Abstract
Human pluripotent stem cells can differentiate into disease-relevant cell types, which capture the unique genome of an individual patient and provide insight into pathological mechanisms of human disease. Recently, human stem cell models for Alzheimer's disease (AD), the most common neurodegenerative dementia, have been described. Stem cell-derived neurons from patients with familial and sporadic AD and Down's syndrome recapitulate human disease phenotypes such as amyloid β peptide production, hyperphosphorylation of tau protein and endosomal abnormalities. Treatment of human neurons with small molecules can modulate these phenotypes, demonstrating the utility of this system for drug development and screening. This review will highlight the current AD stem cell models and discuss the remaining challenges and potential future directions of this field.
INTRODUCTION
A consequence of an aging population is the increased prevalence of neurodegenerative disease. Alzheimer's disease (AD) is incurable and is the most common neurodegenerative dementia, with over 5 million cases in the USA and ∼30 million patients affected worldwide (1). The direct cost of AD in the USA is estimated to be greater than $170 billion/year, making AD a considerable public health issue (2,3). Because AD is a disease of the central nervous system (CNS), obtaining relevant patient tissue before death is challenging. Vertebrate and non-vertebrate models have provided important insights into AD; however, these models often involve the substantial overexpression of proteins, which by itself can cause abnormal phenotypes. Unfortunately, mutations introduced into the endogenous mouse genes do not recapitulate all of human AD pathology (4).
Human pluripotent stem cells (hPSCs) have great potential in disease research because they can differentiate into all cell types and genes of interest are expressed at endogenous levels. Recent advances in reprogramming technology have led to the ability to express defined factors in a somatic cell from an individual patient and induce a pluripotent stem cell state (5). These induced pluripotent stem cells (iPSCs) can be differentiated into multiple cell types, including neurons, capturing the unique genetic background of the individual. Although iPSCs may have an increased incidence of genetic and epigenetic abnormalities (6–8), this technology allows investigations of human-specific phenotypes and behaviors that cannot be evaluated in other organisms. As discussed below, there are now several strategies to differentiate neurons and other CNS cell types from human stem cells. These cells can be isolated using cell sorting strategies and biochemical phenotypes measured on relatively pure populations. Additionally, these approaches may be adaptable to high-throughput methods for therapeutic screens, a promising strategy for AD as there is currently no drug that alters the course of disease.
Familial and sporadic AD
There are two main forms of AD: early-onset, familial AD (FAD) and late-onset, sporadic AD (SAD). Clinically and pathologically FAD and SAD are similar, with both types of patients exhibiting progressive cognitive dementia, senile plaques consisting of amyloid β peptide (Aβ) and neurofibrillary tangles (NFTs) consisting of phosphorylated tau protein (9). FAD and SAD appear to share other cellular phenotypes including axonal transport defects, synapse loss and selective neuronal death (2,10).
FAD accounts for <5% of all AD cases and is primarily due to rare autosomal dominant mutations in the amyloid precursor protein (APP) gene and in two presenilin genes, PS1 and PS2 (2). The APP protein has multiple functions and is highly expressed in the CNS where it is thought to have roles in the formation of synapses, neurogenesis, axonal transport, signaling and plasticity (2,11–13). APP is a type I transmembrane protein, normally cleaved by α, β or γ secretases. When APP is sequentially cleaved first by β and then γ secretase enzymes, the Aβ fragments are generated. PS1 and 2 are transmembrane protein components of the gamma secretase enzyme which cleaves multiple substrates including APP (2). In addition to γ secretase activity, PS1/2 is also involved in the regulation of the endosome/lysosome pathway and autophagy (14).
Extensive studies of APP and PS1/2 in cellular and animal models of AD have led to a proposed pathological pathway in which the cleavage of APP to Aβ plays a central role. The longer forms of Aβ, Aβ42 and higher, become increasingly aggregate prone and are the main components of senile plaques in the AD brain. FAD mutations in the presenilins increase the ratio of the Aβ42 to 40 peptides either by decreasing the production of Aβ40 or increasing Aβ42 peptides (15). FAD mutations in APP increase amyloidogenic processing as well, either by enhancing β and γ secretase cleavage or by increasing gene dosage through duplications of the gene or chromosome (2,15). This evidence has lead to the idea of the amyloid cascade hypothesis (16), which centers around Aβ as the primary toxic molecule in AD. Indeed, peptide levels and plaque formation remain a primary phenotype analyzed in AD studies, although it is still unclear whether senile plaques are a cause or a consequence of AD and how they are connected to tau and NFTs. Tau is a microtubule-stabilizing protein and although there are no known tau mutations in AD, hyperphosphorylation of the protein may reduce its function and contribute to axonal degeneration and neuron loss (9). Interestingly, NFTs correlate more strongly with AD than senile plaques (17,18).
