Abstract
Patients with developmental disorders often harbour sub-microscopic deletions or duplications that lead to a disruption of normal gene expression or perturbation in the copy number of dosage-sensitive genes. Clinical interpretation for such patients in isolation is hindered by the rarity and novelty of such disorders. The DECIPHER project (https://decipher.sanger.ac.uk) was established in 2004 as an accessible online repository of genomic and associated phenotypic data with the primary goal of aiding the clinical interpretation of rare copy-number variants (CNVs). DECIPHER integrates information from a variety of bioinformatics resources and uses visualization tools to identify potential disease genes within a CNV. A two-tier access system permits clinicians and clinical scientists to maintain confidential linked anonymous records of phenotypes and CNVs for their patients that, with informed consent, can subsequently be shared with the wider clinical genetics and research communities. Advances in next-generation sequencing technologies are making it practical and affordable to sequence the whole exome/genome of patients who display features suggestive of a genetic disorder. This approach enables the identification of smaller intragenic mutations including single-nucleotide variants that are not accessible even with high-resolution genomic array analysis. This article briefly summarizes the current status and achievements of the DECIPHER project and looks ahead to the opportunities and challenges of jointly analysing structural and sequence variation in the human genome.
INTRODUCTION
The accurate clinical diagnosis of genetic disorders requires the assessment of a patient's clinical features and their family history supplemented by genetic testing in a diagnostic laboratory. While phenotype assessment alone may often provide clues to an underlying genetic problem, genetic data are essential to make an accurate diagnosis. However, for very rare disorders seen in isolation, establishing causality remains a challenge. Traditional clinical genomic analysis has relied upon cytogenetic techniques such as karyotype-banding and fluorescence in situ hybridization (FISH). These methods are useful for the detection of large, multi-megabase CNVs in the genome of affected patients such as those that result in Down's syndrome (1), Prader–Willi syndrome (2) etc. These techniques do not lend themselves to the identification of smaller CNVs that involve only a few genes or just a single gene. Genomic array analysis, on the other hand, is a powerful and accurate molecular cytogenetic diagnostic tool to identify large and small CNVs. Rapid advances in the genomic technology have seen the resolution of genomic array analysis increase a 1000-fold, from 1 Mb to <1 kb, enabling the precise identification and mapping of CNVs on the reference sequence of the human genome. Genomic microarray analysis using array-comparative genomic hybridization (aCGH) or single-nucleotide polymorphism genotyping arrays has been particularly useful in the identification and management of patients with severe development delay, intellectual disability and congenital structural anomalies (3–7). This has facilitated the identification of hitherto unknown chromosomal syndromes (8–11) and improved the clinical management of patients with such disorders (12,13). With the resolution of cytogenetic analysis methods approaching that of molecular genetics, clinicians and laboratory scientists can now employ methods ranging from FISH to aCGH to genome sequencing to identify sequence and structural variations in patients displaying features suggestive of a genetic disorder.
The biggest challenge arising from the clinical application of genomic analysis remains one of being able to accurately distinguish between causal (pathogenic) and non-causal (often termed ‘benign’) variation. Highly penetrant variants causing rare genetic diseases are, by definition, rare in the population, whereas most variants observed within any human genome are common in the population and therefore likely to be benign. Thus, the advent of genome-wide maps of CNVs that are common in different populations has proven to be an effective strategy to filter out the large fraction of variants that are too common to be plausibly causal. Large-scale genome studies have identified the existence of a number of insertions, deletions and duplications of varying lengths in the normal human population (14–16). The Database of Genomic Variation (14) currently contains over 179 000 published, unique, copy-number variants (CNVs) spread over several thousand loci. Initiatives such as the International HapMap Project (17) and the 1000 Genomes Project (18) continue to enlarge our understanding of common genomic variation (allele frequency >1%) within populations of diverse ancestries. Successful clinical interpretation of sequence and structural variants shares similar requirements: weeding out of common variants by using a good population variation resource and comparison with known pathogenic variants with up-to-date functional annotations of the genome.
