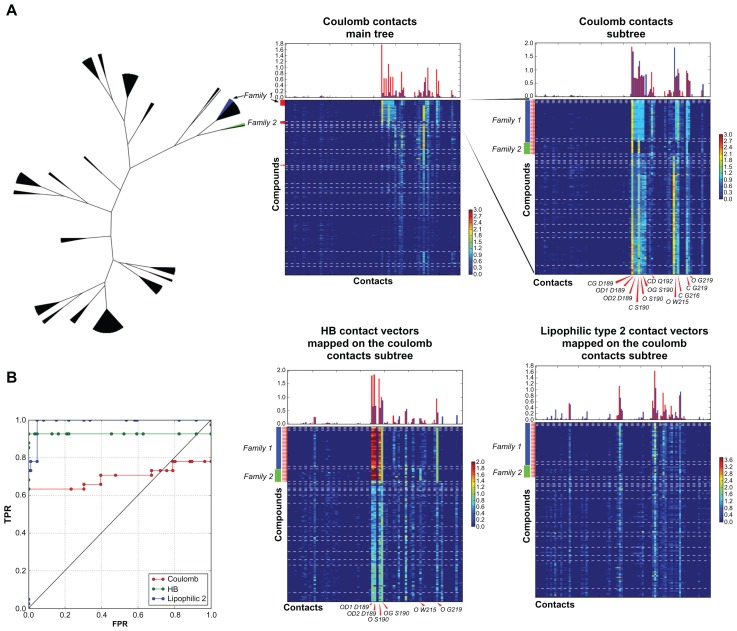
Construction of the subtree for the thrombin coulomb contact set led to the identification of 49% of the active compounds without decoys (Figure 3). The trypsin coulomb contact clustering defined two families of active compounds with different contact fingerprints, yielding two leaves containing 69% and 25% of all active compounds respectively (Figure 4A). The subtree for vectors of these two leaves provided excellent clustering, with six leaves containing 94% of all active compounds without decoys (Figure 4A and B). A high rate of similarity between the contacts for decoys and active compounds meant that the coulomb contact subtree represented a difficult dataset for scoring. Meanwhile, the highest score was assigned to the best leaf, and 63% of all active compounds were identified without decoys (Figure 4B). In order to increase the efficiency of the ligands selection, the coulomb contact subtree was implemented for mapping of the HB and lipophilic type 2 contact vectors. As a result, vectors for HB, coulomb, and hydrophobic contacts were clustered in one tree, providing convenient data organization for scoring cross validation and comparison of the different types of contacts (Figure 4A). However, HB and lipophilic type 2 contacts of decoys and ligands differed significantly, as the selection of the compounds to the subtree was based on the similarity between coulomb contacts. This provided efficient scoring for these contact selections and a corresponding improvement of the ROC curves (Figure 4B). HB scoring selected four leaves with 86% of the active compounds without decoys. Lipophilic contacts scoring yielded a selection of 68% of ligands without decoys, 94% of the active compounds identified together with 0.3% of the decoys.
Figure 3.
Construction of the subtree for the best leaf of thrombin Coulomb contact dataset.
Notes: Number of compounds in the main tree: 2357, in the subtree: 212. (A) Fingerprints for the vectors of the main tree (left) and of the subtree for the best leaf of the main tree (right). (B) AuPosSOM scoring ROC curve for the Coulomb contact subtree dataset.
Figure 4.
Construction of the subtree for two best leaves of the trypsin coulomb contact dataset.
Notes: Number of compounds in the main tree: 1588, in the subtree: 127. (A) Tree representation of the results for the clustering of the full dataset and fingerprints. Top panels: fingerprints for the vectors of the main tree and subtree of the coulomb contact dataset. Bottom panels: fingerprints for the HB and lipophilic contacts 2 vectors mapped to the coulomb contact selection subtree. Key contacts are pointed out. (B) AuPosSOM scoring ROC curves for the coulomb contact subtree scored using three types of contact vectors.
In contrast to coulomb contact sets, the construction of subtrees for the HB contact sets of trypsin and thrombin did not result in an increase in clustering quality. This is due to the high similarity of HB contacts between active compounds and decoys clustered in the best leaf. In addition, the subtree for the DHFR HB data set yielded efficient clustering (data not shown).