SAD accounts for over 95% of all AD cases. Although there are no clear dominant or recessive SAD mutations, many genetic variants have been identified and there is clearly a strong heritable component to the disorder (19). In fact, twin studies have estimated that heritability for SAD is between 60 and 80% (20). Elucidating how genetic risk variants contribute is a key issue in understanding SAD. Dozens of genome-wide association studies (GWASs) have now been performed for AD and hundreds of variants have been implicated in the disorder. Among these, the apolipoprotein E (APOE) isoform E4 has been the most consistent, although recent GWASs have identified other genes such as SORL1, Clusterin, CR1 and Picalm (21–23). Because of the lack of a clear genetic pattern, there are no animal models of SAD and thus attempts to model SAD in the laboratory are challenging.
Currently available drugs for AD, such as Aricept and Memantine, boost neurotransmission or protect from glutamate excitotoxicity, respectively, and provide only modest, short-term symptomatic relief (24). Most studies that have identified possible therapeutics for AD have focused on molecules that reduce the Aβ peptide, but thus far these have failed to alter disease course in patients, which underscores the need for new models to identify more successful strategies (2,25). Development of new human stem cell models for AD might identify novel drug targets with the additional possibility that patient-specific neurons will recapitulate variability in individual genetic backgrounds.
Stem cell models of AD
The first neurological diseases to be modeled using the iPSC technology were either monogenic disorders or versions of complex diseases caused by known mutations. These disorders include Parkinson's disease (PD) (26–29), amyotrophic lateral sclerosis (30,31), smooth muscle atrophy (32) and familial dysautonomia (33), and this work has been recently reviewed (34). IPSC models of neurodevelopmental disorders and psychiatric disorders such as Retts syndrome and schizophrenia have also been described (35,36). Despite the advances in this technology and the need for a human model, stem cell models from AD have only recently been developed, likely because of the complex nature of the disease.
Presenilin point mutation models
Due to the challenges of modeling SAD, most current AD stem cell models are from presenilin FAD point mutation patients. IPSCs have been generated from patient fibroblasts with PS1 A246E and PS2 N141I mutation (37). Independently, these fibroblasts have been used to generate induced neuronal cells (iNs) (38), a method which uses forebrain transcription factors to directly covert fibroblasts to neurons. First described in rodents (39), this stragegy has the advantage of by-passing the time-consuming and patentially mutagenic IPSC reprogramming process, but has the disadvantage that it does not generate a self-renewing, stable progenitor population. In both the IPSC and the iN studies, there was no difference in the neuronal differentiation ability when wild-type and presenilin point mutations were compared.
To test whether FAD presenilin mutant human neurons exhibited AD phenotypes, both groups looked at the production of Aβ peptides. The FAD IPSC-derived neurons and the iNs both showed increased Aβ42/40 ratios compared with controls, suggesting that these human neurons recapitulate a typical patient phenotype. Overexpression of WT PS1 and an FAD PS1 mutation has also recently been described in both iPSCs and human embryonic stem cells (hESCs) (40). This work capitalized on the development of long-term neuroepithelial cell cultures, derived from either iPSCs or hESCs, which can be cultured for over 150 passages and efficiently differentiate into neurons (41). This study used a lentiviral strategy to overexpress the PS1 L166P mutation, which causes a very aggressive form of early-onset FAD (42), along with WT PS1 or PS1 D385N a catalytically inactive form of the protein (40). PS1 L166P mutant neurons had an increase in the Aβ42/40 ratio compared with neurons overexpressing WT PS1, due to a large decrease in Aβ40 peptides in the L166P neurons. Cells expressing the D385N mutation had low to undetectable levels of both peptides, suggesting that this mutation suppresses γ-secretase activity in a dominant-negative fashion (40). While this strategy is useful to compare the effects of these mutations in the same genetic background, the levels of expression of these transgenes are not endogenous and in some FAD cases, such as APP duplication or trisomy 21, overexpression per se may contribute to phenotype (43,44).
Although increased Aβ production is considered pathological in AD, it is unclear whether this is a cause or consequence of disease. Other reported cellular phenotypes include synapse loss, axonal degeneration and endosomal/lysosomal compartment alterations (2,45) all of which may contribute to neuronal stress and cell death before, or independently of, Aβ accumulations. In fact, enlarged endosomes and accumulation of multi-vesicular bodies have been repeatedly described in Down's syndrome (DS) and SAD patient neurons post-mortem, while PS FAD brains show abnormal lysosome pathology (46–48). Interestingly, presenilin itself has been implicated in lysosomal acidification and FAD mutations may disrupt this process, contributing to abnormal lysosomes (49). In PS1/2 FAD iN cells, there was a marked increase in APP positive puncta, colocalizing with the early endosome marker EEA-1 and a general increase in both early and late endosomes. Overexpression of WT PS rescued this phenotype, suggesting that a loss of PS1 function may contribute to abnormal endosomes in this system.
