Abstract
Objectives:
The aim of this study was to define the composition of the gut bacterial flora in Norwegian patients with early stage Crohn’s disease (CD).
Methods:
By using a nonselective metagenomics approach, the general bacterial composition in mucosal biopsies from the ileum and the colon of five subjects, four patients with different phenotypes of CD, and one noninflammatory bowel disease control, was characterized. After partial 16S ribosomal RNA (rRNA) gene sequencing, BLAST homology searches for species identification and phylogenetic analysis were performed.
Results:
An overall biodiversity of 106 different bacterial operational taxonomic units (OTUs) was detected in the cloned libraries. Nearly all OTUs belonged to the phylae Bacteroidetes (42% in CD, 71% in the control) or Firmicutes (42% in CD, 28% in the control), except for some OTUs that belonged to the phylum Proteobacteria (15% in CD, 0% in the control) and a few OTUs that could not be assigned to a phylum (2% in CD, 1% in the control).
Conclusion:
Based on the high incidence of inflammatory bowel disease (IBD) in Norway, this pilot study represents a relevant determination of the gut microbiota in Norwegian patients compared to previous findings in other countries. The bacterial profile of Norwegian CD patients was found to be similar to that of CD patients in other countries. The findings do not support a particular bacterial composition as a predominant causative factor for the high incidence of IBD that exists in some countries.
Keywords: Crohn’s disease, gut microbiota composition, inflammatory bowel disease, IBD, metagenomics, 16S rRNA gene sequences
Introduction
Inflammatory bowel disease (IBD) refers to Crohn’s disease (CD) and ulcerative colitis, which have different clinical phenotypes. However, both are considered to evolve through a combination of genetic, environmental, and immunologic factors. In Norway, IBD occurs at a remarkably high incidence,1 although the exact role of gut microbiota in IBD is still unclear.2 Using a variety of samples and methods, previous studies have described various microbial constellations in CD, ulcerative colitis and healthy individuals. A reduction of bacterial diversity in IBD patients has been identified in reports examining fecal as well as mucosal samples, and a shift in the ratio between the anaerobic phylae Firmicutes towards Bacteroidetes, particularly in CD patients, has been suggested.3–6
The bacterial microbiota in Norwegian CD patients has not been described. In a previous study, we demonstrated the paucity of Mycobacterium avium subspecies paratuberculosis (MAP) in mucosal biopsies from the distal ileum and colon of newly diagnosed CD and ulcerative colitis patients, and hypothesized that this microbe does not play a role in eliciting IBD, but rather may occur during the course of disease as a secondary phenomenon in long-standing CD.7 In the search for bacterial signatures as a confounding factor in CD, an important challenge is to assess and characterize the gut microbiota in both healthy and diseased subjects in various geographical settings since most microbial diversity cannot be detected using cultivation-based methods alone. Thus, metagenomics-based methods represent a revolution in their ability to display the complete content of the gut microbiota.8 Tap and colleagues described a phylogenetic human intestinal core content consisting of a limited number of operational taxonomic units (OTUs).9 In a study of three healthy individuals, Eckburg and Relman described ribosomal RNA (rRNA) gene sequences from multiple colonic mucosal sites and feces in which they found significant interindividual differences, as well as differences between mucosal and fecal samples.10 Wang et al, also using 16S rRNA gene sequences, demonstrated that the bacterial composition was different in the jejunum as compared to ileum, colon and rectum, which exhibited similar compositions,11 whereas Green et al demonstrated that mucosa-adherent bacteria from different sites of the colon showed little variation.12
Both the choice of the DNA extraction method and the sequences of the polymerase chain reaction (PCR) primers are critical in the sensitivity and specificity of the PCR-based portion of the procedure. In a recent study, Hong and colleagues demonstrated that a single combination of DNA extraction and PCR primer may miss half of the microbial diversity present, regardless of sampling and sequencing efforts.13 A limitation of the PCR cloning procedure is that it is not a quantitative method, but only semiquantitative. Detecting highly prevalent bacterial entities may represent a more realistic picture of the skewed composition of a particular gut microbiota, but this may also occur due PCR primer and amplification bias, DNA contamination, or PCR inhibitory components.
Phylogenetic arrays are promising high-throughput methods for analyzing the gut microbiota.14,15 The advantage of the phylogenetic arrays is its ability to detect hundreds or thousands of bacteria, whereas the disadvantage is that the potential findings are predefined. A comparison of random sequence reads with 16S rRNA gene sequences presented similar findings when utilizing these two methods.16 Pyrosequencing and a phylogenetic array for community profiling, the Human Intestinal Tract (HIT) Chip, also demonstrated a good correlation of the bacterial profiles between the two latter methods.17
The aim of the present study was to systematically analyze the intestinal bacterial entities present in selected well-defined CD patients. We therefore assessed the bacterial composition in mucosal biopsies from Norwegian CD patients in a nonselective manner using an open-ended metagenomics approach with shotgun cloning of PCR-amplified gut biopsy DNA and subsequent phylogenetic analysis. We characterized the microbial composition and constellation at a species level, or OTUs, in early, treatment-naïve CD patients. For this purpose, we selected five subjects, four patients with different well-defined phenotypes of CD (one with L1 [ileitis], two with L2 [colitis], and one with L3 [ileocolitis] according to the Montreal classification)18 and one non-IBD control to describe the nature of the microbial constellation. Based on the high incidence of IBD in Norway, this pilot study represents a warranted calibration of the gut bacteria in Norwegian patients in comparison to previous findings in similar patients from other parts of the world.
Patients and methods
Patients
Patients were recruited from the prospective Inflammatory Bowel Disease South-Eastern Norway (IBSEN) II study (2005–2007), which investigated genetic, immunological, and environmental factors that contributed to IBD etiology. Inclusion of the four CD patients in the study was based upon a history of abdominal symptoms, including diarrhea and/or blood in the feces for more than 10 days and typical endoscopic and histological findings. A patient with similar symptoms, but without pathologic endoscopy or histology, was defined as a non-IBD control. Detailed phenotypic data regarding the members of this sub-study are available through the IBSEN II study.19
Clinical specimens and processing of bowel biopsies
Two doses of sodium phosphate (Phosphoral® ; Casen-Fleet, Zaragoza, Spain) were used for bowel cleansing; the first dose was given on the day prior to the colonoscopy and the second dose in the morning on the day of the examination. Colonoscopy was performed and mucosal biopsies were retrieved from the terminal ileum and the descending colon. One biopsy from each location was taken using biopsy forceps used during routine biopsies and immediately snap-frozen in liquid nitrogen after the colonoscopy.
Ethical considerations
All patients provided informed written consent to be involved in this study and ethical approval for the study was given by the Regional Ethics Committee in Oslo, Norway (http://www.etikkom.no/REK/regionSorOst, reference number S-04209).
Metagenomics procedure
The biopsy specimens were investigated for the presence of bacterial 16S rRNA gene fragments representing bacterial species entities in an open-ended manner.
DNA isolation
For DNA isolation, 1 mL of Magnapure bacterial lysis buffer (Stratagene, La Jolla, CA) and 20 μL Proteinase K (Sigma-Aldrich, St Louis, MO) were added to the vial with the snap-frozen biopsy sample. Homogenization was performed at 5600 rpm for 26 seconds using the MagNA Lyser Instrument (Roche Diagnostics, Basel, Switzerland). After homogenization, samples were lysed by incubation at 56°C for 30 minutes. Isolation of nucleic acids was performed using a Magnapure Large Volume Kit (Roche Diagnostics).
