Abstract
Background
Genome-wide association studies have identified multiple variants associating with coronary artery disease (CAD) and myocardial infarction (MI). Whether a combined genetic risk score (GRS) is associated with prevalent and incident MI in high risk subjects remains to be established.
Methods and Results
In subjects undergoing cardiac catheterization (n=2597) we identified cases with a history of MI onset at age <70 years and controls ≥70 years old without prior MI, and followed them for incident MI and death. Genotyping was performed for 11 established CAD/MI variants and a GRS calculated based on average number of risk alleles carried at each locus weighted by effect size. Replication of association findings was sought in an independent angiographic cohort (n=2702). The GRS was significantly associated with prevalent MI, occurring before age 70 years, compared to older controls (≥70 years) with no history of MI (p<0.001). This association was successfully replicated in a second cohort yielding a pooled p value of <0.001. The GRS modestly improved the c-statistic in models of prevalent MI with traditional risk factors. However, the association was not statistically significant when elderly controls without MI but with stable angiographic CAD were examined (pooled p=0.11). Finally, during a median 2.5 year follow-up, only a non-significant trend was noted between the GRS and incident events, which was also not significant in the replication cohort.
Conclusions
A GRS of 11 CAD/MI variants is associated with prevalent MI but not near term incident adverse events in two independent angiographic cohorts. These findings have implications for understanding the clinical utility of genetic risk scores for secondary as opposed to primary risk prediction.
Keywords: cardiovascular disease risk factors, coronary artery disease, myocardial infarction, Genetic Risk Score
Introduction
Genetic susceptibility for myocardial infarction (MI), the potentially fatal consequence of coronary artery disease (CAD), is due at least in part, to the cumulative effect of common polymorphisms with each risk variant conferring a small additive effect.1, 2 Recent genome wide association studies (GWAS) have identified multiple single nucleotide polymorphisms (SNPS) associating with both MI and CAD.3-9 Combination of these variants into a risk score may thus permit examination of the cumulative effect of risk SNP burden on prevalent and incident MI.
Kathiresan et al. demonstrated that a genetic risk score (GRS) based on the 9 variants identified in their GWAS, was linearly associated with their primary phenotype of early onset MI.9 However, as the GRS was based on the same discovery cohort the effect size may have been overestimated. Replication in independent cohorts is therefore required to demonstrate that (1) a GRS derived from large GWAS can be applied to other independent populations, (2) such a GRS is also associated with non premature MI and (3) that a GRS that predicts prevalent MI or CAD is also able to predict future incident major adverse events.
Recent large prospective studies in younger low risk community cohorts have either confirmed or refuted the value of GWAS derived GRS in prediction of incident MI over 10-12 years.10-12 This discrepancy may be because these variants associate primarily with CAD and mechanisms underlying development of atherosclerosis, but not with MI per se. Attempts to dissect these two phenotypes require study of cohorts with detailed phenotyping for both MI and CAD.13, 14 However, to date, no studies have examined the association between a GRS and either prevalent or incident MI in the context of detailed CAD phenotyping. Furthermore, little data exists on use of such a score to predict future prospective risk of adverse events in a CAD cohort in whom secondary risk prediction algorithms are currently inadequate.15
We therefore sought to examine the association between a genetic risk score, based on 11 CAD/MI variants discovered through GWAS, with (1) prevalent MI with onset <70 years of age and (2) incident MI and death in a cohort of subjects undergoing coronary angiography for diagnosis or treatment of CAD.
Methods
Study Design
All primary and subgroup association analyses were performed in the Emory Cardiovascular Biobank (Atlanta, Georgia, USA) with replication of key findings sought in a second independent cohort from the Cleveland Clinic Genebank (Cleveland, Ohio, USA). Significant association results were then pooled through meta-analysis.
Study Samples
The Emory Cardiovascular Biobank consists of over 5000 consecutively recruited patients undergoing cardiac catheterization at Emory University and affiliated hospital sites since 2003 with documented demographic characteristics, medical histories, behavioral factors, and fasting blood samples. Specific details regarding risk factor definitions and coronary angiographic phenotyping have been described previously.16, 17 Family history (FH) of CAD was defined as having any first degree relative (parents, siblings) with MI or coronary revascularization prior to the age of 60 years. All subjects were prospectively followed by telephone interview, chart abstraction and through state vital records data to document major cardiovascular events and death. Follow-up was performed at time periods ranging from 1-5 years. Events were considered as outcomes if they occurred greater than 72 hours after date of discharge. Patients with heart transplantation and missing or incomplete phenotype data were also excluded. As most risk variants have not yet been validated in non-Caucasian populations, we excluded those with self reported non-Caucasian ancestry. The study was approved by the Institutional Review Board and all subjects provided written informed consent.
The Cleveland Clinic GeneBank study is a single site sample repository generated from consecutive patients undergoing elective diagnostic coronary angiography or elective cardiac computed tomographic angiography with extensive clinical and laboratory characterization and longitudinal observation. Subject recruitment occurred between 2001 and 2006. The GeneBank cohort has been used previously for discovery and replication of novel genes and risk factors for atherosclerotic disease.18-22 Ethnicity was self-reported and information regarding demographics, medical history, and medication use was obtained by patient interviews and confirmed by chart reviews. Angiographic CAD was determined as evidence of ≥ 50% stenosis of one or more major epicardial vessel. Prospective cardiovascular risk was assessed by the incidence of major adverse cardiac events (MACE) during three years of follow up from the time of enrollment, which included MI, stroke, and all-cause mortality. Adjudicated outcomes ascertained over the ensuing 3 years for all subjects following enrollment were confirmed using source documentation. All patients provided written informed consent prior to being enrolled in GeneBank and the study was approved by the Institutional Review Board of the Cleveland Clinic.