Contact selection
Five types of contact selection were utilized. One of the main goals of this procedure was to probe different types of contacts to find the types of contacts that yielded the best clustering of active compounds. Two types of contacts were described: polar contacts (selections 1 and 2), and lipophilic contacts (selections 3 and 4). All atom contact selection was made as a control for the presence of the specific contacts that were not included in other selections. Atom selection by partial charge was used for selections 2 and 3. This is a robust and flexible way to probe contacts between specific atoms. An additional reason for using this approach was to take into account the electrostatic properties of the molecules, as atoms with the highest partial charges of opposite signs could be expected to come into contact in a space. Atoms with partial charges close to zero could also be expected to group together as a result of hydrophobic interactions.
The best results were obtained for the HB and coulomb contact selections. Lipophilic and all contact evaluation of the interactions appeared to be not specific enough for efficient clustering. The coulomb contact selection yielded the best results for five targets (DHFR, thrombin, HIV RT, HIV protease, COX1, and HSP90) providing the highest enrichment of the best leaf (Table 2). HB was better for PR, CDK2 and trypsin. The better clustering for the trypsin HB contact set can be explained by the fact that coulomb contacts represent more detailed information about binding by providing clustering of active compounds in two leaves. Selections 3 and 4 for lipophilic contacts yielded approximately the same results (Table 2). Even for targets with hydrophobic binding sites, better clustering than polar selections was not achieved.
Filtering technique
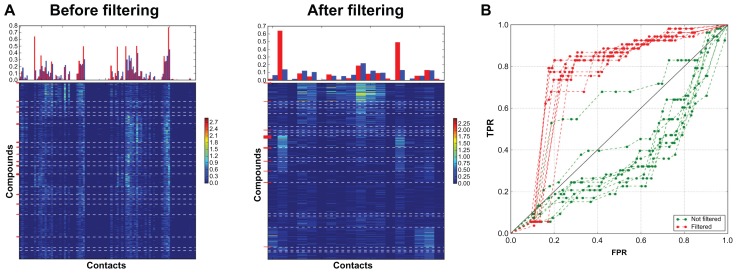
The technique for filtering the contacts that are present in all leaves helps to simplify vectors for the SOM analysis and can increase its efficiency. Additionally, removing equally populated contacts can improve scoring and significantly simplifies visual identification of key contacts on the heat maps. The technique is especially useful when the number of equally populated contacts is much higher than the number of specific contacts. In this case high-populated non selective contacts mask the difference in binding modes and decrease the efficiency of AuPosSOM. Removing these contacts may significantly improve clustering. An example of this is the HIV protease coulomb contact dataset (Figure 5 and Table 1). For good datasets, like DHFR, where a few very specific contacts are present for active compounds and where the number of contacts that are common for all leaves is not higher than the number of specific contacts, filtering decreases the number of decoys in the best leaf (Table 1). However, this may also slightly decrease the number of active compounds. Removing weakly populated contacts does not influence clustering or scoring, but simultaneously simplifies the analysis of the heat maps.
Figure 5.
Clustering for the HIV protease coulomb contact dataset vectors before and after filtering. (A) Fingerprints. (B) AuPosSOM scoring ROC curves for ten runs of clustering with and without filtering.
AuPosSOM scoring function
AuPosSOM scoring function provided good evaluation of the clustering, the scoring consistent with the distribution of the active compounds in the tree for eight datasets. AuPosSOM scoring for the clustered HB contact matrixes without filtering is shown in Figure 6. Scoring function defined the best leaves for eight datasets unambiguously. Heat maps for the corresponding matrixes are displayed in Figure 2. Careful analysis of the contacts revealed that clustering of the best leaf active compounds for the group 1 and progesterone receptor datasets is governed by the presence of selective contacts. For HIV RT, HIV protease, CDK2, and COX1 datasets, the best leaf ligands’ clustering is mainly provided by highly populated contacts. These two types of contacts make a strong positive contribution to scoring, yielding the highest scores for the leaves containing active compounds. The HSP dataset was the most difficult for scoring, as active compounds did not contain remarkably populated HB contacts. This corresponds to a close to zero value of the score for the best leaf.
Figure 6.
AuPosSOM scoring for 20 leaves of the HB contact selection trees for nine datasets.
Notes: Top panels: AuPosSOM scoring for the leaves. Middle panels: average contacts population intensities for the leaves. Bottom panels: distribution of the active compounds in the tree. A 4×5 SOM matrix was used yielding 20 leaves in the tree. Clustering was performed without filtering.
Scoring values provide information about dispersion of the contact populations over the leaves. A negative scoring value indicates that the leaf contains mainly equally populated contacts. The absence of positive scores for all leaves of the tree implies that compounds of the dataset do not exhibit significant differences in the contact sets, and that the probability of efficient clustering is low.
