Abstract
A genetic contribution to develop chronic obstructive pulmonary disease (COPD) is well established. However, the specific genes responsible for enhanced risk or host differences in susceptibility to smoke exposure remain poorly understood. The goal of this review is to provide a comprehensive literature overview on the genetics of COPD, highlight the most promising findings during the last few years, and ultimately provide an updated COPD gene list. Candidate gene studies on COPD and related phenotypes indexed in PubMed before January 5, 2012 are tabulated. An exhaustive list of publications for any given gene was looked for. This well-documented COPD candidate-gene list is expected to serve many purposes for future replication studies and meta-analyses as well as for reanalyzing collected genomic data in the field. In addition, this review summarizes recent genetic loci identified by genome-wide association studies on COPD, lung function, and related complications. Assembling resources, integrative genomic approaches, and large sample sizes of well-phenotyped subjects is part of the path forward to elucidate the genetic basis of this debilitating disease.
Keywords: COPD, genetics, lung function, candidate genes, genome-wide association study
Video abstract
Introduction
Chronic obstructive pulmonary disease (COPD) is the third-leading cause of worldwide mortality and is predicted to remain a major public health problem in the near future.1,2 It is characterized by airflow limitations that occur in approximately 10% of adults aged ≥ 40 years.3 Cigarette smoking is the primary risk factor. However, only a fraction of smokers (~20%) develop the disease, and host differences in susceptibility are thus persuasive. The author has previously reviewed the genetics of COPD and COPD-related phenotypes.4 The current review aims to: (1) update this publication, (2) provide a comprehensive literature overview on the genetics of COPD, (3) highlight the most promising findings during the last few years, and ultimately (4) provide an updated COPD gene list.
Chronic obstructive pulmonary disease candidate-gene studies
A systematic review of the literature was conducted in order to provide a comprehensive overview of genes associated with COPD and related phenotypes. PubMed was searched using the string “genetics and COPD” on January 5, 2012. All titles and abstracts were reviewed for inclusion. The goal was to obtain all publications testing genetic variants in humans for association with COPD and related phenotypes (ie, spirometric measurements, emphysema, chronic bronchitis, lung-function decline, etc). Population-based, case-control, and family studies were included. The author attempted to include all reported articles without quality assessment or exclusion criteria based on sample size or other criteria. The search for relevant publications was complemented using the list of references in relevant manuscripts and the COPD genetic association compendium.5 Readers are welcome to contact the author for any articles missed in the current review.
A large number of candidate gene–association studies were conducted to identify the COPD-susceptibility genes. Table 1 provides a comprehensive overview of the genes associated with COPD and related phenotypes using this genetic approach. Supplementary Table 1 presents additional genes tested but showing lack of association with COPD and related phenotypes. Most genes in these tables were studied because of their potential role in the pathobiology of COPD, but some also represent follow-up genes originally identified from genome-wide linkage and association studies. Genes are presented in alphabetical order. Single studies and metaanalyses testing each gene are indicated. An attempt was made to classify each article as supportive or not of a given gene based on the conclusions provided by the authors. Single genetic markers, haplotypes, or combinations of variants associated with COPD, COPD severity, COPD-related phenotypes, or complications were considered as positives. Table 1 aims to provide an exhaustive list of publications for any given gene.
Table 1.
List of genes associated with chronic obstructive pulmonary disease
| Symbol | Name | Chromosome | References | |||
|---|---|---|---|---|---|---|
|
|
||||||
| Single studies | Meta-analyses | |||||
|
|
|
|||||
| Positive | Negative | Positive | Negative | |||
| A2M | Alpha-2-macroglobulin | 12 | 51 | |||
| ABCC1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | 16 | 52–54 | |||
| ACE | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | 17 | 55–60 | 61,62 | 5 | |
| ADAM33 | ADAM metallopeptidase domain 33 | 20 | 63–68 | 69,70 | ||
| ADRB2 | Adrenergic, beta-2-, receptor, surface | 5 | 71–82 | 83 | 5,83 | |
| ALOX5AP | Arachidonate 5-lipoxygenase-activating protein | 13 | 84 | |||
| AQP5 | Aquaporin 5 | 12 | 85,86 | |||
| BCL2 | B-cell CLL/lymphoma 2 | 18 | 87 | |||
| BDKRB2 | Bradykinin receptor B2 | 14 | 88 | |||
| CASP10 | Caspase 10, apoptosis-related cysteine peptidase | 2 | 89 | |||
| CAT | Catalase | 11 | 90 | 91,92 | ||
| CCL5 (RANTES) | Chemokine (C-C motif) ligand 5 | 17 | 93 | 79 | ||
| CCR2 | Chemokine (C-C motif) receptor 2 | 3 | 94 | |||
| CD14 | CD14 molecule | 5 | 95,96 | |||