The ever reducing cost and speed of whole genome/exome sequencing promises to make it practical for use in a clinical setting for patients with genetic disorders (19), and next-generation sequencing (NGS) is already being used in a research context for the identification of novel Mendelian disease genes (20–23). The proliferation of publicly available genomic variation data including locus-specific databases (LSDB) is also driving the development of tools for gene prioritization and annotation of functional impact (see 24 for a review of all available genome interpretation databases). However, the accurate diagnosis of human genetic disorders in a clinical setting requires the identification of other patients that share the same/similar genomic variants and comparison of their phenotypes. Few data collections correlating genotype with phenotype in human genetic disorders exist.
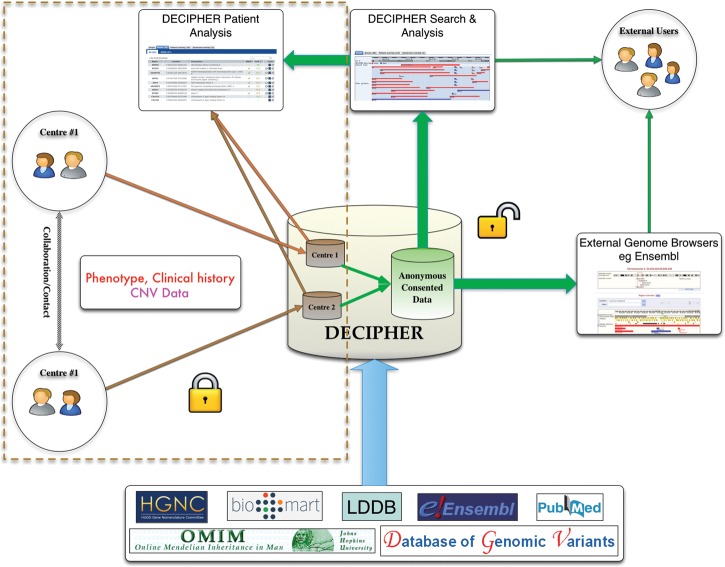
The DECIPHER project (https://decipher.sanger.ac.uk) was initiated in 2004 to address the need for international collaboration in the reporting and cataloguing of genotype–phenotype associations, with the aim of enabling the identification of clusters of patients with developmental disorders sharing similar phenotypes and genomic variants (25). Since its inception, DECIPHER has become a valuable international resource, and now contains variant and phenotype information on over 17 000 patients from more than 200 contributing academic departments of clinical genetics in 30 countries. Each centre maintains full control of its own confidential patient data in DECIPHER until patient consent is given to allow their linked-anonymized genomic and phenotypic data to be shared (Fig. 1). Data are shared with other clinical DECIPHER users for collaboration and comparison and with academic researchers (in the form of anonymized bulk data under data access agreements) to drive the development of interpretative tools. The importance of a collaborative effort as in DECIPHER can be garnered from the fact that the project has been instrumental in the identification of multiple new syndromes (11,26–28). Other worldwide initiatives such as the International Standards for Cytogenomic Arrays (ISCA) Consortium (29) and the European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA) (30) also provide a mechanism to upload and share patient cytogenetic data with other members of the project.
Figure 1.
DECIPHER provides participating clinical genetics centres secure and private area for depositing patient phenotypic and genotypic information. Preliminary analysis and comparison of these data can be carried out against anonymized consented data to aid contact and collaboration with other clinicians with patients with similar variation. Once patient consent for data release has been obtained, these anonymized data are supplemented with information from various bioinformatics resources and shared with the larger DECIPHER community and within external genome browsers such as the Ensembl Genome Browser and the UCSC Browser.
Advances in the technology are blurring the boundaries between classical cytogenetic techniques (FISH and aCGH) and molecular genetics and providing methods for the analysis and discovery of nucleotide-level changes in the genome that result in disease, cancer or inherited and de novo disorders in humans (20,31). There is a pressing need for the development of tools and services that integrate data from both sequence and structural variation to provide a complete understanding of a patient genome as well as delineate causal genes based on comparison of different variants (CNVs and deleterious mutations) with similar functional impact in similar regions of the genome. This new paradigm has critical implications for services like DECIPHER (primarily designed for capturing data from genomic array experiments) not only for data deposition and analysis, but also for the identification, storage and display of sequence variants alongside CNVs. In this review, we describe the current functionality available in DECIPHER and discuss the opportunities and challenges offered by emerging NGS technologies. We discuss the future direction of DECIPHER development, mindful of the ethical issues surrounding the deposition of genomic data derived from patients.