Discussion
We have developed a scoring function for the identification of the active compounds in the AuPosSOM clustered dataset. The results demonstrate that the AuPosSOM contact analysis followed by scoring of the compounds using the scoring function developed can provide a high level of selection for active compounds. For three datasets, the clustering scoring method gave better ROC curves than the best energy scoring functions and for five datasets the efficiency was approximately the same. Thus, this new approach represents an alternative to the energy-based scoring functions and can be more efficient than conventional techniques. Construction of subtrees for the best leaves and cross validation of scoring for different types of contacts proved to be a powerful method for increasing the level of identification of active compounds.
Analysis of the results revealed that clustering efficiency depends strongly on the data set and contact selection. AuPosSOM ROC curves for the filtered coulomb contact selection were better or comparable to those of the conventional scoring functions for eight out of nine datasets, indicating that the clustering approach is more robust than the energy-based one. Score values may be used to estimate the probability for efficient clustering, while information regarding the receptor binding site’s properties can also be helpful.
The clustering was efficient in cases where the difference between contact sets of active compounds and decoys was significant, while the presence of well-populated contacts for active compounds was additionally required for successful scoring. These conditions were satisfied for HB and coulomb contact selections for eight tested datasets. The only exception was the HSP90 target, for which active compounds were characterized by the absence of well-populated, selective HB and coulomb contacts. This may point out the need to search for new types of contact selection. At the same time, energy scoring did not provide good results for this target either. Altogether, these data may indicate that docking failed for HSP90.
Various contact selections were tested for clustering. They characterized two types of contacts: polar and lipophilic. Additionally, all contact selections were probed. Polar contact data sets provided significantly better clustering than lipophilic and all contact selections. Even for hydrophobic active sites, like that of COX-1, lipophilic datasets were not superior. The selection of the atoms by their partial charges appeared to be an efficient method for the evaluation of coulomb interactions, providing the best results for most of the targets. All atom selections failed as a result of masking of specific contacts by a high number of non specific interactions.
An important difference in the AuPosSOM clustering and scoring approach in comparison with the energy scoring approach, is that it takes information about the contacts of all poses of the docked compound simultaneously. This allows for average docking imperfections and avoids errors related to the best pose search. A weakness of this approach might be its inability to evaluate the results correctly when the number of poses with correct contact sets is low. In this setting, the energy scoring-based approach may be used to extract the right pose by energy estimation. Remarkably, in accordance with our results, the scoring functions used in the tests were not efficient for most of the difficult targets. Another important idea is that the contact-based approach does not take the conformation of the pose into consideration. This approach greatly simplifies the analysis, as the main requirement for successful clustering is the presence of a unique set of contacts for active compounds rather than the correct overall conformation of the pose. The latter is often hard to achieve, especially for ligands that were not obtained from the receptor’s crystal structure used for docking.
Fingerprint heat map data representation allows for the identification of key contacts for groups of compounds, as well as easy comparison of contact sets for compounds of different structural families. The examples of the implementation of the AuPosSOM software demonstrate the possibility of its utilization for pharmacophore characterization and its applicability to CAR analysis. The analysis of a contact set of compounds with known activity can directly provide the requirements needed for a search of compounds with the highest binding affinity. The information about key contacts and their populations may also be utilized as a filter for screening large libraries of ligands. One of the recent uses of AuPosSOM clustering is the integrative computational protocol for the discovery of the inhibitors of the Helicobacter pylori nickel response regulator.30
It is necessary to emphasize that the DUD database contains ligands and decoys that have similar physicochemical properties, and thus represents challenging objects for CAR analysis. For AuPosSOM contact-based clustering and scoring, the search for active compounds in libraries containing compounds with highly diverse properties, should be a much easier problem to manage than DUD datasets. Active compounds have a high affinity for the receptor and are expected to form the largest number of high-populated contacts corresponding to the decoys. Consequently, clusters with these vectors will be assigned the highest scores. Additionally, these clusters can be defined by the visual analysis of the heat maps. Good efficiency of clustering for libraries of compounds with highly diverse properties was demonstrated in our previous publication for Thrombin and HIV Protease targets.19
Version 2.0 of AuPosSOM is available online (http://www.aupossom.com). Further improvement of clustering and scoring efficiency is in progress.
Supplementary figures
ROC curves for the Coulomb contact set of protease. (A) Vectors were not filtered, (B) Vectors were filtered.
Notes: Different charges were tested for the contact search. ROC curves obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree.
ROC curves for the coulomb contact sets of protease (A and B) and thrombin (C and D). (A and C) Vectors were not filtered, (B and D) Vectors were filtered.