APP duplication and DS models
Duplication of the APP gene on chromosome 21 causes FAD (43) and individuals with trisomy 21, or DS, show clinical symptoms of AD at ages 40–50 and have characteristic senile plaques post-mortem (44). Recently, our group reported the generation of iPSC neurons from two non-demented controls (NDCs) and two FAD patients with duplications in APP (APPDp) (50). This work took the advantage of a previously established fluorescence-activated cell sorting (FACS) protocol to purify differentiated neural precursor and subsequently neuronal cells (51). FACS selection led to a population of highly purified neurons, which were used to assay the cellular and biochemical AD phenotypes directly. Neurons from both FAD APPDp patients showed higher levels of Aβ1–40 and tau phosphorylation when compared with neurons from non-demented, age-matched individuals. Levels of activated GSK3β, a kinase thought to be involved in the phosphorylation of tau (52), were also increased in neurons from both FAD APPDp patients, pointing to a potential mechanism for the tau pathology in these cells. Interestingly, while γ-secretase inhibition lowered the levels of Aβ40, the increased activated GSK3β and phospho-tau levels were only reduced when the cells were treated with β-secretase inhibitors, which reduce the β-C-terminal fragment (β-CTF) of APP. APP CTFs have been implicated in mouse models of APP duplications as contributing to axonal transport defects (53), and these data in human neurons suggest that the β-CTF, prior to γ-cleavage and Aβ generation, may be a toxic entity and therefore an important molecule to focus on in terms of therapeutics. Finally, the phenotypic characterization was extended to examine endosome pathology and synaptic loss. As with the Aβ and tau phenotypes, FAD APPDp neurons had increased numbers of Rab5+ endosomes, a phenotype observed in SAD patient tissue as well as the brains and fibroblasts from DS patients (54,55). In FAD APPDp neurons, however, despite a 12-day culture period, no loss of the presynaptic protein synapsin-I was observed. Thus, endosomal abnormalities may be an early phenotype in AD and a longer culture period may be necessary to detect synapse loss and other signs of neuron degeneration, such as axonal defects, which have been shown to be independent of Aβ (56).
Complementary work demonstrated AD phenotypes in neurons derived from DS iPSCs (57). This study used a new protocol to generate human cortical neurons, a major cell type affected in AD (58). Cortical neurons from DS iPSCs secreted increased levels of both Aβ40 and 42 peptides compared with control cells and, similar to other studies, treatment with γ-secretase inhibitors blocked the synthesis of these peptides. Additionally, this group reported the formation of extracellular Aβ aggregates in DS patient neurons after 2 months in culture from the initial cortical differentiation, which is the first report of these extracellular accumulations in human neurons in vitro. Similar to FAD APPDp neurons, these studies detected hyperphosphorylation of the tau protein in DS neuronal cells. Intracellularly, the phosphorylated tau was aberrantly localized and they were also able to detect extracellular accumulations of tau (57), consistent with findings seen in the CSF analysis of AD patients (59). This finding correlated with increased cell death in these cultures.
SAD models
One main challenge of studying AD is that most of the cases have no clear genetic lesion, although it is assumed that SAD phenotypes will be similar to FAD. Generating iPSCs for SAD by reprogramming patients with sporadic disease captures the unique genome of each patient. Neurons derived from these iPSCs can then be used to directly test whether an SAD genome will confer similar cellular phenotypes to those seen in FAD. To investigate this question, our group recently reported the comparison of iPSC-derived neurons from two SAD patients, NDCs and the two FAD patients with APPDp described above (50). Neurons from one of the two SAD patients showed higher levels of Aβ1-40, increased GSK3β activity and increased tau phosphorylation compared with neurons from age-matched, NDC individuals. Similar to FAD APPDp neurons, cells from one SAD patient had increased numbers of Rab5+ endosomes after 12 days in culture, but no loss of the presynaptic protein synapsin-1. Therefore, while the numbers are thus far too small to draw general conclusions, it can be suggested that there are at least some genomes in the human population that generate neuronal phenotypes similar to those induced by an APPDp mutation. Whether this type of stem cell-based phenotypic analysis of human neurons carrying genomes from SAD patients can be used as a prospective diagnostic or clinical trial stratification strategy remains to be determined.
Drug screening
Screening candidate drugs in human disease-specific cells is an important goal of a human stem cell system. Forebrain neurons derived from iPSCs have been used to perform a proof-of-principle experiment demonstrating the efficiency of these cells to serve a platform to screen disease-modifying drugs (60). This study specifically assayed for components related to Aβ production and demonstrated increased levels of APP, APP-cleavage products and β/γ secretase enzymes upon neuronal differentiation. Treatment of cultures with β and γ secretase inhibitors and the non-steroidal anti-inflammatory drug (NSAID) sulindac sulfide all inhibited Aβ production (60). Chemicals that block γ-secretase reduce Aβ peptide levels by inhibiting the PS enzyme complex. Four of the above studies, the PS FAD iPSCs, PS FAD iNs, PS1 overexpressing iPSCs and the FAD APPDP and SAD iPSCs, all report lowering of Aβ in stem cell-derived neuronal cultures upon treatment with γ-secretase inhibiting compounds. However, γ-secretase inhibitors reduce both 40 and 42 peptides and may also impede Aβ-independent γ-secretase functions, such as cleavage of Notch (61). In addition to γ-secretase inhibitors, the data from the FAD APPDP iPSCs suggest that inhibiting β-secretase may be protective against GSK3β activation and phosphorylation of tau, implicating a pathway independent of the Aβ peptide for this phenotype.