DECIPHER TODAY
The original aims of DECIPHER were to:
provide a secure deposition interface to enable the upload and storage of patient genomic data from microarray analysis with accompanying phenotypes before linked-anonymized data sharing (following informed consent);
identify genes involved in a specific microduplication, microdeletion, inversion or translocation using coordinates from human genome assemblies;
aid in the interpretation of uploaded data by comparison of both genomic and phenotypic information from consented patients already in DECIPHER, as well as genome annotation resources;
encourage collaboration between clinicians and molecular cytogeneticists, both within and between centres, to facilitate the identification of new syndromes and gene function.
DECIPHER provides a simple, password-protected secure interface for entering patient genomic and phenotype data, which the clinician and clinical scientist can enter and modify. DECIPHER uses restricted phenotype vocabulary from the Baraitser-Winter Neurogenetics Database (BWDB) (32) in order to ensure consistent and accurate representation of patient phenotypes. Until explicit patient consent is granted for sharing linked-anonymous data, all deposited studies are only accessible to the depositing centre. Shared consented data are made available to other DECIPHER members and is also accessible without password protection via the Ensembl and UCSC genome browsers. DECIPHER offers a powerful search functionality that allow consented data to be interrogated over many different types of query. Search results are presented in an intuitive and sortable interface that allows for quick visual comparison between different patients (Fig. 2). For a summary of features in DECIPHER including data content and visualization, see Table 1.
Figure 2.
Search and analysis. (A) DECIPHER provides a simple search interface that supports many different types of queries (highlighted: search DECIPHER by chromosomal position 17p11.2). (B) List of overlapping DECIPHER patients with additional detail and annotated syndromes found matching query. (C) DECIPHER patient page showing variant detail (position, mean ratio, genes involved and inheritance), with external genome browser links (highlighted). Also shown is a graphical comparison of patient CNV overlaid on other DECIPHER patients having an overlapping variant. (D) DECIPHER Gene Data: description for every gene affected by the CNV, additional data including Online Mendelian Inheritance in man (OMIM), OMIM Morbid, and Haploinsufficiency Scores and external links to gene resources (highlighted). (E) DECIPHER Patient Overlap: other consented DECIPHER patients that have an overlapping CNV with the query patient, common phenotypes are shown in bold.
Table 1.
DECIPHER Functionality
| Registered depositors | Data deposition | Password-protected secure interface for patient genotype and phenotype data |
| Data analysis | Comparison with all patient data in own centre and all consented data in DECIPHER | |
| Data retrieval | Patient ID, phenotype, location, karyotype etc. search all patient data in own centre and all consented data in DECIPHER | |
| Collaboration | Contact clinicians of consented patients directly from within DECIPHER | |
| All Users | Data retrieval | Patient ID, phenotype, location, karyotype etc. search all consented patient data in DECIPHER |
| Data content and visualization | Location, size and nature of the variation observed (deletion/duplication) | |
| Number of genes affected and any inheritance information | ||
| Approved gene symbols and description of the genes involved, highlighting genes present in OMIM and OMIM Morbid databases with predicted haploinsufficiency scores (42) | ||
| Other patients that share variants in the same region of the chromosome shared phenotypes highlighted | ||
| Known syndromes that overlap with the recorded variant | ||
| Colour-coded visualization of CNVs for quick identification of the other overlapping patient | ||
| Visualization of genes that are in the affected region coloured by haploinsufficiency scores | ||
| Interactive visualization of consented CNV data in external genome browsers (Ensembl and UCSC) | ||
| Syndrome views |
DECIPHER curated ‘syndrome’ pages provide a single point of reference and links for a given syndrome. The syndrome pages provide salient features including expert-reviewed clinical synopsis, size and nature of deletion or duplication, a list of genes contained with the aberration, literature references and links to appropriate support groups. The syndrome pages can also be used to query all consented patients in DECIPHER who have similar affected regions (Fig. 3).
Figure 3.