| CD40 | CD40 molecule, TNF receptor superfamily member 5 | 20 | 97 | |||
| CD63 | CD63 molecule | 12 | 98 | |||
| CD86 | CD86 molecule | 3 | 99 | |||
| CDC6 | Cell division cycle 6 homolog (S cerevisiae) | 17 | 100 | |||
| CDKN1A (p21) | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 6 | 101 | |||
| CFTR | Cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | 7 | 102–108 | 109,110 | ||
| CHI3L1 | Chitinase 3-like 1 (cartilage glycoprotein-39) | 1 | 111 | |||
| CHRNA3 | Cholinergic receptor, nicotinic, alpha 3 (neuronal) | 15 | 26,30,31, 112,113 | |||
| CHRNA5 | Cholinergic receptor, nicotinic, alpha 5 (neuronal) | 15 | 26,30,31, 112,113 | |||
| CLCA1 | Chloride channel accessory 1 | 1 | 114 | |||
| COL4A3 | Collagen, type IV, alpha 3 (Goodpasture antigen) | 2 | 115 | |||
| CRP | C-reactive protein, pentraxin-related | 1 | 116 | 117–119 | ||
| CSF2 | Colony stimulating factor 2 (granulocyte-macrophage) | 5 | 120 | 121 | ||
| CSF3 | Colony stimulating factor 3 (granulocyte) | 17 | 121 | |||
| CTLA4 | Cytotoxic T-lymphocyte-associated protein 4 | 2 | 99,122,123 | |||
| CTSS | Cathepsin S | 1 | 124 | |||
| CYBA | Cytochrome b-245, alpha polypeptide | 16 | 125 | |||
| CYP1A1 | Cytochrome P450, family 1, subfamily A, polypeptide 1 | 15 | 125–128 | 129,130 | ||
| CYP1A2 | Cytochrome P450, family 1, subfamily A, polypeptide 2 | 15 | 129,131 | 125,128 | ||
| CYP2E1 | Cytochrome P450, family 2, subfamily E, polypeptide 1 | 10 | 127,132 | 130 | ||
| CYP2F1 | Cytochrome P450, family 2, subfamily F, polypeptide 1 | 19 | 133 | |||
| CYP3A5 | Cytochrome P450, family 3, subfamily A, polypeptide 5 | 7 | 134 | |||
| DEFB1 | Defensin, beta 1 | 8 | 135,136 | 137 | ||
| DEFB4A | Defensin, beta 4A | 8 | 138 | |||
| EDN1 | Endothelin 1 | 6 | 139–141 | 142,143 | ||
| EDNRB | Endothelin receptor type B | 13 | 143 | |||
| ELN | Elastin (supravalvular aortic stenosis, Williams–Beuren syndrome) | 7 | 144,145 | 146,147 | ||
| EPHX1 | Epoxide hydrolase 1, microsomal (xenobiotic) | 1 | 77,83,130, 146–167 | 127,168–174 | 175 | 5,8,176 |
| ESR1 | Estrogen receptor 1 | 6 | 177 | |||
| FAM13A | Family with sequence similarity 13, member A | 4 | 26 | |||
| FGF10 | Fibroblast growth factor 10 | 5 | 178 | |||
| GC | Group-specific component (vitamin D binding protein) | 4 | 179–186 | 146,147,151, 155,187 | ||
| GCLC | Glutamate-cysteine ligase, catalytic subunit | 6 | 188 | 172,189 | ||
| GCLM | Glutamate-cysteine ligase, modifier subunit | 1 | 190 | 172,188 | ||
| GSTCD | Glutathione S-transferase, C-terminal domain containing | 4 | 191 | |||
| GSTM1 | Glutathione S-transferase M1 | 1 | 127,148,161, 164,165, 192–202 | 90,130,146,147, 151,169,203–206 | 5,7,8 | |
| GSTO1 | Glutathione S-transferase omega 1 | 10 | 207 | |||
| GSTO2 | Glutathione S-transferase omega 2 | 10 | 207 | |||
| GSTP1 | Glutathione S-transferase pi 1 | 11 | 77,90,146, 148,151,152, 157,164,165, 193,194,196, 204,208–210 | 69,127,130,147, 149,159,171,185, 197,203,211,212 | 8,213 | 5,214 |
| GSTT1 | Glutathione S-transferase theta 1 | 22 | 127,165,193, 196–198, 204–206 | 90,130,148,161, 164,169,194, 199–201,203 | 5,7,8 | |
| HCK | Hemopoietic cell kinase | 20 | 215 | |||
| HHIP | Hedgehog interacting protein | 4 | 26,191,216 | |||
| HLA | Classical class 11 subregion of the MHC | 6 | 217,218 | 219,220 | ||
| HMOX1 | Heme oxygenase (decycling) 1 | 22 | 130,151,166, 221–224 | 69,147,185, 196,225 | ||
| HTR4 | 5-hydroxytryptamine (serotonin) receptor 4 | 5 | 191 | |||
| IFNG | Interferon, gamma | 12 | 226–228 | |||
| IL1A | Interleukin 1, alpha | 2 | 227 | |||
| IL1B | Interleukin 1, beta | 2 | 227,229–233 | 120,228, 234–238 | 5,8 | |
| IL1RN | Interleukin 1 receptor antagonist | 2 | 231,232, 234,235 | 228,230, 236–238 | 8 | |
| IL2 | Interleukin 2 | 4 | 227 | |||
| IL27 | Interleukin 27 | 16 | 239 | |||
| IL4 | Interleukin 4 | 5 | 71,227,240 | 120,241,242 | 5 | |
| IL4R | Interleukin 4 receptor | 16 | 227,243 | 79,241 | ||
| IL5 | Interleukin 5 (colony-stimulating factor, eosinophil) | 5 | 244 | |||
| IL6 | Interleukin 6 | 7 | 118,228,234, 245–247 | 116,233, 236,248 | 5,8 | |
| IL8 | Interleukin 8 | 4 | 120 | 234,235,238, 249,250 | ||
| IL8RA | Interleukin 8 receptor, alpha | 2 | 251 | 120,146,147 | ||
| IL8RB (CXCR2) | Interleukin 8 receptor, beta | 2 | 250 | 120,146,147 | ||
| IL10 | Interleukin 10 | 1 | 149,227,235, 248,252–254 | 120,234,255 | ||
| IL12B | Interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) | 5 | 227 | 239 | ||
| IL13 | Interleukin 13 | 5 | 79,241,242, 256–261 | 71,120,238, 243,262 | 5 | |
| IL13RA1 | Interleukin 13 receptor, alpha 1 | X | 241 | |||
| IL17F | Interleukin 17F | 6 | 263 | |||
| IREB2 | Iron-responsive element binding protein 2 | 15 | 26,30,47 | |||
| KCNMB1 | Potassium large conductance calcium-activated channel, subfamily M, beta member 1 | 5 | 264 | |||
| KEAP1 | Kelch-like ECH-associated protein 1 | 19 | 265 | |||
| LEP | Leptin | 7 | 266 | |||
| LEPR | Leptin receptor | 1 | 267 | |||
| LTA | Lymphotoxin alpha (TNF superfamily, member 1) | 6 | 234,268–272 | 120,233,248, 273–275 | 5 | |
| LTA4H | Leukotriene A4 hydrolase | 12 | 84 | |||
| LTBP4 | Latent transforming growth factor beta binding protein 4 | 19 | 146,147 | |||
| MBL2 | Mannose-binding lectin (protein C) 2, soluble | 10 | 276,277 | |||
| MICB | MHC class I polypeptide-related sequence B | 6 | 278 | |||
