Abstract
Genetic association and linkage studies can provide insights into complex disease biology, guiding the development of new diagnostic and therapeutic strategies. Over the past decade, genetic association studies have largely focused on common, easy to measure genetic variants shared between many individuals. These common variants typically have subtle functional consequence and translating the resulting association signals into biological insights can be challenging. In the last few years, exome sequencing has emerged as a cost-effective strategy for extending these studies to include rare coding variants, which often have more marked functional consequences. Here, we provide practical guidance in the design and analysis of complex trait association studies focused on rare, coding variants.
INTRODUCTION
Over the past decade, genome-wide association studies have identified hundreds of common risk alleles for complex human diseases (1–9). These studies were enabled by a combination of the availability of large well-characterized sample collections (6–8, 10–13), advances in genotyping technologies (14–16) and advances in methods for the analysis of the resulting data (17–20). These studies have provided several biological insights, highlighting the role of the complement genes in age-related macular degeneration (21–23), of autophagy in Crohn's disease (24–26) or of specific regulatory proteins in blood lipid levels (6), among others. Still, translating the resulting signals into function has been challenging because most common variants have only subtle functional consequences.
Over the past several years, great advances have been made in sequencing and capture technologies, enabling accurate determination of nearly all protein-coding sequence variants in an individual (27–29). These exome-sequencing technologies have already accelerated genetic studies of Mendelian disorders (30) and there is great interest in extending them to complex traits (31). To support this goal, many methods for the design, analysis and interpretation of exome-sequencing studies have been proposed (32–34) and focused candidate gene-sequencing studies have been undertaken, with promising results (35–43).
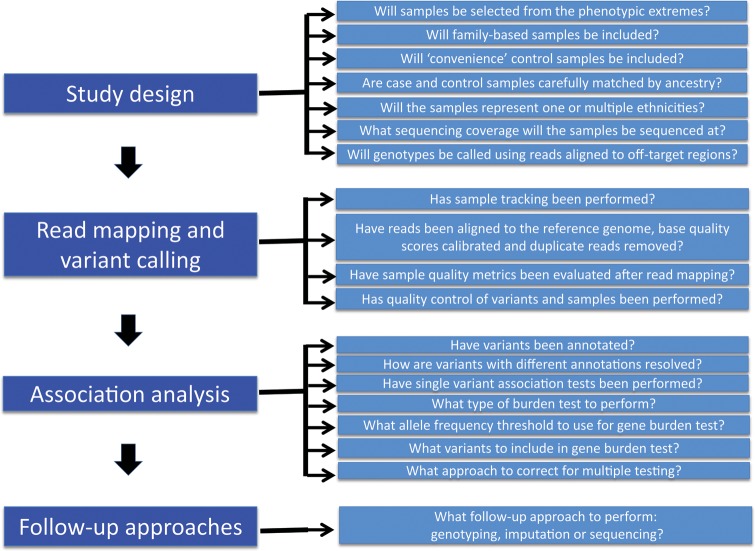
We have been involved in the planning, execution and analysis of several exome-sequencing studies encompassing information on >10 000 individuals. In this review, we focus on the practical aspects of such studies, highlighting important issues to consider when undertaking or evaluating exome-sequencing studies to dissect complex trait genetics. Given the rapidly changing nature of the field, we have tried not to be prescriptive. Rather, we encourage readers to carefully consider a series of key questions when evaluating alternatives for study design, generation of sequence data and variant calling, quality control of the resulting data, rare variant association analysis and follow-up approaches (Fig. 1).
Figure 1.
Key questions and considerations for different stages of an exome sequencing study of complex disease.
STUDY DESIGN: SAMPLE SELECTION
Perhaps the most important step in any exome-sequencing study is the choice of samples to sequence. As with any genetic study, we encourage researchers to start by clearly stating their objectives at the outset (is the objective to survey the range of variation in normal individuals, to find variants that predispose to risk of a specific disease, like diabetes or myocardial infarction, to find variants that influence a specific quantitative trait, like glucose or lipid levels, or to simultaneously investigate a wide-range of quantitative outcomes?) and to systematically inventory all samples in which the traits of interest might be examined (these might include population samples, case and control series, and even families that might be segregating Mendelian forms of disease).
