Abstract
The worldwide dog population is fragmented into >350 domestic breeds. Breeds share a common ancestor, the gray wolf. The intense artificial selection imposed by humans to develop breeds with particular behaviors and phenotypic traits has occurred primarily in the last 200–300 years. As a result, the number of genes controlling the major differences in body size, leg length, head shape, etc. that define each dog is small, and genetically tractable. This is in comparison to many human complex traits where small amounts of variance are controlled by literally hundreds of genes. We have been interested in disentangling the genetic mechanisms controlling breed-defining morphological traits in the domestic dog. The structure of the dog population, comprised large numbers of pure breeding populations, makes this task surprisingly doable. In this review, we summarize recent work on the genetics of body size, leg length and skull shape, while setting the stage for tackling other traits. It is our expectation that these results will contribute to a better understanding of mammalian developmental processes overall.
INTRODUCTION
Most common dog breeds were developed recently, within the last 200–300 years in Europe and were optimized for both appearance and behavior (1,2). As a result, domestic dog breeds today display a dazzling array of variations in terms of leg length, skull shape, coat color, bone width, body size, etc. However, the number of genes that are likely to control each trait is small, compared with the large numbers assigned to many human phenotypes (3,4). Why is this? The observation likely reflects several aspects of canine population structure. Fanciers have historically placed dogs under strong selection to create breeds with specific phenotypes. In addition, many dog breeds are descended from small numbers of founders and, additionally, many underwent bottlenecks early in their development. As a result, the number of genes controlling major traits that have slipped through the bottleneck is limited (5,6). So, rather than having a large number of genes of small effect, as is observed in humans, a small number of genes of large effect predominate in dogs. Many such loci are marked by the presence of selective sweeps, or regions of reduced heterozygosity, indicating loci that breeders have been selecting for the formation of particular breed-defining traits (7).
We have been interested in finding the genes that control fixed morphological traits in the domestic dog and then determining the importance of those genes in humans. In the United States, today, there are 167 dog breeds recognized by the American Kennel Club (AKC) and worldwide, over 350 breeds are recognized. In this review, we discuss current findings, highlighting the advances in canine genomics that have contributed to our growing understanding of mammalian morphologic genetics.
ORIGINS OF THE DOMESTIC DOG
The dog is the only large carnivore that has been domesticated (8), and several studies show that the wolf is the direct ancestor of today's dog (9–11). However, the timing and geographical region where key domestication events took place are controversial. Archeological data suggest that dogs first appeared 15 000 up to 100 000 years ago in multiple locations, including Europe, the Middle East and East Siberia (10,12–15). Studies of the canine nuclear genome support this statement, suggesting that South West Asia and/or Europe were principle regions of domestication (16). However, separate studies analyzing mitochondrial and the Y-chromosome DNA suggested that a South East Asian origin is more likely (17–19). The debate continues to this day (8,20).
BREED RELATIONSHIPS
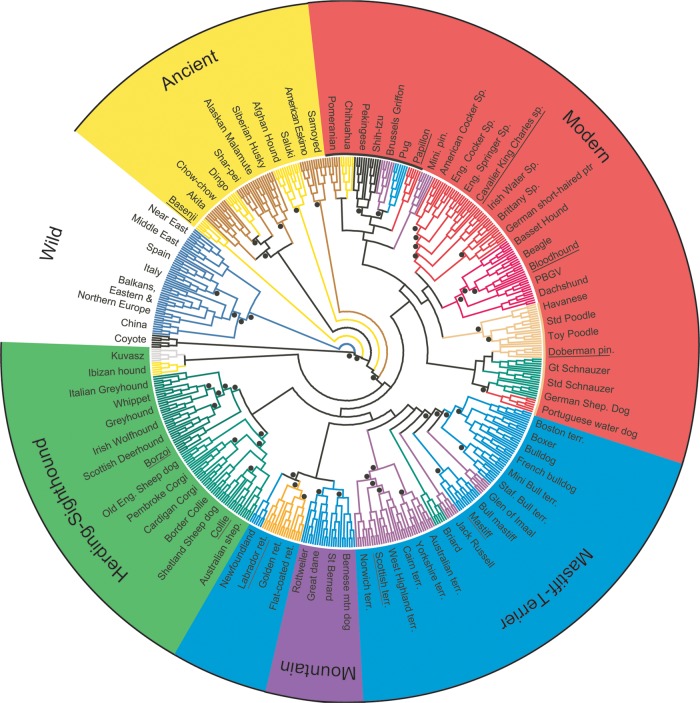
Several studies have addressed the issue of how dog breeds relate one to another. The first by Parker et al. asked how well each dog, in a blinded analysis, was correctly assigned to its breed (21). She found that most breeds had a distinct polymorphic signature: individual dogs were accurately assigned to their breed >99% of the time. Expansion of the dataset to 132 breeds both validated these findings and provided insights regarding how dog breeds are related one to another (22). Using clustering analysis, it was possible to divide the 132 breeds into five groups that disproportionately shared at the DNA level (Fig. 1). Most recently vonHoldt et al. published the definitive study in this area using a dataset of >50 000 single-nucleotide polymorphisms (SNPs) to build neighbor joining trees from 80 common dog breeds (16). Their trees demonstrate how the various breeds relate to one another, and highlight likely common ancestors (Fig. 1).