| MIR196A2 | MicroRNA 196a-2 | 12 | 279 | |||
| MIR499A | MicroRNA 499a | 20 | 279 | |||
| MMP1 | Matrix metallopeptidase 1 (interstitial collagenase) | 11 | 146,280,281 | 69,128,147, 151,282–285 | ||
| MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72 kDa gelatinase, 72 kDa type IV collagenase) | 16 | 285 | 69,281 | ||
| MMP3 | Matrix metallopeptidase 3 (stromelysin 1, progelatinase) | 11 | 286 | 128,287 | ||
| MMP9 | Matrix metallopeptidase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) | 20 | 128,202,281, 282,284, 288–290 | 69,147,151, 280,283,285,287 | 5,8 | |
| MMP12 | Matrix metallopeptidase 12 (macrophage elastase) | 11 | 280,283, 291,292 | 69,146,147, 282,284, 285,287 | ||
| MMP14 | Matrix metallopeptidase 14 (membrane-inserted) | 14 | 293 | |||
| MSR1 | Macrophage scavenger receptor 1 | 8 | 137,294 | |||
| NAT2 | N-acetyltransferase 2 (arylamine N-acetyltransferase) | 8 | 132 | |||
| NFE2L2 | Nuclear factor (erythroid-derived 2)-like 2 | 2 | 265,295 | |||
| NFKBIB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | 19 | 185 | |||
| NOS3 | Nitric oxide synthase 3 (endothelial cell) | 7 | 57,62, 296,297 | 149 | ||
| NQO1 | NAD(P)H dehydrogenase, quinone 1 | 16 | 90 | |||
| NR3C1 | Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | 5 | 298 | 299 | ||
| OGG1 | 8-oxoguanine DNA glycosylase | 3 | 300 | 189 | ||
| OR4X1 | Olfactory receptor, family 4, subfamily X, member 1 | 11 | 301 | |||
| PDE4D | Phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, drosophila) | 5 | 302 | |||
| PLAUR | Plasminogen activator, urokinase receptor | 19 | 303,304 | |||
| PPARG | Peroxisome proliferator-activated receptor gamma | 3 | 163 | |||
| PTEN | Phosphatase and tensin homolog | 10 | 14 | |||
| PTGDR | Prostaglandin D2 receptor (DP) | 14 | 305 | |||
| PTGS2 (COX2) | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 1 | 306,307 | |||
| SERPINA1 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | 14 | 76,308–325 | 326–336 | ||
| SERPINA3 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 14 | 337–343 | 146,147,149, 151,310,314, 326,332,344 | 5 | |
| SERPINE2 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 | 2 | 77,146,149, 326,345–348 | 147,152,171, 349,350 | ||
| SFTPA1 | Surfactant protein A1 | 10 | 69,351 | |||
| SFTPA2 | Surfactant protein A2 | 10 | 69 | |||
| SFTPB | Surfactant protein B | 2 | 147,151,171, 351–354 | 69,77,146, 149,152,355 | ||
| SFTPC | Surfactant protein C | 8 | 356 | 357 | ||
| SFTPD | Surfactant protein D | 10 | 69,358,359 | 151,351 | ||
| SIRT2 | Sirtuin 2 | 19 | 185 | |||
| SLC6A4 | Solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 17 | 360 | |||
| SLC11A1 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | 2 | 361 | |||
| SMAD3 | SMAD family member 3 | 15 | 362 | |||
| SMOC2 | SPARC related modular calcium binding 2 | 6 | 363 | |||
| SOD2 | Superoxide dismutase 2, mitochondrial | 6 | 364–366 | 91,92,271 | 8 | |
| SOD3 | Superoxide dismutase 3, extracellular | 4 | 90,91,364, 367–370 | 5 | 8 | |
| SOX5 | SRY (sex determining region Y)-box 5 | 12 | 371 | |||
| STAT1 | Signal transducer and activator of transcription 1, 91 kDa | 2 | 185 | |||
| STAT3 | Signal transducer and activator of transcription 3 (acute-phase response factor) | 17 | 372 | |||
| STAT6 | Signal transducer and activator of transcription 6, interleukin-4 induced | 12 | 79 | 241 | ||
| STIP1 | Stress-induced-phosphoprotein 1 | 11 | 373 | |||
| TBXA2R | Thromboxane A2 receptor | 19 | 244,374 | |||
| TGFB1 | Transforming growth factor, beta 1 | 19 | 69,77,146, 147,238, 375–382 | 30,149,151, 171,383 | 5,8 | 384 |
| TGFBR3 | Transforming growth factor, beta receptor III | 1 | 190 | |||
| TIMP1 | TIMP metallopeptidase inhibitor 1 | X | 285 | 69 | ||
| TIMP2 | TIMP metallopeptidase inhibitor 2 | 17 | 146,385,386 | 147,151,387 | 5 | |
| TLR4 | Toll-like receptor 4 | 9 | 388,389 | 96,271 | ||
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | 6 | 11,149,151, 234,238,250, 262,268, 270–272, 390–398 | 83,120,146, 147,155,230, 233,235–237, 248,269, 273–275, 399–403 | 5,8–11 | |
| TNS1 | Tensin 1 | 2 | 191 | |||
| TP53 (p53) | Tumor protein p53 | 17 | 101,307 | |||
| TRPV4 | Transient receptor potential cation channel, subfamily V, member 4 | 12 | 404 | |||
| TSLP | Thymic stromal lymphopoietin | 5 | 405 | |||
| VDR | Vitamin D (1,25-dihydroxyvitamin D3) receptor | 12 | 406–408 | 409 | ||
| VEGFA | Vascular endothelial growth factor A | 6 | 410 | 411 | ||
| XRCC1 | X-ray repair complementing defective repair in Chinese hamster cells 1 | 19 | 300 | |||
| XRCC5 | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | 2 | 412 | |||
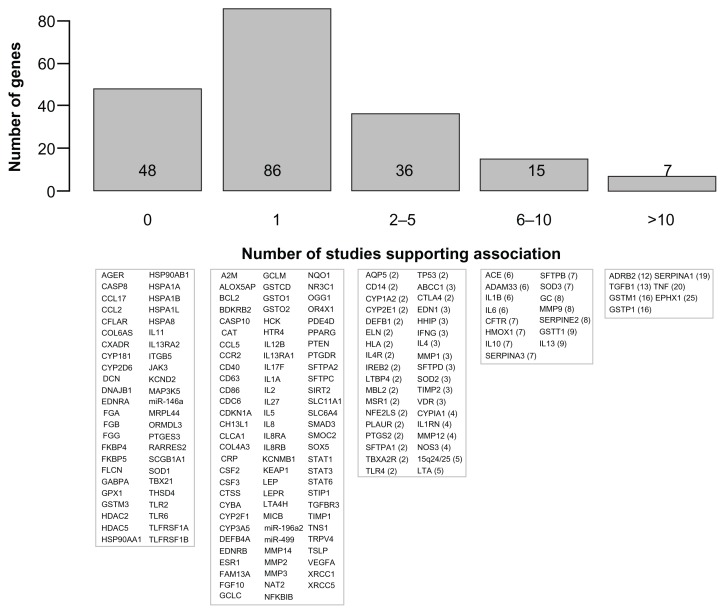
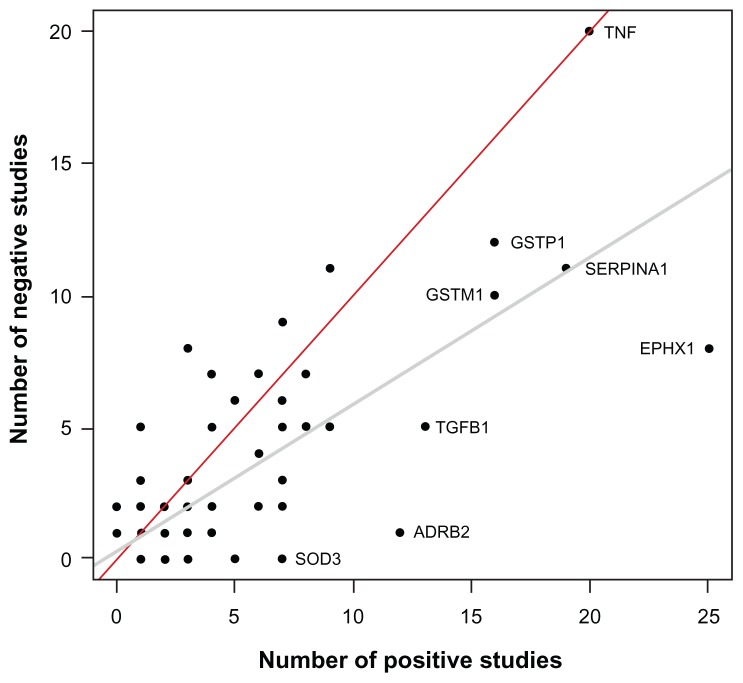