Nearly always, the range of potentially informative samples exceeds the available sequencing budget. Therefore, careful consideration of which samples to sequence will be extremely important. In most instances, it will be fruitful to focus on samples with an extreme outcome (44–46)—for a quantitative trait, these are naturally defined as samples at the extremes of the trait distribution after accounting for known modifiers, which might include age, sex and diet but also previously identified genetic risk factors. For a discrete trait, these are samples whose outcomes are ‘unusual’ after accounting for previously known risk factors (46)—for example, individuals who present with myocardial infarction at an unusually young age. Another general strategy for increasing power is to focus on samples whose relatives have similarly extreme phenotypes (such as high lipid levels) or a history of disease (such as myocardial infarction) (47).
Although selecting individuals with phenotypes that appear extreme or unusual based on known risk factors is important, other considerations can also greatly impact outcome of the study. For example, if a role for de novo mutation events is suspected, it will be extremely useful to sequence related individuals (48–50) and, if the identification of individuals who are homozygous for rare loss-of-function alleles is desired, sequencing of individuals with evidence of inbreeding will be appealing (27).
It is expected that many rare variants will have a very restricted geographic distribution (51,52) so that careful matching of case and control ancestries is likely to be extremely important. In contrast to genome-wide association studies of common variants, where methods for removing artifacts due to mismatches between case and control ancestries are mature (18,53) and the use of ‘convenience’ control samples is relatively widespread, we expect that extreme care will be needed when using convenience controls in exome-sequencing studies because of the potential for false signals to be introduced by small differences in ancestry. As with genome-wide association studies, when these concerns can be overcome, convenience controls can provide for greatly increased sample sizes and power (54).
Most protein-coding variants are extremely rare, previously undescribed and with a geographically restricted segregation pattern (52,55,56). Often, interesting and informative variants will segregate in a population-specific manner. For example, Y142X, a nonsense variant in PCSK9 that demonstrates that knockout of the gene results in greatly reduced low-density lipoprotein cholesterol levels and decreased coronary heart disease risk has frequency of 0.8% in African-ancestry individuals but is virtually absent from European-ancestry samples (44). For this reason, the most complete exome-sequencing studies will examine individuals from a variety of ancestries—with the expectation that segregating variants will provide insights about different (but potentially overlapping) subset of genes in each population. In this context, founder populations—where it may be possible to observe multiple copies of alleles that are otherwise extremely rare—may prove very useful for exome-sequencing studies [just as they were for earlier studies of Mendelian disorders (57,58)].
STUDY DESIGN: SEQUENCING STRATEGY
Standards for generation of high-quality exome sequence data are rapidly emerging. There are several good summaries of raw data quality, but it is common to aim for coverage with high-quality bases to reach 20× or greater in 80–95% of the protein-coding sequences in each genome, after removal of ambiguously mapped reads and of duplicated reads (4,55). With this level of coverage, it should be possible to identify the vast majority of protein-coding variants with high specificity (55). Because the efficiency of enrichment protocols exhibits great local variation, achieving this level of coverage requires sequencing the protein-coding regions of each individual to an average depth of 60–80×.
Most protocols for targeted exome sequencing also result in relatively light coverage of the rest of the genome, typically on the range of 0.2–2.0× on average. Although these ‘off-target’ reads are sometimes discarded in analyses, in our view, they can be extremely useful, particularly in samples that have not been genotyped with whole genome arrays. These off-target reads can be used to estimate the local or global ancestry of each sample (enabling improved case–control matching in association analyses or admixture mapping analyses), can be combined with a panel of reference haplotypes to estimate genotypes across the genome (59–61) and can facilitate detection of large structural variants (such as deletions of entire genes) (62).
VARIANT CALLING
Once sequence data are generated, there are several steps required to process raw short read sequences into high-quality genotypes for each individual. Typically, we first check whether DNA samples have been contaminated and, if DNA fingerprints are available, also check whether samples were tracked correctly during processing (63,64). Next, the process proceeds to the alignment of short sequence reads to the reference genome (65–67), calibration of base-quality scores (68) and removal of duplicate reads (69). After this initial processing, it is useful to examine per sample quality metrics—which might include the fraction of the exome covered at various depths, after removal of duplicates and poorly mapped reads, evaluating the distribution of empirical base quality scores, and the relationship between coverage and GC content. Data for samples with outlier properties such as a low fraction of the genome covered or low base quality scores can be excluded, flagged and/or reprocessed.