Prevalent MI
MI was consistently defined according to standard criteria from medical records as documented history of acute (at enrollment) or remote MI.23 Age at first MI was recorded to the closest year. Subjects with MI onset at age <70 years were defined as cases. We also specified a-priori subgroup analyses to explore association with more premature MI defined as cases with MI <60 and <50 years of age. To mitigate case misclassification bias, whereby younger control subjects could become cases by developing MI in later life, we selected subjects ≥70 years at enrollment without a history of prior MI (or during the study period), as controls. These controls included those without ever experiencing an MI, but who may have had significant angiographic CAD. Given that our statistical test of MI versus no MI would also test the hypothesis of CAD versus no CAD, we utilized previously described methods to examine the effect of the risk score on odds of MI in the presence of significant CAD determined as angiographic CAD ≥50% luminal stenosis in any major epicardial vessel (MI+/CAD+ versus MI-/CAD+).13, 14
Incident MI
All patients in both the Emory and Cleveland Clinic cohorts were prospectively followed for adverse events, as described above. The primary endpoint for incident event analysis in this study was defined as incident MI, using standard criteria as described above. The composite outcome of all cause death and MI was defined as a secondary endpoint.
SNP Selection and Genotyping
We selected CAD/MI risk variants discovered through GWAS and published before June 2009, whose primary phenotype was MI or a combination of CAD/MI and which satisfied the genome wide association threshold of 5×10−8, (reflecting a million tests of association). The selected SNPs are listed in Table 1 along with risk allele frequencies for each cohort with published effect sizes. The LPA locus was not included for logistical reasons despite publication in 2009.24, 25
Table 1.
Details of the selected myocardial infarction loci and their risk variants in each cohort
| Emory CV Biobank |
Cleveland Clinic Genebank |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Locus | SNP ID | Nearest Gene | Risk allele | Published risk allele frequency |
Observed frequency |
HWE p value |
Observed frequency |
HWE p value |
Reported allelic MI risk (OR) |
Ref* |
| 1p13 | rs646776 | CELSR2-PSRC1- SORT1 |
T | 0.81 | 0.80 | 0.18 | 0.80 | 0.80 | 1.19 (1.13-1.26) | 9 |
| 1p32 | rs11206510 | PCSK9 | T | 0.81 | 0.82 | 0.19 | 0.83 | 0.12 | 1.15 (1.10-1.21) | 9 |
| 1q41 | rs17465637 | MIA3 | C | 0.72 | 0.73 | 0.89 | 0.73 | 0.26 | 1.14 (1.10-1.19) | 9 |
| 2q33 | rs6725887 | WDR12 | C | 0.14 | 0.14 | 0.33 | 0.15 | 0.83 | 1.17 (1.11-1.23) | 9 |
| 3q22.3 | rs9818870 | MRAS | T | 0.15 | 0.16 | 0.89 | 0.16 | 0.53 | 1.15 (1.11-1.19) | 7 |
| 6p24 | rs12526453 | PHACTR1 | C | 0.65 | 0.67 | 0.09 | 0.67 | 0.21 | 1.12 (1.08-1.17) | 9 |
| 9p21 | rs10757278 | CDKN2A/B | G | 0.50 | 0.50 | 0.50 | 0.53 | 1.00 | 1.29 (1.22-1.35) | 3 |
| 10q11 | rs1746048 | CXCL12 | C | 0.84 | 0.87 | 0.66 | 0.86 | 0.64 | 1.17 (1.11-1.24) | 9 |
| 12q24 | rs3184504 | SH2B3 | T | 0.47 | 0.49 | 0.30 | 0.52 | 0.03 | 1.13 (1.08-1.18) | 8 |
| 19p13 | rs1122608 | LDLR | G | 0.75 | 0.75 | 1.0 | 0.76 | 0.22 | 1.15 (1.10-1.20) | 9 |
| 21q22 | rs9982601 | SLC5A3-MRPS6-KCNE2 | T | 0.13 | 0.13 | 0.25 | 0.15 | 0.66 | 1.20 (1.14-1.27) | 9 |
References relate to studies utilizing myocardial infarction as the primary phenotype, from which values in the table are derived, except 3q22.3 for which a combined phenotype was used.
Genotyping for Emory Cardiovascular Biobank Samples was performed using the Centaurus (Nanogen) platform at deCODE genetics, Reykjavik, Iceland or with the SNPstream (Beckman Coulter) platform at Emory University, Atlanta, GA. 26, 27 Genotyping for Cleveland Clinic GeneBank samples was performed using the Affymetrix Genome-Wide Human SNP Array 6.0 platform at the University of Ottawa with imputation for untyped SNPs using MACH 1.0 software. Full details are provided in the Supplementary Methods.
Genetic Risk Score (GRS)
A weighted GRS was calculated based on the average number of risk alleles (0, 1 or 2) in the SNPs genotyped weighted by the effect size published in larger scale studies (natural log of the OR x no. of risk alleles for each SNP), using the “Profile Scoring” option in PLINK.28 Thus, 2 copies of the risk allele 9p21 (published OR per risk allele 1.3) contributed proportionately more to the overall GRS for an individual than two copies of the risk allele 6p24 (OR 1.1). For the Emory cohort, missing genotype data for individual SNPs was accounted for by mean imputation based on overall risk allele frequency. Details of missingness are provided in Supplementary Table 1. We performed validation analysis using only data from subjects with the full complement of genotyped variants to ensure imputation did not adversely influence our results. Cleveland Clinic genotype data was derived from imputation of a GWAS dataset (Supplementary Methods).