Figure 1.
Genetic clustering of domestic dogs and gray wolves. The inner circle is a neighbor-joining tree representing the clustering of domestic dog breeds into ∼10 groups based on the comparison of 10-SNP haplotypes (tree originally published in (16)). The outer circle shows the five groups previously found from a microsatellite analysis of 130 breeds (22). Reproduced with permission from Mammalian Genome (45).
These results suggested a way to use dog breeds for genetic mapping. One could reasonably hypothesize that breeds within a cluster or branch share ancestral mutations for both morphological traits and disease. Thus, the most efficient way to localize a gene of interest is to select cases and controls from among closely related breeds that share a common trait (6,23). This would allow sufficient power to correctly identify a locus, and then reduce the region of interest by taking advantage of the unique meiotic events that characterized each breed.
CANINE GENOMICS AND IMPLICATIONS FOR ASSOCIATION STUDIES
In 2005, a female boxer dog was sequenced to 7.5× and several other breeds were sequenced to a lesser extent (11). Based on the analysis of these data, the authors proposed that ancestral dogs started out with short linkage disequilibrium (LD) blocks, as would be expected from their more ancient history. The development of breeds introduced tight and breed-specific bottlenecks, with modern breeds retaining only a small percentage of the original haplotype pool. Surprisingly, the tight bottlenecks did not result in massive random fixation of individual haplotypes within specific breeds. In fact, <15% of the small ancestral haplotypes identified proved monomorphic within any single breed (11). There was also significant sharing of large haplotypes across breeds, with 60% of 100 kb haplotypes observed in multiple breeds, albeit at varying distributions (11,24).
This information provided a blueprint for how to set up canine genome-wide association studies (GWAS) (25). There is sufficient homogeneity within breeds that a large number of dogs are not required for any single GWAS (11). It is important, however, to develop maximally unrelated populations among both the case and control populations to improve power. Since LD in dogs extends long distances, sometimes up to megabases (Mb), only tens of thousands of SNPs would be needed to complete a GWAS in dogs, compared with the hundreds of thousands required for comparable human studies (11,24).
Today, a successful canine study, seeking to disentangle a 1–2 gene trait, uses 100–200 cases and controls, each, for the initial GWAS and ∼50 000 SNPs. A variety of related breeds are then used to reduce the region of interest to <0.5 Mb. Fine mapping or next generation sequencing is then used to find the mutation. The dog system can, therefore, be used to circumvent many of the problems faced by human geneticists who have limited numbers of adequately sized families for linkage, small datasets for association studies and who frequently face the problem of phenocopy. Regarding the latter, since all dogs of a breed share the same genetic background, there is far less variation in the presentation of disease phenotypes in dogs than in humans (26).