Previous studies have suggested that NSAIDs may have a therapeutic effect for AD by selectively reducing Aβ42 (62). Stem cell-derived neurons overexpressing either WT or mutant PS1 were treated with NSAIDS and a reduction in Aβ42 levels were noted in the WT PS1 cells but not the L166P mutant (40). This is consistent with reports that some FAD mutations may be immune to this type of treatment (63); however, additional FAD mutations should be screened for responsiveness to test for the generality of this conclusion. Taken together, however, these studies demonstrate the possible feasibility of stem cell models for therapeutic screens.
CONCLUSIONS AND CHALLENGES
The different AD models, differentiation methods and AD relevant phenotypes described are summarized in Table 1. From this first set of stem cell-derived AD models, there are several positive findings. First, none of the FAD mutations described seemed to influence the differentiation ability of the parental cell types into neurons, demonstrating that various stem cell systems can generate disease-specific cell types in good quantities, even though the differentiation protocols are different from lab-to-lab. Second, a main disease phenotype, secretion or altered processing of Aβ peptides, can be detected in a neuron-specific fashion as most of these studies reported low to undetectable Aβ levels in parent fibroblasts. Third, the Aβ levels reported in these systems could be modulated by treatment with different molecules, demonstrating that this system provides a relevant platform for screening disease-modifying drugs. Finally, and perhaps most significantly, three of the studies report phenotypes in addition to Aβ generation, including tau phosphorylation (50,57), GSK3β activation (50) and abnormal endosomes (38,50), highlighting pathways that can be investigated alongside of APP processing. This is important as Aβ-lowering strategies, such as γ-secretase inhibition, have not yet been shown to alter the course of AD (64).
Table 1.
Summary of hPSC models generated for AD
| Model | Mutation | Differentiation method | Neuronal markers | AD-relevant phenotype | Lines |
|---|---|---|---|---|---|
| iPSC (37,76) | PS1 A246E PS2 N141I |
|
|
|
Seven lines:Mutant: PS1-2, PS1-4, PS2-1, PS2-1Control: IPS201B7, PD01-25 and 26 |
| iPSC (60) | Wild-type |
|
|
|
One line: hIPS 253G4 |
| iN (38) | PS1 A246E PS2 N141I |
Direct conversion: expression of Brn2, Myt1, Olig2 and Ascl1 in fibroblasts |
|
|
Three fibroblast cultures per group |
| iPSC (50) | APPDp sporadic |
|
|
|
20 lines: NDC1.1-3, NDC2.1-3, SAD1.1-3, SAD2.1-5, APPDp1.1-3 and APPDp2.1-3 |
| iPSC (57,58) | Trisomy 21 (DS) |
|
|
|
|
| hESC iPSC (40,41) | PS1 L166P PS1 D385N Overexpression |
|
|
|
Two lines: hESC line I3 and iPSC line PKa |
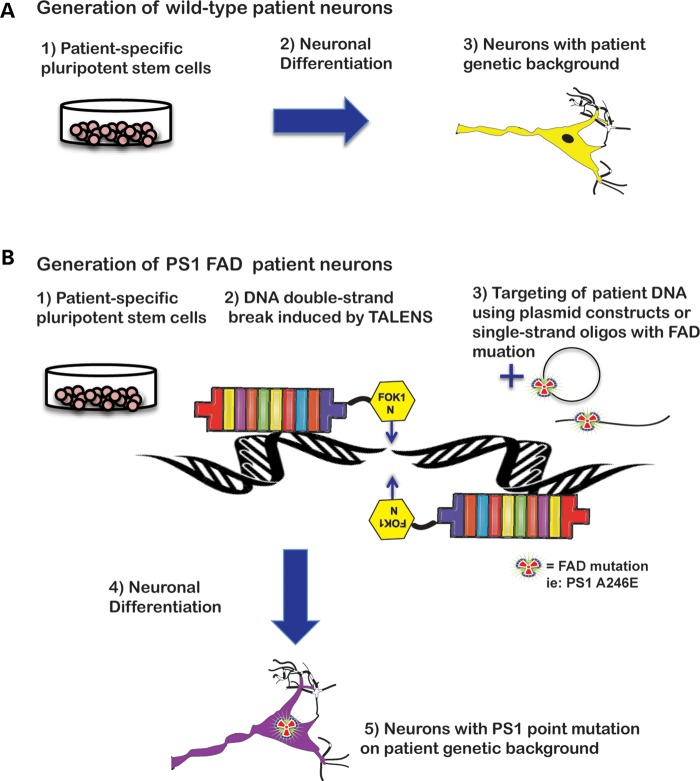
How can we build on these early studies to develop a more complete human model for AD? One prospect is to use the powerful emerging technology of engineered nucleases to modify individual genomes. Zinc-finger nucleases (ZFNs) and, more recently, tal-effector nucleases (TALENs) take the advantage of the fact that a DNA double-strand break will trigger a cell's endogenous recombination machinery. Arrayed domains, which bind to a specific nucleotide sequence, are fused to the FOK1 nuclease and resulting DNA breaks are repaired either by non-homologous end-joining (NHEJ) or by homologous recombination, in the presence or the absence of a donor sequence (65,66). Using TALENs or ZFNs, point mutations can be introduced into wild-type cells, creating isogenic lines in which mutant and normal genes are expressed endogenously in an identical genetic background (Fig. 1). This technology has recently been applied to early-onset PD in which a PD point mutation from patient-derived iPSCs was corrected and several PD mutations were introduced into WT hESCs (67). For stem cell models of AD, this strategy holds great promise. First of all, a direct comparison of isogenic cell lines differing only in the presence or the absence of a defined FAD point mutation, either in APP or presenilin, can reduce the intrinsic variability that comes from comparing cells from different patients. Second, the targeted mutations will be under endogenous control of expression, which will help to precisely determine how the mutation affects the cell. This is particularly important for AD as there is some debate as to whether presenilin mutations constitute a gain or a loss of function of the protein (68,69) and given the unclear connection between Aβ and NFTs.