Notes: Different distances were tested for the contact search. ROC curves were obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree.
ROC curves for the selection 4 contact set of thrombin. (A) Vectors were not filtered, (B) Vectors were filtered.
Notes: Different distances were tested for the contact search. ROC curves were obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree.
Data for the selection 4 contact set of COX1. (A) Fingerprint before filtering, (B) Fingerprint after filtering, (C) ROC curves.
Note: ROC curves were obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree.
ROC curves for the AuPosSOM clustering and conventional scoring functions results.
Notes: ROC curves for AuPosSOM were obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree. ROC curves for the lipophilic contacts 2 selection is not shown as the results are close to those of lipophilic contacts 1. ROC curves for the clustering of the filtered contact matrixes are presented for AuPosSOM.
AuPosSOM scoring.
Notes: ROC curves for ten runs of the AuPosSOM clustering. HB contact dataset. Filled circles: clustering without filtering, blank circles: clustering with filtering.
AuPosSOM scoring.
Notes: ROC curves for ten runs of the AuPosSOM clustering. Coulomb contact dataset. Filled circles: clustering without filtering, blank circles: clustering with filtering.
Acknowledgments
We thank the Language Centre at Paris Descartes University, for English corrections in this manuscript.
Footnotes
Disclosure
The authors report no conflict of interest in this work.
References
- 1.Warren GL, Andrews CW, Capelli A-M, et al. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 2.Moitessier N, Englebienne P, Lee D, Lawandi J, Corbeil CR. Towards the development of universal, fast and highly accurate docking/ scoring methods: a long way to go. Br J Pharmacol. 2008;153(1):S7–S26. doi: 10.1038/sj.bjp.0707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hevener KE, Zhao W, Ball DM, et al. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model. 2009;49:444–460. doi: 10.1021/ci800293n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellenberger E, Rodrigo J, Muller P, Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins. 2004;57:225–242. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 5.Pencheva T, Soumana OS, Pajeva I, Miteva MA. Post-docking virtual screening of diverse binding pockets: comparative study using DOCK, AMMOS, X-Score and FRED scoring functions. Eur J Med Chem. 2010;45:2622–2628. doi: 10.1016/j.ejmech.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 6.Giganti D, Guillemain H, Spadoni J-L, Nilges M, Zagury J-F, Montes M. Comparative evaluation of 3D virtual ligand screening methods: impact of the molecular alignment on enrichment. J Chem Inf Model. 2010;50:992–1004. doi: 10.1021/ci900507g. [DOI] [PubMed] [Google Scholar]
- 7.Kinnings SL, Liu N, Tonge PJ, Jackson RM, Xie L, Bourne PE. A machine learning-based method to improve docking scoring functions and its application to drug repurposing. J Chem Inf Model. 2011;51:408–419. doi: 10.1021/ci100369f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng W, Breneman C, Embrechts MJ. Predicting protein-ligand binding affinities using novel geometrical descriptors and machine-learning methods. J Chem Inf Comput Sci. 2004;44:699–703. doi: 10.1021/ci034246+. [DOI] [PubMed] [Google Scholar]
- 9.Ballester PJ, Mitchell JBO. A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics. 2010;26:1169–1175. doi: 10.1093/bioinformatics/btq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charifson PS, Corkery JJ, Murcko MA, Walters WP. Consensus scoring: A method for obtaining improved hit rates from docking databases of three-dimensional structures into proteins. J Med Chem. 1999;42:5100–5109. doi: 10.1021/jm990352k. [DOI] [PubMed] [Google Scholar]
- 11.Wang R, Wang S. How does consensus scoring work for virtual library screening? An idealized computer experiment. J Chem Inf Comput Sci. 2001;41:1422–1426. doi: 10.1021/ci010025x. [DOI] [PubMed] [Google Scholar]
- 12.Yang JM, Chen YF, Shen TW, Kristal BS, Hsu DF. Consensus scoring criteria for improving enrichment in virtual screening. J Chem Inf Model. 2005;45:1134–1146. doi: 10.1021/ci050034w. [DOI] [PubMed] [Google Scholar]
- 13.Deng Z, Chuaqui C, Singh J. Structural interaction fingerprint (SIFt): a novel method for analyzing three-dimensional protein-ligand binding interactions. J Med Chem. 2004;47:337–344. doi: 10.1021/jm030331x. [DOI] [PubMed] [Google Scholar]
- 14.Renner S, Derksen S, Radestock S, Mörchen F. Maximum common binding modes (MCBM): consensus docking scoring using multiple ligand information and interaction fingerprints. J Chem Inf Model. 2008;48:319–332. doi: 10.1021/ci7003626. [DOI] [PubMed] [Google Scholar]
- 15.Boström J, Hogner A, Schmitt S. Do structurally similar ligands bind in a similar fashion? J Med Chem. 2006;49(23):6716–6725. doi: 10.1021/jm060167o. [DOI] [PubMed] [Google Scholar]
- 16.Fujita S, Orita M. Method of searching for ligand. [Accessed Jul 7, 2012]. pp. 1–19. Pub. No.: WO/2008/035729 International Application No.: PCT/JP2007/068245 [cited 2012 Jul] http://patentscope.wipo.int.