PCR analysis
For detection of bacterial DNA, PCR amplification targeting the 16S rRNA gene was employed. PCR primers 16S PA_f (AGAGTTTGATCCTGGCTCAG) and16S PD_r (GTATTAC-CGCGGCTGCTG) were used to amplify a 529 bp fragment of the 16S rRNA gene. The PCR mixture was composed of 5 μL of DNA sample extract in a final volume of 50 μL containing 2 μM each of the primers and PCR buffer containing 2.5 mM MgCl2, 0.2 mM dNTPs, and 1 U Taq polymerase (New England Biolabs, Ipswich, MA). The PCR cycling conditions were: initial denaturation for 2 minutes at 95°C, followed by denaturation for 30 seconds at 95°C, annealing for 40 seconds at 61°C, and extension for 40 seconds at 72°C for 36 cycles, with final extension for 10 minutes at 72°C. The sensitivity of the 16S rRNA gene-specific PCR reaction was determined using dilutions of a known Escherichia coli DNA-positive control until estimated to contain 10–20 of fewer 16S rRNA gene copies. The PCR amplification products of 529 bp were excised from the gel after electrophoresis, placed into separate Eppendorf tubes, and purified using a Qiaex II Agarose Gel Purification Kit (Strata-gene) according to the manufacturer’s instructions. The positive control material for mycobacteria was a mucosal bowel biopsy from a MAP-infected goat. MAP detection was performed using IS900-specific PCR as previously described.7
Cloning of PCR products with the TOPO TA Cloning®Kit
The PCR products were cloned into competent E. coli cells using the pCR®2.1-TOPO® vector (Invitrogen; Life Technologies, Carlsbad, CA) for sequencing according to the manufacturer’s instructions. For the DNA from each biopsy, 96 white E. coli colonies were picked and subjected to PCR-based confirmation of bacterial inserts.
DNA sequencing
The purity of the PCR products was verified using gel electrophoresis, and DNA sequence analysis of the 16S rRNA gene clone inserts was performed in a semi-automated capillary sequencer (Applied Biosciences, Foster City, CA).
Sequence preparation and homology search for species identification/BLAST analysis
Sequences were exported from the original ABI files using Chromas Lite (Technelysium Pty Ltd, Helensvale, Australia; 2012. See http://www.technelysium.com.au/chromas_lite.html), converted into FASTA format, and vector and primer sequences were removed. The 16S rRNA gene-specific sequences were compiled and used for bacterial taxonomic entity identification by BLAST analysis on the NCBI website using search results of at least 96% sequence similarity to define the OTUs. Alignment of the nucleotide sequences was conducted with ClustalX20 with the default program settings. Phylogenetic trees were generated using the Dendroscope software21 and manually edited using Inkscape (GNU GPL v2; Albert M, et al; 2010. See http://www.inkscape.org).
Statistics
Comparison of the distribution of the bacterial species between the CD patients and the non-IBD control was conducted using Fisher’s exact test.
Results
Bacterial composition of the bowel biopsies
Patients were grouped according to their CD status: CD or non-IBD control (Table 1). Bowel biopsies from each patient were subjected to global 16S rRNA gene-specific PCR amplification and cloning. Assembled gene sequences were assessed by BLAST analysis. A simplified overview of the bacterial clones detected in this open-ended search in all the study subjects is shown in Table 2 and dendrograms for the respective patient categories and biopsy sites are presented in Figures 1 and 2. IS900-specific PCR was negative for all bowel biopsies, demonstrating no occurrence of MAP.
Table 1.
Characteristics of the study subjects
| ID | Non-IBDa | CD1b | CD2 | CD3 | CD4 |
|---|---|---|---|---|---|
| Diagnosis | Normal | Colitis | Colitis | Ileitis | Ileocolitis |
| Symptom duration (months) | 3 | 5 | 2 | 7 | 2 |
| Age | 41 | 52 | 44 | 38 | 39 |
| Sex | M | M | M | F | M |
| Smoking | N | N | Y | Y | N |
| NOD2 mutation | – | – | – | – | – |
| HBI | 3 | 6 | 8 | 5 | 9 |
| Calprotectinc | 5 | 477 | 32 | 29 | 354 |
| CRPd | 7 | 7 | 7 | 7 | 21 |
| Symptoms | Abdominal pain, temporary loose stools | Loose stools, no abdominal pain or bloody stools | Loose stools, no abdominal pain or bloody stools | Abdominal pain in lower right quadrant, no loose stools | Episodic abdominal pain with loose, bloody stools |
| Macroscopic findings | Normal | Continuous diffuse inflammation with erythema, edema, and fragile mucosa throughout the colon | Aphtous lesions from the ascending colon to the sigmoid, but not in the ileum or the rectum | Aphtous lesions in the terminal ileum | Stenosis of the ileocoecal valve |
| Microscopic findings | Normal | Patchy chronic active inflammation | Light grade of patchy inflammation throughout the colon | Light chronic active inflammation | Chronic active inflammation of the ileocoecal valve, widespread light chronic active inflammation throughout the colon |
| Granuloma | No | Yes | No | No | No |
Notes:
Non-IBD control;
Crohn’s disease.
Table 2.
Simplified overview of bacterial operational taxonomic units (OTUs) detected in the study subjects
|
ID
|
Non-IBDa
|
CD1b
|
CD2
|
CD3
|
CD4
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Segment | Ileum | Colon | Ileum | Colon | Ileum | Colon | Ileum | Colon | Ileum | Colon |
| Bacteroidetes clones, uncultured | 2 | 1 | 1 | 1 | ||||||
| Bacteroidales clones, uncultured | 2 | 4 | ||||||||
| Bacteroides fragilis | 7 | 5 | 1 | 1 | 6 | 1 | 1 | 5 | ||
| Bacteroides thetaiotaomicron | 1 | 1 | 1 | 1 | 1 | |||||
| Bacteroides uniformis | 1 | 3 | 3 | 2 | 3 | 2 | ||||
| Bacteroides vulgatus | 30 | 50 | 6 | 14 | 54 | 5 | 40 | 22 | 1 | |
| Bacteroides sp. | 6 | 2 | 1 | 7 | 4 | 3 | 2 | |||
| Parabacteroides sp. | 1 | 2 | ||||||||
| Sum Bacteroidetes | 45 | 61 | 15 | 24 | 62 | 12 | 48 | 27 | 1 | 7 |
| Pseudomonas frederiksbergensis | 1 | |||||||||
| Citrobacter freundii | 51 | |||||||||
| Citrobacter brakii | 6 | |||||||||
| Enterobacter hormachei | 2 | |||||||||
| Eschericia coli | 1 | |||||||||
| Haemophilus parainfluenzae | 1 | |||||||||
| Klebsiella sp. | 1 | 1 | ||||||||
| Parasutterella excrementihominis | 3 | 1 | ||||||||
| Sutterella sp. | 2 | |||||||||
| Sum Proteobacteria | 0 | 0 | 4 | 1 | 0 | 58 | 4 | 0 | 0 | 3 |
| Clostridiales bacterium | 3 | 2 | 1 | 1 | ||||||
| Clostridium sp. | 4 | 1 | 1 | 76 | 45 | |||||
| Coprococcus sp. | 1 | 1 | ||||||||
| Lachnospiraceae clones, unc. | 3 | 1 | 1 | 1 | ||||||
| Lachnospira pectinoschiza | 1 | |||||||||
| Ruminococcus sp. | 2 | 4 | 4 | 2 | 2 | 4 | 6 | 5 | ||
| Faecalibacterium prausnitzii | 1 | 1 | ||||||||
| Faecalibacterium sp. | 4 | 1 | 3 | 10 | 3 | 1 | 13 | |||
| Roseburia sp. | 3 | 5 | 1 | 6 | ||||||
| Eubacterium halii | 1 | |||||||||
| Firmicutes bacterium clones, unc. | 11 | 1 | 1 | |||||||
| Sum Firmicutes | 29 | 13 | 9 | 14 | 14 | 4 | 19 | 2 | 83 | 51 |
| Bacterium clones, uncultured | 2 | 0 | 0 | 1 | 0 | 1 | 3 | 1 | 0 | 2 |
| Total number of clones | 76 | 74 | 28 | 40 | 76 | 75 | 74 | 30 | 84 | 63 |
| 150 | 68 | 151 | 104 | 147 | ||||||
| Total number of OTUs | 42 | 22 | 27 | 30 | 12 | |||||
Notes:
Non-IBD control;
Crohn’s disease.