Figure 1.
Genome editing of hPSCs. (A) Patient-specific pluripotent stem cells can be differentiated to neurons with the unique genetic background of the individual. (B) Modification of an individual genome with TALENs can introduce a point mutation in a disease gene of interest. The resulting isogenic cells can then be studied to determine the exact effect of the mutation in an otherwise identical genetic background. 215 × 279 mm (300 × 300 DPI).
Stem cell-derived neurons will also provide an important tool to dissect the contribution of a mutation or a risk factor to a specific cell type. For example, experiments with different types of human CNS cells derived from individuals of different genotypes may shed light on cell-autonomous factors. This strategy could be used to understand the role of APOE, a protein involved in cholesterol metabolism, in AD. The APOE4 isoform has been consistently implicated as an AD risk factor with people carrying the E4 allele at either a 3-fold (heterozygous) or a 12-fold (homozygous) higher chance of developing AD, while individuals with the E2 isoform have a decreased risk (70). APOE production varies greatly in the different cell types of the CNS, with glia producing more than neurons (71). Thus, co-culture experiments of human neurons of one APOE genotype with human glia of a different genotype will yield insights into the mechanisms and cell autonomy of the APOE contribution to AD phenotypes.
Elucidating the factors that result in SAD also remains a primary challenge. Although lacking a defined genetic mechanism, there is clearly a heritable component to SAD and each individual's unique genetic background contains variants that may predispose to, or protect from, disease. This raises several key questions: What is the genetic contribution to disease risk in an individual? What are the phenotypic consequences of harboring genetic risk variants in an individual genetic background? A potential strategy is to evaluate variants identified by GWASs. Identification of molecular or biochemical phenotypes caused by variants, and their impact in different genetic backgrounds could yield important new information about interactions of genetic variants in individual genomes and their contributions to neuronal phenotypes and AD risk. This line of attack will surely find novel pathways that can be used to test new possible therapeutics. How to decide which variants to study? Analysis of the top-scoring GWASs from Alzgene has yielded a pattern of common cellular pathways in which the genes carrying these variants are involved, including inflammation, endocytosis and cholesterol metabolism (72). In particular, the endo/lysosome pathway appears to play a prominent role in AD as the main components of the APP processing machinery reside in vesicles trafficked through this system and pathological abnormalities are present in AD patient tissue (54). GWASs hits such as SORL1 and PICALM play a direct role in endocytic and vesicular processes. SORL1, identified in 2007 as a risk factor for AD (21) directly interacts with APP and sequesters the full-length protein away from late endosomes where amyloidogenic processing can occur (73). PICALM is involved in clathrin-mediated endocytosis and may be involved in trafficking important components of synapses, a process disrupted in AD that correlates strongly with cognitive deficits (74,75). Due to limited success of current AD therapies, perhaps investigating these cellular pathways using human, patient-specific cells will yield new targets for drug discovery.
Although human stem cell research and its application to AD is still a field in its infancy, the basic biology of these cells is being intensively studied and protocols are being rapidly developed, validated and improved upon. As the first AD studies show, it is possible to study neurons from a patient with AD in a dish and observe relevant phenotypes and behaviors. Our challenge now is to use this system to build on decades of previous AD work so that we can and more fully understand and treat this devastating disorder.
FUNDING
The authors are funded by the California Institute of Regenerative Medicine (CIRM) (RT2-01927) and the National Institutes of Health/National Institutes of Aging (R01AG032180). J.E.Y. is supported by the A.P. Giannini Foundation for Medical Research.