Statistical Analysis
Continuous variables are presented as means (SD) and categorical variables as proportions (%) with one way ANOVA and chi-squared tests used to determine differences between groups.
Prevalent MI association analysis
Logistic regression models were constructed to first test the effect of the individual SNPs (under an additive model, with risk allele coding 0,1,2) and then the weighted GRS, as a continuous variable on prevalent MI risk (case= MI<70yrs; controls = no MI≥70yrs). We further examined the association with the GRS in younger cases, defined as MI<60 years and MI<50 years against the same control group. Replication analyses were performed in the Cleveland Clinic Genebank, with a random effects meta-analysis to combine results in order to derive a final pooled estimate. For illustrative purposes, we then categorized into quintiles of GRS to derive estimates for effect size (OR) for the top quintile compared to the first quintile as the referent category. We also tested for association between the continuous GRS and MI in all patients with CAD (MI+/CAD+ versus MI-/CAD+), again with attempted replication in Cleveland Clinic Genebank. This was performed using the same cases (MI<70yrs) but selecting controls (≥70 yrs) without prior MI but with significant stable angiographic CAD (defined as ≥50% stenosis). Analyses were performed before and after adjusting for gender, body mass index, diabetes, hypertension, hyperlipidemia, smoking and family history (FH) of premature MI/CAD. Interaction terms were tested for association between the risk score and risk factors including FH.
Incremental predictive value of the GRS for prevalent MI occurring before the age of 70 in the Emory cohort was tested using ROC analysis and estimated through the c-statistic. Models were constructed with and without the risk score using (1) traditional risk factors (gender, BMI, diabetes, hypertension, hyperlidemia, smoking and FH), and (2) with gender and FH only, which are the only variables likely to be known in early life, when risk prediction could be employed.
Incident MI risk analysis
Cox regression analyses were applied to identify associations between the individual SNPs (under an additive model, with risk allele coding 0,1,2) and then the GRS with risk of incident MI and combined death and MI during follow-up. Models were constructed using both a continuous and a categorized GRS, with and without traditional risk factors and presence of baseline angiographic CAD (≥50%). Hazard Ratios (HR) were estimated for the top quintile of the GRS referenced to the first quintile as before. Subgroup analyses included stratifying by history of MI at enrollment to examine risk of recurrent and first MI during follow up and examining incident risk in younger subjects <60 years. The proportional hazards assumption was evaluated graphically using log-log plots.
A 2-tailed P value <0.05 was considered significant. PLINK statistical software was used to estimate Hardy-Weinberg equilibrium and minor allele frequencies for all SNPs.28 All remaining statistical analyses were performed using SAS (Cary, NC) and SPSS 17.0 (Chicago, IL).
Results
For the 11 SNPs genotyped in the Emory and Cleveland Clinic cohorts, the observed frequencies for each risk allele were similar to published and Hap-Map data. All SNPs were in Hardy Weinberg equilibrium (HWE) with p-value threshold of 0.01 considering multiple testing. Only one SNP, rs3184504, in the Cleveland Clinic Genebank had a nominal p < 0.05 (Table 1). A total of 2597 self-reported Caucasians (mean age 63.9 ± 11.1 years and 67% male) were genotyped from the Emory Cardiovascular Biobank and 2702 (mean age63.0 ± 11.2 years and 72% male) from the Cleveland Clinic Genebank. Although the two cohorts were generally similar, differences were noted including for example a greater prevalence of treated hyperlipidaemia and smoking in the Cleveland Clinic Genebank compared to those in the Emory Cardiovascular Biobank (Table 2).
Table 2.
Details of demographics for each cohort and by case-control status for prevalent MI analysis
| Emory Cardiovascular Biobank | Cleveland Clinic Genebank | Overall Comparison |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Overall N=2597 |
Cases (MI<70 yrs) N=889 |
Controls (no MI≥70 yrs) N=449 |
P value | Overall N=2702 |
Cases (MI<70 yrs) |
Controls (no MI≥70 yrs) |
P value | P value | |
|
|
|||||||||
| Age, yrs | 63.9 (11.3) | 62.9 (10.0) | 75.7 (4.20) | <0.001 | 63.0 (11.2) | 60.4 (10.4) | 76.0 (4.3) | <0.001 | 0.998 |
| Male Gender | 67.6 | 76.2 | 59.0 | <0.001 | 71.7 | 77.8 | 52.5 | <0.001 | 0.001 |
| Body Mass Index, kg/m2 | 28.9 (5.7) | 29.7 (5.8) | 27.6 (5.3) | <0.001 | 29.2 (5.6) | 29.5 (5.7) | 27.8 (5.2) | <0.001 | 0.023 |
| Diabetes | 30.2 | 35.9 | 27.2 | 0.001 | 20.1 | 18.8 | 20.0 | 0.572 | <0.001 |
| Hypertension | 70.6 | 70 | 73.9 | 0.148 | 72.4 | 74.2 | 71.0 | 0.181 | 0.155 |
| Hyperlipidemia | 68.2 | 77.8 | 69.0 | 0.001 | 88.2 | 93.8 | 75.7 | <0.001 | <0.001 |
| Ever Smoked | 61.2 | 67.8 | 55.6 | 0.001 | 70.8 | 78.5 | 58.8 | <0.001 | <0.001 |
| Family History CAD <60yrs | 15.3 | 20.5 | 8.7 | <0.001 | N/A | NA | NA | NA | N/A |
Mean and standard deviations or % presented unless otherwise indicated. Overall comparison for Emory versus Cleveland Clinic cohorts
Prevalent MI association with a genetic risk score
For prevalent MI (cross sectional) analysis, we examined 889 cases of MI<70 years and 449 controls ≥70 years without prior MI from the Emory Cardiovascular Biobank (Table 2). The replication analysis included 1154 cases and 495 controls from Cleveland Clinic Genebank. Cases were significantly younger than controls by design, and expected significant differences in other risk factors were also observed (Table 2). Individual SNP associations with prevalent MI are presented in Supplementary Table 2. While trends were noted for association with most of the SNPS, statistically significant associations were noted only for SNPs at loci 1q41 (rs17465637), 6p24 (rs12526453), 9p21 (rs10757278) and 21q22 (rs9982601) in the Emory cohort and only for 9p21 (rs10757278) in the Cleveland Clinic Genebank.