Expert-reviewed syndrome resource in DECIPHER. (A) Karyotype view showing the location and nature of all DECIPHER syndromes. (B) DECIPHER expert overview for 17q21.31 deletion syndrome (11) with associated citations and external links to support groups and additional sources of information. (C) Detail for 22q11 deletion syndrome (65) showing genes involved in the syndrome with associated haploinsufficiency scores and external links to OMIM, OMIM Morbid and other data sources.
DECIPHER continues to play a critical role in the delineation and identification of new syndromes (11,26–28), as well as providing crucial data for the analysis of known disorders (33–37). By providing mechanisms to contact and collaborate with other clinicians having patients with similar disorders, DECIPHER facilitates knowledge exchange pertaining to patient advice and long-term care.
DECIPHER collaborative framework
Collaboration and information sharing are the cornerstones of scientific discovery, and DECIPHER provides an invaluable tool for clinical geneticists, clinical laboratory scientists and researchers to share and analyse their findings. The different levels at which DECIPHER facilitates collaboration are:
Peer-to-Peer: Registered members (people who also deposit data to DECIPHER) have access to the contact details of other members who have shared their data enabling direct contact for information and/or collaboration resulting in joint findings in literature (38–41).
External User to Peer: Unregistered users have anonymized access to all consented patient data. DECIPHER facilitates collaboration between these groups by acting as an intermediary and forwarding all genuine requests for information/collaboration to depositing clinicians. On average, DECIPHER receives ∼10 such contact requests every month.
Anonymized Bulk Metadata: DECIPHER provides anonymized consented data under a Data Access Agreement to individuals seeking to undertake large-scale analysis. These bulk data have been used to catalyse research into improved interpretative tools, most notably in the prediction of haploinsufficiency (42), mouse–human phenotype association studies (33) and the characterization of copy-number stable regions in the human genome (43).
LOOKING AHEAD
DECIPHER and other peer databases
At the research level, national and international initiatives are underway to identify and catalogue the genomic basis of developmental disorders (deciphering developmental disorders, DDD) (44), cancer (Cancer Genome Project) (45), Mendelian diseases (Centres for Mendelian Genomics) (46), learning disabilities (Genetics of Learning Disability, GOLD) (47), rare genetic variants in health and disease (UK10K) to name a few. Large-scale genome studies in the population like the 1000 Genomes Project (18) have provided a detailed map of normal genetic variation in the population. Individual clinicians and laboratories are also generating a wealth of structural and sequence variation data on specific genetic disorders that are deposited in a myriad of LSDB (48), making data access or comparison difficult. Structural variants are deposited with DECIPHER and other projects like ISCA Consortium (29) and ECARUCA (30). The spread of patient variant information across multiple (and sometimes protected) collections not only poses a genuine problem in the overall retrieval, comparison and analysis, but also slows down the discovery of new syndromes and disease-causing genes.
In order to provide a comprehensive resource for all forms of human genetic variation, it is desirable for these initiatives to work together and share data. This has traditionally been difficult due to funding models, legislative hurdles, ethical issues and specialization (49,50). Initiatives such as the Human Variome Project (HVP) (51) seek to redress this imbalance by setting up a public catalogue of curated variation data for each gene with associated phenotypes by using standardized methods and standards for data sharing. It is not far fetched to envisage a collaborative competitive environment, where the underlying data are shared using an agreed mechanism and format, but the different projects develop different tools and services for interrogation of these data. This will not only help to identify and weed out duplicate submissions and enhance the quality of data, but also enforce data uniformity in terms of phenotype ontologies and variant descriptors. The collaborative competitive model is popular in industry and has been shown to work successfully for scientific consortia like the worldwide Protein Data Bank (wwPDB) (52) and the Unified Protein Resource (UniProt) (53). DECIPHER has led the way in defining an ethical and collaborative framework for the sharing of human CNV data. In the future, DECIPHER and similar initiatives such as HVP, ISCA Consortium and others will need to pool their data resources to provide an integrated view of human genotype–phenotype correlations in genetic disorders.