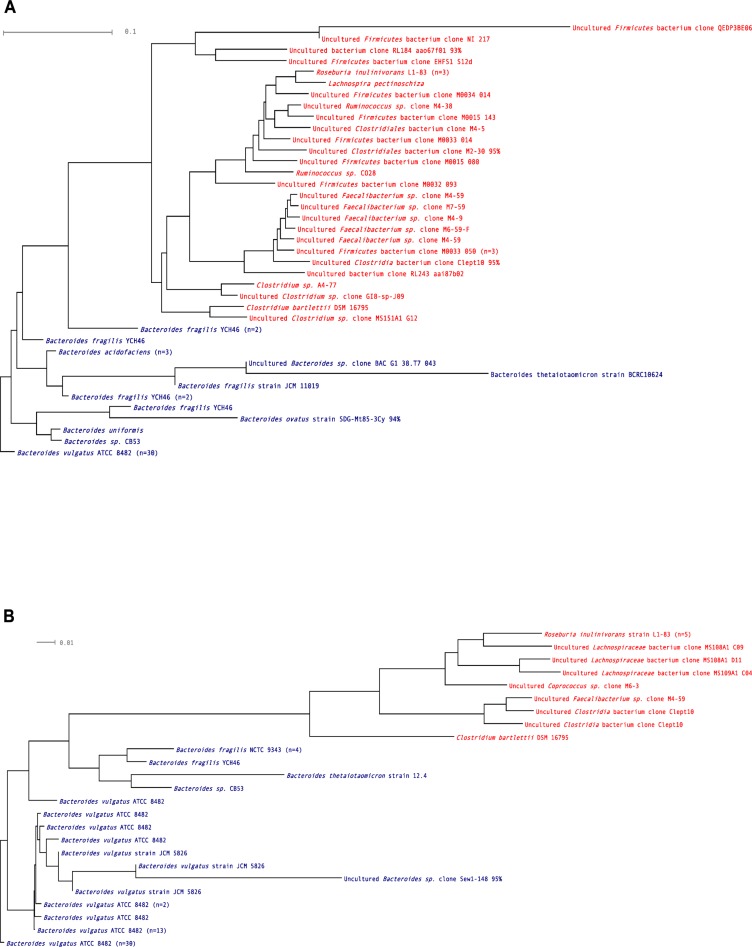
Figure 1.
Operational taxonomic units (OTUs) found in the non-IBD control.
Notes: (A) Ileum; (B) colon. Red = Firmicutes; blue = Bacteroidetes. n = number of clones.
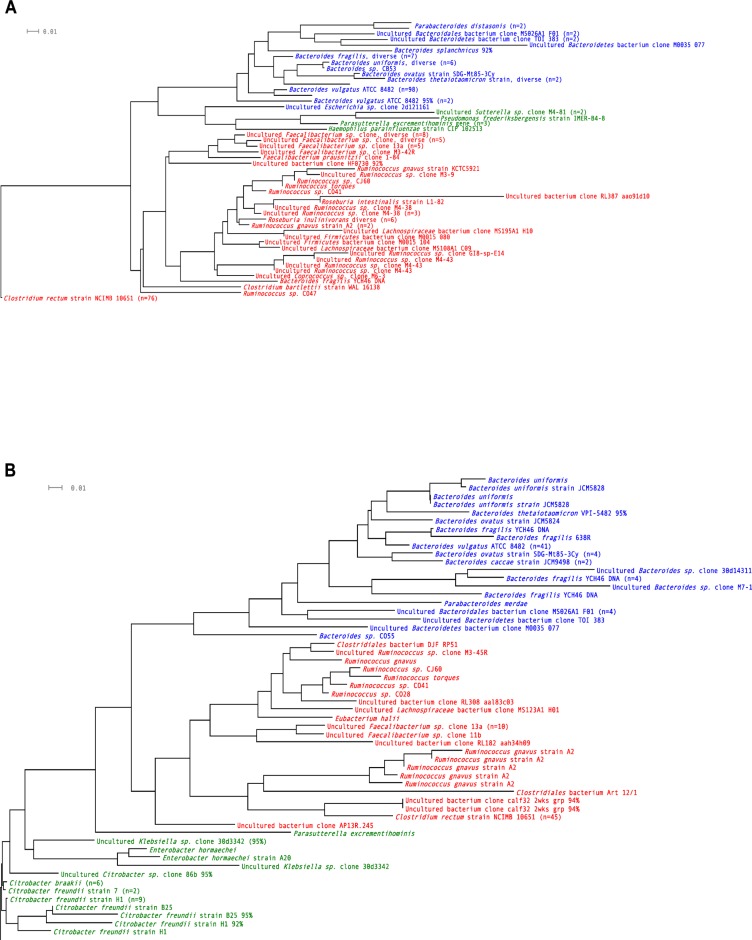
Figure 2.
Operational taxonomic units (OTUs) found in patients with Crohn’s disease.
Notes: (A) Ileum; (B) colon. Blue = Bacteroidetes; green = Proteobacteria; red = Firmicutes. n = number of clones.
The total number of OTUs detected was 106 (Table S1). Out of these, 82 (77%) were present in the ileal biopsies and 52 (49%) were present in the colonic biopsies, while 55 (52%) were present only in ileal biopsies, 31 (29%) only in colonic biopsies, and 20 (19%) were present in both ileal and colonic biopsies. Notably, 74 (70%) of the OTUs identified were found in single biopsies only.
The number of taxonomic entities found concomitantly in both ileal and colonic biopsies differed significantly among the five individuals; six of 42 (14%) in the non-IBD control, eight of 22 (36%) in CD1, one of 27 (4%) in CD2, three of 30 (10%) in CD3, and two of 12 (17%) in CD4.
A predominant intraindividual clustering of OTUs was observed when pairing ileum and colon, whereas a clustering of OTUs was weak when comparing ileum and colon interindividually. In 16 of the 20 cases for which the same OTU was present in both ileal and colonic biopsies, it was within the same individual or individuals. Due to the limited amount of data, a rigorous statistical comparison regarding clustering of the OTUs was not possible.