- 17.Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 18.Heberlé G, de Azevedo WF., Jr Bio-inspired algorithms applied to molecular docking simulations. Curr Med Chem. 2011;18(9):1339–1352. doi: 10.2174/092986711795029573. [DOI] [PubMed] [Google Scholar]
- 19.Bouvier G, Evrard-Todeschi N, Girault J-P, Bertho G. Automatic clustering of docking poses in virtual screening process using self-organizing map. Bioinformatics. 2010;26:53–60. doi: 10.1093/bioinformatics/btp623. [DOI] [PubMed] [Google Scholar]
- 20.Kohonen T. Self-Organizing Maps. Heidelberg, Germany: Springer Series in Information Sciences; 2001. [Google Scholar]
- 21.Samsonova EV, Kok JN, Ijzerman AP. TreeSOM: Cluster analysis in the self-organizing map. Neural Netw. 2006;19(6–7):935–949. doi: 10.1016/j.neunet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain AN. Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput Aided Mol Des. 2007;21(5):281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 24.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 25.Lindorff-Larsen K, Piana S, Palmo K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Evnin LB, Vásquez JR, Craik CS. Substrate specificity of trypsin investigated by using a genetic selection. Proc Natl Acad Sci U S A. 1990;87:6659–6663. doi: 10.1073/pnas.87.17.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briand L, Chobert JM, Gantier R, et al. Impact of the lysine-188 and aspartic acid-189 inversion on activity of trypsin. FEBS Lett. 1999;442:43–47. doi: 10.1016/s0014-5793(98)01611-1. [DOI] [PubMed] [Google Scholar]
- 29.Presnell SR, Patil GS, Mura C, et al. Oxyanion-mediated inhibition of serine proteases. Biochemistry. 1998;37:17068–17081. doi: 10.1021/bi981636u. [DOI] [PubMed] [Google Scholar]
- 30.Segura-Cabrera A, Guo X, Rojo-Domínguez A, Rodríguez-Pérez MA. Integrative computational protocol for the discovery of inhibitors of the Helicobacter pylori nickel response regulator (NikR) J Mol Model. 2011;17(12):3075–3084. doi: 10.1007/s00894-011-0962-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ROC curves for the Coulomb contact set of protease. (A) Vectors were not filtered, (B) Vectors were filtered.
Notes: Different charges were tested for the contact search. ROC curves obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree.
ROC curves for the coulomb contact sets of protease (A and B) and thrombin (C and D). (A and C) Vectors were not filtered, (B and D) Vectors were filtered.
Notes: Different distances were tested for the contact search. ROC curves were obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree.
ROC curves for the selection 4 contact set of thrombin. (A) Vectors were not filtered, (B) Vectors were filtered.
Notes: Different distances were tested for the contact search. ROC curves were obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree.
Data for the selection 4 contact set of COX1. (A) Fingerprint before filtering, (B) Fingerprint after filtering, (C) ROC curves.
Note: ROC curves were obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree.
ROC curves for the AuPosSOM clustering and conventional scoring functions results.
Notes: ROC curves for AuPosSOM were obtained using preliminary knowledge of the best leaf position in the tree and distances between the leaves in the tree. ROC curves for the lipophilic contacts 2 selection is not shown as the results are close to those of lipophilic contacts 1. ROC curves for the clustering of the filtered contact matrixes are presented for AuPosSOM.
AuPosSOM scoring.
Notes: ROC curves for ten runs of the AuPosSOM clustering. HB contact dataset. Filled circles: clustering without filtering, blank circles: clustering with filtering.
AuPosSOM scoring.
Notes: ROC curves for ten runs of the AuPosSOM clustering. Coulomb contact dataset. Filled circles: clustering without filtering, blank circles: clustering with filtering.