Abstract
Many eukaryotic membrane proteins have a single C-terminal transmembrane domain that anchors them to a variety of organelles in the secretory and endocytic pathways. These tail-anchored (TA) proteins are post-translationally inserted into the endoplasmic reticulum by molecular mechanisms that have long remained mysterious. This review describes how, in just the last five years, intense research by a handful of labs has led to the identification of all the key components of one such mechanism: the guided entry of TA proteins (GET) pathway, which is conserved from yeast to man. The GET pathway is both surprisingly complicated and yet more experimentally tractable than most other membrane insertion mechanisms, and is rapidly revealing new fundamental concepts in membrane protein biogenesis.
Membrane Protein Targeting to the Endoplasmic Reticulum
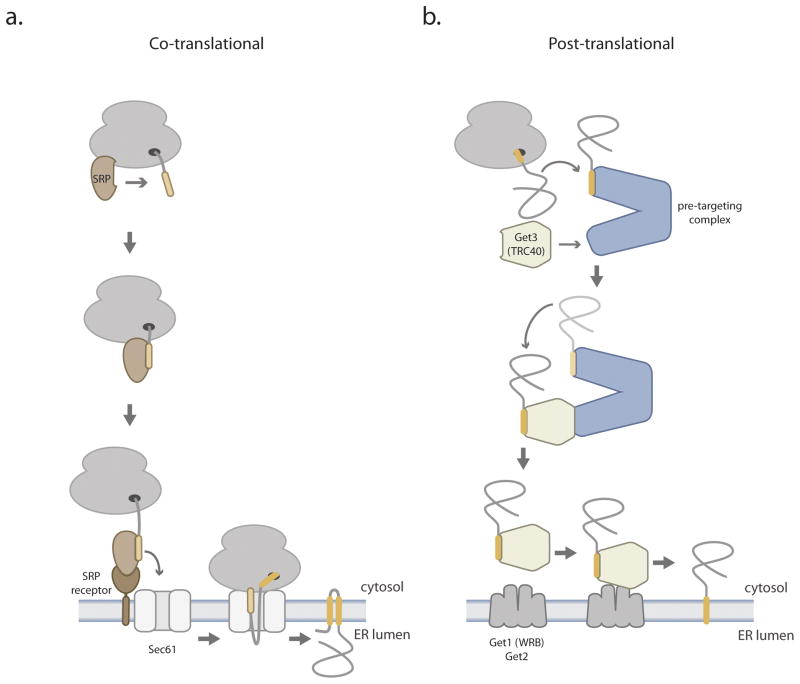
Most integral proteins are embedded in membranes by hydrophobic, alpha helical sequences (~20 amino acids long) called transmembrane domains (TMDs). Eukaryotic cells have a variety of mechanisms that shield TMDs from the moment they emerge out of the ribosome until they are stably inserted into the target organelle. When normal protein targeting is disrupted, membrane proteins can form cytosolic protein aggregates, become targeted to incorrect membranes, or be prematurely destroyed by the proteasome. For most membrane proteins in the secretory pathway, exposure of TMDs to the cytosol is minimized by a mechanism that physically couples protein synthesis to insertion into the endoplasmic reticulum (ER) membrane [1] (Figure 1). This is achieved by co-translational recognition of a hydrophobic sequence on the substrate (either a cleavable signal sequence or the first TMD) by the signal recognition particle (SRP), which is docked near the nascent chain exit tunnel on the ribosome. Signal sequence binding to the SRP causes translational pausing and enhances recruitment to the SRP receptor in the ER membrane. The ribosome and signal sequence are then transferred to the Sec61 channel, protein synthesis resumes, and nascent chains translocate into the ER lumen while allowing TMDs to partition into the lipid bilayer through a lateral gate.
Figure 1. Co-translational and post-translational biogenesis of ER membrane proteins.
(a) Signal sequence (yellow) or transmembrane domain (not shown) recognition by SRP (signal recognition particle) enables coupling of protein translation with translocation across the ER membrane. This is achieved by SRP binding to the SRP receptor, followed by substrate transfer from SRP to Sec61. Consequently, most transmembrane domains pass directly from the ribosome exit tunnel into the Sec61 channel without exposure to the cytosol. Signal sequence cleavage by signal peptidase is not illustrated.