The total number of OTUs in the non-IBD control was 42 out of 150 clones. Among the specimens from the four individuals with CD, the numbers of OTUs detected were as follows: CD1 22 out of 68 clones, CD2 27 out of 151 clones, CD3 30 out of 104 clones, and CD4 12 out of 147 clones. The complexity of the gut microbiota was lower in CD2 and CD4 than in the non-IBD control (P = 0.004 and P < 0.001, respectively; Table S2) due to a high prevalence of the Citrobacter sp. in CD2 and Clostridium rectum in CD4.
Main taxonomic entities detected
Nearly all OTUs belonged to the anaerobic phylae Bacteroidetes (42% in CD and 71% in the control, P ≤ 0.0001) and Firmicutes (42% in CD and 28% in the control, P = 0.0028), except for some OTUs that belonged to the aerobic phylum Proteobacteria (15% in CD and 0% in the control, P ≤ 0.001) and a few OTUs that could not be assigned to a phylum (2% in CD and 1% in the control, P = 1.0). When distinguishing between the four CD patients, the distribution was as follows: CD1; Bacteroidetes 57%, Firmicutes 34%, Proteobacteria 7%, CD2; Bacteroidetes 49%, Firmicutes 12%, Proteobacteria 38%, CD3; Bacteroidetes 72%, Firmicutes 20%, Proteobacteria 4%, CD4; Bacteroidetes 5%, Firmicutes 91%, Proteobacteria 2%.
The most predominant bacterial genera present in all or multiple biopsies were Bacteroides in 100% of the biopsies, Ruminococcus in 80%, Faecalibacterium in 70% and Clostridium in 50% of the biopsies (Figures S1 and S2). The genus Bacteroides made up a large proportion of the bacterial microbiota in the non-IBD control and in three of the four CD patients. We detected a number of different Bacteroides spp.; in particular, Bacteroides vulgatus, as represented by the clone ATCC 8482, was highly represented in many of the biopsies and present in all except the biopsy from the colon of patient CD4. Faecalibacterium sp. was found in seven out of 10 biopsies, whereas Faecalibacterium prausnitzii, an anaerobic bacterium suggested to be an anti-inflammatory commensal,22 was found only in two biopsies, one from the ileum of patient CD3, the only female, who also had the lowest clinical activity, discrete endoscopic findings and a low level of inflammatory markers in serum and feces, and, as expected, one from the ileum of the non-IBD control.
Bacteria in CD
All aerobic Proteobacteria entities detected, which included E. coli, Citrobacter freundii, and C. brakii, Pseudomonas frederiksbergensis, Enterobacter hormacheii, Haemophilus parainfluenzae, Sutterella sp., and Parasutterella excrementihominis, were found only in biopsies from CD patients. This was also the case for the anaerobic Parabacteroides sp. belonging to the Bacteroidetes and Eubacterium sp. belonging to the Firmicutes. Thus, the bacterial microbiota in Norwegian CD patients was found to be very diverse. However, no clear bacterial signature constellations of predominant OTUs were detected in either of the patient groups.
Discussion
Based on the high incidence of IBD in Norway, it was important to perform a pilot study to determine bacterial profile in the small and large bowels of CD patients and compared these findings to those from other parts of the world. Previously, newly diagnosed CD in Norway was not found to be associated with the occurrence of MAP in distal ileal or colon biopsies.7 Here, extending this data, we performed an open-ended metagenomics search for gut bacterial OTUs or constellations useful towards determining factors that may or may not elicit IBD. No previous studies have been able to identify a specific bacterial pathogen or entity, or a particular constellation of gut bacteria, as a hallmark of CD. It remains unclear whether gut bacteria induce or elicit CD or fuel the disease once it is established. In this study, we assessed the bacterial profile in treatment-naïve CD patients who had not been subjected to immunomodulating treatment and had a short duration of time from the onset of symptoms to diagnosis. We employed a general approach using nonspecific bacterial 16S rRNA gene sequences with the primary goal of identifying the between five and ten most prevalent bacterial species in four CD patients with different phenotypes and a non-IBD control.
A recent study proposed three different enterotypes of the normal human gut microbiome based on fecal metagenomes from four different countries.23 Mucosa-associated bacteria in bowel biopsies appear to be a more relevant target to study in the IBD setting compared to luminal bacteria in fecal samples since previous studies have shown a different bacterial composition in samples from these two sites24 and most significant host–microbial crosstalk takes place at the mucus–epithelial interface.
The ratio between the bacterial phylae Bacteroidetes, Firmicutes, and Proteobacteria in the CD patients in this study was similar to the findings of Bibiloni et al, who investigated intestinal biopsies from newly diagnosed, untreated Canadian patients.5 We also demonstrate a similar depletion of Bacteroidetes and Firmicutes with a parallel increase of Proteobacteria in CD patients as demonstrated by Frank et al in a study where rRNA gene sequence analysis was performed on surgical specimens from US patients.25
Bacterial species of the genus Bacteroides are likely the most abundantly occurring gut OTUs, both in health and disease, and were present in all biopsies investigated in this study. They generally prevent pathogens from colonizing the gut and contribute to food digestion by breaking down complex molecules. There are conflicting observations concerning the possible role of Bacteroides in the setting of CD. Swidsinski et al reported that a primary feature of IBD is the presence of a Bacteroides fragilis-predominant biofilm.26
Ruminococcus gnavus and R. torques, which are species that belong to the Clostridium group XIVa, are known to degrade cellulose in ruminants but are also found in the human gut, where they are able to ferment glucose and xylose. They are important mucus-degrading anaerobic bacterial species that were detected in 80% of the gut biopsies in our study.
Among the most important bacteria normally found in the colon, Bacteroides, Eubacterium, Enterobacteriaceae, and Clostridia were all present, while Bifidobacteria, Enterococci, Fusobacteria, and Lactobacilli were not detected. Furthermore, among the two pathogens that have been most widely discussed with regard to their role as causative agents of CD, MAP7,27 was not detected and adhesive and invasive E. coli (AIEC)28 could also not be distinguished from E. coli using 16S rRNA gene-specific PCR, despite the prevailing overall detection of other Proteobacteria in all CD patients.
We identified Faecalibacterium sp. in 70% of the gut biopsies; however, the signature OTU F. prausnitzii was identified in only 20% of CD biopsies, the healthy control, and the least-affected CD patient, corroborating previous findings. Sokol et al detected an under-representation of F. prausnitzii in active IBD and infectious colitis29 and a reduction of F. prausnitzii counts was found in cases of postoperative recurrence of ileal CD,22 while the presence of F. prausnitzii was associated with a healthy bowel. Willing et al described changes specific to twin patients with ileal CD, including the disappearance of Faecalibacterium and Roseburia and increased amounts of Enterobacteriaceae and Ruminococcus gnavus, for instance.30 A less abundant finding was that one patient (CD2) was heavily colonized with Citrobacter freundii, an opportunistic pathogen, indicative of a predominant proteobacterial contingent in the apparently relatively aerobic gut microenvironment of this patient. Another patient (CD4) exhibited a predominance of Clostridium rectum in both the ileal and colonic segments.