A total of 192 genes are summarized in Table 1 and Supplementary Table 1. Figure 1 illustrates these genes based on the number of publications supporting the association with COPD phenotypes. Briefly, 86 genes are supported by one study, 36 genes by two to five studies, 15 genes by six to ten studies, and seven genes by more than ten studies. The latter seven genes include ADRB2, TGFB1, TNF, GSTM1, GSTP1, SERPINA1, and EPHX1. Note that Figure 1 must be interpreted with caution. Replication of genotype–phenotype associations is the gold standard to identify genes conferring susceptibility.6 However, the number of supportive studies is not necessarily an indication that a gene is consistently replicated. Figure 2 illustrates the relationship between the number of studies supporting and not supporting the list of COPD genes. It seems that genes replicated many times in COPD are simply the most popular genes studied. For example, the author found 20 studies supporting TNF as a COPD-susceptibility gene. However, lack of association between this gene and COPD phenotypes was found in 20 other studies (Table 1). Considering publication bias, candidate genes associated with COPD are not consistently replicated and the overall results are rather inconclusive. In fact, excluding SERPINA1 (encoding the alpha-1 antitrypsin protein), none of the other genes are well-proven susceptibility genes for COPD. Perhaps the most convincing candidate COPD genes up to now are those less studied but consistently replicated, such as SOD3. Many of the most studied COPD genes have now been investigated in meta-analyses.
Figure 1.
Candidate genes associated with chronic obstructive pulmonary disease (COPD) or related phenotypes.
Notes: The upper part shows a histogram of the number of COPD susceptibility genes based on the number of publications supporting a significant genetic association. The lower part shows the corresponding genes in each bar. Official gene symbols are indicated. The number of publications that are supportive is indicated in parentheses. References are provided in Table 1 for genes supported by at least one publication and in Supplementary Table 1 for genes tested but not supported.
Figure 2.
Scatter plot showing the number of studies supporting and not supporting candidate genes for chronic obstructive pulmonary disease.
Notes: A total of 192 genes are illustrated. Note that many genes overlap in the lower-left corner and the 192 dots cannot be visualized on this display. The gray and red lines are the regression and identity lines, respectively. Genes studied many times or more consistently replicated are illustrated.
Meta-analyses
A number of meta-analyses have been conducted to identify genes robustly associated with COPD and lung function. So far, meta-analyses have been conducted for genes involved in the following pathways: inflammation (IL4, IL6, IL13, IL1B, IL1RN, LTA, TNF, and TGFB1), protease/antiprotease (MMP9, TIMP2, and SERPINA3), oxidative stress (GSTM1, GSTP1, GSTT1, EPHX1, SOD2, and SOD3), and others (ACE and ADRB2). These studies and their main outcomes are summarized by gene in Table 1. Among these genes, GSTM1 was consistently associated with COPD in more than one meta-analysis.5,7,8 This is also true for TNF, but only in Asian populations.5,8–11 In contrast, other genes have not been supported in meta-analyses conducted so far, including GSTT1,5,7,8 IL1B,5,8 IL6,5,8 and MMP9.5,8 The other genes considered in meta-analyses were either reported in only one study or showed conflicting results across studies (Table 1).
As genetic data accumulates, more genes and polymorphisms will be considered in meta-analyses. Combining the findings of an increasing number of studies will allow pooled analyses in more homogenous subgroups based on ethnicity, smoking history, emphysema vs airway type of COPD, and others. These subgroup analyses are likely to be important in finding susceptibility genes for COPD. Ongoing activities gathering genetic data in the field of COPD are important. For example, a web application summarizing candidate-gene studies was recently established.5 At the time of publication, this database included 108 genetic-association studies, including population-based and case-control studies but excluding family-based studies. Seventy-two genes were studied, focusing strictly on single-marker biallelic polymorphisms. A total of 27 genetic variants were found to be reported in three or more independent study populations and summarized into a meta-analysis. Four genes were found to carry a single genetic variant significantly associated with COPD, being GSTM1, TGFB1, TNF, and SOD3. It should be noted that this COPD genetic-association compendium has not been updated since April 2010 and does not included more recent genetic studies on COPD. Updating this type of resource is important to draw reliable conclusions about the contribution of genes. The number of studies for most COPD-susceptibility genes is currently insufficient to reach firm conclusions.
Multi-gene-association studies
A systematic replication study of genes associated with lung function was recently conducted in the SpiroMeta Consortium.12 A literature search identified 104 publications reporting a positive association with lung-function traits in the general populations of diverse origins or in cohorts of patients with respiratory diseases. A total of 130 genes and 48 intergenic regions were studied in 20,288 individuals. Among the 16,936 genotyped or imputed single-nucleotide polymorphisms (SNPs) in these loci, none was significantly associated with forced expiratory volume in one second (FEV1) or FEV1/forced vital capacity (FVC) ratio after correction for multiple testing. The strongest genetic association signals with FEV1 were observed in ever-smokers in the SERPINA1 and PDE4D genes.