After this step, the reads overlapping each position are inspected to identify variant sites. Typically, these sites will be covered by many reads that differ from the reference genome (68,70). The initial list of variant sites is then inspected by a machine-learning-based classifier that tries to separate variants likely to be polymorphic from those that might be calling artifacts (lists of known variants and common artifacts generated by the 1000 Genomes Project can often be used to train these classifiers) (4,68,71). To distinguish true and false positive variants, the machine learning classifiers typically evaluate metrics like the mapping quality of reads supporting each allele, the fraction of reads supporting the alternate allele in putative heterozygotes and sequencing depth. In very small data sets, it may not be practical to tune machine-learning-based classifiers, and it may be necessary to manually review each of these quality metrics to determine appropriate quality cut-offs for each quantity (31). Note that, while variant calls can be generated across the entire genome, producing accurate genotypes in regions that are not deeply covered typically requires an additional post-processing step—using a haplotype aware genotype caller (59,72,73). These haplotype aware callers are quite useful for variants shared across many individuals but are not useful for the rarest variants (including private variants). We also note that calling of insertion–deletion polymorphisms remains especially challenging and that improved analysis of these important variants will likely require a new generation of sequence analysis tools.
At this stage in the process, it is again common to generate a series of quality metrics—these might include the number of variants per individual (typically, we expect 10 000–12 500 synonymous variants, 9500–12 000 non-synonymous variants and 100–200 stop or splice altering variants per individual), the fraction of variants in each category that is unique to each sequenced sample (typically, we expect that nearly all the variants in each sample have been previously described), the fraction of heterozygous sites per sample and the fraction of coding indels that result in a frameshift. Samples with unusual profiles can be flagged, reprocessed or excluded from downstream analyses (55). Within each of these categories, it is also common to compare the transition–transversion ratio of new and previously described variants (74). The transition–transversion ratio is a useful diagnostic metric because, in nature, transitions (A<-> G and C <-> T) occur much more often than transversions (A <-> C, A <-> T, G <-> C or G <-> T). For the exome, we expect the ratio to be a little above 2.0 for non-synonymous variants and above 5.0 for synonymous variants (55, 71). It is often a good idea to manually review the evidence supporting a random subset of the sites—for example, using the integrative genomics viewer (75,76)—and this review should always be carried out for the key variants supporting a manuscript or novel finding. If sufficient resources are available, genotyping or Sanger sequencing of putative carriers can validate a subset of newly identified variants.
Although it is not yet standard to do so, we recommend that the depth of coverage with high-quality bases and the fraction of samples reaching coverage of 20× or greater at each position should also be recorded for each position. These quantities facilitate comparisons between exome-sequencing studies, helping distinguish regions where one study found variation and another study had poor coverage from regions where there truly are differences in the rate of variation across studies.
While there are many reasonable choices for these steps (ranging from the choice of read mapper, specific criteria for filtering poorly mapped reads, criteria for declaring variant calls to be high quality), we note that these choices—just like choices of sequencing and exome capture technology and protocol—do have a small impact on results and can make it difficult to directly contrast samples analyzed with different protocols. In particular, in a few hard to interpret regions or genes, different analytical protocols (or variations on the same protocol) can result in markedly different lists of variants. A welcome development in this area is the development of standards for storing sequence data (69) and resulting variant calls (77), which make it easy for tools developed in different groups to interoperate.
ASSOCIATION ANALYSIS
The final step before association analysis is annotation of functional effects for each variant. There are now reliable, widely used tools for this purpose (78–81). According to their impact on protein-coding transcripts, these tools can identify single nucleotide variants that result in synonymous, missense, nonsense, splice site alterations [typically defined as within 2 bp of an intron–exon boundary, as supported by empirical analyses (82)] or read-through alleles; indels are typically annotated according to whether or not they result in a frameshift or not. Typically, they also assign each variant a score, based on analysis of protein structure or evolutionary conservation, to separate variants with little functional impact from those more likely to damage protein function (83,84). A strategy must be selected for dealing with variants that have multiple annotations—for example, a variant might alter the protein-coding sequence for one transcript but not for other overlapping transcripts. These annotation conflicts can be resolved by focusing only on canonical transcripts for each gene (for example, RefSeqGene), by focusing on the longest transcript in each gene, or by using the most deleterious prediction from all available transcripts.
We recommend that every analysis of exome sequence data should start with single variant association tests. While these tests are typically not well powered for rare variants (most of which will be seen only once or twice, even in very large datasets), they provide a convenient opportunity to quality check the data—by verifying that previously reported common variant signals are reproduced and by inspecting genome-wide QQ plots to ensure samples are adequately matched and results are not unduly influenced by population structure (85).