(b) In both the yeast and the mammalian GET pathways (generically illustrated to emphasize commonalities between them), related pre-targeting complexes mediate the transfer of newly synthesized TA proteins from the ribosome to a conserved ER targeting factor (Get3 in yeast, TRC40 in mammals). Get1/2 are two ER membrane proteins that interact with Get3 and comprise the minimal membrane insertion machinery for the yeast GET pathway. WRB is a mammalian Get1 homolog.
Approximately 5% of membrane proteins in the secretory pathway have a single TMD near the C terminus, which also serves as an ER membrane signal sequence. These tail-anchored (TA) proteins are functionally diverse and many are essential. For example, a large fraction of SNAREs (soluble NSF attachment protein receptors, which are proteins that mediate vesicle fusion) are tail-anchored. Historically, a key operational problem of TA protein biogenesis was defined by the discovery of a protein-assisted mechanism that can post-translationally insert TA proteins into the ER membrane independently of Sec61 [2]. After the identification of a TA protein targeting factor [4,5], a flurry of complementary genetic, biochemical, and structural studies have in a short time identified most, if not all, of the components for a novel protein targeting pathway [6,7]. This review presents my synthetic view of the conserved GET (guided entry of TA proteins) pathway in yeast and mammalian cells (Figure 1) with emphasis on fundamental mechanistic insights of general relevance to membrane protein targeting and discusses the critical unanswered questions in the field.
The pre-targeting steps of the GET pathway
Yeast
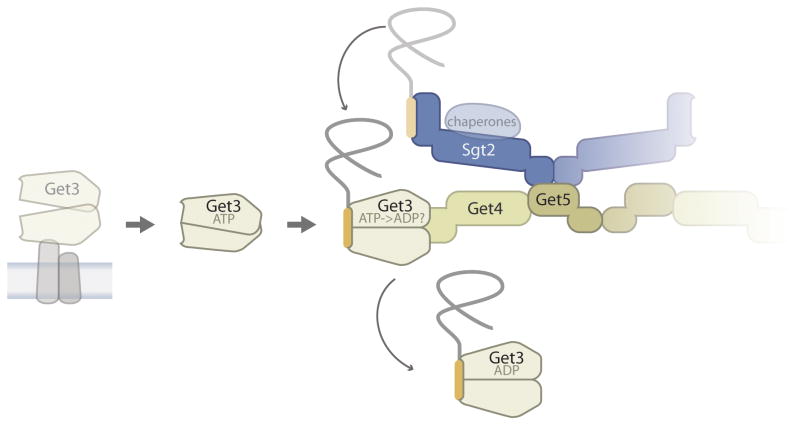
Entry of newly synthesized TA proteins into the GET pathway in Saccharomyces cerevisiae begins with efficient TMD capture by Sgt2 (a small glutamine-rich tetratricopeptide repeat-containing protein) [8] (Figure 2). This chaperone shields TMDs after they are released from the ribosome to prevent TA protein aggregation in the cytosol or mistargeting to mitochondria [9,10,11]. Sgt2 is in a complex with Get4 and Get5, two pathway components that facilitate TA protein transfer from Sgt2 to Get3, a homodimeric ATPase that is the ER membrane targeting factor of the GET pathway [8,11]. This is achieved by a dual mechanism (Figure 2). First, ATP stimulates binding of Get3 to Get4 [12,13,14], and this increases the local concentration of Get3 near TA proteins because of the Get4-Get5-Sgt2 bridge [8]. Second, Get4 increases the intrinsic rate of Get3-TA protein complex formation, most likely by making the Get3 dimer conformation receptive for TMD binding [8] (Figure 2). These biochemical insights agree with the dominant structural view of TMD recognition by Get3 [15]: ATP binding converts the Get3 dimer from an open to a semi-closed state; ATP hydrolysis fully closes the Get3 conformation, creating a composite, hydrophobic groove that most likely cradles the TMD. In addition, there is some evidence for a competing view that tail anchors are sandwiched inside a tetrameric Get3, which has a head-to-head arrangement of hydrophobic grooves [16]. Regardless of the precise structural details, the sum of biochemical and structural evidence has revealed a surprisingly elaborate pre-targeting mechanism that facilitates TMD recognition by Get3.