Smaller-scale studies testing multiple genes were also conducted in China. First, 170 asthmatic cases and 347 controls were evaluated for 119 SNPs in 98 genes for association with lung function.13 After correction for multiple testing, none of the SNPs was significantly associated with lung function (ie, FEV1, FVC, or FEV1/FVC). The strongest association was observed between rs320995 (Phe309Phe) in CYSLTR1 and FEV1/FVC (P = 0.0004). Second, 1,261 SNPs in 380 candidate genes for cancer or other human diseases were tested for association with COPD in 53 cases and 107 controls with in-home coal exposure.14 A total of 22 genes were associated with COPD risk, but only PTEN was significant after correction for multiple testing. Considering the small sample sizes, the results of these studies must be replicated before reaching firm conclusions.
Genome-wide association studies on COPD
Table 2 summarizes COPD susceptibility loci identified by genome-wide association (GWA) studies. The results of the first GWA study on COPD were published in 2009.15 The GWA study was conducted in a case-control cohort of Norway (823 COPD cases and 810 controls), and the top 100 SNPs were followed up in the family-based International COPD Genetics Network (ICGN). Two susceptibility loci were identified. The most definitive evidence of association was found with two SNPs at the α-nicotinic acetylcholine receptor locus on chromosome 15q25, the same locus implicated in the risk of lung cancer.16–18 Two SNPs at the hedgehog interacting protein (HHIP) locus on chromosome 4q31 also showed strong associations.
Table 2.
Susceptibility loci for chronic obstructive pulmonary disease (COPD) and related phenotypes identified by genome-wide association studies
| Reference | Study* | Sample size (cases/controls) | Disease/trait | Platform (# SNPs) | Region (size) | Gene | Key SNPs |
|---|---|---|---|---|---|---|---|
| Pillai et al15 | Norway | 823/810 | COPD | Illumina (Human Hap550) | 15q25 | CHRNA3 | rs8034191 |
| ICGN | 1891 | CHRNA5 | rs1051730 | ||||
| NETT-NAS | 389/472 | ||||||
| EOCOPD | 949 | ||||||
| 4q31 | HHIP | rs1828591 | |||||
| rs13118928 | |||||||
| Cho et al19 | Norway | 2940/1380 | COPD | Illumina (Human Hap550 or Quad610) | 4q22 | FAM13A | rs7671167 |
| NETT-NAS | rs1903003 | ||||||
| ECLIPSE | |||||||
| COPDGene | 502/504 | ||||||
| EOCOPD | 949 | ||||||
| ICGN | 2859 | ||||||
| 15q25 | CHRNA3 | rs1062980 | |||||
| CHRNA5 | |||||||
| IREB2 | |||||||
| 4q31 | HHIP | rs1828591 | |||||
| Cho et al20 | ECLIPSE | 1764/178 | COPD | Illumina (Human Hap550, Quad610, or Omni1 Quad) | 19q13 | RAB4B | rs7937 |
| NAS-NETT | 373/435 | EGLN2 | rs2604894 | ||||
| GenKOLS | 863/808 | MIA | |||||
| COPDGene | 499/501 | CYP2A6 | |||||
| ICGN | 983 probands/ 1876 siblings | ||||||
| 4q22 | FAM13A | rs1964516 | |||||
| rs7671167 | |||||||
| 4q31 | HHIP | rs13141641 | |||||
| rs13118928 | |||||||
| 15q25 | CHRNA3 | rs11858836 | |||||
| CHRNA5 | rs13180 | ||||||
| IREB2 | |||||||
| Wilk et al37 | FHS | 1059–1222 | Ten spirometry phenotypes | Affymetrix (70,987) | 10q25 | GSTO2 | rs156697 |
| Wilk et al38 | FHS | 7691 | FEV1/FVC | Affymetrix (500 K + 50 K) | 4q31 | HHIP | rs13147758 |
| Family heart study | 835 | ||||||
| Repapi et al40 | SpiroMeta Consortium | 20,288 | FEV1 and FEV1/FVC | Affymetrix and Illumina (2.5 million) | 4q31 | HHIP | rs12504628 |
| CHARGE consortium | 32,184 | ||||||
| 21,209 | |||||||
| Health 2000 survey | 883 | ||||||
| FEV1 | 2q35 | TNS1 | rs2571445 | ||||
| 4q24 | GSTCD | rs10516526 | |||||
| 5q33 | HTR4 | rs3995090 | |||||
| FEV1/FVC | 6p21 | AGER | rs2070600 | ||||
| 15q23 | THSD4 | rs12899618 | |||||
| Hancock et al39 | CHARGE Consortium | 20,890 | FEV1/FVC | Affymetrix and Illumina (2,515,866) | 2q36 | PID1 | rs1435867 |
| SpiroMeta consortium | 16,178 | ||||||
| 4q22 | FAM13A | rs2869967 | |||||
| 4q31 | HHIP | rs1980057 | |||||
| 5q33 | HTR4 | rs11168048 | |||||
| 5q33 | ADAM19 | rs2277027 | |||||
| 6p21 | AGER-PPT2 | rs2070600 | |||||
| 6q24 | GPR126 | rs3817928 | |||||
| 9q22 | PTCH1 | rs16909898 | |||||
| FEV1 | 4q24 | INTS12 | rs17331332 | ||||
| GSTCD | |||||||
| NPNT | |||||||
| Soler Artigas et al41,** | 23 studies | 48,201 | FEV1 | Illumina and Affymetrix (2,706,349) | 3q26 | MECOM | rs134555 |
| 17 studies | 46,411 | ||||||
| 6p22 | ZKSCAN3 | rs6903823 | |||||
| 10q22 | C10orf11 | rs11001819 | |||||
| FEV1/FVC | 1p36 | MFAP2 | rs2284746 | ||||
| 1q41 | TGFB2- LYPLAL1 | rs993925 | |||||
| 2q37 | HDAC4- FLJ43879 | rs12477314 | |||||
| 3p24 | RARB | rs1529672 | |||||
| 5q15 | SPATA9- RHOBTB3 | rs153916 | |||||
| 6q21 | ARMC2 | rs2798641 | |||||
| 6p21 | NCR3-AIF1 | rs2857595 | |||||
| 12q13 | LRP1 | rs11172113 | |||||
| 12q22 | CCDC38 | rs1036429 | |||||
| 16q13 | MMP15 | rs12447804 | |||||
| 16q23 | CFDP1 | rs2865531 | |||||
| 21q22 | KCNE2- LINC00310 | rs9978142 | |||||
| FEV1 and FEV1/FVC | 10p23 | CDC123 | rs7068966 | ||||
| Imboden et al42 | SAPALDIA | 2677 nonasthmatic, 1441 asthmatic | FEV1 decline in nonasthmatic | Illumina | 13q14 | DLEU7 | rs9316500 |
| ECRHS | Human | ||||||
| EGEA | 610quad | ||||||
| FHS | 10,858 nonasthmatic, 1138 asthmatic | ||||||
| ARIC | |||||||
| B58C | |||||||
| Dutch asthma study | |||||||
| FEV1/FVC decline in asthmatic | 8p22 | TUSC3 | rs4831760 | ||||
| Kong et al43 | ECLIPSE | 1557 | Emphysema (qualitative) | Illumina | 12q11 | BICD1 | rs10844154 |
| Norway | 432 | Human | rs161976 | ||||
| Hap550 (499,578) | |||||||
| Wan et al44 | ECLIPSE | 1734 | Cachexia-related phenotypes (BMI and fat-free mass index) | Illumina | 16q12 | FTO | rs8050136 |
| Norway | 851 | ||||||
| NETT | 365 | ||||||
| COPDGene | 502 |
Notes:
Bold entries indicates replication cohorts;
only the new loci are identified for this study, but ten loci previously reported by Hancock et al39 and Repapi et al40 were also detected.