ACKNOWLEDGEMENTS
The authors would like to thank A. Almenar-Queralt and G. Woodruff for critical feedback and editorial comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Barnes D.E., Yaffe K. The projected effect of risk factor reduction on Alzheimer's disease prevalence. Lancet Neurol. 2011;10:819–828. doi: 10.1016/S1474-4422(11)70072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holtzman D.M., Morris J.C., Goate A.M. Alzheimer's disease: the challenge of the second century. Sci. Transl. Med. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lunn J.S., Sakowski S.A., Hur J., Feldman E.L. Stem cell technology for neurodegenerative diseases. Ann. Neurol. 2011;70:353–361. doi: 10.1002/ana.22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duff K., Suleman F. Transgenic mouse models of Alzheimer's disease: how useful have they been for therapeutic development. Brief Funct. Genomic. Proteomic. 2004;3:47–59. doi: 10.1093/bfgp/3.1.47. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Hussein S.M., Batada N.N., Vuoristo S., Ching R.W., Autio R., Narva E., Ng S., Sourour M., Hamalainen R., Olsson C., et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 7.Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E., et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O'Malley R., Castanon R., Klugman S., et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee V.M., Brunden K.R., Hutton M., Trojanowski J.Q. Developing therapeutic approaches to tau, selected kinases, and related neuronal protein targets. Cold Spring Harb. Perspect. Med. 2011;1:a006437. doi: 10.1101/cshperspect.a006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein L.S. Axonal transport and neurodegenerative disease: can we see the elephant? Prog. Neurobiol. 2012 doi: 10.1016/j.pneurobio.2012.03.006. http://dx.doi.org/10.1016/j.pneurobio.2012.03.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunawardena S., Goldstein L.S. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001;32:389–401. doi: 10.1016/s0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- 12.Lazarov O., Demars M.P. All in the family: how the APPs regulate neurogenesis. Front Neurosci. 2012;6:81. doi: 10.3389/fnins.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunholz S., Sisodia S., Lorenzo A., Deyts C., Kins S., Morfini G. Axonal transport of APP and the spatial regulation of APP cleavage and function in neuronal cells. Exp. Brain Res. 2012;217:353–364. doi: 10.1007/s00221-011-2870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nixon R.A., Yang D.S. Autophagy failure in Alzheimer's disease—locating the primary defect. Neurobiol. Dis. 2011;43:38–45. doi: 10.1016/j.nbd.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selkoe D.J. Alzheimer's disease. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy J.A., Higgins G.A. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 17.Buerger K., Ewers M., Pirttila T., Zinkowski R., Alafuzoff I., Teipel S.J., DeBernardis J., Kerkman D., McCulloch C., Soininen H., et al. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer's disease. Brain. 2006;129:3035–3041. doi: 10.1093/brain/awl269. [DOI] [PubMed] [Google Scholar]
- 18.Buerger K., Teipel S.J., Zinkowski R., Blennow K., Arai H., Engel R., Hofmann-Kiefer K., McCulloch C., Ptok U., Heun R., et al. CSF tau protein phosphorylated at threonine 231 correlates with cognitive decline in MCI subjects. Neurology. 2002;59:627–629. doi: 10.1212/wnl.59.4.627. [DOI] [PubMed] [Google Scholar]
- 19.Avramopoulos D. Genetics of Alzheimer's disease: recent advances. Genome Med. 2009;1:34. doi: 10.1186/gm34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatz M., Reynolds C.A., Fratiglioni L., Johansson B., Mortimer J.A., Berg S., Fiske A., Pedersen N.L. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 21.Rogaeva E., Meng Y., Lee J.H., Gu Y., Kawarai T., Zou F., Katayama T., Baldwin C.T., Cheng R., Hasegawa H., et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M.J., Tavernier B., et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 23.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tayeb H.O., Yang H.D., Price B.H., Tarazi F.I. Pharmacotherapies for Alzheimer's disease: beyond cholinesterase inhibitors. Pharmacol. Ther. 2012;134:8–25. doi: 10.1016/j.pharmthera.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Reitz C. Alzheimer's disease and the amyloid cascade hypothesis: a critical review. Int. J. Alzheimers Dis. 2012;2012:369808. doi: 10.1155/2012/369808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soldner F., Hockemeyer D., Beard C., Gao Q., Bell G.W., Cook E.G., Hargus G., Blak A., Cooper O., Mitalipova M., et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen H.N., Byers B., Cord B., Shcheglovitov A., Byrne J., Gujar P., Kee K., Schule B., Dolmetsch R.E., Langston W., et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seibler P., Graziotto J., Jeong H., Simunovic F., Klein C., Krainc D. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J. Neurosci. 2011;31:5970–5976. doi: 10.1523/JNEUROSCI.