Figure 2. The pre-targeting steps of the yeast GET pathway.
ATP facilitates the release of Get3 from the membrane. Further destabilization of membrane association probably comes from competitive binding to the pre-targeting complex (containing Get4, Get5, Sgt2, and chaperones) which preferentially transfers TA proteins from Sgt2 to ATP-bound Get3 [13]. This cartoon interprets the stimulatory effect of Get4/5 on the intrinsic rate of substrate handoff [8] as an induced fit in the Get3 conformation. It is not clear if TMD binding promotes ATP hydrolysis by Get3, however, the resulting Get3-TA protein complex is shown as ADP because an assembled hydrophobic groove for substrate binding has only been visualized in the post-hydrolysis state [28,50]. Fade out of one set of Sgt2, Get5, Get4 subunits indicates that the precise stoichiometry of the pre-targeting complex is not known. For a more detailed discussion of how existing structural data fit substrate handoff models of different stoichiometries the reader is referred to a recent review [51]. The chaperones bound to the TPR domain of Sgt2 are shown as a transparent oval because their contribution to the GET pathway is unclear.
Mammalian
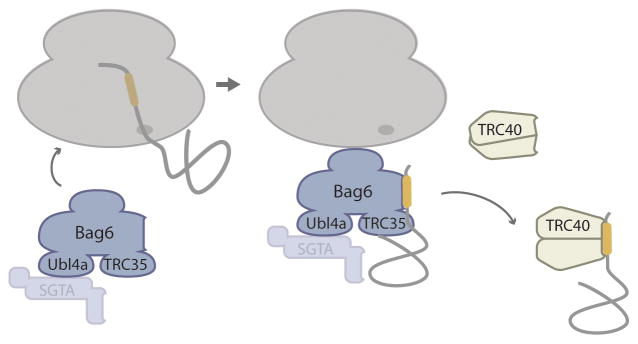
A similar pre-targeting mechanism is also required for delivering mammalian TA proteins to TRC40 (the Get3 homolog; originally named the 40kDa component of the TMD recognition complex). In this case, Bag6 (a protein encoded by BCL2-associated athanogene 6, which has no apparent yeast homolog) captures newly synthesized TA proteins [17,18] as part of a stable protein complex with the mammalian Get4 and Get5 homologs, TRC35 (C7orf20) and Ubl4A, respectively [17,19] (Figure 3). The mammalian homolog of Sgt2 (SGTA) also recognizes TMDs, but it appears to associate weakly with Bag6 [18,20]. Remarkably, even though TMD recognition by the Bag6 complex is post-translational, it can still occur on the ribosome [17]. The precise molecular details remain to be worked out, but in vitro nascent TMD sequences inside the exit tunnel enhance recruitment of the Bag6 complex to the ribosome (Figure 3). Moreover, TMD synthesis slows down mRNA translation, thus presumably allowing enough time for the Bag6 complex to be recruited before translation terminates. More concretely, following completion of proteins synthesis, TA proteins can remain ribosome-associated via the Bag6 complex and then get transferred to TRC40 (Figure 3). It would be satisfying to find that a conserved mechanism in yeast selectively recruits Sgt2 to ribosomes with nearly completed TA protein nascent chains. Certainly, this would strengthen the view that the very hydrophobic nature of most tail anchors necessitated a mechanistic convergence between the GET and SRP/Sec61 pathways, to create distinct substrate channeling mechanisms that minimize TMD exposure to the cytosol during transit from the ribosome to the ER membrane.
Figure 3. The pre-targeting steps of the mammalian GET pathway.
Bag6 (a mammalian protein with no apparent yeast homolog) scaffolds the formation of a stable ternary complex with Ubl4a and TRC35 (the mammalian homologs of Get5 and Get4, respectively). The TMD (yellow) in the exit tunnel somehow recruits this complex to the ribosome to presumably minimize competing off-GET pathway interactions. The Bag6 complex can handoff substrates to TRC40 (the mammalian Get3 homolog) while still associated with the ribosome. SGTA (the mammalian Sgt2 homolog) is shown as a transparent shape to de-emphasize its role, but it probably also recognizes newly synthesized TA proteins and delivers them to TRC40 by dynamically associating with the ternary Ubl4a-Bag6-TRC35 complex.