In our primary analysis there was a significant association between the overall weighted GRS and prevalent MI, when modeled as a continuous variable (p=9.74×10−5) (Table 3). A modest but statistically significant association was identified with prevalent MI using the same definitions and analysis methods (p=0.04). When combined in the pooled analysis, there was a significant overall association for the GRS with prevalent MI (p=4.5×10−5) (Table 3). These findings persisted after adjustment for risk factors (pooled p value=1.4×10−4)
Table 3.
Association between weighted risk scores and prevalent MI <70 years with and without consideration of CAD
| Emory Cardiovascular Biobank |
Cleveland Clinic Genebank | Pooled Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| Beta | SE | P | Beta | SE | P | Effect | SE | P | P adj | Direction | |
| MI <70yrs v no MI≥70 yrs (All) |
17.05 | 4.37 | 9.74×10−5 | 7.63 | 3.76 | 0.04 | 11.64 | 2.85 | 4.51×10−5 | 1.4×10−4 | ++ |
| MI<70 yrs v no MI≥70 yrs (CAD+)* |
11.73 | 4.96 | 0.02 | 0.69 | 4.95 | 0.89 | 6.20 | 3.50 | 0.08 | 0.11 | ++ |
See methods. Adjusted in both groups for gender, BMI, diabetes, hypertension, hyperlipidaemia, smoking
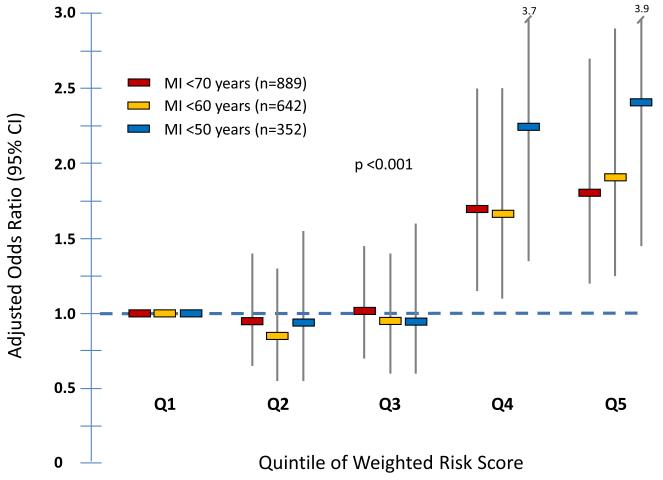
To illustrate this association further, we grouped the Emory cohort by quintile of GRS. In comparison to the lowest quintile, the adjusted OR for MI for an individual in the highest quintile of GRS was 1.81 (95% CI 1.21-2.70) (Figure 1). Furthermore, the effect sizes were greater in cases with MI <60 years or <50 years of age compared to older controls. Thus, the adjusted OR for MI for the highest quintile of GRS compared to the lowest was 1.90 (95% CI 1.24-2.92) for MI<60 years and 2.37 (95% CI 1.44-3.91) for MI<50 years (Figure 1). Interestingly, it appears that a significant increase in odds of MI was only observed in the highest quintiles, with no apparent increase between the first 3 quintiles (Figure 1).
Figure 1.
Association between quintiles of genetic risk score and MI in different age groups within the Emory Cardiovascular Biobank. Adjusted Odds Ratios and 95% confidence intervals are presented. Covariates included gender, BMI, diabetes, hypertension, hyperlipidemia, smoking and family history of CAD.
We did not observe significant interactions between the GRS and covariates in the association with MI <70 years. Most pertinent was the lack of interaction for family history (FH) of CAD (p for interaction 0.63). Overall, the mean weighted GRS was marginally higher in subjects with a positive FH compared to those with a negative or unknown FH (0.085±0.01 v 0.083±0.01 respectively, p=0.05).
Given the significant association with prevalent MI, we examined the additive value of the GRS when modeled with existing risk factors for prevalent MI risk at <70 years using the area under the curve statistic. There was a statistically significant increase in the c-statistic when the GRS was added to a model containing gender, BMI, traditional risk factors and FH (0.681 (0.015) v 0.693 (0.015), p=0.012) (Table 4). Similarly, when modeled for MI at younger ages, there was again a significant increase in the c-statistic (<60yrs 0.699 (0.016) v 0.712 (0.016) p=0.03; <50yrs 0.716 (0.018) v 0.733 (0.018), p=0.04) (Table 4). The difference between the AUC was even greater when the GRS was modeled with the non modifiable risk factors of FH and gender (difference in AUC = 0.027; 0.030; 036 for MI <70; <60 and <50yrs respectively, all p<0.005) (Table 4).
Table 4.