We also found that Bacteroidetes was the most predominant phylum detected in general, while we detected excessive Proteobacteria in CD patients only. Hatoori and Taylor previously found that adult gut-enriched bacterial genes aggregated to the three species Bacteroides, Eubacterium, and Ruminococcus.31 Bacteroides sp. and Ruminococcus sp. were the most abundant OTUs in our study. Although similar in the distribution of the bacterial divisions represented, our findings of bacterial species differed significantly from the biopsy based findings of Bibiloni et al in newly diagnosed untreated CD patients, likely since they targeted specific bacterial groups using defined phylogenetic arrays,5 while we used an open-ended search. The findings of Ott et al, who performed quantification of 20 dominant bacterial species using quantitative real-time PCR for German CD patients,3 and our findings showed clear similarities, particularly regarding the presence of several Bacteroides species, such as B. acidifaciens, B. fragilis, B. ovatus, B. thetaiotaomicron, and B. ovatus, and also Ruminococcus gnavus. Gophna et al compared Canadian CD and UC patients and healthy controls using a metagenomics approach.6 They found an increase of Proteobacteria in CD, Bacteroidetes as the most dominant phylum in general and increased in CD, leading to an associated decrease in Clostridium sp. When searching for signature bacteria in Spanish CD patients, Martinez-Medina et al found that E. coli, Clostridium sp. and R. torques together may serve as indicators of CD, whereas F. prausnitzii may serve as an indicator for a healthy colon,32 corroborating our study.
A number of limitations exist in metagenomics approaches, including the method employed here. Regarding sampling of the material, the quality and representative nature of the gut mucosa biopsies taken are critical, while the procedures for enrichment and isolation of nucleic acids may lead to a loss of nucleic acids from many of the bacteria localized in the mucous surface layer, therefore predominantly displaying the more mucosal epithelium-adherent microbiota. Generally, the submucosa, and not only the mucosa, is the source of marked inflammatory hyperplastic and exudative changes in IBD. As a result, the bowel wall becomes thickened. Therefore, the mucosa may not reveal the cause of CD, but rather only elucidate secondary changes.33 This may, however, be significant if searching for invasive bacteria. Additionally, the procedure of lysis is easier to perform on Gram-negative than on Gram-positive bacteria, which may lead to a selection of Gram-negative bacteria.
The bacterial profile in Norwegian CD patients was found to have a profile similar to that of CD patients in other countries such as Canada, Germany, Spain, and the US. Despite the small sample size in the present study and the limitations of open-ended metagenomics procedures, the results presented are very consistent with those of studies from other locations with a high incidence of IBD, indicating that the material analyzed is representative for CD patients in Norway. Considering the particularly high incidence of IBD in Norway, the constellation of gut OTUs in the CD patients we studied did not reveal significant uniqueness or differences in microbial composition from other parts of the Western world. We also did not identify a predominance of special commensal or pathogen species. The results do not support the hypothesis that a particular bacterial composition is a main driver or causative factor for the high incidence of IBD in Norway, although Bibiloni et al showed different bacterial composition in different geographical regions when comparing patients from the high-incidence country Canada with the IBD low-incidence country Mexico.34 In a microbe-host interaction context, Smith and Garrett suggest that specific bacterial species modulate the development and function of mucosal T-cells, and that not only the presence but also the absence of specific bacterial species may increase host susceptibility to chronic inflammation.35
The microbes inhabiting the human gut provide additional metabolic capacities to their host and regulate expression of genes involved in lipid and carbohydrate metabolism, thereby influencing nutrient supply, energy balance, and body weight. The gut microbiota is also a critical stimulus for adequate maturation of the immune system, which contributes to reducing infections and aberrant immune responses. Thus, the microbiota inhabiting the intestinal tract has an array of physiologic roles within the human body, influencing both metabolic and immune functions, particularly when they are altered. Several mechanisms connect the gut microbiota to host energy metabolism: increased energy harvesting from the diet, regulation of appetite through gut peptide secretion, regulation of tissue-free fatty acid composition and uptake, storage, and oxidation, modulation of intestinal barriers by glucagon-like peptide-2 secretion, activation of innate immunity, and hepatic fibrogenesis through the lipopolysaccharide-toll-like receptor-4 axis.36 Gut microbiota manipulation through antibiotics, prebiotics, and probiotics yields encouraging results for treating obesity and diabetes.36 Additionally, addressing multifactorial diseases, our findings here examining IBD and CD development indirectly indicate that endogenous or environmental factors other than the bacterial profile alone should be characterized. Future studies examining gut microbiota in larger, different populations should be conducted to determine the genetics and factors of socioeconomic origin that affect mucosal barriers and their (dys)function, which may be of biological, epigenetic, or geographical nature.
Supplementary material
Table S1.
Identity of 16S rRNA gene sequences subjected to BLAST-based searches for species identification
| Non-IBD | CD1 Colitis | CD2 Colitis | CD3 Ileitis | CD4 Ileocolitis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||
| Ileum 76 | Colon 74 | Ileum 28 | Colon 40 | Ileum 76 | Colon 75 | Ileum 74 | Colon 30 | Ileum 84 | Colon 63 | |
| Bacteroides acidofaciens | 3 | |||||||||
| B. caccae strain JCM 9498 | 2 | |||||||||
| B. fragilis YCH46 | 6 | 1 | 3 | 1 | 1 | 5 | ||||
| B. fragilis NCTC 9343 | 4 | 1 | 3 | |||||||
| B. fragilis strain JCM 11019 | 1 | |||||||||