The membrane-associated steps of the GET pathway
Yeast
Historically, the membrane requirements for TA protein targeting and insertion have been difficult to pin down. A pioneering cell-free study established that insertion of a tail-anchored SNARE protein into mammalian microsomes (ER-derived membranes) was abolished by prior protease treatment of the membrane [2]. Sec61 was excluded as a possible protease-sensitive candidate but the identities of the relevant membrane proteins were not established. Subsequently, this picture was complicated by the discovery that cytochrome b5 (Cb5) is a TA protein that inserts efficiently into liposomes (protein-free lipid bilayers) [21,22]. This could be reconciled with selective Cb5 targeting to the ER in vivo because sterols block insertion into liposomes: because the ER membrane has a relatively low sterol content compared to the other membranes in the secretory and endocytic pathways, it should be selectively permissive for Cb5 insertion. This is, however, difficult to test in vivo because the tools for manipulation of ER lipid composition are still rudimentary.
With the biochemical identification of TRC40 came the exciting possibility of using it as a molecular handle to identify the missing membrane components [23]. Indeed, Get3 had already been genetically and physically linked to two ER membrane proteins, Get1 and Get2 [24,25]. It was not long before a powerful combination of yeast genetics, cell microscopy, and cell-free insertionassays explained the strong ER selectivity of TA proteins [26]: most TA proteins interact with Get3 and are targeted to the ER through the Get3 interaction with its ER receptor, Get1/2 (Figure 1). Thus, after some historical uncertainty, the targeting of most TA proteins fell in line with the tenets of the signal hypothesis [27]: a cytosolic targeting factor recognizes a hydrophobic signal on the substrate and delivers it to the appropriate membrane by interacting with a receptor.
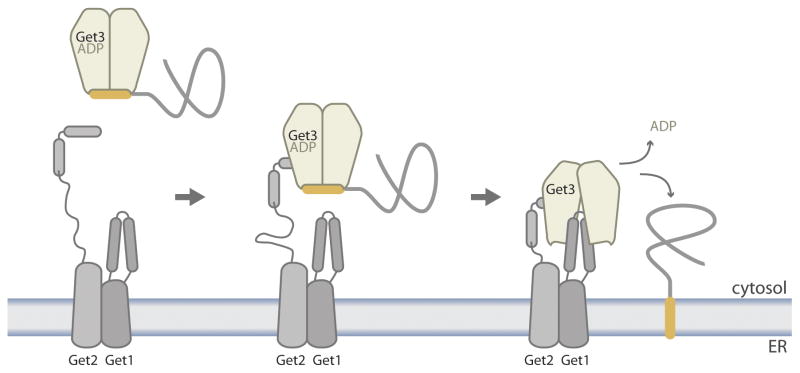
Three recent studies of Get1/2 interactions with Get3 have provided a clear mechanistic insight into one of the least understood steps of the signal hypothesis: how the targeting factor lets go of its substrate. Get1 and Get2 are three-pass proteins that interact via their transmembrane regions and use distinct cytosolic domain (CD) structures to bind individually to Get3 [13,28,29]. Specifically, X-ray crystallography revealed that the very N-terminal region of Get2, which is part of an otherwise intrinsically disordered cytosolic region, forms a complex with a Get3 dimer that is stabilized in the closed conformation by the presence of bound nucleotide [28,29]. This interaction is mediated by a pair of short, charged Get2 alpha helices, separated by a linker, which interact symmetrically with residues largely restricted to one or the other subunit of the Get3 dimer. Notably, Get2 binding is compatible with an assembled hydrophobic groove on Get3, which suggests that Get2 captures Get3-TA protein complexes from the cytosol with a long-reaching tether (Figure 4). This view also agrees with the conformational state of Get3 in the crystal structures of the Get3-Get1CD complex. In those studies, despite inclusion of nucleotide, the Get3 dimer is either fully or partially open and lacks nucleotide [28,29]. This is because the two Get1 coiled coils progressively shuck open the Get3 dimer and, as a result, break apart the substrate-binding groove and nucleotide-binding sites (Figure 4).
Figure 4. Membrane-associated steps of the GET pathway in yeast.