DECIPHER: integrating CNV and sequence variation
The traditional approaches to identifying genetic disorders caused by copy-number variation involve the use of techniques such as G-banded karyotyping and FISH. These methods are useful in the identification of relatively large-scale variations in the genome ranging from ∼5 Mb (G-banding) to between 2 Mb and 500 kb depending on the FISH method (54). The increasing resolution of genomic array analysis now offers resolutions sufficient for the identification of many microdeletion or microduplication events. However, monogenic disorders resulting from even smaller changes in dosage-sensitive genes often fall below the detection threshold of detection by classical molecular cytogenetic techniques. Massively parallel exome sequencing, combined with statistical filtering methodologies (55) to exclude benign or unrelated variants, is a powerful technique to identify very small causative mutations in a single gene, as shown for Miller syndrome (56), Charcot–Marie–Tooth disease (21) and Kabuki syndrome (57). With the falling costs of whole genome and exome sequencing, it is not unrealistic to assume that the coming years will see a huge increase in the diagnosis and discovery of rare diseases and their causal genes in individual patients and lead to improvements in personalized medical care.
As the disciplines of cytogenetics, molecular genetics and genomics gradually merge to provide a comprehensive view of all forms of human genetic variation, services like DECIPHER have an even more important role to play in providing an integrated phenotype-linked structural and sequence variation analysis platform. Phenotype associations in sequence variation, as with statistical filtering against control populations, are critical in the identification of pathogenic variants (58,59). DECIPHER is already working on extending its scope to provide a platform for accepting and displaying sequence variant data as well as using controlled phenotype ontologies from the Human Phenotype Ontology (HPO) project (60). With the availability of phenotype-linked sequence variation data, DECIPHER has the potential to give access to genotype-driven and phenotype-driven interpretative tools and resources ranging from differential diagnosis based on phenotypes (Phenomizer) (60) and variant effect predictor in Ensembl (61), to identifying affected protein families and domains using Pfam (62) and InterPro (63) or interaction networks using IntAct (64). The inclusion of sequence variant data in a project originally designed for storing and displaying data from genomic microarray analysis poses significant challenges, not least in the scale and detail of the information, but also in the meaningful visualization of relevant information in a genomic context. Sequence variant display in DECIPHER is currently being developed in partnership with the DDD project (44) and is scheduled to be available by the end of 2012.
CONCLUSION
The field of medical genetics is poised at the cusp of a revolution. Exponential advances in sequencing technologies have made it practical to carry out exome and/or whole genome sequencing of patients with rare disorders; however, the fundamental interpretative challenge encountered with novel or extremely rare variants remains. The increasing availability of data from large-scale genome studies of normal populations is facilitating the discrimination between normal and pathogenic variation. At the same time, improvements in aCGH technologies have made it possible to detect ever-smaller CNVs in the human genome and narrow the gap between classical cytogenetics and molecular genetics.
DECIPHER continues to be an essential international resource for submitting and accessing phenotype-linked patient copy-number variation data and plans to extend its scope and usability by accepting sequence variation data. The incorporation of sequence variation data in DECIPHER poses significant technological, visualization and ethical challenges. Building on the existing global network of members, DECIPHER is strategically positioned to integrate sequence variation data with structural variation data and phenotype thus becoming an invaluable and essential service for clinical diagnosis and genomic research.
FUNDING
The authors are grateful to the Wellcome Trust for their financial support (WT077008).
ACKNOWLEDGEMENTS
The authors thank the patients and their families for permission to include their information in the DECIPHER database. Finally, the authors would like to thank all of the DECIPHER Consortium Centres, a list of which can be found on the DECIPHER website. The DECIPHER project has UK Research Ethics Committee approval (04/MRE05/50, granted by the Cambridge South REC).
Conflict of Interest statement: None declared.