Because most variants are individually rare, achieving adequate statistical power requires a design where additional copies of the variant of interest can be sampled (perhaps in a family study or in a founder population) or the ability to combine and evaluate groups of variants likely to have similar function (86). The basic idea behind most rare variant association tests is to group variants likely to have an impact on the function of a specific gene and to compare the distribution of these variant groupings to the distribution of the trait of interest.
There are two major categories of association tests for groups of rare variants. In one type of test, the total number of rare alleles across a gene is tabulated in each individual and these totals are compared between cases and controls, for a discrete trait, or correlated with trait values, for a quantitative trait (32). These tests can be carried out by assigning all variants the same weight or they can be designed to place more weight on rarer variants and other variants that are expected to have more severe functional consequences (87,88). While early versions of these tests require explicit allele frequency cut-offs for defining rare variants, newer versions use adaptive thresholds whose choice is guided by available data (89).
Another type of test allows for the situation where a gene might harbor both deleterious and protective variants. Instead of comparing the total number of variants per individual, these tests examine whether the number of variants with non-zero effect sizes (whether positive or negative) exceeds chance expectations (33,89,90). In general, we recommend that at least one test from each category (that is, one burden test assuming all alleles impact the trait in the same direction and one burden test allowing for alleles with opposite directions of effect in each gene) should be considered and that variable threshold implementations of these tests should be used. When it is not practical to use variable threshold methods, we recommended that a variety of frequency cut-offs should be considered (for example, 0.05, 0.01 and 0.001). An additional analysis, focused on individuals who are homozygous or compound heterozygous for deleterious variants in a gene, might eventually become a useful complement to these tests—because it focuses explicitly in individuals where gene function might be ablated.
A number of packages under active development now implement a variety of these tests (89–91, http://genome.sph.umich.edu/wiki/EPACTS, http://atgu.mgh.harvard.edu/plinkseq). In addition to implementing multiple tests, these packages make it simple to consider different subsets of the data for analysis. For example, an initial analysis might include all missense, splice or stop altering variants, excluding only synonymous and non-coding variants. Since many missense variants will not significantly impact protein function (92,93), a second analysis might focus on the subset of these variants that are predicted to have deleterious consequences. And an even more restricted analysis might focus only on splice, frame and stop-altering variants among this later set (94).
We expect there will be no optimal statistical test, filtering strategy or frequency cut-off for gene-based tests. The spectrum of functional variants and their characteristics will likely differ between genes, depending on the importance of the gene's function for the organisms overall function and the luck of the evolutionary draw. Given the multiplicity of statistical tests (and of filtering strategies used to decide which variants are proposed as input for these tests), permutation-based approaches should be used for evaluating statistical significance. Permutations can naturally account for the fact that some genes have very few rare alleles (and thus can never produce a significant burden test result) and that multiple correlated tests might have been undertaken (31). In the absence of permutation-based significance thresholds, a good rule of thumb is that burden test results from exome-sequencing studies should reach P-values on the order of 5 × 10−7 or less before being declared significant (this stringent threshold accounts for the number of genes tested but also for the variety of tests that must be considered and the choice of variants to test inherent in the analysis of these studies). Just as with single variant tests, we recommend generating QQ plots to summarize association results across the genome and ensure test statistics are well behaved. We note that it is valid to combine results for all the tests considered (single variant, burden tests using different frequency thresholds and/or aggregation strategies, etc.) into a single QQ plot.
APPROACHES FOR FOLLOW-UP OF PROMISING SIGNALS
In some rare cases, exome sequencing of a single large sample will be sufficient to demonstrate association (perhaps after technical validation of key genotypes, to show that they are not genotyping artifacts). More often, it will be necessary to examine the most promising variants in additional samples (95). A range of approaches are available for follow-up, ranging from in silico approaches (based on genotype imputation) to targeted genotyping or targeted sequencing.