The nucleotide state of the Get3-TA protein complex upon encountering the N-terminal helix-linker-helix motif of Get2 is uncertain. For simplicity, however, it is shown as ADP because ATP hydrolysis is required for substrate release from Get3. The Get1 coiled coil forces ADP dissociation from Get3 and forms a stable complex with an open Get3 dimer lacking nucleotide. This cartoon illustrates the lowest stoichiometry of Get1/2 interactions consistent with biochemical studies of substrate release from Get3. The fact that the cytosolic domains of Get1 and Get2 by themselves form 2:2 complexes with the Get3 dimer has also lead to models with more complex stoichiometries that are discussed elsewhere [7,51,52].
Biochemical reconstitution experiments support the substrate-release mechanism gleaned from the structures and further demonstrate that Get1 and Get2 are the minimal membrane machinery for TA protein insertion [13,28]. Namely, Get3 can still hold its substrate while interacting with Get2’s N-terminal helix-linker-helix motif, but Get1CD binding to Get3-TA protein complexes causes substrate release. Importantly, both in vivo and under physiologically relevant in vitro conditions, efficient TA protein insertion requires Get3 binding to both cytosolic domains of the Get1/2 receptor. This is because, in the absence of the Get2CD tether, the local concentration of a Get3-TA protein complex near Get1CD is not high enough to cause substrate release.
Lastly, the role of the Get3 ATP cycle in coordinating the pre-targeting and membrane-associated steps of the GET pathway is slowly coming into light. There are two crucial observations. First, a Get3-TA protein complex formed in the presence of ATP has to undergo hydrolysis before TA protein release can occur [13,28]. Second, ATP induces dissociation of Get3 from the membrane, which resets the GET pathway to its starting point [13,28] (Figure 3). Consistent with this latter finding, ATP (but not ADP or AMP) competes with Get1CD for Get3 binding. The intimate nature of this competition is illustrated by the conserved loop at the tip of Get1’s coiled coil that extends into the nucleotide-binding pocket of Get3 [28,29]. A fuller understanding of how the GET pathway presumably avoids futile cycles of ATP hydrolysis in the absence of substrate targeting (or whether it uses energy for substrate proofreading) will require a more detailed kinetic analysis. Nonetheless, we have already reached a good understanding of how an ATPase molecular switch coordinates the targeting and insertion stages of the GET pathway.
Mammalian
We know comparatively little about how TRC40 delivers TA proteins to the ER of mammalian cells, but several lines of evidence point to a conserved mechanism. First, WRB (tryptophan-rich basic protein) has weak sequence homology and a similar predicted topology to Get1 [26]. This protein localizes to the ER membrane, where it functions as the TRC40 receptor [30]. Second, the isolated WRB coiled coil forms a complex with TRC40 and inhibits TA protein insertion in vitro [30]. Third, native microsomes recruit but fail to induce substrate release from TRC40-TA protein complexes when ATP hydrolysis is blocked [4]. Last, ATP stimulates dissociation of TRC40 that co-purifies with microsomes [4].
An important piece of evidence that is still missing is whether WRB is necessary for TA protein biogenesis in vivo. Deletion of the GET1 gene in yeast cells results in cytosolic aggregation or mitochondrial mislocalization of secretory pathway TA proteins [26]. Future studies should establish if WRB RNAi knockdown leads to a similar phenotype or if increased TA protein degradation dominates under these conditions [19,31]. Furthermore, the Get2 homolog remains elusive. This is perhaps not surprising given that the majority of the Get2 cytosolic domain is intrinsically disordered and poorly conserved even among closely related yeast species. Thus, the functional requirements for the mammalian equivalent might be unimposing: a TRC40 tether (ie., a helix-linker-helix motif at the end of an intrinsically disordered cytosolic region), and transmembrane domains that interact with WRB. More sophisticated bioinformatic approaches and purification of native WRB complexes should facilitate identification of mammalian Get2, which would nail the point that the GET pathway is conserved from beginning to end.
Another issue is the recent connection between the mammalian GET pathway and ER targeting of secretory peptides. Because of their short length (<100 amino acids), these proteins are synthesized faster than the SRP pathway can target them to the ER membrane, but they still depend on Sec61 for translocation into the ER lumen [32,33]. Surprisingly, TRC40 can recognize secretory peptides in vitro and promote their ER translocation by a WRB-dependent mechanism [34]. These findings raise the intriguing possibility that the GET pathway can feed certain substrates to the Sec61 channel.