| B. fragilis 638R | 1 | |||||||||
| B. ovatus SDG-Mt85-3Cy | 1 | 4 | 1 | |||||||
| B. ovatus JCM5824 | 1 | |||||||||
| B. splanchnicus 92% | 1 | |||||||||
| B. thetaiotaomicron, div. | 1 | |||||||||
| B. thetaiotaomicron 12.4 | 1 | |||||||||
| B. thetaiotaomicron 17.4 | 1 | |||||||||
| B. thetaiotaomicron VPI-5482 95% | 1 | |||||||||
| B. thetaiotaomicron BCRC 10624 | 1 | |||||||||
| B. uniformis | 1 | 2 | 3 | |||||||
| B. uniformis JCM 5826 | 3 | 2 | 2 | |||||||
| B. uniformis JCM 5828 | 1 | |||||||||
| B. vulgatus ATCC 8482 | 30 | 50 | 6 | 14 | 54 | 5 | 39 | 22 | 1 | |
| B. sp. CB53 | 1 | 1 | 1 | |||||||
| B. sp. CO55 | 1 | |||||||||
| B. sp., uncultured | 1 | |||||||||
| B. sp. 30d14311, uncultured | 1 | |||||||||
| B. sp. M7-1, uncultured | 1 | |||||||||
| B. sp. BAC G1 38.T7 043 | 1 | |||||||||
| Parabacteroides johnsonii | 1 | |||||||||
| Parabacteroides distasonis | 2 | |||||||||
| Bacteroidales clone uncultured MS026 A1F01 | 2 | 4 | ||||||||
| Bacteroidetes clone uncultured TOI 383 | 2 | 1 | ||||||||
| Bacteroidetes clone uncultured M0035 077 | 1 | 1 | ||||||||
| Bacteroidetes | 45 | 61 | 15 | 24 | 62 | 12 | 48 | 27 | 1 | 7 |
| Pseudomonas frederiksbergensis strain IMER-B4-8 | 1 | |||||||||
| Citrobacter freundii strain 7 | 37 | |||||||||
| Citrobacter freundii H1 | 11 | |||||||||
| Citrobacter freundii B25 | 2 | |||||||||
| Citrobacter braakii 167 | 6 | |||||||||
| Citrobacter sp. clone 86b 95% | 1 | |||||||||
| Enterobacter hormachei | 2 | |||||||||
| Escherichia sp. clone 2d121161, uncultured | 1 | |||||||||
| Haemophilus parainfluenzae strain CIP 102513 | 1 | |||||||||
| Klebsiella sp. clone 30d3342, uncultured | 1 | 1 | ||||||||
| Parasutterella excrementihominis | 3 | 1 | ||||||||
| Sutterella sp. uncultured clone M4-81 | 2 | |||||||||
| Proteobacteria | 0 | 0 | 4 | 1 | 0 | 58 | 4 | 0 | 0 | 3 |
| Cl. bartletii DSM 16795 | 1 | 1 | ||||||||
| Cl. bartletii WAL 16138 | 1 | |||||||||
| Cl. rectum NCIMB 10651 | 76 | 45 | ||||||||
| Clostridium sp. A4-77 | 1 | |||||||||
| Clostridium sp. clone GI8-sp-J09, uncultured | 1 | |||||||||
| Clostridium sp. clone MS151A1 G12, uncultured | 1 | |||||||||
| Clostridiales bacterium DJF RP51 | 1 | |||||||||
| Clostridiales bacterium Art 12/1 | 1 | |||||||||
| Clostridiales bacterium clone M4-5, uncultured | 1 | |||||||||
| Clostridiales bacterium clone M2-30, uncultured | 1 | |||||||||
| Clostridia bacterium clone Clept10, uncultured | 1 | 2 | ||||||||
| Coprococcus sp. clone M6-3, uncultured | 1 | 1 | ||||||||
| Lachnospira pectinoschiza | 1 | |||||||||
| Lachnospiraceae clone uncultured, div. | 3 | |||||||||
| Lachnospiraceae bacterium clone MS 108A1 C09, uncultured | 1 | |||||||||
| Lachnospiraceae bacterium clone MS 123A1 H01, uncultured | 1 | |||||||||
| Lachnospiraceae bacterium clone MS 195A1 H10, uncultured | 1 | |||||||||
| Ruminococcus gnavus | 1 | |||||||||
| R. gnavus KCTC5921 | 1 | |||||||||
| R. gnavus A2 | 2 | 5 | ||||||||
| R. torques | 1 | 1 | ||||||||
| R. sp. clone M3-9 | 1 | |||||||||
| R. sp. CJ60 | 1 | 1 | ||||||||
| R. sp. CO28 | 1 | 1 | ||||||||
| R. sp. CO41 | 1 | 1 | ||||||||
| R. sp. CO47 | 1 | |||||||||
| R. sp. clone GI8-sp-E14, uncultured | 1 | |||||||||
| Ruminococcus sp. clone M3-45R | 1 | |||||||||
| Ruminococcus sp. clone M4-38, uncultured | 1 | 4 | ||||||||
| Ruminococcus sp. clone M4-43, uncultured | 2 | 1 | ||||||||
| Faecalibacterium prausnitzii clone 1-84 | 1 | |||||||||
| Faecalibacterium sp. clone GI5-003-A10, uncultured | 1 | |||||||||
| Faecalibacterium sp. clone M3-42R, uncultured | 4 | |||||||||
| Faecalibacterium sp. clone M4-59, uncultured | 2 | 1 | ||||||||
| Faecalibacterium sp. clone M4-69, uncultured | 1 | |||||||||
| Faecalibacterium sp. clone M7-59, uncultured | 1 | 2 | ||||||||
| Faecalibacterium sp. clone M4-9, uncultured | 1 | 3 | ||||||||
| Faecalibacterium sp. clone M6-59-F, uncultured | 1 | |||||||||
| Faecalibacterium sp. clone PP071-b29, uncultured | 2 | |||||||||
| Faecalibacterium sp. clone 13a, uncultured | 3 | 10 | 2 | |||||||
| Faecalibacterium sp. clone 11b, uncultured | 1 | |||||||||
| Faecalibacterium sp. clone M4-59, uncultured | 1 | |||||||||
| Roseburia inulinivorans strain L1-83 | 3 | 5 | 5 | |||||||
| Roseburia inulinivorans type strain A2-194T | 1 | |||||||||
| Roseburia intestinalis strain L1-82 | 1 | |||||||||
| Eubacterium halii | 1 | |||||||||
| Firmicutes bacterium clone QEDP3BE06, uncultured | 1 | |||||||||
| Firmicutes bacterium clone NI 217, uncultured | 1 | |||||||||
| Firmicutes bacterium clone EHFS1 S12d, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0034 014, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0015 143, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0033 014, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0015 080, uncultured | 1 | 1 | ||||||||
| Firmicutes bacterium clone M0015 104, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0032 093, uncultured | 1 | |||||||||
| Firmicutes | 29 | 13 | 9 | 14 | 14 | 4 | 19 | 2 | 83 | 51 |
| Bacterium clone AP13R.245, uncultured | 1 | |||||||||
| Bacterium clone HF0730, uncultured 92% | 1 | |||||||||
| Bacterium clone RL 305 aal87f07, uncultured | 1 | |||||||||
| Bacterium clone RL 182 aah34h09, uncultured | 1 | |||||||||
| Bacterium clone RL 184 aao67f01, uncultured | 1 | |||||||||
| Bacterium clone RL 243 aai87b02, uncultured | 1 | |||||||||
| Bacterium clone RL308 aal83c03, unculturded | 1 | |||||||||
| Bacterium clone RL 387 aao91d10, uncultured | 1 | |||||||||
| Bacterium clone calf32 2 wks grp 94% | 2 | |||||||||
| Bacterium clone, uncultured | 2 | 0 | 0 | 1 | 0 | 1 | 3 | 1 | 0 | 2 |
Abbreviation: IBD, inflammatory bowel disease.
Main findings in ileal biopsies.
Main findings in colonic biopsies.
Table S2.
2 × 2 contigency tables comparing differences in complexity of the bacterial OTUs detected in each of the study subjects (Fischer’s exact test)
| Total OTU variants | OTU variants identified > 1 | Sum | |
|---|---|---|---|
| P = 0.52 | |||
| Non-IBD | 42 | 108 | 150 |
| CD1 | 22 | 46 | 68 |
| Sum | 64 | 154 | 218 |
| P = 0.04 | |||
| Non-IBD | 42 | 108 | 150 |
| CD2 | 27 | 124 | 151 |
| Sum | 69 | 232 | 301 |
| P = 0.89 | |||
| Non-IBD | 42 | 108 | 150 |
| CD3 | 30 | 74 | 104 |
| Sum | 72 | 182 | 254 |
| P < 0.001 | |||
| Non-IBD | 42 | 108 | 150 |
| CD4 | 12 | 135 | 147 |
| Sum | 54 | 243 | 297 |
Abbreviations: IBD, inflammatory bowel disease; OTUs, operational taxonomic units.