REFERENCES
- 1.Roizen N.J., Patterson D. Down's syndrome. Lancet. 2003;361:1281–1289. doi: 10.1016/S0140-6736(03)12987-X. [DOI] [PubMed] [Google Scholar]
- 2.Cassidy S.B., Schwartz S., Miller J.L., Driscoll D.J. Prader-Willi syndrome. Genet. Med. 2012;14:10–26. doi: 10.1038/gim.0b013e31822bead0. [DOI] [PubMed] [Google Scholar]
- 3.Shaw-Smith C., Redon R., Rickman L., Rio M., Willatt L., Fiegler H., Firth H., Sanlaville D., Winter R., Colleaux L., et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J. Med. Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu X., Shaw C.A., Patel A., Li J., Cooper M.L., Wells W.R., Sullivan C.M., Sahoo T., Yatsenko S.A., Bacino C.A., et al. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS One. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stankiewicz P., Beaudet A.L. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr. Opin. Genet. Dev. 2007;17:182–192. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Veltman J.A. Genomic microarrays in clinical diagnosis. Curr. Opin. Pediatr. 2006;18:598–603. doi: 10.1097/MOP.0b013e3280105417. [DOI] [PubMed] [Google Scholar]
- 7.Vissers L.E., de Vries B.B., Osoegawa K., Janssen I.M., Feuth T., Choy C.O., Straatman H., van der Vliet W., Huys E.H., van Rijk A., et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am. J. Hum. Genet. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballif B.C., Hornor S.A., Jenkins E., Madan-Khetarpal S., Surti U., Jackson K.E., Asamoah A., Brock P.L., Gowans G.C., Conway R.L., et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat. Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 9.Ballif B.C., Theisen A., McDonald-McGinn D.M., Zackai E.H., Hersh J.H., Bejjani B.A., Shaffer L.G. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin. Genet. 2008;74:469–475. doi: 10.1111/j.1399-0004.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- 10.Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 11.Shaw-Smith C., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 12.Shaffer L.G., Bejjani B.A., Torchia B., Kirkpatrick S., Coppinger J., Ballif B.C. The identification of microdeletion syndromes and other chromosome abnormalities: cytogenetic methods of the past, new technologies for the future. Am. J. Med. Genet. C Semin. Med. Genet. 2007;145C:335–345. doi: 10.1002/ajmg.c.30152. [DOI] [PubMed] [Google Scholar]
- 13.Shevell M.I., Bejjani B.A., Srour M., Rorem E.A., Hall N., Shaffer L.G. Array comparative genomic hybridization in global developmental delay. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:1101–1108. doi: 10.1002/ajmg.b.30730. [DOI] [PubMed] [Google Scholar]
- 14.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 15.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 16.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium T.G. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzo J.M., Buck M.J. Key principles and clinical applications of ‘next-generation’ DNA sequencing. Cancer Prev. Res. 2012;5:887–900. doi: 10.1158/1940-6207.CAPR-11-0432. [DOI] [PubMed] [Google Scholar]
- 20.Ku C.S., Cooper D.N., Polychronakos C., Naidoo N., Wu M., Soong R. Exome sequencing: dual role as a discovery and diagnostic tool. Ann. Neurol. 2012;71:5–14. doi: 10.1002/ana.22647. [DOI] [PubMed] [Google Scholar]
- 21.Montenegro G., Powell E., Huang J., Speziani F., Edwards Y.J., Beecham G., Hulme W., Siskind C., Vance J., Shy M., et al. Exome sequencing allows for rapid gene identification in a Charcot-Marie-Tooth family. Ann. Neurol. 2011;69:464–470. doi: 10.1002/ana.22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnefond A., Durand E., Sand O., De Graeve F., Gallina S., Busiah K., Lobbens S., Simon A., Bellanne-Chantelot C., Letourneau L., et al. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS One. 2010;5:e13630. doi: 10.1371/journal.pone.0013630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilissen C., Hoischen A., Brunner H.G., Veltman J.A. Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capriotti E., Nehrt N.L., Kann M.G., Bromberg Y. Bioinformatics for personal genome interpretation. Brief Bioinform. 