Outstanding questions about the GET pathway
Is the ribosome a conserved component of the GET pathway? Proteomic analysis identified Get5 as a ribosome-associated protein [35] but more directed biochemical experiments should establish if the yeast GET pathway, like the mammalian one, also begins on the ribosome. Evidence for a conserved ribosome surveillance mechanism by the GET pathway would be exciting because yeast genetics could then be used to hunt for ribosomal get mutants. More concretely, a mammalian in vitro system should be used to establish whether TA proteins are mistargeted when the connection between the ribosome and the Bag6 complex is severed. Lastly, cryoelectron microscopy studies should look for meaningful structural changes in the ribosome that track with the ability of nascent chain sequences in the exit tunnel to generate a recruitment signal for the Bag6 complex.
What is the role of Sgt2/SGTA-tethered heat shock proteins (Hsps)? Prior to the discovery that SGTA directly recognizes tail anchors, many studies established that it also co-chaperones Hsp-mediated protein folding of non-TA protein substrate [36,37,38]. The interaction between Sgt2 and Hsps, however, is not apparently crucial for efficient TA protein capture and handoff [8,11]. Nonetheless, a possible connection between Hsps and the GET pathway might still exist. Specifically, protein localization by fluorescence cell microscopy of get1/2 mutants revealed that Get3 is recruited to TA protein aggregates in a Get4/5-dependent manner [39]. Thus, establishing if Sgt2/SGTA and/or the associated Hsps go after TA protein aggregates to maintain protein homeostasis is an important future goal.
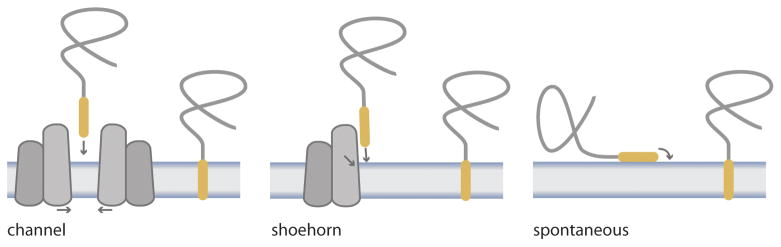
What is the mechanism of the insertion step? It is not known how tail anchors get inserted into the membrane after they are released from Get3. One proposal is that this occurs spontaneously [21] (Figure 5). In this view, the GET pathway is done with the tail anchor once it deposits it on the cytosolic surface of the ER membrane. Indeed, moderately hydrophobic peptides spontaneously insert into liposomes all on their own [40]. Most tail anchors are significantly more hydrophobic, however, which is why the GET pathway is necessary to maintain them in a soluble state and, at the same time, confer selectivity for the ER membrane. Thus, an alternative hypothesis is that the GET pathway assists the insertion step [4] (Figure 5) presumably to reduce a kinetic barrier to spontaneous insertion. Spontaneous insertion models for protein targeting pathways are intrinsically difficult to prove and have historically been in decline [41]. Future studies of the structure and function of Get1/2 transmembrane domains should reveal whether another spontaneous insertion model bites the dust.
Figure 5. Three hypothetical mechanisms for the insertion step of the GET pathway.
The ‘channel’ and ‘shoehorn’ evoke more or less elaborate mechanisms, respectively, by which Get1/2 (shown as the two grey membrane protein shapes) could be chaperoning tail anchors (yellow) into the lipid bilayer. Any future structure/function studies of Get1/2 in support of a chaperoned insertion mechanism should attempt to explain the apparent thermodynamic membrane partitioning of the anchor following release from TRC40 [54]. For simplicity, Get3 is not illustrated, but it could also play a more active role during the insertion step. In the ‘spontaneous’ model, following release from Get3, the tail anchor first associates with the cytosolic leaflet of the ER membrane and then flips the C terminus across. The thermodynamics of this kind of mechanism are well-established for model membrane-inserting peptides [48].
How will the GET pathway grow up?