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R., et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 31.Mitne-Neto M., Machado-Costa M., Marchetto M.C., Bengtson M.H., Joazeiro C.A., Tsuda H., Bellen H.J., Silva H.C., Oliveira A.S., Lazar M., et al. Downregulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum. Mol. Genet. 2011;20:3642–3652. doi: 10.1093/hmg/ddr284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebert A.D., Yu J., Rose F.F., Jr., Mattis V.B., Lorson C.L., Thomson J.A., Svendsen C.N. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A., et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchetto M.C., Brennand K.J., Boyer L.F., Gage F.H. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum. Mol. Genet. 2011;20:R109–R115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennand K.J., Simone A., Jou J., Gelboin-Burkhart C., Tran N., Sangar S., Li Y., Mu Y., Chen G., Yu D., et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yagi T., Ito D., Okada Y., Akamatsu W., Nihei Y., Yoshizaki T., Yamanaka S., Okano H., Suzuki N. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 38.Qiang L., Fujita R., Yamashita T., Angulo S., Rhinn H., Rhee D., Doege C., Chau L., Aubry L., Vanti W.B., et al. Directed conversion of Alzheimer's disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch P., Tamboli I.Y., Mertens J., Wunderlich P., Ladewig J., Stuber K., Esselmann H., Wiltfang J., Brustle O., Walter J. Presenilin-1 L166P mutant human pluripotent stem cell-derived neurons exhibit partial loss of gamma-secretase activity in endogenous amyloid-beta generation. Am. J. Pathol. 2012;180:2404–2416. doi: 10.1016/j.ajpath.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Koch P., Opitz T., Steinbeck J.A., Ladewig J., Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl Acad. Sci. USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moehlmann T., Winkler E., Xia X., Edbauer D., Murrell J., Capell A., Kaether C., Zheng H., Ghetti B., Haass C., et al. Presenilin-1 mutations of leucine 166 equally affect the generation of the Notch and APP intracellular domains independent of their effect on Abeta 42 production. Proc. Natl Acad. Sci. USA. 2002;99:8025–8030. doi: 10.1073/pnas.112686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rovelet-Lecrux A., Hannequin D., Raux G., Le Meur N., Laquerriere A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M., et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 44.Lemere C.A., Blusztajn J.K., Yamaguchi H., Wisniewski T., Saido T.C., Selkoe D.J. Sequence of deposition of heterogeneous amyloid beta-peptides and APO E in Down syndrome: implications for initial events in amyloid plaque formation. Neurobiol. Dis. 1996;3:16–32. doi: 10.1006/nbdi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 45.Nixon R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 46.Cataldo A.M., Hamilton D.J., Barnett J.L., Paskevich P.A., Nixon R.A. Abnormalities of the endosomal-lysosomal system in Alzheimer's disease: relationship to disease pathogenesis. Adv. Exp. Med. Biol. 1996;389:271–280. [PubMed] [Google Scholar]
- 47.Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., Cuervo A.M. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 48.Cataldo A.M., Peterhoff C.M., Schmidt S.D., Terio N.B., Duff K., Beard M., Mathews P.M., Nixon R.A. Presenilin mutations in familial Alzheimer disease and transgenic mouse models accelerate neuronal lysosomal pathology. J. Neuropathol. Exp. Neurol. 2004;63: 821–830. doi: 10.1093/jnen/63.8.821. [DOI] [PubMed] [Google Scholar]
- 49.Lee J.H., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.M., Martinez-Vicente M., Massey A.C., Sovak G., et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Israel M.A., Yuan S.H., Bardy C., Reyna S.M., Mu Y., Herrera C., Hefferan M.P., Van Gorp S., Nazor K.L., Boscolo F.S., et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan S.H., Martin J., Elia J., Flippin J., Paramban R.I., Hefferan M.P., Vidal J.G., Mu Y., Killian R.L., Israel M.A., et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho J.H., Johnson G.V. Primed phosphorylation of tau at Thr231 by glycogen synthase kinase 3beta (GSK3beta) plays a critical role in regulating tau's ability to bind and stabilize microtubules. J. Neurochem. 2004;88:349–358. doi: 10.1111/j.1471-4159.2004.02155.x. [DOI] [PubMed] [Google Scholar]