2012;13:495–512. doi: 10.1093/bib/bbr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahir F., Firth H.V., Baross A., Delaney A.D., Eydoux P., Gibson W.T., Langlois S., Martin H., Willatt L., Marra M.A., et al. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J. Med. Genet. 2007;44:556–561. doi: 10.1136/jmg.2007.050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malan V., Raoul O., Firth H.V., Royer G., Turleau C., Bernheim A., Willatt L., Munnich A., Vekemans M., Lyonnet S., et al. 19q13.11 deletion syndrome: a novel clinically recognisable genetic condition identified by array comparative genomic hybridisation. J. Med. Genet. 2009;46:635–640. doi: 10.1136/jmg.2008.062034. [DOI] [PubMed] [Google Scholar]
- 28.Engels H., Wohlleber E., Zink A., Hoyer J., Ludwig K.U., Brockschmidt F.F., Wieczorek D., Moog U., Hellmann-Mersch B., Weber R.G., et al. A novel microdeletion syndrome involving 5q14.3-q15: clinical and molecular cytogenetic characterization of three patients. Eur. J. Hum. Genet. 2009;17:1592–1599. doi: 10.1038/ejhg.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaminsky E.B., Kaul V., Paschall J., Church D.M., Bunke B., Kunig D., Moreno-De-Luca D., Moreno-De-Luca A., Mulle J.G., Warren S.T., et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feenstra I., Fang J., Koolen D.A., Siezen A., Evans C., Winter R.M., Lees M.M., Riegel M., de Vries B.B., Van Ravenswaaij C.M., et al. European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA); an online database for rare chromosome abnormalities. Eur. J. Med. Genet. 2006;49:279–291. doi: 10.1016/j.ejmg.2005.10.131. [DOI] [PubMed] [Google Scholar]
- 31.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 32.Winter R.M., Baraitser M. The London dysmorphology database. J. Med. Genet. 1987;24:509–510. doi: 10.1136/jmg.24.8.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boulding H., Webber C. Large-scale objective association of mouse phenotypes with human symptoms through structural variation identified in patients with developmental disorders. Hum. Mutat. 2012;33:874–883. doi: 10.1002/humu.22069. [DOI] [PubMed] [Google Scholar]
- 34.Santen G.W., Aten E., Sun Y., Almomani R., Gilissen C., Nielsen M., Kant S.G., Snoeck I.N., Peeters E.A., Hilhorst-Hofstee Y., et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat. Genet. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- 35.Dharmadhikari A.V., Kang S.H., Szafranski P., Person R.E., Sampath S., Prakash S.K., Bader P.I., Phillips J.A., 3rd, Hannig V., Williams M., et al. Small rare recurrent deletions and reciprocal duplications in 2q21.1, including brain-specific ARHGEF4 and GPR148. Hum. Mol. Genet. 2012;21:3345–3355. doi: 10.1093/hmg/dds166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Priest J.R., Girirajan S., Vu T.H., Olson A., Eichler E.E., Portman M.A. Rare copy number variants in isolated sporadic and syndromic atrioventricular septal defects. Am. J. Med. Genet. A. 2012;158A:1279–1284. doi: 10.1002/ajmg.a.35315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davidson T.B., Sanchez-Lara P.A., Randolph L.M., Krieger M.D., Wu S.Q., Panigrahy A., Shimada H., Erdreich-Epstein A. Microdeletion del(22)(q12.2) encompassing the facial development-associated gene, MN1 (meningioma 1) in a child with Pierre-Robin sequence (including cleft palate) and neurofibromatosis 2 (NF2): a case report and review of the literature. BMC Med. Genet. 2012;13:19. doi: 10.1186/1471-2350-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thevenon J., Callier P., Andrieux J., Delobel B., David A., Sukno S., Minot D., Mosca Anne L., Marle N., Sanlaville D., et al. 12p13.33 microdeletion including ELKS/ERC1, a new locus associated with childhood apraxia of speech. Eur. J. Hum. Genet. 2012 doi: 10.1038/ejhg.2012.116. doi:10.1038/ejhg.2012.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez E., Callier P., Cormier-Daire V., Lacombe D., Moncla A., Bottani A., Lambert S., Goldenberg A., Doray B., Odent S., et al. Search for a gene responsible for Floating-Harbor syndrome on chromosome 12q15q21.1. Am. J. Med. Genet. A. 2012;158A:333–339. doi: 10.1002/ajmg.a.34401. [DOI] [PubMed] [Google Scholar]