The emergence of the GET pathway as the dominant mechanism for targeting TA proteins to the ER raises a nagging question: why is the GET pathway not essential in budding yeast (nota bene: mice lacking TRC40 die during early embryogenesis [32])? One obvious interpretation is that other targeting pathways work redundantly with the GET pathway to either help carry the load, or split the clientele. This could be mediated through tail anchor interactions with the SRP, protein folding chaperones, and/or calmodulin, which have all been observed in vitro [33,34]. It has been difficult, however, to mechanistically build alternative pathways from these observations or establish their physiological significance. A related possibility is that get mutants turn on the heat shock response, similar to the yeast srp mutants [35], to compensate by providing an inefficient TA protein delivery mechanism to Sec61. Conversely, disruption of the GET pathway might indirectly perturb other ER targeting pathways (or more generally, protein homeostasis) by engenderinga competition between TA proteins and other hydrophobic substrates. Recent technological advances in ribosome profiling [36] point a way out of this morass, at least in mammalian cells. Specifically, because the Bag6 complex is selectively recruited to ribosomes that are synthesizing TA proteins, footprinting of ‘Bag6-marked’ ribosomes should reveal the natural flux of substrates through the GET pathway in unperturbed cells.
Our mechanistic understanding of the GET pathway has gotten sophisticated over the past five years. Biochemical reconstitution approaches carried out in yeast and mammalian cell-free systems have lead to a coherent view of how newly synthesized TA proteins hop from a pre-targeting complex to Get3 or TRC40, respectively. In yeast, we even know the structures of many of the pre-targeting components and have a basic understanding of how they work. More broadly, substrate handoff between two chaperones is a recurring mechanism in biology. For example, the folding of many regulatory proteins in eukaryotes depends on their transfer from Hsp70 to Hsp90 as part of a single complex organized by a scaffolding protein [37]. The two significant technical challenges associated with studying this mechanism are the biochemically complex nature of Hsp90 and the poorly characterized and multivalent hydrophobic epitopes on the substrate. In comparison, the tail anchor is a well-defined substrate that moves through a more elementary chaperone machine. Nonetheless, it is remarkable that Sgt2 and Get3 do not apparently incur the energetic cost of exposing empty substrate-binding sites to the cytosol during substrate transfer. Future structural and single-molecule studies of the role of Get4/5 during TA protein handoff should extend our understanding of chaperone trapeze acts in cell biology.
Arguably, the hardest conceptual problem in all membrane insertion pathways is visualizing TMD insertion into the lipid bilayer (Figure 5). Structural studies of the Sec61 translocon have revealed an elegant solution to this: a protein translocation channel with a lateral gate [1]. Nonetheless, this insertion mechanism is difficult to probe further because both Sec61 and ribosome-associated substrates are biochemically complex and staging the insertion reaction for detailed kinetic analysis is challenging. By contrast, the insertion step of the GET pathway is specialized for insertion of a single hydrophobic epitope without coupling to biosynthesis and extensive protein translocation. Consequently, it has relatively few moving parts and they can all be made recombinantly. Staging will still be a challenge but the accessibility of the TMD C terminus offers hope that conditional roadblocks can be attached to it. As such, structural and functional studies of the Get1/2 complex are promising to move us one step closer towards a fundamental understanding of how cells exploit chaperones to put transmembrane domains into lipid bilayers.