- 53.Salehi A., Delcroix J.D., Belichenko P.V., Zhan K., Wu C., Valletta J.S., Takimoto-Kimura R., Kleschevnikov A.M., Sambamurti K., Chung P.P., et al. Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron. 2006;51:29–42. doi: 10.1016/j.neuron.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Cataldo A.M., Peterhoff C.M., Troncoso J.C., Gomez-Isla T., Hyman B.T., Nixon R.A. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer's disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am. J. Pathol. 2000;157:277–286. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cataldo A.M., Mathews P.M., Boiteau A.B., Hassinger L.C., Peterhoff C.M., Jiang Y., Mullaney K., Neve R.L., Gruenberg J., Nixon R.A. Down syndrome fibroblast model of Alzheimer-related endosome pathology: accelerated endocytosis promotes late endocytic defects. Am. J. Pathol. 2008;173:370–384. doi: 10.2353/ajpath.2008.071053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stokin G.B., Almenar-Queralt A., Gunawardena S., Rodrigues E.M., Falzone T., Kim J., Lillo C., Mount S.L., Roberts E.A., McGowan E., et al. Amyloid precursor protein-induced axonopathies are independent of amyloid-beta peptides. Hum. Mol. Genet. 2008;17:3474–3486. doi: 10.1093/hmg/ddn240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y., Kirwan P., Smith J., MacLean G., Orkin S.H., Livesey F.J. A human stem cell model of early Alzheimer's disease pathology in Down syndrome. Sci. Transl. Med. 2012;4:124ra129. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Y., Kirwan P., Smith J., Robinson H.P., Livesey F.J. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 2012;15:477–486. doi: 10.1038/nn.3041. S471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holtzman D.M. CSF biomarkers for Alzheimer's disease: current utility and potential future use. Neurobiol. Aging. 2011;32(Suppl. 1):S4–S9. doi: 10.1016/j.neurobiolaging.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yahata N., Asai M., Kitaoka S., Takahashi K., Asaka I., Hioki H., Kaneko T., Maruyama K., Saido T.C., Nakahata T., et al. Anti-Abeta drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer's disease. PLoS One. 2011;6:e25788. doi: 10.1371/journal.pone.0025788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geling A., Steiner H., Willem M., Bally-Cuif L., Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gasparini L., Ongini E., Wenk G. Non-steroidal anti-inflammatory drugs (NSAIDs) in Alzheimer's disease: old and new mechanisms of action. J. Neurochem. 2004;91:521–536. doi: 10.1111/j.1471-4159.2004.02743.x. [DOI] [PubMed] [Google Scholar]
- 63.Czirr E., Leuchtenberger S., Dorner-Ciossek C., Schneider A., Jucker M., Koo E.H., Pietrzik C.U., Baumann K., Weggen S. Insensitivity to Abeta42-lowering nonsteroidal anti-inflammatory drugs and gamma-secretase inhibitors is common among aggressive presenilin-1 mutations. J. Biol. Chem. 2007;282:24504–24513. doi: 10.1074/jbc.M700618200. [DOI] [PubMed] [Google Scholar]
- 64.Talan J. In a large clinical trial, gamma-secreatase inhibitor Tarenflurbil is found ineffective for mild Alzheimer disease. Neurology Today. 2010;10:16–17. [Google Scholar]
- 65.Cathomen T., Joung J.K. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 66.Mussolino C., Cathomen T. TALE nucleases: tailored genome engineering made easy. Curr. Opin. Biotechnol. 2012 doi: 10.1016/j.copbio.2012.01.013. doi:10.1016/j.copbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 67.Soldner F., Laganiere J., Cheng A.W., Hockemeyer D., Gao Q., Alagappan R., Khurana V., Golbe L.I., Myers R.H., Lindquist S., et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfe M.S. When loss is gain: reduced presenilin proteolytic function leads to increased Abeta42/Abeta40. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:136–140. doi: 10.1038/sj.embor.7400896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen J., Kelleher R.J., 3rd The presenilin hypothesis of Alzheimer's disease: evidence for a loss-of-function pathogenic mechanism. Proc. Natl Acad. Sci. USA. 2007;104:403–409. doi: 10.1073/pnas.0608332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guerreiro R.J., Hardy J. Alzheimer's disease genetics: lessons to improve disease modelling. Biochem. Soc. Trans. 2011;39:910–916. doi: 10.1042/BST0390910. [DOI] [PubMed] [Google Scholar]
- 71.Vance J.E., Hayashi H., Karten B. Cholesterol homeostasis in neurons and glial cells. Semin. Cell Dev. Biol. 2005;16:193–212. doi: 10.1016/j.semcdb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 72.Olgiati P., Politis A.M., Papadimitriou G.N., De Ronchi D., Serretti A. Genetics of late-onset Alzheimer's disease: update from the alzgene database and analysis of shared pathways. Int. J. Alzheimers Dis. 2011;2011:832379. doi: 10.4061/2011/832379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pallesen L.T., Vaegter C.B. Sortilin and SorLA regulate neuronal sorting of trophic and dementia-linked proteins. Mol. Neurobiol. 2012;45:379–387. doi: 10.1007/s12035-012-8236-2. [DOI] [PubMed] [Google Scholar]
- 74.Harel A., Wu F., Mattson M.P., Morris C.M., Yao P.J. Evidence for CALM in directing VAMP2 trafficking. Traffic. 2008;9:417–429. doi: 10.1111/j.1600-0854.2007.00694.x. [DOI] [PubMed] [Google Scholar]
- 75.Masliah E., Mallory M., Alford M., DeTeresa R., Hansen L.A., McKeel D.W., Jr, Morris J.C. Altered expression of synaptic proteins occurs early during progression of Alzheimer's disease. Neurology. 2001;56:127–129. doi: 10.1212/wnl.56.1.127. [DOI] [PubMed] [Google Scholar]
- 76.Miura K., Okada Y., Aoi T., Okada A., Takahashi K., Okita K., Nakagawa M., Koyanagi M., Tanabe K., Ohnuki M., et al. Variation in the safety of induced pluripotent stem cell lines. Nat. Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]