THE CANMAP STUDY
In 2008, we initiated a dataset that would allow us to map multiple breed-defining traits simultaneously. The study, termed CanMap, had at its heart a population of 915 dogs from 80 domestic breeds as well as 83 wild canids (3). Each breed is represented by ∼12 highly unrelated individuals. Each dog was genotyped with a 127 000 SNP chip, and the data analyzed for 57 morphological traits, allowing us to identify 51 regions of the dog genome associated with phenotypic variation. This includes traits as varied as ear position to tail curl, leg length/width ratios to skull measurements (3). As predicted (27,28), a small number of loci (≤3) appear to underlie most morphological features (3). We explore several of these below.
BODY SIZE
One of the most interesting things to come out of the CanMap project was further elucidation of the study of body size in dogs. A microsatellite linkage study of Portuguese Water Dogs (PWD) (27) and a multi-breed SNP study (28) had previously reported that about six QTLs are important in regulating canine body size, with the most predominant on canine chromosome 15 (CFA15).
Building on that work, Sutter et al. fine-mapped the 15 Mb region on CFA15 (29) by first reducing the region using data from very large and small PWD. Then, analyzing 14 small dog breeds, he found evidence for a selective sweep that corresponded precisely with the insulin-like growth factor 1 (IGF1) gene (Fig. 2A). He analyzed dogs of all sizes from over 80 breeds, ultimately demonstrating a 20 SNP haplotype spanning part of IGF1. While several haplotypes were theoretically present, the ‘B’ haplotype was found on the majority of chromosomes from small dogs, suggesting that the contribution that IGF1 makes to small dogs uses alleles that have been in existence since early in breed development.
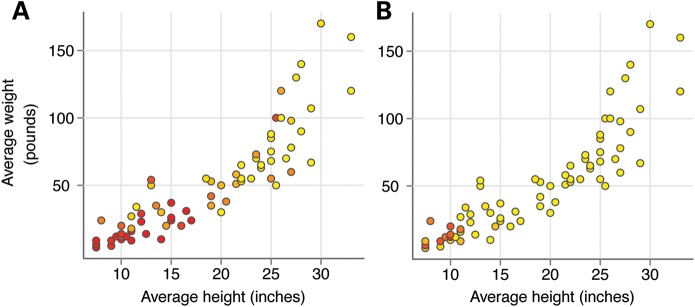
Figure 2.
Correlation of the allele frequency and size at IGF1 and IGF1R loci. The average height and weight of breeds from the CanMap dataset are plotted (3). Each circle represents one breed. The ancestral allele frequency is represented as a gradient from red (low) to yellow (high). SNPs depicted include CFA15:44,226,659 (A) and CFA3:43,756,620 (B). These SNPs are highly associated with canine body size at IGF1 and IGF1R loci, respectively (3,31).
However, finding the associated mutation has proven difficult. A ∼10 kb interval spanning intron 2 was most strongly associated with size variation. Both a short interspersed SINEC_Cf element and a SNP that were in strong LD were reasonable candidates. Gray et al. extended the IGF1 studies by considering these two polymorphisms using gray wolves (30). Both of these markers were part of the ‘B’ haplotype, and the alleles were fixed in the nearly all the small breeds tested. However, in wolves the ‘derived’ or canine allele was rarely present. Indeed, it was found only in Middle Eastern wolves, suggesting that the dog variant arose, perhaps, coincident with dog domestication, in the Middle East. However, neither the SINE nor the SNP appear to be trait-associated, and the variant remains to be found.
Recently, work on the other loci has matured (Rimbault et al., personal communication). We have been especially interested in loci that control extremes of size, i.e. very large and very small dogs. Toward that end, Hoopes et al. has shown that the IGF1 receptor (IGF1R) is strongly associated with very small skeletal size in dogs (31) (Fig. 2B). We again made use of the CanMap dataset and found a strong signal on CFA3 (P = 1.9 × 10−70) when comparing tiny dogs with the rest. Fine mapping revealed a non-synonymous SNP at CFA3:44 706 389 that changed a highly conserved amino acid (R204H) in the IGF1R gene. This particular mutation is likely functional, as it is predicted to block formation of multiple hydrogen bonds within the cysteine-rich domain of the receptor's ligand-binding extracellular subunit. Further evidence from SIFT (32) and Polyphen-2 (33) indicate that the R204H substitution is ‘not tolerated’ and ‘probably damaging’.