Abbreviations: ARIC, Atherosclerosis Risk in Communities; B58C, British 1958 Birth Cohort; EOCOPD, Boston Early-Onset COPD Study; BMI, body mass index; COPDGene, COPDGene study; ECLIPSE, Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; ECRHS, European Community Respiratory Health Survey; EGEA, Epidemiological study on the Genetics and Environment of Asthma; FEV1, forced expiratory volume in 1 second; FHS, Framingham Heart Study; FVC, forced vital capacity; GenKOLS, Bergen, Norway COPD Cohort; ICGN, International COPD Genetics Network study; NAS-NETT, Normative Aging Study and National Emphysema Treatment Trial; SAPALDIA, Swiss Cohort Study on Air Pollution and Lung and Heart Disease in Adults; SNPs, single-nucleotide polymorphisms.
The case-control cohort of Norway was then combined with the COPD cases from the National Emphysema Treatment Trial (NETT) and unaffected individuals from the Normative Aging Study (NAS), as well as cases and controls from the multicenter Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Study.19 A total of 2940 cases and 1380 controls were considered. Loci 15q25-CHRNA3/CHRNA5/IREB2 and 4q31-HHIP were replicated in this study. A third locus was also identified at 4q22.1 harboring the FAM13A gene. The latter was followed up and validated in the COPDGene study and the ICGN. A trend was also observed in the Boston Early-Onset COPD Study (EOCOPD). The latest GWA study on COPD was performed using 3499 cases and 1922 controls regrouping the ECLIPSE, NETT-NAS, Norway, and COPDGene studies. 20 The three GWA-nominated COPD-susceptibility loci (ie, CHRNA3/CHRNA5/IREB2, HHIP, and FAM13A) were confirmed in this extended GWA study. In addition, a new COPD locus was identified on chromosome 19q13, which harbored the RAB4B, EGLN2, MIA, and CYP2A6 genes. It was estimated that the four GWA-nominated COPD loci accounted for ~5% of the total variance of the sibling relative risk of COPD.20
Two of the four genome-wide associated loci found in COPD – 15q25 and 19q13 – were previously associated with cigarettes smoked per day and cotinine levels,21–25 suggesting that the risk alleles are acting through smoking behavior. Further studies support this hypothesis on 15q25. In fact, previous studies suggested that sequence variants on chromosome 15q25 confer risk of smoking-related lung diseases (ie, COPD and lung cancer) through its effect on tobacco addiction.17,26 This is consistent with the lack of association between the 15q25 locus and lung cancer among never-smokers.27–29 In contrast, other evidence argues against this hypothesis, showing weak or no evidence that the 15q25 locus directly influences smoking behavior,15,16 no appreciable variation in the risk of lung cancer across smoking categories,18 and significant effect of the 15q25 locus on smoking-related diseases after adjustment for smoking exposure.30,31 Multiple distinct loci affecting both smoking behavior24,31 and lung cancer32 were reported on 15q25. It is still unknown whether genes located at any of these loci are causally involved in the pathogenesis of COPD and lung cancer or the effect is mediated by changing smoking behavior. Risk alleles on chromosome 15q25 were shown to modulate the mRNA expression levels of the CHRNA5 gene in the brain33,34 and lung35 tissues as well as the expression of CHRNA5 and IREB2 genes in sputum.36 The regulation of genes in primary disease tissues, such as lung and sputum, suggests a direct effect of 15q25 genes on COPD susceptibility. More functional studies are needed to find the causal alleles and genes on 15q25 as well as to disentangle their impact on correlated traits associated with this chromosomal region.
GWA studies on lung function
In 2007, Wilk et al37 reported the first GWA study on lung function in approximately 1200 individuals. The study was conducted as part of the Framingham Heart Study. Association tests were performed on 70,987 autosomal SNPs and for ten spirometry phenotypes. No SNP was associated with lung-function phenotypes using stringent criteria for genome-wide significance, but suggestive evidence of association was provided for a nonsynonymous coding SNP in exon 5 of the GSTO2 gene. In 2009, a larger GWA study from the Framingham Heart Study was performed in 7691 participants.38 Interestingly, the 4q31-HHIP COPD locus was associated with percent predicted FEV1/FVC ratio. This locus was confirmed in a second set of participants from the Family Heart Study (n = 835).
In January 2010, two articles reported GWA studies for lung function.39,40 First, Repapi et al40 performed a GWA study on FEV1 and FEV1/FVC ratio in the SpiroMeta consortium (20,288 individuals of European ancestry). They have also followed up the best associated SNPs in 32,184 additional individuals. Overall, they have identified five novel genome-wide significant loci for pulmonary function, being 2q35 (TNS1), 4q24 (GSTCD), and 5q33 (HTR4) for FEV1, and 6p21 (AGER) and 15q23 (THSD4) for FEV1/FVC. Second, Hancock et al39 conducted a GWA study on the same two clinically important pulmonary function measures in the CHARGE consortium consisting of 20,890 participants of European ancestry. They identified significant associations with FEV1/FVC ratio for SNPs located in seven previously unrecognized loci: 6q24 (GPR126), 5q33 (ADAM19), 6p21 (AGER and PPT2), 4q22 (FAM13A), 9q22 (PTCH1), 2q36 (PID1), and 5q33 (HTR4). For FEV1, one new locus annotated by three genes (INTS12, GSTCD, and NPNT) on 4q24 was identified. 4q24 (GSTCD), 5q33 (HTR4) and 6p21 (AGER) were common in both consortia, ie, SpiroMeta and CHARGE. The previously reported 4q31 locus located upstream of the HHIP gene associated with FEV1 and FEV1/ FVC ratio was also confirmed in these consortia.