Discriminatory analysis using AUC for prevalent MI in the Emory Cardiovascular Biobank
| Traditional RFs |
Traditional RFs + GRS |
Improvement in C statistic |
P value | Early RFs | Early RFs + GRS |
Improvement in C statistic |
P value | |
|---|---|---|---|---|---|---|---|---|
| MI<70 | 0.681(0.015) | 0.693(0.015) | 0.012 | 0.032 | 0.612 | 0.639 | 0.027 | 0.002 |
| MI<60 | 0.699(0.016) | 0.712(0.016) | 0.013 | 0.016 | 0.625 | 0.655 | 0.030 | 0.002 |
| MI<50 | 0.716(0.018) | 0.733(0.018) | 0.017 | 0.040 | 0.639 | 0.675 | 0.036 | 0.001 |
Traditional Risk Factors (RFs) = gender, BMI, diabetes, hypertension, hyperlipidaemia, smoking and family history (FH) CAD; Early RFs = Gender and FH CAD only; GRS = Genetic Risk Score; MI<70/60/50 = myocardial infarction occurring before respective age compared to older controls (≥70) without prior MI; P values for comparison of AUC.
Finally, to determine if these variants confer risk of MI (by plaque rupture or thrombosis) over and above the risk of developing CAD, we repeated the main analysis using older CAD+ controls (methods). We examined 889 cases (MI<70yrs) and 370 controls (no MI≥70 years with CAD+; Supplementary Table 3). Overall the results were significantly attenuated with a modest association identified only in the Emory cohort (p=0.02). This finding however was not replicated in the Cleveland Clinic cohort (1154 cases and 241 controls; Supplementary Table 3), with a final non significant association in the pooled adjusted analysis (p=0.11) (Table 3).
Incident MI: Association with a genetic risk score
For incident MI (prospective) analysis we followed the entire Emory cohort of 2597 subjects for adverse outcomes. Demographic data by quintile of risk score for the Emory cohort are presented in Supplementary Table 4, for which there was a statistically significant difference in age with increasing risk score, whereby those with lower scores were older than those with higher scores (p trend =0.001). Also, those with the highest scores tended to have higher incidence of hyperlipidemia.
After a median of 2.5 years follow-up, 101 subjects experienced an MI and 358 the composite endpoint of death or MI. For the primary endpoint of incident MI there was no significant association for either the individual SNPS (Supplementary Table 2) or the cumulative GRS (adjusted p=0.68). When grouped into quintiles of GRS, compared to the lowest quintile, subjects in the highest quintile trended towards more events with an adjusted HR of 1.08 (95% CI 0.57-1.99) for incident MI (Table 5). In subgroup analysis, there was no association between the GRS and future recurrent or first MI during follow up (p= 0.67 and p=0.21 respectively)
Table 5.
Prospective follow up events and association with weighted risk score by quintiles
| Emory Cardiovascular Biobank | Cleveland Clinic Genebank | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||
| Events by quintile | Top v Bottom Quintile | Events by quintile | Top v Bottom Quintile | |||||||||||
| Q1 | Q2 | Q3 | Q4 | Q5 | HR (95% CI) | HR (95% CI) Adjusted |
Q1 | Q2 | Q3 | Q4 | Q5 | HR (95% CI) | HR (95% CI) Adjusted |
|
|
|
||||||||||||||
| All Subjects * | N=2597 | N=2702 | ||||||||||||
| Non Fatal MI | 18 | 20 | 17 | 20 | 26 | 1.24 (0.67-2.29) | 1.08 (0.57-1.99) | 21 | 21 | 23 | 29 | 30 | 1.27 (0.84-1.91) | 1.28 (0.85-1.93) |
| Death/MI | 82 | 74 | 73 | 63 | 66 | 0.72 (0.51-0.99) | 0.65 (0.46-0.91) | 56 | 55 | 54 | 61 | 58 | 1.03 (0.77-1.37) | 1.07 (0.80-1.44) |
| < 60 yrs | N=970 | N=1108 | ||||||||||||
| Non fatal MI | 6 | 6 | 8 | 3 | 14 | 2.25 (0.86-5.88) | 2.02 (0.75-5.40) | 9 | 6 | 12 | 10 | 8 | 0.74 (0.35-1.60) | 0.76(0.35-1.65) |
| Death/ MI | 18 | 12 | 20 | 19 | 22 | 1.16 (0.62-2.17) | 1.02 (0.54-1.93) | 13 | 15 | 19 | 17 | 14 | 0.76 (0.43-1.36) | 0.80 (0.45-1.43) |
All Subjects includes all those enrolled and prospectively followed irrespective of age at enrollment
Adjusted for age at enroll, gender, BMI, hyperlipidaemia, hypertension, diabetes, ever smoked, baseline angiographic CAD ≥50%
For the composite endpoint of Death/MI however, there was a significant, paradoxically, protective association with GRS (adjusted β= −11.4(3.98), p=0.004), with an adjusted HR of 0.68 (95% CI 0.49-0.94) for top v bottom quintile. Other significant multivariate predictors in the latter model included age (HR 1.03 (1.02-1.04)), diabetes (HR 1.51 (1.20-1.90)), hypertension (HR 1.28 (1.00-1.65)), smoking (HR 1.39 (1.10-1.75)) and baseline significant angiographic CAD (HR 1.31 (1.0-1.73)). A history of hyperlipidaemia was also noted to be protective (HR 0.78 (0.61-0.99)).