In addition to IGF1 and its receptor, many other genes remain to be investigated. While it is unlikely that dogs will require 180 or so genes that have been reported in human GWAS studies of height (34–38), several papers (3,4,27,28) highlight common loci, including good candidates like HMGA2 (38) and the growth hormone receptor, both of which have been implicated in studies of human height. Our challenge is to find the underlying variants, determine how they account for the continuum of body size in dogs and then use that data to try and help organize the human GWAS findings.
LEG LENGTH
Chondrodysplastic breeds have disproportionately short, curved limbs and heavy bones (39) and include the Basset Hound, Corgi, Pekingese and Dachshunds among others. Using AKC standards, a multi-breed GWAS compared 95 dogs from eight chondrodysplastic breeds with a control group of 64 non-chondrodysplastic breeds (40), revealing a locus on CFA18. Sequencing showed that all short-legged breeds have a 5 kb insertion containing a conserved retrogene of the fibroblast growth factor 4 (FGF4) gene. The retrogene includes a complete coding sequence as well as the 3′ untranslated region and a polyadenylated tail, but is lacking the 5′ regulatory sequences of the original canine FGF4 gene. The retrogene is expressed in the articular cartilage of the long bones from chondrodysplastic dogs. We proposed that this insertion could result in the increased levels of FGF4 protein, leading to premature closure of growth plates in the long bones of carrier breeds. The fgf4 retrogene was absent from all wolves tested, suggesting that this duplication event probably occurred after domestication but before radiation of early dogs into modern breeds. Therefore, not only does this study suggest a novel type of mutation for mammals, but it also provides a new candidate gene for unaccounted cases of human dwarfism.
SKULL SHAPE
One of the most interesting breed fixed differences observed between breeds is in skull shape (Fig. 3). The literature describes two approaches for finding genes associated with differences in skull shape that characterize the various breeds. One, involving cloning candidates genes associated with diseases such as Treacher Collins syndrome, has been fruitful (41), but is limiting because it only evaluates one gene at a time and relies entirely on known information (42).
Figure 3.
Skull shape variation in the dog. Above are shown extreme examples of brachycephalic breeds (on top, from left to right: Pug, Japanese Chin, Brussels Griffon, French Bulldog, Neapolitan Mastiff) and dolichocephalic breeds (on bottom, from left to right: German Shepherd Dog, Bull Terrier, Saluki, Ibizan Hound, Collie) (images: Mary Bloom, AKC).
Using a GWAS approach, Schoenebeck et al. collected skull phenotypes from 533 dogs representing 120 breeds and 4 wolf subspecies sampled from all over the United States and Europe (42). Fifty-one stereotyped landmarks were recorded using a microscribe digitizer, which allowed them to later perform principal component analyses and determine which sets of measurements were likely to be coordinately regulated. The data showed that four PCs controlled over 75% of the variance in the canine skull, including changes in rostrum length and angle, palate and zygomatic arch width, and depth of the neurocranium. These are essentially the cranioskeletal features that describe the continuum between dolichocephalic (hounds) and brachycephalic breeds (i.e. Bulldogs) (Fig. 3).
One locus had been explored previously. Bannasch et al. mapped the locus on CFA1 to an interval of ∼300 kb (43). However, no gene or mutations were ever identified, although several are plausible and presumably studies are continuing.
Schoenebeck chose to focus on CFA32, as it was highly associated with PC1, displayed showed strong evidence of selection, and it had not been previously examined (42). The initial locus was 190 kb and spanned two genes, one of which was bone morphogenesis protein 3 (BMP3), an excellent candidate. Analysis of additional SNPs contributed from the sequence of 11 breeds other than the reference Boxer reduced the interval to 85 kb and 48 critical variants. Only one of those 48 was compelling in terms of its conservation, association with the phenotype and likely functional significance: a phenylalanine to leucine change at amino acid 452 (F452L).
The argument was strengthened by making zebrafish morpholinos which, when injected, reduced the expression of the nascent BMP3 transcript in the zebrafish embryo. We then assessed the head phenotype of the fish at various times post fertilization. This loss of function experiments demonstrated a clear role of BMP3 in craniofacial development with loss of jaw structures and frontal bossing apparent in both severely and mildly affected embryos. Thus, these brachycephalic-like zebrafish demonstrate the importance of BMP3 expression in skull formation, particularly at about 48 h post fertilization.