Acknowledgments
We wish to thank Jorunn Bratlie at Oslo University Hospital (Rikshospitalet) for expert technical assistance, Gunn Seim Ekeland at Akershus University Hospital for excellent assistance with the protocol, and Ellen Gussiås, Elseber Jørgensen and Toril Sandvold at Akershus University Hospital for their invaluable assistance at the Endoscopic Unit. This work was supported by grants from HelseSør RHF (the IBSEN II study), an Akershus University Hospital strategic grant and a Centre of Excellence grant from the Research Council of Norway to the Centre of Molecular Biology and Neuroscience (CMBN).
Footnotes
Disclosure
There was no conflict of interest between the authors and the host institutions. The authors declare that no financial or other conflict of interest exists in relation to the content of this article.
References
- 1.Shivananda S, Lennard-Jones J, Logan R, et al. Incidence of inflammatory bowel disease across Europe: is there a difference between north and south? Results of the European Collaborative Study on Inflammatory Bowel Disease (EC-IBD) Gut. 1996;39:690–697. doi: 10.1136/gut.39.5.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibiloni R, Mangold M, Madsen KL, Fedorak RN, Tannock GW. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn’s disease and ulcerative colitis patients. J Med Microbiol. 2006;55:1141–1149. doi: 10.1099/jmm.0.46498-0. [DOI] [PubMed] [Google Scholar]
- 6.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricanek P, Lothe SM, Szpinda I, et al. IBSEN II study group. Paucity of mycobacteria in mucosal bowel biopsies from adults and children with early inflammatory bowel disease. J Crohns Colitis. 2010;4:561–566. doi: 10.1016/j.crohns.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Ventura M, Turroni F, Canchaya C, Vaughan EE, O’Toole PW, van Sinderen D. Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci. 2009;14:3214–3221. doi: 10.2741/3445. [DOI] [PubMed] [Google Scholar]
- 9.Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 10.Eckburg PB, Relman DA. The role of microbes in Crohn’s disease. Clin Infect Dis. 2007;44:256–262. doi: 10.1086/510385. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Ahrne S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Green GL, Brostoff J, Hudspith B, et al. Molecular characterization of the bacteria adherent to human colorectal mucosa. J Appl Microbiol. 2006;100:460–469. doi: 10.1111/j.1365-2672.2005.02783.x. [DOI] [PubMed] [Google Scholar]
- 13.Hong S, Bunge J, Leslin C, Jeon S, Epstein SS. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009;3:1365–1373. doi: 10.1038/ismej.2009.89. [DOI] [PubMed] [Google Scholar]
- 14.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 15.Paliy O, Kenche H, Abernathy F, Michail S. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microbiol. 2009;75:3572–3579. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manichanh C, Chapple CE, Frangeul L, Gloux K, Guigo R, Dore J. A comparison of random sequence reads versus 16S rDNA sequences for estimating the biodiversity of a metagenomic library. Nucleic Acids Res. 2008;36:5180–5188. doi: 10.1093/nar/gkn496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claesson MJ, O’Sullivan O, Wang Q, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–753. doi: 10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricanek P, Brackmann S, Perminow G, et al. Evaluation of disease activity in IBD at the time of diagnosis by the use of clinical, biochemical, and fecal markers. Scand J Gastroenterol. 2011;46:1081–1091. doi: 10.3109/00365521.2011.584897. [DOI] [PubMed] [Google Scholar]
- 20.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoetendal EG, von WA, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feller M, Huwiler K, Stephan R, et al. Mycobacterium avium subspecies paratuberculosis and Crohn’s disease: a systematic review and meta-analysis. Lancet Infect Dis. 2007;7:607–613. doi: 10.1016/S1473-3099(07)70211-6. [DOI] [PubMed] [Google Scholar]
- 28.Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 29.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 30.Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 31.Hattori M, Taylor TD. The human intestinal microbiome: a new frontier of human biology. DNA Res. 2009;16:1–12. doi: 10.1093/dnares/dsn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 33.Crohn BB, Ginzberg L, Oppenheimer GD. Regional ileitis. JAMA. 1932;99:1323–1328. [Google Scholar]
- 34.Bibiloni R, Tandon P, Vargas-Voracka F, et al. Differential clustering of bowel biopsy-associated bacterial profiles of specimens collected in Mexico and Canada: what do these profiles represent? J Med Microbiol. 2008;57:111–117. doi: 10.1099/jmm.0.47321-0. [DOI] [PubMed] [Google Scholar]
- 35.Smith PM, Garrett WS. The gut microbiota and mucosal T cells. Front Microbio. 2011:2. doi: 10.3389/fmicb.2011.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musso G, Gambino R, Cassader M. Gut microbiota as a regulator of energy homeostasis and ectopic fat deposition: mechanisms and implications for metabolic disorders. Curr Opin Lipidol. 2010;21:76–83. doi: 10.1097/MOL.0b013e3283347ebb. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Identity of 16S rRNA gene sequences subjected to BLAST-based searches for species identification
| Non-IBD | CD1 Colitis | CD2 Colitis | CD3 Ileitis | CD4 Ileocolitis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||
| Ileum 76 | Colon 74 | Ileum 28 | Colon 40 | Ileum 76 | Colon 75 | Ileum 74 | Colon 30 | Ileum 84 | Colon 63 | |
| Bacteroides acidofaciens | 3 | |||||||||
| B. caccae strain JCM 9498 | 2 | |||||||||
| B. fragilis YCH46 | 6 | 1 | 3 | 1 | 1 | 5 | ||||
| B. fragilis NCTC 9343 | 4 | 1 | 3 | |||||||
| B. fragilis strain JCM 11019 | 1 | |||||||||
| B. fragilis 638R | 1 | |||||||||
| B. ovatus SDG-Mt85-3Cy | 1 | 4 | 1 | |||||||
| B. ovatus JCM5824 | 1 | |||||||||
| B. splanchnicus 92% | 1 | |||||||||
| B. thetaiotaomicron, div. | 1 | |||||||||
| B. thetaiotaomicron 12.4 | 1 | |||||||||
| B. thetaiotaomicron 17.4 | 1 | |||||||||