SNPs with frequencies >1% can usually be tested in thousands of samples by direct genotyping or imputation since these SNPs are frequent enough to be tested individually. A recent Crohn's disease-sequencing study illustrates the possibilities (96): after analysis of sequence data for 350 cases and 350 controls, 70 variants were examined in >16 000 additional cases of Crohn's, >12 000 cases of ulcerative colitis and 17 000 controls—resulting in a clear association for a splice variant in CARD9 (allele frequency = 0.2–0.7%, odds ratio = 0.29, P < 1 × 10−16). An important extension of this approach are studies that attempt to examine essentially all, or most, of the variants discovered in a sequencing experiment in very large numbers of additional samples. One notable set of these experiments, currently underway, are the exome chip experiments. These experiments use arrays designed to include >250 000 non-synonymous variants identified by sequencing >12 000 individuals and are being genotyped on >1 000 000 individuals to explore genetic contributions to a great variety of traits. A limitation of exome chips is that they will miss a significant fraction (∼15–20%) of variants because their genomic context is incompatible with array-based genotyping, variants highly specific to non-European populations (∼10 000 of the 12 000 sequenced individuals considered for the design of exome chip were of European ancestry) as well as the rarest variants in any population. Still, because of their focus on very rare coding variation (the vast majority of variants on the exome chip have frequency <0.5%), the analyses of exome chip experiments will be more similar to the analysis of exome-sequencing studies than to the analysis of genome-wide association studies—requiring careful attention to ancestry matching and the consideration of tests that consider many coding variants in a gene, for example. While these exome chip studies will only provide an imperfect approximation to the results of sequencing studies, we hope they will provide a preview of the discoveries that will be possible when exome sequencing is applied to 100 000 s of samples.
When a very large number of individuals with exome sequence data and whole genome genotypes is available, statistical imputation can also provide an effective strategy for extending sample sizes (97,98). The approach can be relatively fast and economical. Currently, sufficiently large reference panels that can support imputation of very rare variants are not available for most cosmopolitan populations. However, several examples of the success of this approach exist, many from the isolated population of Iceland. There, relatively limited genetic diversity, a panel of sequenced Icelanders, and the availability of 10 000 s of genotyped individuals have enabled recent discoveries using imputation. MYH6 L721W (a variant with allele frequency of 0.4%) was evaluated in 38 000 individuals and associated with the risk for sick sinus syndrome (odds ratio = 12.5, P = 2 × 10−29) (99) and of variant APP A673T (allele frequency 0.1%) was evaluated in 71 000 individuals and associated with the risk for Alzheimer's disease (odds ratio = 5.29 and P = 5 × 10−27) (100).
When targeted genotyping and imputation are not possible or when the association signal is driven by a burden of very rare mutations (101), it will be necessary to undertake targeted sequencing of genes prioritized on the basis of initial analyses. While current methods for sequencing 50–200 genes in 10 000 s of samples are cumbersome, this is an area of active technology development where we expect important advances will soon be available. These advances should perform at a fraction of the cost of traditional Sanger sequencing and will allow follow-up of exome-sequencing studies to explore promising signals due to a burden of rare variants.
THE ROLE OF FUNCTIONAL ASSAYS IN INTERPRETING EXOME-SEQUENCING STUDIES
Genetic analyses that consider groups of rare variants will improve in power if functional variants can be separated from those that have no impact on function so that association tests and follow-up experiments can focus on the functional variants. In this context, functional or computational assays that identify variants most likely to impact gene function—particularly when they can be carried out on a genomic scale—could play a very important role in the successful interpretation of exome-sequencing studies. As these functional assays are expanded to the rest of the genome, they will likely play a critical role in expanding studies of rare variation beyond the exome and to the rest of the genome—where identifying, aggregating and grouping functional variants remain much harder.
Functional characterization of non-synonymous changes will also help interpret rare variant association signals and help transform genetic findings into precise mechanistic insights. Functional studies can reveal the specific molecular changes consequences of coding variation on gene products, as well as the molecular mechanisms by which genes produce disease (102). However, such functional data, when used to support statistical signals that cannot stand on their own, are susceptible to many biases (94). The historical example of candidate gene association studies is informative—in that setting, the widespread use of functional information to support marginal genetic association signals produced a situation where many published findings were irreproducible and most such studies are now discounted. In our view, claims of significance for marginal statistical signals based on modest functional evidence should be considered only when generating additional genetic data is impossible.
We encourage human geneticists to carefully plan and consider the functional experiments that will follow identification of robust, rare variant association signals. However, in most cases, these experiments should only be undertaken when the initial association signal is clearly established. As noted above, we do make an exception for high-throughput assays that attempt to separate variants that are likely to be functional from those are that are likely to be neutral—for example, so as to focus burden analysis on the most deleterious variants. These analyses can be productively used in the discovery process—however, if not used judiciously, they will require the use of even more stringent thresholds because they imply an additional round of statistical tests and potential false discoveries.