Interestingly, several cases have been reported in the literature of children with microdeletions at 4q21, and the deletion always includes at least part of the BMP3 gene (44). In each case, the deletion is associated with craniofacial abnormalities that are reminiscent of those seen in our zebrafish experiments. This argues for the value of studying naturally occurring breed phenotypes as a way of further understanding rare human syndromes. It also suggests that BMP3 dysfunction could form the basis of cephalic conditions in humans whose genetic cause remains, to this day, unknown.
SUMMARY
The domestic dog is an entirely unique experiment in human history. Never before, or since, have humans had the chance to determine the physical and behavioral attributes of such an important member of the animal kingdom. As a result, dogs serve not only as our closest companion, but they aid us as hunters, guide dogs, herders, drafters, etc.
In this review, we have considered how genomics has influenced the types of experiments that are now possible in model organisms. We now have in hand the tools to understand the genetic basis of that variation in a unique way. In doing so we are learning about new roles of old genes, roles of genes we knew nothing about and new ways to think about how variation occurs.
In the world of science, it is common to love what we do and take our work home with us. But it is rare that we let it crawl in bed with us, feed it off our plate and with expectant eyes ask it ‘what kind of a day did you have?’. It is our considered good luck to be working in a time where genomics and genetics make it possible to do experiments that 20 years ago were unimaginable. It is our absolute privilege to be doing them with our best friend.
FUNDING
E.A.O. and M.R. are funded by the Intramural Program of the National Human Genome Research Institute (NHGRI) at NIH.
ACKNOWLEDGEMENTS
We thank Ostrander lab colleagues for reading this manuscript and their helpful comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Fogel B. The Encyclopedia of the Dog. New York, NY: DK Publishing, Inc; 1995. [Google Scholar]
- 2.Wilcox B., Walkowicz C. Atlas of Dog Breeds of the World. Neptune City, NJ: T.F.H. Publications; 1995. [Google Scholar]
- 3.Boyko A., Quignon P., Li L., Schoenenbeck J., Degenhardt J., Lohmueller K., Zhao K., Brisbin A., Parker H.G., vonHoldt B.M., et al. Simplified genetic architecture underlies morphological variation in dogs. PLoS Biology. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. doi:10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaysse A., Ratnakumar A., Derrien T., Axelsson E., Rosengren Pielberg G., Sigurdsson S., Fall T., Seppälä E.H., Hansen M.S., Lawley C.T., et al. Identification of genomic regions associated with phenotypic variation between dog Breeds using selection mapping. PLoS Genet. 2011;7:e1002316. doi: 10.1371/journal.pgen.1002316. doi:10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostrander E.A., Kruglyak L. Unleashing the canine genome. Genome Res. 2000;10:1271–1274. doi: 10.1101/gr.155900. doi:10.1101/gr.155900. [DOI] [PubMed] [Google Scholar]
- 6.Parker H.G., Shearin A.L., Ostrander E.A. Man's best friend becomes biology's best in show: genome analyses in the domestic dog. Annu. Rev. Genet. 2010;44:309–336. doi: 10.1146/annurev-genet-102808-115200. doi:10.1146/annurev-genet-102808-115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollinger J.P., Bustamante C.D., Fledel-Alon A., Schmutz S., Gray M.M., Wayne R.K. Selective sweep mapping of genes with large phenotypic effects. Genome Res. 2005;15:1809–1819. doi: 10.1101/gr.4374505. doi:10.1101/gr.4374505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wayne R.K., vonHoldt B.M. Evolutionary genomics of dog domestication. Mamm. Genome. 2012;23:3–18. doi: 10.1007/s00335-011-9386-7. doi:10.1007/s00335-011-9386-7. [DOI] [PubMed] [Google Scholar]
- 9.Wayne R.K. Molecular evolution of the dog family. Trends Genet. 1993;9:218–224. doi: 10.1016/0168-9525(93)90122-x. doi:10.1016/0168-9525(93)90122-X. [DOI] [PubMed] [Google Scholar]