- 40.van Bon B.W., Gilissen C., Grange D.K., Hennekam R.C., Kayserili H., Engels H., Reutter H., Ostergaard J.R., Morava E., Tsiakas K., et al. Cantu syndrome is caused by mutations in ABCC9. Am. J. Hum. Genet. 2012;90:1094–1101. doi: 10.1016/j.ajhg.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker K., Di Donato N., Holder-Espinasse M., Andrieux J., Cuisset J.M., Vallee L., Plessis G., Jean N., Delobel B., Thuresson A.C., et al. De novo microdeletions of chromosome 6q14.1-q14.3 and 6q12.1-q14.1 in two patients with intellectual disability—further delineation of the 6q14 microdeletion syndrome and review of the literature. Eur. J. Med. Genet. 2012;55:490–497. doi: 10.1016/j.ejmg.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson A.C., Feuk L. Characterization of copy number-stable regions in the human genome. Hum. Mutat. 2011;32:947–955. doi: 10.1002/humu.21524. [DOI] [PubMed] [Google Scholar]
- 44.Firth H.V., Wright C.F. The deciphering developmental disorders (DDD) study. Dev. Med. Child Neurol. 2011;53:702–703. doi: 10.1111/j.1469-8749.2011.04032.x. [DOI] [PubMed] [Google Scholar]
- 45.Cox C., Bignell G., Greenman C., Stabenau A., Warren W., Stephens P., Davies H., Watt S., Teague J., Edkins S., et al. A survey of homozygous deletions in human cancer genomes. Proc. Natl Acad. Sci. USA. 2005;102:4542–4547. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bamshad M.J., Shendure J.A., Valle D., Hamosh A., Lupski J.R., Gibbs R.A., Boerwinkle E., Lifton R.P., Gerstein M., Gunel M., et al. The centers for Mendelian genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am. J. Med. Genet. A. 2012;158A:1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raymond F.L., Whibley A., Stratton M.R., Gecz J. Lessons learnt from large-scale exon re-sequencing of the X chromosome. Hum. Mol. Genet. 2009;18:R60–R64. doi: 10.1093/hmg/ddp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Celli J., Dalgleish R., Vihinen M., Taschner P.E., den Dunnen J.T. Curating gene variant databases (LSDBs): toward a universal standard. Hum. Mutat. 2012;33:291–297. doi: 10.1002/humu.21626. [DOI] [PubMed] [Google Scholar]
- 49.Whitworth J. Data sharing: reaching consensus. Bull. World Health Organ. 2010;88:467–468. doi: 10.2471/BLT.10.079202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pisani E., AbouZahr C. Sharing health data: good intentions are not enough. Bull. World Health Organ. 2010;88:462–466. doi: 10.2471/BLT.09.074393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cotton R.G. Collection of variation causing disease—the Human Variome Project. Hum. Genomics. 2009;3:301–303. doi: 10.1186/1479-7364-3-4-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berman H., Henrick K., Nakamura H., Markley J.L. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Apweiler R., Bairoch A., Wu C.H. Protein sequence databases. Curr. Opin. Chem. Biol. 2004;8:76–80. doi: 10.1016/j.cbpa.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Heiskanen M., Peltonen L., Palotif A. Visual mapping by high resolution FISH. Trends Genet. 1996;12:379–382. doi: 10.1016/0168-9525(96)30083-8. [DOI] [PubMed] [Google Scholar]
- 55.Ionita-Laza I., Makarov V., Yoon S., Raby B., Buxbaum J., Nicolae D.L., Lin X. Finding disease variants in Mendelian disorders by using sequence data: methods and applications. Am. J. Hum. Genet. 2011;89:701–712. doi: 10.1016/j.ajhg.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C., et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 59.Schork N.J., Wessel J., Malo N. DNA sequence-based phenotypic association analysis. Adv. Genet. 2008;60:195–217. doi: 10.1016/S0065-2660(07)00409-9. [DOI] [PubMed] [Google Scholar]
- 60.Robinson P.N., Mundlos S. The human phenotype ontology. Clin. Genet. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 61.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T.K., Bateman A., Bernard T., Binns D., Bork P., Burge S., et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., Duesbury M., Dumousseau M., Feuermann M., Hinz U., et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perez E., Sullivan K.E. Chromosome 22q11.2 deletion syndrome (DiGeorge and velocardiofacial syndromes) Curr. Opin. Pediatr. 2002;14:678–683. doi: 10.1097/00008480-200212000-00005. [DOI] [PubMed] [Google Scholar]