FORWARD GENETICS
Instead of characterizing function in model systems, exome sequencing potentially allows for evaluating the functional consequence of pathogenic mutations directly in humans. It is now possible to envision an era of ‘forward genetics’ involving humans. The concept involves understanding gene function by identifying patients harboring specific mutations and characterizing the physiologic and clinical consequences of these mutations. Direct study of rare, human ‘knock-out’ variants may be particularly illustrative (103–105). For example, humans heterozygous or homozygous for knockout alleles at several plasma lipid genes have been identified and detailed study of these individuals has led to new biologic insights.
WHERE DO WE GO FROM HERE?
Exome sequencing has already been successful at identifying the genetic cause of many Mendelian disorders. While applications of exome sequencing to common, complex diseases will be more challenging, we expect that the continued availability of high-quality phenotyped samples, combined with advances in sequencing technology and analytical methods, will soon allow >10 000 s of individuals to be examined for many common outcomes and quantitative traits. As large numbers of sequenced individuals become available, a particular challenge will be the development of appropriate methods for combining information (or results) across studies that might have used different sequencing platforms or analytical approaches for converting sequence data into genotypes. In the context of common variant association studies, such approaches have been instrumental in the rapid rate of discovery of the past few years. In the context of rare variant studies, we believe that new protocols and statistical methods that allow rare variant burden tests to be reconstructed through meta-analysis of study specific summary statistics will be extremely useful.
As larger exome-sequencing studies become common place, and the barriers to cross study analyses are surmounted, perhaps a harvest of specific biological insights will arrive—producing a great need for cellular and model organism systems where these hypotheses can be evaluated. As for human geneticists, we predict they will then be ready to continue their systematic exploration of the genome—proceeding from common variants, to rare coding variants, to a systematic evaluation of all variation (including rare non-coding variation) using whole genome-sequencing approaches.
Conflict of Interest statement. None declared.
FUNDING
This work was supported in part by research grants from the US National Institutes of Health. R.D. is funded by a CIHR Banting Fellowship. Funding to pay the Open Access publication charges for this article was provided by the University of Michigan and the National Human Genome Research Institute.
REFERENCES
- 1.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hindorff L.A., MacArthur J., Wise A., Junkins H.A., Hall P.N., Klemm A.K., Manolio T.A. 2012. www.genome.gov/gwastudies. [accessed August 21, 2012]
- 3.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altshuler D., Daly M.J., Lander E.S. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stranger B.E., Stahl E.A., Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhernakova A., Stahl E.A., Trynka G., Raychaudhuri S., Festen E.A., Franke L., Westra H.J., Fehrmann R.S., Kurreeman F.A., Thomson B., et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ripke S., Sanders A.R., Kendler K.S., Levinson D.F., Sklar P., Holmans P.A., Lin D.Y., Duan J., Ophoff R.A., Andreassen O.A., et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voight B.F., Kang H.M., Ding J., Palmer C.D., Sidore C., Chines P.S., Burtt N.P., Fuchsberger C., Li Y., Erdmann J., et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaFramboise T. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res. 2009;37:4181–4193. doi: 10.1093/nar/gkp552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunderson K.L., Kruglyak S., Graige M.S., Garcia F., Kermani B.G., Zhao C., Che D., Dickinson T., Wickham E., Bierle J., et al. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuzaki H., Dong S., Loi H., Di X., Liu G., Hubbell E., Law J., Bernsten T., Chadha M., Hui H., et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 17.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 18.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., Sangiovanni J.P., Mane S.M., Mayne S.T., et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards A.O., Ritter R., 3rd, Abel K.J., Manning A., Panhuysen C., Farrer L.A. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 23.Haines J.L., Hauser M.A., Schmidt S., Scott W.K., Olson L.M., Gallins P., Spencer K.L., Kwan S.Y., Noureddine M., Gilbert J.R., et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 24.Franke A., McGovern D.P., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42: 1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkes M., Barrett J.C., Prescott N.J., Tremelling M., Anderson C.A., Fisher S.A., Roberts R.G., Nimmo E.R., Cummings F.R., Soars D., et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P., Nayir A., Bakkaloglu A., Ozen S., Sanjad S., et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gnirke A., Melnikov A., Maguire J., Rogov P., LeProust E.M., Brockman W., Fennell T., Giannoukos G., Fisher S., Russ C., et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 31.Kiezun A., Garimella K., Do R., Stitziel N.O., Neale B.M., McLaren P.J., Gupta N., Sklar P., Sullivan P.F., Moran J.L., et al. Exome sequencing and the genetic basis of complex traits. Nat. Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neale B.M., Rivas M.A., Voight B.F., Altshuler D., Devlin B., Orho-Melander M., Kathiresan S., Purcell S.M., Roeder K., Daly M.J. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris A.P., Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet. Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo G.S., Farooqi I.S., Aminian S., Halsall D.J., Stanhope R.G., O'Rahilly S. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 1998;20:111–112. doi: 10.1038/2404. [DOI] [PubMed] [Google Scholar]