In the Cleveland Clinic cohort, we also observed a trend towards decreasing age with increasing quintile of risk score (p=0.01) (Supplementary Table 5). After 3 years of follow up in 2702 subjects there were 124 incident MIs and 284 composite end points of Death/MI. There was again no significant association with individual SNPs (Supplementary Table 2) or the cumulative GRS (adjusted p=0.40) with incident MI, although there was again a non significant trend with more events in the top quintile yielding an adjusted HR of 1.28 (95% CI 0.85-1.93) compared to the lowest quintile. For the composite endpoint of Death/MI there was also no significant association (p=0.91) with an adjusted HR of 1.07 (0.80-1.44).
Analysis of those <60 years of age at enrollment revealed no significant associations in either cohort for incident MI or Death/MI (Table 5).
Finally, we repeated the analyses for prevalent and incident MI using only subjects with full genotype data for all 11 variants in the Emory cohort (Supplementary Table 1) and identified similar findings, thereby confirming that mean imputation did not appreciably influence our results (data not shown).
Discussion
In a study of high risk angiographically phenotyped patients, we demonstrate that a cumulative risk score generated from published GWAS variants is: (1) significantly associated with MI before the age of 70 years in two Caucasian cohorts, independent from the discovery samples; (2) not associated with MI when both cases and controls have established CAD; and (3) not a strong predictor of incident near term adverse events.
Although genetic risk scores using variants identified through genome wide linkage scans and candidate gene studies have been previously studied, the first attempt to devise a cumulative risk score from GWAS variants was by Kathiresan et al, using 9 risk variants for premature MI. 9 They found that subjects within the highest quintile of a weighted risk score had two-fold higher odds of premature MI than those in the lowest quintile. Our study validates the use of such a score in two independent cohorts after addition of two extra variants that have since been identified.7, 8 In comparison, our cohorts were significantly older, but we still showed that subjects within the highest quintile of risk score also had an almost two fold greater odds of MI prior to the age of 70 years than those in the lowest quintile, with an even greater risk in subjects with more premature disease (MI <60 and <50 years), thereby confirming that hereditary effects are greatest in younger age. Interestingly the association observed for prevalent MI in our study was non-linear, with only the highest quintile of risk score significantly associating with MI. These findings may reflect possible gene-gene interactions or simply more extensive CAD prone to complications, such as a greater lipid core or thin cap fibro-atheroma arising as a consequence of inheriting a greater than average number of these atherosclerosis predisposing risk variants.
Use of the broad phenotype of “coronary heart disease” by early large scale GWAS has raised concerns as to whether these risk variants confer risk of MI or just CAD, since the processes involved in atherosclerosis may differ from those that lead to MI. We and others showed that 9p21, for example, associates primarily with CAD and atherosclerosis.16, 29, 30 Moreover in subjects with angiographically significant CAD, the 12 risk variants individually do not associate with MI.14 Thus, at the pathophysiologic level, these variants are unlikely to be associated with thrombosis or plaque rupture, and MI risk observed in prior studies is likely secondary to development of greater CAD burden. This study extends these observations by demonstrating that the association between the cumulative risk score and MI is considerably attenuated and not replicated in a second cohort. In contrast to the study of Reilly et al, where controls were younger and thus subject to possible case-misclassification, use of older controls is a major strength of this study, given the critical time sensitive nature of the MI phenotype. Although this finding is of major interest for mechanistic understanding, MI is a more tangible clinical endpoint and as such association of a risk score with MI, even by proxy may still be clinically relevant.
Although subjects reporting a positive FH of CAD/MI had a marginally higher risk score, the cross sectional association between the risk score and MI was independent of FH, and furthermore, there was no interaction with FH, suggesting that genetic risk is mediated both in those with and without a FH of premature CAD. Despite the challenges related to accurate documentation of FH, this serves to highlight the complexity of heritability of MI in relation to common variants. Identification of rarer variants with more Mendelian characteristics through next generation sequencing may yield further insights into the role of FH in relation to complex diseases.31
Genetic risk score and incident MI
Three large studies have examined the value of genetic risk scores in predicting first MI in asymptomatic populations. In a prospective study of 19,000 community based women, no association was observed between a panel of 12 MI risk variants and future adverse CV events after 12 year follow-up.10 However, Ripatti et al found that a risk score of 13 variants was associated with prevalent and incident CV events in a community based population of 31,000 men and women; subjects in the highest quintile were at 1.66-fold greater risk of adverse events during a 10-year follow-up period compared to those in the lowest quintile.11 An association between incident MI risk and 13 SNPs was also observed in the Framingham cohort.12 In contrast to our population who had established CAD or were at high risk of CAD where future risk prediction is particularly challenging, all of these studies examined primary risk of MI.
In this study, although we observed a very marginal trend towards more incident MI in those with the highest quintile of risk score, the final association results were not statistically significant in both cohorts. This is likely to reflect the fact that the primary phenotype for most of these variants is atherosclerosis, which had already developed by the time of enrollment into a cardiac catheterization cohort. Interestingly, we noted that the distribution of risk scores in both the Emory and Cleveland Clinic cohorts was related to age with older subjects trending towards lower scores, potentially indicating that those with high scores were at greater risk for premature fatal events. This finding may help to explain the paradoxically protective association for those with the highest risk scores with the composite outcome of Death/MI in the Emory cohort. Given that a history of hyperlipidemia was also protective in the same model, it is plausible that unmeasured treatment effects such as intensive statin use may also be responsible. Overall, the observation of a lack of association between genetic variants and incident events in a diseased cohort has also been described in other studies. For example, the 9p21 risk is associated with incident risk prediction in primary prevention populations but not in those with diagnosed CAD.32
Finally, we also tested the discriminatory value of the genetic risk score for identifying cases of prevalent MI, which may be greater than for single SNPS as advocated by Davies et al.33 Indeed, after addition of the genetic risk score to traditional risk factors, we found a modest, but statistically significant improvement in the c-statistic for prevalent MI <70 yrs as well as more premature MI. However, genetic data is unique and provides risk information at birth, unlike the majority of acquired risk factors that impact later in life. When the genetic risk score was added to a model containing the non modifiable risk factors of gender and FH (the only ones likely to be known in adolescence), we found that it markedly enhanced the c-statistic, with the greatest discriminatory value for more premature MI. Use of genomic data in this manner would represent a shift in genetic biomarker assessments and requires further exploration.