- 10.Vilà C., Savolainen P., Maldonado J.E., Amorim I.R., Rice J.E., Honeycutt R.L., Crandall K.A., Lundeberg J., Wayne R.K. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. doi:10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 11.Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., 3rd, Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. doi:10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 12.Sablin M., Khlopachev G. The earliest ice age dogs: evidence from Eliseevichi I. Curr. Anthropol. 2002;43:795–798. doi:10.1086/344372. [Google Scholar]
- 13.Germonpré M., Sablin M.V., Stevens R.E., Hedges R.E., Hofreiter M., Stiller M., Després V.R. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. J. Archaeol. Sci. 2009;36:473–490. doi:10.1016/j.jas.2008.09.033. [Google Scholar]
- 14.Ovodov N.D., Crockford S.J., Kuzmin Y.V., Higham T.F., Hodgins G.W., van der Plicht J. A 33,000-year-old incipient dog from the Altai mountains of Siberia: evidence of the earliest domestication disrupted by the last glacial maximum. PloS One. 2011;6:e22821. doi: 10.1371/journal.pone.0022821. doi:10.1371/journal.pone.0022821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ardalan A., Kluetsch C.F., Zhang A.B., Erdogan M., Uhlén M., Houshmand M., Tepeli C., Ashtiani S.R., Savolainen P. Comprehensive study of mtDNA among Southwest Asian dogs contradicts independent domestication of wolf, but implies dog-wolf hybridization. Ecol. Evol. 2011;1:373–385. doi: 10.1002/ece3.35. doi:10.1002/ece3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.vonHoldt B.M., Pollinger J.P., Lohmueller K.E., Han E., Parker H.G., Quignon P., Degenhardt J.D., Boyko A.R., Earl D.A., Auton A., et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. doi:10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savolainen P., Zhang Y.P., Luo J., Lundeberg J., Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. doi:10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 18.Pang J.F., Kluetsch C., Zou X.J., Zhang A.B., Luo L.Y., Angleby H., Ardalan A., Ekström C., Skollermo A., Lundeberg J., et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol. Biol. Evol. 2009;26:2849–2864. doi: 10.1093/molbev/msp195. doi:10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Z.L., Oskarsson M., Ardalan A., Angleby H., Dahlgren L.G., Tepeli C., Kirkness E., Savolainen P., Zhang Y.P. Origins of domestic dog in southern East Asia is supported by analysis of Y-chromosome DNA. Heredity. 2012;108:507–514. doi: 10.1038/hdy.2011.114. doi:10.1038/hdy.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larson G., Karlsson E.K., Perri A., Webster M.T., Ho S.Y., Peters J., Stahl P.W., Piper P.J., Lingaas F., Fredholm M., et al. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc. Natl Acad. U.S.A. 2012;109:8878–8883. doi: 10.1073/pnas.1203005109. doi:10.1073/pnas.1203005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker H.G., Kim L.V., Sutter N.B., Carlson S., Lorentzen T.D., Malek T.B., Johnson G.S., DeFrance H.B., Ostrander E.A., Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. doi:10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 22.Parker H.G., Kukekova A.V., Akey D.T., Goldstein O., Kirkness E.F., Baysa K.C., Mosher D.S., Aguirre G.D., Acland G.M., Ostrander E.A. Breed relationships facilitate fine-mapping studies: A 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 2007;17:1652–1671. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlsson E.K., Baranowska I., Wade C.M., Salmon Hillbertz N.H.C., Zody M.C., Anderson N., Biagi T.M., Patterson N., Pielberg G.R., Kulbokas E.J., et al. Efficient mapping of Mendelian traits in dogs through genome-wide association. Nat. Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. doi:10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- 24.Sutter N.B., Eberle M.A., Parker H.G., Pullar B.J., Kirkness E.F., Kruglyak L., Ostrander E.A. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. doi:10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlsson E.K., Lindblad-Toh K. Leader of the pack: gene mapping in dogs and other model organisms. Nat. Rev. Genet. 2008;9:713–724. doi: 10.1038/nrg2382. doi:10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- 26.Ostrander E.A. Franklin H. Epstein Lecture. Both ends of the leash the human links to good dogs with bad genes. N. Engl. J. Med. 2012;367:636–646. doi: 10.1056/NEJMra1204453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chase K., Carrier D.R., Adler F.R., Jarvik T., Ostrander E.A., Lorentzen T.D., Lark K.G. Genetic basis for systems of skeletal quantitative traits: Principal component analysis of the canid skeleton. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9930–9935. doi: 10.1073/pnas.152333099. doi:10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones P., Chase K., Martin A., Davern P., Ostrander E.A., Lark K.G. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics. 2008;179:1033–1044. doi: 10.1534/genetics.108.087866. doi:10.1534/genetics.108.087866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutter N.B., Bustamante C.D., Chase K., Gray M.M., Zhao K., Zhu L., Padhukasahasram B., Karlins E., Davis S., Jones P.G., et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. doi:10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray M.M., Sutter N.B., Ostrander E.A., Wayne R.K. The IGF1 small dog haplotype is derived from Middle Eastern grey wolves. BMC Biol. 2010;8:16. doi: 10.1186/1741-7007-8-16. doi:10.1186/1741-7007-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoopes B.C., Rimbault M., Liebers D., Ostrander E.A., Sutter N.B. The insulin-like growth factor 1 receptor (IGF1R) gene contributes to reduced size in dogs. Mamm. Genome. 2012 doi: 10.1007/s00335-012-9417-z. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. doi:10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 33.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. doi:10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. doi:10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 35.Lango A.H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. doi:10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J.-J., Lee H.-I., Park T., Kim K., Lee J.-E., Cho N.H., Shin C., Cho Y.S., Lee J.-Y., Han B.-G., et al. Identification of 15 loci influencing height in a Korean population. J. Hum. Genet. 2009;55:27–31. doi: 10.1038/jhg.2009.116. doi:10.1038/jhg.2009.116. [DOI] [PubMed] [Google Scholar]
- 37.Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. doi:10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weedon M.N., Lettre G., Freathy R.M., Lindgren C.M., Voight B.F., Perry J.R.B., Elliott K.S., Hackett R., Guiducci C., Shields B., et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. doi:10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The American Kennel Club. The Complete Dog Book. New York, NY: Howell Book House; 1998. [Google Scholar]
- 40.Parker H.G., vonHoldt B.M., Quignon P., Margulies E.H., Shao S., Mosher D.S., Spady T.C., Elkahloun A., Cargill M., Jones P.G., et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;315:995–998. doi: 10.1126/science.1173275. doi:10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haworth K.E., Islam I., Breen M., Putt W., Makrinou E., Binns M., Hopkinson D., Edwards Y. Canine TCOF1; cloning, chromosome assignment and genetic analysis in dogs with different head types. Mamm. Genome. 2001;12:622–629. doi: 10.1007/s00335-001-3011-0. doi:10.1007/s00335-001-3011-0. [DOI] [PubMed] [Google Scholar]
- 42.Schoenebeck J.J., Hutchinson S.A., Byers A., Beale H.C., Carrington B., Faden D.L., Rimbault M., Decker B., Kidd J.M., Sood R., et al. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 2012;8:e1002849. doi: 10.1371/journal.pgen.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bannasch D., Young A., Myers J., Truve K., Dickinsn P., Gregg J., Davis R., Bongcam-Rudlff E., Webster M.T., Lindblad-Toh K., Pedersen N. Localization of canine brachycephaly using an across breed mapping approach. PLoS One. 2010;10:e9632. doi: 10.1371/journal.pone.0009632. doi:10.1371/journal.pone.0009632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonnet C., Adrieux J., Béri-Dexheimer M., Leheup B., Boute O., Manouvrier S., Delobel B., Copin H., Receveur A., Mathieu M., et al. Microdeletion at chromosome 4q21 defines a new emerging syndrome with marked growth restriction, mental retardation and absent or severely delayed speech. J. Med. Genet. 2010;47:377–384. doi: 10.1136/jmg.2009.071902. doi:10.1136/jmg.2009.071902. [DOI] [PubMed] [Google Scholar]
- 45.Parker H.G. Genomic analyses of modern dog breeds. Mamm. Genome. 2012;23:19–27. doi: 10.1007/s00335-011-9387-6. doi:10.1007/s00335-011-9387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]