More recently, a larger GWA study of FEV1 and FEV1/FVC ratio was reported, comprising more than 48,000 individuals of European ancestry and followed up for replication in more than 46,000 individuals.41 Ten out of eleven loci previously reported by the SpiroMeta and CHARGE consortia were replicated in this extended GWA study. Only PID1 on 2q36 was not replicated. More interestingly, 16 new loci were identified, including twelve loci for FEV1/FVC, three for FEV1, and one for both traits. Thus, 26 loci were associated with lung function in this GWA study. Together, these loci explain 3.2% of the additive polygenic variance for FEV1/ FVC and 1.5% of the variance for FEV1.
The first GWA study on lung-function decline was recently reported.42 Briefly, genome-wide analyses on FEV1 and FEV1/FVC decline were conducted in 2677 nonasthmatics and 1441 asthmatics separately. The top hits were then replicated in 10,858 nonasthmatic and 1138 asthmatic participants. Decline of FEV1 and FEV1/FVC ratio was evaluated during a follow-up examination period of roughly 10 years in these participants. No SNP reached genome-wide significance in the discovery set. However, one locus on chromosome 13q14.3 containing the DLEU7 gene was strongly associated with FEV1 decline in nonasthmatics from the discovery set and confirmed in the replication set. A strong association signal was also reported on 8p22 harboring the TUSC3 gene for FEV1/FVC decrease in asthmatics, but not validated in the replication set. Many loci previously associated with cross-sectional lung function in GWA studies described above were replicated with baseline lung function in either asthmatic or nonasthmatic subjects. However, few GWAS-nominated lung-function loci were associated with lung-function decline, suggesting different genetic mechanisms governing baseline lung function and decline with age. In addition, this study showed the genetic heterogeneity of lung-function decline between subjects with and without asthma. Table 2 summarizes lung-function susceptibility loci identified by GWA studies.
GWA studies on COPD-related phenotypes
Other GWA studies were reported on COPD-related phenotypes. Emphysema is an important feature of COPD and varies considerably between patients. A recent GWA study was performed on emphysema measures by computed tomography scan and defined by radiologist qualitative scores and quantitative assessments of low-attenuation areas.43 The qualitative scores obtained in 1557 patients from the ECLIPSE study and 432 subjects from the Norway cohort led to the identification of an emphysema locus on chromosome 12p11.2. The most strongly associated SNP is located in the BICD1 gene, known to be involved in regulating telomere length. The ECLIPSE, Norway, and NETT studies were also used to perform a GWA study on COPD-related cachexia phenotypes, including body mass index and fat-free mass index.44 Cachexia occurs in approximately 10% of patients with COPD and is associated with increased mortality. The GWA study on body mass index and fat-free mass index in patients with COPD identified a single susceptibility locus that harbored the FTO gene, the most robust gene associated with obesity. Whether FTO acts through obesity or directly affects lung function remains to be elucidated.
GWA studies on COPD, lung function, and related phenotypes provided strong and consistent evidence of genetic susceptibility loci. These studies also highlight the large number of participants required to identify reproducible genetic loci. So far, GWA studies have identified only a small fraction of the genetic variants contributing to COPD risk, related complications, and lung-function variability. GWA studies on larger sample sizes, especially for COPD, will be required to identify the genetic factors underpinning COPD and related phenotypes. Large international efforts are under way to increase sample sizes and use more comprehensive molecular phenotyping (eg, gene expression in the lung) to elucidate the genetic component of COPD.45,46 It should be emphasized that the causal genes and genetic variants of all these newly discovered loci by GWA studies remain to be identified. More integrative genomic approaches will be required for these purposes. Different study designs testing rare and copy-number variants as well as gene-smoking interaction are also needed.
Integrative genomic approaches
More studies are being conducted using integrative genomic approaches in order to identify COPD susceptibility genes. For example, the IREB2 gene was identified by combining gene expression in human lungs and genetic association in COPD cohorts.47 In this study, lung specimens were obtained from patients undergoing lung nodule resection, and gene expression was compared between 15 COPD and 18 non-COPD patients using whole-genome gene-expression arrays. A total of 889 SNPs found in the 62 genomic regions containing genes differentially expressed between patients with or without COPD were tested for association with COPD and lung function. Seventy-one SNPs nominally associated (P ≤ 0.05) with COPD in the NETT-NAS study were followed up for replication in the EOCOPD study. A gene-based replication was then completed to confirm genetic association between genetic variants in the IREB2 gene and lung function. Overall, the IREB2 gene was shown to be upregulated in lung specimens of COPD patients and to contain genetic variants associated with COPD. Gene expression in a larger number of lung specimens will be required to test whether COPD-associated SNPs in the IREB2 gene influence the expression of its gene product.
Although Table 2 shows the major susceptibility loci identified by GWA studies, many additional loci were borderline significant in these studies. Many true positives are likely to be missed by this approach owing to the stringent threshold used to control for false-discovery rates. Different weighting methods and SNP-prioritization strategies are currently used to find true-positive signals from previous GWA studies. For example, the FGF7 gene was recently identified as a COPD susceptibility locus by weighting GWA analysis on regions of conserved homozygosity haplotype in subjects affected with COPD compared to unaffected subjects.48 As mentioned previously,49 further studies reanalyzing genome-wide SNP datasets with weighting methods based on function annotations (eg, coding variants or regions) or prior knowledge (eg, candidate genes or genome-wide linkage studies) will be required. Similarly, ongoing lung expression quantitative trait loci (eQTLs) mapping data36,46 are likely to leverage the impact of previous GWA studies on COPD by providing a list of SNPs that regulate gene expression in relevant tissues. SNPs associated with gene expression will provide crucial functional information to understand the molecular changes introduced by the susceptibility DNA variants. The identification of SNPs associated with both disease traits and quantitative transcript levels of one or more genes in relevant tissues will highlight the most likely causal gene within the susceptibility loci and the functional SNPs that are prime candidates to be directly involved in the pathogenesis of COPD.