Acknowledgments
The author would like to thank members of the Denic lab for critical reading of the manuscript and B. Toyama for help with graphics. The work in the author’s lab is supported by the NIH (RO1GM0999943-01), the Ellison Medical Foundation, and Harvard University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 2.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 4.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 5.Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12:787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battle A, Jonikas MC, Walter P, Weissman JS, Koller D. Automated identification of pathways from quantitative genetic interaction data. Mol Syst Biol. 2010;6:379. doi: 10.1038/msb.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohl C, Tessarz P, von der Malsburg K, Zahn R, Bukau B, et al. Cooperative and independent activities of Sgt2 and Get5 in the targeting of tail-anchored proteins. Biol Chem. 2011;392:601–608. doi: 10.1515/BC.2011.066. [DOI] [PubMed] [Google Scholar]
- 12.Chartron JW, Suloway CJ, Zaslaver M, Clemons WM., Jr Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci U S A. 2010;107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Whynot A, Tung M, Denic V. The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell. 2011;43:738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang YW, Chuang YC, Ho YC, Cheng MY, Sun YJ, et al. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2010;285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson PJ, Schwappach B, Dohlman HG, Isaacson RL. Structures of Get3, Get4, and Get5 provide new models for TA membrane protein targeting. Structure. 2010;18:897–902. doi: 10.1016/j.str.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suloway CJ, Rome ME, Clemons WM., Jr Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue. EMBO J. 2012;31:707–719. doi: 10.1038/emboj.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leznicki P, Clancy A, Schwappach B, High S. Bat3 promotes the membrane integration of tail-anchored proteins. J Cell Sci. 2010;123:2170–2178. doi: 10.1242/jcs.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, et al. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell. 2011;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winnefeld M, Grewenig A, Schnolzer M, Spring H, Knoch TA, et al. Human SGT interacts with Bag-6/Bat-3/Scythe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp Cell Res. 2006;312:2500–2514. doi: 10.1016/j.yexcr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Borgese N, Brambillasca S, Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, et al. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005;24:2533–2542. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandon EC, Gilmore R. The tail end of membrane insertion. Cell. 2007;128:1031–1032. doi: 10.1016/j.cell.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Auld KL, Hitchcock AL, Doherty HK, Frietze S, Huang LS, et al. The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae. Genetics. 2006;174:215–227. doi: 10.1534/genetics.106.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 26.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blobel G, Walter P, Chang CN, Goldman BM, Erickson AH, et al. Translocation of proteins across membranes: the signal hypothesis and beyond. Symp Soc Exp Biol. 1979;33:9–36. [PubMed] [Google Scholar]
- 28.Mariappan M, Mateja A, Dobosz M, Bove E, Hegde RS, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefer S, Reitz S, Wang F, Wild K, Pang YY, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333:758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vilardi F, Lorenz H, Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J Cell Sci. 2011;124:1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, et al. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay R, Ho YS, Swiatek PJ, Rosen BP, Bhattacharjee H. Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett. 2006;580:3889–3894. doi: 10.1016/j.febslet.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Rabu C, Schmid V, Schwappach B, High S. Biogenesis of tail-anchored proteins: the beginning for the end? J Cell Sci. 2009;122:3605–3612. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassdenteufel S, Schauble N, Cassella P, Leznicki P, Muller A, et al. Ca2+-calmodulin inhibits tail-anchored protein insertion into the mammalian endoplasmic reticulum membrane. FEBS Lett. 2011;585:3485–3490. doi: 10.1016/j.febslet.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mutka SC, Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol Biol Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh E, Becker AH, Sandikci A, Huber D, Chaba R, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- 40.Chirico WJ, Waters MG, Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- 41.Plath K, Rapoport TA. Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J Cell Biol. 2000;151:167–178. doi: 10.1083/jcb.151.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matlack KE, Misselwitz B, Plath K, Rapoport TA. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- 43.Shao S, Hegde RS. A calmodulin-dependent translocation pathway for small secretory proteins. Cell. 2011;147:1576–1588. doi: 10.1016/j.cell.2011.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Neil KT, DeGrado WF. How calmodulin binds its targets: sequence independent recognition of amphiphilic alpha-helices. Trends Biochem Sci. 1990;15:59–64. doi: 10.1016/0968-0004(90)90177-d. [DOI] [PubMed] [Google Scholar]
- 45.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tobaben S, Thakur P, Fernandez-Chacon R, Sudhof TC, Rettig J, et al. A trimeric protein complex functions as a synaptic chaperone machine. Neuron. 2001;31:987–999. doi: 10.1016/s0896-6273(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 47.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reshetnyak YK, Andreev OA, Segala M, Markin VS, Engelman DM. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc Natl Acad Sci U S A. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981;23:411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 50.Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chartron JW, Clemons WM, Jr, Suloway CJ. The complex process of GETting tail-anchored membrane proteins to the ER. Curr Opin Struct Biol. 2012;22:217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinning I, Bange G, Wild K. It takes two to Get3. Structure. 2011;19:1353–1355. doi: 10.1016/j.str.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Lyman SK, Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- 54.Leznicki P, Warwicker J, High S. A biochemical analysis of the constraints of tail-anchored protein biogenesis. Biochem J. 2011;436:719–727. doi: 10.1042/BJ20101737. [DOI] [PMC free article] [PubMed] [Google Scholar]