- 36.Vaisse C., Clement K., Guy-Grand B., Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet. 1998;20:113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 37.Cohen J.C., Kiss R.S., Pertsemlidis A., Marcel Y.L., McPherson R., Hobbs H.H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 38.Morita H., Larson M.G., Barr S.C., Vasan R.S., O'Donnell C.J., Hirschhorn J.N., Levy D., Corey D., Seidman C.E., Seidman J.G., et al. Single-gene mutations and increased left ventricular wall thickness in the community: the Framingham Heart Study. Circulation. 2006;113:2697–2705. doi: 10.1161/CIRCULATIONAHA.105.593558. [DOI] [PubMed] [Google Scholar]
- 39.Kotowski I.K., Pertsemlidis A., Luke A., Cooper R.S., Vega G.L., Cohen J.C., Hobbs H.H. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji W., Foo J.N., O'Roak B.J., Zhao H., Larson M.G., Simon D.B., Newton-Cheh C., State M.W., Levy D., Lifton R.P. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat. Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nejentsev S., Walker N., Riches D., Egholm M., Todd J.A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raychaudhuri S., Iartchouk O., Chin K., Tan P.L., Tai A.K., Ripke S., Gowrisankar S., Vemuri S., Montgomery K., Yu Y., et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat. Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansen C.T., Wang J., McIntyre A.D., Martins R.A., Ban M.R., Lanktree M.B., Huff M.W., Peterfy M., Mehrabian M., Lusis A.J., et al. Excess of rare variants in non-genome-wide association study candidate genes in patients with hypertriglyceridemia. Circ. Cardiovasc. Genet. 2012;5:66–72. doi: 10.1161/CIRCGENETICS.111.960864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen J.C., Boerwinkle E., Mosley T.H., Jr, Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 45.Li D., Lewinger J.P., Gauderman W.J., Murcray C.E., Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet. Epidemiol. 2011;35:790–799. doi: 10.1002/gepi.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guey L.T., Kravic J., Melander O., Burtt N.P., Laramie J.M., Lyssenko V., Jonsson A., Lindholm E., Tuomi T., Isomaa B., et al. Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genet. Epidemiol. 2011;35:236–246. doi: 10.1002/gepi.20572. [DOI] [PubMed] [Google Scholar]
- 47.Li M., Boehnke M., Abecasis G.R. Efficient study designs for test of genetic association using sibship data and unrelated cases and controls. Am. J. Hum. Genet. 2006;78:778–792. doi: 10.1086/503711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neale B.M., Kou Y., Liu L., Ma'ayan A., Samocha K.E., Sabo A., Lin C.F., Stevens C., Wang L.S., Makarov V., et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D., et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L., et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathieson I., McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat. Genet. 2012;44:243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson M.R., Wegmann D., Ehm M.G., Kessner D., St Jean P., Verzilli C., Shen J., Tang Z., Bacanu S.A., Fraser D., et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirschhorn J.N., Daly M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 55.Tennessen J.A., Bigham A.W., O'Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keinan A., Clark A.G. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de la Chapelle A. Disease gene mapping in isolated human populations: the example of Finland. J. Med. Genet. 1993;30:857–865. doi: 10.1136/jmg.30.10.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de la Chapelle A., Wright F.A. Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc. Natl Acad. Sci. USA. 1998;95:12416–12423. doi: 10.1073/pnas.95.21.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y., Sidore C., Kang H.M., Boehnke M., Abecasis G.R. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasaniuc B., Rohland N., McLaren P.J., Garimella K., Zaitlen N., Li H., Gupta N., Neale B.M., Daly M.J., Sklar P., et al. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat. Genet. 2012;44:631–635. doi: 10.1038/ng.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flannick J., Korn J.M., Fontanillas P., Grant G.B., Banks E., Depristo M.A., Altshuler D. Efficiency and power as a function of sequence coverage, SNP array density, and imputation. PLoS Comput. Biol. 2012;8:e1002604. doi: 10.1371/journal.pcbi.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cibulskis K., McKenna A., Fennell T., Banks E., DePristo M., Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27: 2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jun G., Flickinger M., Hetrick K.N., Romm J.M., Doheny K.F., Abecasis G.R., Boehnke M., Kang H.M. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 2012 doi: 10.1016/j.ajhg.2012.09.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Metzker M.L. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 66.Li H., Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.The 1000 Genomes Project. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 doi: 10.1038/nature11632. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Browning B.L., Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am. J. Hum. Genet. 2009;85:847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le S.Q., Durbin R. SNP detection and genotyping from low-coverage sequencing data on multiple diploid samples. Genome Res. 2011;21:952–960. doi: 10.1101/gr.113084.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics, 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012 doi: 10.1093/bib/bbs017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27: 2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yandell M., Huff C., Hu H., Singleton M., Moore B., Xing J., Jorde L.B., Reese M.G. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w (1118) ; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Habegger L., Balasubramanian S., Chen D.Z., Khurana E., Sboner A., Harmanci A., Rozowsky J., Clarke D., Snyder M., Gerstein M. VAT: a computational framework to functionally annotate variants in personal genomes within a cloud-computing environment. Bioinformatics. 2012;28:2267–2269. doi: 10.1093/bioinformatics/bts368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calvo S.E., Tucker E.J., Compton A.G., Kirby D.M., Crawford G., Burtt N.P., Rivas M., Guiducci C., Bruno D.L., Goldberger O.A., et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat. Genet. 2010;42:851–858. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sunyaev S., Hanke J., Aydin A., Wirkner U., Zastrow I., Reich J., Bork P. Prediction of nonsynonymous single nucleotide polymorphisms in human disease-associated genes. J. Mol. Med. 1999;77:754–760. doi: 10.1007/s001099900059. [DOI] [PubMed] [Google Scholar]
- 84.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 86.Kryukov G.V., Shpunt A., Stamatoyannopoulos J.A., Sunyaev S.R. Power of deep, all-exon resequencing for discovery of human trait genes. Proc. Natl Acad. Sci. USA. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zawistowski M., Gopalakrishnan S., Ding J., Li Y., Grimm S., Zollner S. Extending rare-variant testing strategies: analysis of noncoding sequence and imputed genotypes. Am. J. Hum. Genet. 2010;87:604–617. doi: 10.1016/j.ajhg.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Magi R., Kumar A., Morris A.P. Assessing the impact of missing genotype data in rare variant association analysis. BMC Proc. 2011;5(Suppl. 9):S107. doi: 10.1186/1753-6561-5-S9-S107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin D.Y., Tang Z.Z. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 2011;89:354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li B, Wang G., Leal S.M. SimRare: a program to generate and analyze sequence-based data for association studies of quantitative and qualitative traits. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts499. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gorlov I.P., Gorlova O.Y., Sunyaev S.R., Spitz M.R., Amos C.I. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am. J. Hum. Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kryukov G.V., Pennacchio L.A., Sunyaev S.R. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am. J. Hum. Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Skol A.D., Scott L.J., Abecasis G.R., Boehnke M. Optimal designs for two-stage genome-wide association studies. Genet. Epidemiol. 2007;31:776–788. doi: 10.1002/gepi.20240. [DOI] [PubMed] [Google Scholar]
- 96.Rivas M.A., Beaudoin M., Gardet A., Stevens C., Sharma Y., Zhang C.K., Boucher G., Ripke S., Ellinghaus D., Burtt N., et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43: 1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holm H., Gudbjartsson D.F., Sulem P., Masson G., Helgadottir H.T., Zanon C., Magnusson O.T., Helgason A., Saemundsdottir J., Gylfason A., et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat. Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jonsson T., Atwal J.K., Steinberg S., Snaedal J., Jonsson P.V., Bjornsson S., Stefansson H., Sulem P., Gudbjartsson D., Maloney J., et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 101.Kathiresan S., Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ioannidis J.P., Thomas G., Daly M.J. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 2009;10:318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao Z., Tuakli-Wosornu Y., Lagace T.A., Kinch L., Grishin N.V., Horton J.D., Cohen J.C., Hobbs H.H. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 2006;79:514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pollin T.I., Damcott C.M., Shen H., Ott S.H., Shelton J., Horenstein R.B., Post W., McLenithan J.C., Bielak L.F., Peyser P.A., et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Musunuru K., Pirruccello J.P., Do R., Peloso G.M., Guiducci C., Sougnez C., Garimella K.V., Fisher S., Abreu J., Barry A.J., et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]