Limitations & Strengths
There are some limitations of our study. Firstly, both our discovery and replication cohorts are selected populations with CAD and thus our findings may be susceptible to unmeasured bias. Secondly, our controls were not healthy, although risk allele frequency would likely be even lower in healthier controls and would strengthen the observed statistical associations. Thirdly, we used imputation to estimate risk scores for those with missing data. However, an analysis using non imputed data revealed similar overall results. Fourthly, while we may have been underpowered for analysis of incident events, our study was powered sufficiently to be able to detect the effects of age/10 yr to a HR of 1.16. Finally, we only included 11 MI SNPs in our score and have not included variants in relevant metabolic pathways or newer and other recently discovered CAD/MI variants.24, 25, 34 However, the recent study by Thannassoulis et al demonstrates that addition of 16 newer variants to a 13 SNP model did not impact on their association findings.12 Moreover, some of the SNPs examined may not to be true risk SNPs in a particular locus, possibly attenuating the results.35
Major strengths of our study include (1) study of independent discovery and replication cohorts (2) the use of elderly controls in the cross sectional analysis to mitigate the important problem of case-misclassification bias (3) examination of the association with the risk score in the context of proven CAD to determine if there was MI risk over and above association with atherosclerosis, which to our knowledge has not been performed previously and is important given the overlap in phenotypes and (4) study of both prevalent and incident MI in the same cohorts to avoid concerns including those of population stratification.
Conclusion
In conclusion, in two independent CAD cohorts, we demonstrate that a high burden of CAD/MI risk variants is associated with prevalent MI but not with near term incident MI or death. We propose that cumulatively these variants confer risk of MI through development of CAD, but once disease has developed the association with future risk of adverse events is attenuated. As such the clinical utility of a GRS is of limited value in secondary risk prediction. These findings have implications for understanding the clinical utility of genetic risk scores for secondary as opposed to primary risk prediction
Supplementary Material
Supplementary Table 1: Details of genotype missingness in relation to subject numbers for the Emory Cardiac Biobank
Supplementary Table 2: Individual SNP associations with prevalent and incident MI
Supplementary Table 3: Demographics for Analysis of MI+/CAD+ v MI-/CAD+ in both cohorts
Supplementary Table 4: Patient characteristics by quintile of risk score for the Emory Cardiovascular Biobank
Supplementary Table 5: Patient characteristics by quintile of risk score for Cleveland Clinic Genebank
Over the last 5 years, genome wide association studies or GWAS have identified multiple novel variants associating with coronary artery disease and myocardial infarction (MI). Given the small effect sizes individually, there is interest in combining these multiple variants into genetic risk scores for predicting events and determining an individual’s personalized risk. With increasing direct-to-consumer testing and growing physician requested testing for genetic variants; this approach is likely to be of increasing relevance. In this paper, we sought to explore the association between a cumulative genetic risk score composed of 11 of the earliest and most replicated variants with both prevalent and incident MI risk in two independent populations of subjects at high risk for or with established CAD. We demonstrate robust association with prevalent MI occurring before the age of 70, confirming the cumulative impact of genetic burden over a lifetime. However we only demonstrate non significant trends for association with near term MI, which is in contrast to some population studies which have shown significant association with incident MI. We propose that cumulatively these CAD/MI variants confer risk of MI through development of CAD, but once disease has developed the association with future risk of adverse events is likely attenuated. These findings therefore have implications for understanding the clinical utility of genetic risk scores for secondary as opposed to primary risk prediction.
Acknowledgments
We would like to thank all our research coordinators, nursing staff and volunteers for help with patient recruitment and sample collection for the Emory Cardiovascular Biobank. We would also like to thank the attendings and fellows from the Cardiology Division of Emory University as well as the cardiac catheterization laboratory staff for their support and assistance in recruiting patients for this study.
Funding Sources: Emory: This work was supported by the American Heart Association (Postdoctoral Fellowship for RSP), National Institutes of Health R01 HL89650-01, Robert W. Woodruff Health Sciences Center Fund, Emory Heart and Vascular Center Funds and supported in part by NIH Grant UL1 RR025008 from the Clinical and Translational Science Award program and NIH grant R24HL085343. Cleveland Clinic: GeneBank has been supported by National Institutes of Health grants P01HL087018-020001, P01HL076491-055328, 1R01HL103866, 1RO1HL103931, and the Cleveland Clinic Clinical Research Unit of the Case Western Reserve University CTSA (1UL1RR024989). SLH has also been supported in part by a grant from the LeDucq Foundation, and a gift from the Leonard Kreiger Fund.