| B. thetaiotaomicron VPI-5482 95% | 1 | |||||||||
| B. thetaiotaomicron BCRC 10624 | 1 | |||||||||
| B. uniformis | 1 | 2 | 3 | |||||||
| B. uniformis JCM 5826 | 3 | 2 | 2 | |||||||
| B. uniformis JCM 5828 | 1 | |||||||||
| B. vulgatus ATCC 8482 | 30 | 50 | 6 | 14 | 54 | 5 | 39 | 22 | 1 | |
| B. sp. CB53 | 1 | 1 | 1 | |||||||
| B. sp. CO55 | 1 | |||||||||
| B. sp., uncultured | 1 | |||||||||
| B. sp. 30d14311, uncultured | 1 | |||||||||
| B. sp. M7-1, uncultured | 1 | |||||||||
| B. sp. BAC G1 38.T7 043 | 1 | |||||||||
| Parabacteroides johnsonii | 1 | |||||||||
| Parabacteroides distasonis | 2 | |||||||||
| Bacteroidales clone uncultured MS026 A1F01 | 2 | 4 | ||||||||
| Bacteroidetes clone uncultured TOI 383 | 2 | 1 | ||||||||
| Bacteroidetes clone uncultured M0035 077 | 1 | 1 | ||||||||
| Bacteroidetes | 45 | 61 | 15 | 24 | 62 | 12 | 48 | 27 | 1 | 7 |
| Pseudomonas frederiksbergensis strain IMER-B4-8 | 1 | |||||||||
| Citrobacter freundii strain 7 | 37 | |||||||||
| Citrobacter freundii H1 | 11 | |||||||||
| Citrobacter freundii B25 | 2 | |||||||||
| Citrobacter braakii 167 | 6 | |||||||||
| Citrobacter sp. clone 86b 95% | 1 | |||||||||
| Enterobacter hormachei | 2 | |||||||||
| Escherichia sp. clone 2d121161, uncultured | 1 | |||||||||
| Haemophilus parainfluenzae strain CIP 102513 | 1 | |||||||||
| Klebsiella sp. clone 30d3342, uncultured | 1 | 1 | ||||||||
| Parasutterella excrementihominis | 3 | 1 | ||||||||
| Sutterella sp. uncultured clone M4-81 | 2 | |||||||||
| Proteobacteria | 0 | 0 | 4 | 1 | 0 | 58 | 4 | 0 | 0 | 3 |
| Cl. bartletii DSM 16795 | 1 | 1 | ||||||||
| Cl. bartletii WAL 16138 | 1 | |||||||||
| Cl. rectum NCIMB 10651 | 76 | 45 | ||||||||
| Clostridium sp. A4-77 | 1 | |||||||||
| Clostridium sp. clone GI8-sp-J09, uncultured | 1 | |||||||||
| Clostridium sp. clone MS151A1 G12, uncultured | 1 | |||||||||
| Clostridiales bacterium DJF RP51 | 1 | |||||||||
| Clostridiales bacterium Art 12/1 | 1 | |||||||||
| Clostridiales bacterium clone M4-5, uncultured | 1 | |||||||||
| Clostridiales bacterium clone M2-30, uncultured | 1 | |||||||||
| Clostridia bacterium clone Clept10, uncultured | 1 | 2 | ||||||||
| Coprococcus sp. clone M6-3, uncultured | 1 | 1 | ||||||||
| Lachnospira pectinoschiza | 1 | |||||||||
| Lachnospiraceae clone uncultured, div. | 3 | |||||||||
| Lachnospiraceae bacterium clone MS 108A1 C09, uncultured | 1 | |||||||||
| Lachnospiraceae bacterium clone MS 123A1 H01, uncultured | 1 | |||||||||
| Lachnospiraceae bacterium clone MS 195A1 H10, uncultured | 1 | |||||||||
| Ruminococcus gnavus | 1 | |||||||||
| R. gnavus KCTC5921 | 1 | |||||||||
| R. gnavus A2 | 2 | 5 | ||||||||
| R. torques | 1 | 1 | ||||||||
| R. sp. clone M3-9 | 1 | |||||||||
| R. sp. CJ60 | 1 | 1 | ||||||||
| R. sp. CO28 | 1 | 1 | ||||||||
| R. sp. CO41 | 1 | 1 | ||||||||
| R. sp. CO47 | 1 | |||||||||
| R. sp. clone GI8-sp-E14, uncultured | 1 | |||||||||
| Ruminococcus sp. clone M3-45R | 1 | |||||||||
| Ruminococcus sp. clone M4-38, uncultured | 1 | 4 | ||||||||
| Ruminococcus sp. clone M4-43, uncultured | 2 | 1 | ||||||||
| Faecalibacterium prausnitzii clone 1-84 | 1 | |||||||||
| Faecalibacterium sp. clone GI5-003-A10, uncultured | 1 | |||||||||
| Faecalibacterium sp. clone M3-42R, uncultured | 4 | |||||||||
| Faecalibacterium sp. clone M4-59, uncultured | 2 | 1 | ||||||||
| Faecalibacterium sp. clone M4-69, uncultured | 1 | |||||||||
| Faecalibacterium sp. clone M7-59, uncultured | 1 | 2 | ||||||||
| Faecalibacterium sp. clone M4-9, uncultured | 1 | 3 | ||||||||
| Faecalibacterium sp. clone M6-59-F, uncultured | 1 | |||||||||
| Faecalibacterium sp. clone PP071-b29, uncultured | 2 | |||||||||
| Faecalibacterium sp. clone 13a, uncultured | 3 | 10 | 2 | |||||||
| Faecalibacterium sp. clone 11b, uncultured | 1 | |||||||||
| Faecalibacterium sp. clone M4-59, uncultured | 1 | |||||||||
| Roseburia inulinivorans strain L1-83 | 3 | 5 | 5 | |||||||
| Roseburia inulinivorans type strain A2-194T | 1 | |||||||||
| Roseburia intestinalis strain L1-82 | 1 | |||||||||
| Eubacterium halii | 1 | |||||||||
| Firmicutes bacterium clone QEDP3BE06, uncultured | 1 | |||||||||
| Firmicutes bacterium clone NI 217, uncultured | 1 | |||||||||
| Firmicutes bacterium clone EHFS1 S12d, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0034 014, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0015 143, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0033 014, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0015 080, uncultured | 1 | 1 | ||||||||
| Firmicutes bacterium clone M0015 104, uncultured | 1 | |||||||||
| Firmicutes bacterium clone M0032 093, uncultured | 1 | |||||||||
| Firmicutes | 29 | 13 | 9 | 14 | 14 | 4 | 19 | 2 | 83 | 51 |
| Bacterium clone AP13R.245, uncultured | 1 | |||||||||
| Bacterium clone HF0730, uncultured 92% | 1 | |||||||||
| Bacterium clone RL 305 aal87f07, uncultured | 1 | |||||||||
| Bacterium clone RL 182 aah34h09, uncultured | 1 | |||||||||
| Bacterium clone RL 184 aao67f01, uncultured | 1 | |||||||||
| Bacterium clone RL 243 aai87b02, uncultured | 1 | |||||||||
| Bacterium clone RL308 aal83c03, unculturded | 1 | |||||||||
| Bacterium clone RL 387 aao91d10, uncultured | 1 | |||||||||
| Bacterium clone calf32 2 wks grp 94% | 2 | |||||||||
| Bacterium clone, uncultured | 2 | 0 | 0 | 1 | 0 | 1 | 3 | 1 | 0 | 2 |
Abbreviation: IBD, inflammatory bowel disease.
Main findings in ileal biopsies.
Main findings in colonic biopsies.
Table S2.
2 × 2 contigency tables comparing differences in complexity of the bacterial OTUs detected in each of the study subjects (Fischer’s exact test)
| Total OTU variants | OTU variants identified > 1 | Sum | |
|---|---|---|---|
| P = 0.52 | |||
| Non-IBD | 42 | 108 | 150 |
| CD1 | 22 | 46 | 68 |
| Sum | 64 | 154 | 218 |
| P = 0.04 | |||
| Non-IBD | 42 | 108 | 150 |
| CD2 | 27 | 124 | 151 |
| Sum | 69 | 232 | 301 |
| P = 0.89 | |||
| Non-IBD | 42 | 108 | 150 |
| CD3 | 30 | 74 | 104 |
| Sum | 72 | 182 | 254 |
| P < 0.001 | |||
| Non-IBD | 42 | 108 | 150 |
| CD4 | 12 | 135 | 147 |
| Sum | 54 | 243 | 297 |
Abbreviations: IBD, inflammatory bowel disease; OTUs, operational taxonomic units.