Footnotes
Conflict of Interest Disclosures: Dr. Tang received research grant support from Abbott Laboratories. Dr. Hazen reports being listed as co-inventor on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics. Dr. Hazen reports having been paid as a consultant for the following companies: AstraZeneca Pharmaceuticals LP, Cleveland Heart Lab, Eli Lilly, Esperion, Liposciences, Inc., Merck & Co., Inc., and Pfizer Inc. Dr. Hazen reports receiving research funds from Abbott, Cleveland Heart Lab, Esperion, and Liposcience. Dr. Hazen reports having the right to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics from the following companies: Abbott Laboratories, Inc., Cleveland Heart Lab., Inc., Frantz Biomarkers, LLC, Liposciences, Inc., and Siemens.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Hemminki K, Forsti A, Bermejo JL. The ‘common disease-common variant’ hypothesis and familial risks. PLoS One. 2008;3:e2504. doi: 10.1371/journal.pone.0002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 4.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samani NJ, Deloukas P, Erdmann J, Hengstenberg C, Kuulasmaa K, McGinnis R, et al. Large scale association analysis of novel genetic loci for coronary artery disease. Arterioscler Thromb Vasc Biol. 2009;29:774–780. doi: 10.1161/ATVBAHA.108.181388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 9.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paynter NP, Chasman DI, Pare G, Buring JE, Cook NR, Miletich JP, et al. Association between a literature-based genetic risk score and cardiovascular events in women. Jama. 2010;303:631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, et al. A multilocus genetic risk score for coronary heart disease: Case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, et al. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium - the framingham heart study. Circ Cardiovasc Genet. 2012 doi: 10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horne BD, Carlquist JF, Muhlestein JB, Bair TL, Anderson JL. Association of variation in the chromosome 9p21 locus with myocardial infarction versus chronic coronary artery disease. Circ Cardiovasc Genet. 2008;1:85–92. doi: 10.1161/CIRCGENETICS.108.793158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, et al. Identification of adamts7 as a novel locus for coronary atherosclerosis and association of abo with myocardial infarction in the presence of coronary atherosclerosis: Two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner SD, Rabbani LE. Secondary prevention strategies for coronary heart disease. J Thromb Thrombolysis. 2010;29:8–24. doi: 10.1007/s11239-009-0381-8. [DOI] [PubMed] [Google Scholar]
- 16.Patel RS, Su S, Neeland IJ, Ahuja A, Veledar E, Zhao J, et al. The chromosome 9p21 risk locus is associated with angiographic severity and progression of coronary artery disease. Eur Heart J. 2010;31:3017–3023. doi: 10.1093/eurheartj/ehq272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gensini G. Coronary arteriography. Futura Publishing Co; New York: 1975. [Google Scholar]
- 18.Hartiala J, Li D, Conti DV, Vikman S, Patel Y, Tang WH, et al. Genetic contribution of the leukotriene pathway to coronary artery disease. Hum Genet. 2011;129:617–627. doi: 10.1007/s00439-011-0963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang WH, Wu Y, Hartiala J, Fan Y, Stewart AF, Roberts R, et al. Clinical and genetic association of serum ceruloplasmin with cardiovascular risk. Arterioscler Thromb Vasc Biol. 2012;32:516–522. doi: 10.1161/ATVBAHA.111.237040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Nicholls SJ, Rodriguez ER, Kummu O, Horkko S, Barnard J, et al. Protein carbamylation links inflammation, smoking, uremia and atherogenesis. Nat Med. 2007;13:1176–1184. doi: 10.1038/nm1637. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya T, Nicholls SJ, Topol EJ, Zhang R, Yang X, Schmitt D, et al. Relationship of paraoxonase 1 (pon1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. Jama. 2008;299:1265–1276. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thygesen K, Alpert JS, White HD. Universal definition of myocardial infarction. Eur Heart J. 2007;28:2525–2538. doi: 10.1093/eurheartj/ehm355. [DOI] [PubMed] [Google Scholar]
- 24.Tregouet DA, Konig IR, Erdmann J, Munteanu A, Braund PS, Hall AS, et al. Genome-wide haplotype association study identifies the slc22a3-lpal2-lpa gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 25.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, et al. Genetic variants associated with lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 26.Kutyavin IV, Milesi D, Belousov Y, Podyminogin M, Vorobiev A, Gorn V, et al. A novel endonuclease iv post-pcr genotyping system. Nucleic Acids Res. 2006;34:e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell PA, Chaturvedi S, Gelfand CA, Huang CY, Kochersperger M, Kopla R, et al. Snpstream uht: Ultra-high throughput snp genotyping for pharmacogenomics and drug discovery. Biotechniques. 2002;(Suppl):70–72. 74, 76–77. [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dandona S, Stewart AF, Chen L, Williams K, So D, O’Brien E, et al. Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2010;56:479–486. doi: 10.1016/j.jacc.2009.10.092. [DOI] [PubMed] [Google Scholar]
- 30.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 32.Gong Y, Beitelshees AL, Cooper-Dehoff RM, Lobmeyer MT, Langaee TY, Wu J, et al. Chromosome 9p21 haplotypes and prognosis in caucasian and african american patients with coronary artery disease. Circ Cardiovasc Genet. 2011;4:169–178. doi: 10.1161/CIRCGENETICS.110.959296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies RW, Dandona S, Stewart AF, Chen L, Ellis SG, Tang WH, et al. Improved prediction of cardiovascular disease based on a panel of single nucleotide polymorphisms identified through genome-wide association studies. Circ Cardiovasc Genet. 2010;3:468–474. doi: 10.1161/CIRCGENETICS.110.946269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, et al. From noncoding variant to phenotype via sort1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Details of genotype missingness in relation to subject numbers for the Emory Cardiac Biobank
Supplementary Table 2: Individual SNP associations with prevalent and incident MI
Supplementary Table 3: Demographics for Analysis of MI+/CAD+ v MI-/CAD+ in both cohorts
Supplementary Table 4: Patient characteristics by quintile of risk score for the Emory Cardiovascular Biobank
Supplementary Table 5: Patient characteristics by quintile of risk score for Cleveland Clinic Genebank