Conclusion
Elucidating the genetic component of COPD and lung function turned out to be a challenging task. Major resources and collaborative efforts will be required to achieve our goal. In this review, the author provides an updated list of COPD genes and a summary of GWAS results conducted during the last few years. It is hoped that the gene list can be used by investigators to replicate or refute susceptibility genes of COPD. As eluded above, this gene list can also be used to reanalyze GWA data by prioritizing genes previously associated with COPD or related phenotypes or enter into more global gene network and causality analyses. Owing to the challenge faced by the genetic community, large collections of patients well characterized for COPD phenotypes are ongoing to identify the genuine COPD genes. A lumping and splitting strategy is an old idea in the field of genetics of complex traits50 that will certainly be essential in the field of COPD. Pooling resources (ie, lumping) is required to obtain proper sample sizes, but is likely to increase heterogeneity. These larger sample sizes, however, provide the opportunity to subdivide (ie, splitting) the pooled data into more homogeneous subgroups where the molecular defects are more likely to be similar. Accordingly, not only the genetic community but the entire spectrum of experts managing and treating patients with COPD will be required to provide samples, precise phenotypes, and expertise to search for the underlying genetic mechanisms. In parallel, complementary multidimensional genomic data in relevant tissues (eg, lung eQTLs) will be crucial to uncover causal genes and genetic variants that contribute to COPD and to discover new molecular targets for prevention, diagnosis, and treatment.
Supplementary materials
Table S1.
Genes tested but showing lack of association with chronic obstructive pulmonary disease
| Symbol | Name | Chromosome | References | |||
|---|---|---|---|---|---|---|
|
|
||||||
| Single studies | Meta-analyses | |||||
|
|
|
|||||
| Positive | Negative | Positive | Negative | |||
| AGER | Advanced glycosylation end product-specific receptor | 6 | 1,2 | |||
| CASP8 | Caspase 8, apoptosis-related cysteine peptidase | 2 | 3 | |||
| CCL17 (TARC) | Chemokine (C-C motif) ligand 17 | 16 | 4 | |||
| CCL2 | Chemokine (C-C motif) ligand 2 | 17 | 5 | |||
| CFLAR | CASP8 and FADD-like apoptosis regulator | 2 | 3 | |||
| COL6A5 | Collagen, type VI, alpha 5 | 3 | 6 | |||
| CXADR | Coxsackie virus and adenovirus receptor | 21 | 7 | |||
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 2 | 8,9 | |||
| CYP2D6 | Cytochrome P450, family 2, subfamily D, polypeptide 6 | 22 | 10 | |||
| DCN | Decorin | 12 | 11 | |||
| DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 19 | 12 | |||
| EDNRA | Endothelin receptor type A | 4 | 13 | |||
| FGA | Fibrinogen alpha chain | 4 | 14 | |||
| FGB | Fibrinogen beta chain | 4 | 14,15 | |||
| FGG | Fibrinogen gamma chain | 4 | 14 | |||
| FKBP4 | FK506 binding protein 4, 59 kDa | 12 | 12 | |||
| FKBP5 | FK506 binding protein 5 | 6 | 12 | |||
| FLCN | Folliculin | 17 | 16 | |||
| GABPA | GA binding protein transcription factor, alpha subunit 60 kDa | 21 | 17 | |||
| GPX1 | Glutathione peroxidase 1 | 3 | 18,19 | |||
| GSTM3 | Glutathione S-transferase mu 3 (brain) | 1 | 20 | |||
| HDAC2 | Histone deacetylase 2 | 6 | 1 | |||
| HDAC5 | Histone deacetylase 5 | 17 | 1 | |||
| HSP90AA1 (HSPCA) | Heat shock protein 90 kDa alpha (cytosolic), class A member 1 | 14 | 12 | |||
| HSP90AB1 (HSPCB) | Heat shock protein 90 kDa alpha (cytosolic), class B member 1 | 6 | 12 | |||
| HSPA1A | Heat shock 70 kDa protein 1A | 6 | 21 | |||
| HSPA1B | Heat shock 70 kDa protein 1B | 6 | 21 | |||
| HSPA1L | Heat shock 70 kDa protein 1-like | 6 | 21 | |||
| HSPA8 | Heat shock 70 kDa protein 8 | 11 | 12 | |||
| IL11 | Interleukin 11 | 19 | 1 | |||
| IL13RA2 | Interleukin 13 receptor, alpha 2 | X | 22 | |||
| ITGB5 | Integrin, beta 5 | 3 | 7 | |||
| JAK3 | Janus kinase 3 | 19 | 1 | |||
| KCND2 | Potassium voltage-gated channel, Shal-related subfamily, member 2 | 7 | 1 | |||
| MAP3K5 | Mitogen-activated protein kinase kinase kinase 5 | 6 | 1 | |||
| MIR146a | MicroRNA 146a | 5 | 23 | |||
| MRPL44 | Mitochondrial ribosomal protein L44 | 2 | 24 | |||
| ORMDL3 | ORM1-like 3 (S cerevisiae) | 17 | 25 | |||
| PTGES3 | Prostaglandin E synthase 3 (cytosolic) | 12 | 12 | |||
| RARRES2 | Retinoic acid receptor responder (tazarotene induced) 2 | 7 | 1 | |||
| SCGB1A1 (CC16) | Secretoglobin, family 1A, member 1 (uteroglobin) | 11 | 26 | |||
| SOD1 | Superoxide dismutase 1, soluble | 21 | 18,27 | |||
| TBX21 | T-box 21 | 17 | 28 | |||
| THSD4 | Thrombospondin, type I, domain containing 4 | 15 | 2 | |||
| TLR2 | Toll-like receptor 2 | 4 | 29,30 | |||
| TLR6 | Toll-like receptor 6 | 4 | 31 | |||
| TNFRSF1A | Tumor necrosis factor receptor superfamily, member 1A | 12 | 32 | |||
| TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B | 1 | 32 | |||
References
- 1.Bakke PS, Zhu G, Gulsvik A, et al. Candidate genes for COPD in two large data sets. Eur Respir J. 2011;37(2):255–263. doi: 10.1183/09031936.00091709. [DOI] [PubMed] [Google Scholar]
- 2.Soler Artigas M, Wain LV, Repapi E, et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med. 2011;184(7):786–795. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith AK, Lange LA, Ampleford EJ, Meyers DA, Bleecker ER, Howard TD. Association of polymorphisms in CASP10 and CASP8 with FEV(1)/FVC and bronchial hyperresponsiveness in ethnically diverse asthmatics. Clin Exp Allergy. 2008;38(11):1738–1744. doi: 10.1111/j.1365-2222.2008.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung TF, Chan IH, Wong GW, et al. Association between candidate genes and lung function growth in Chinese asthmatic children. Clin Exp Allergy. 2007;37(10):1480–1486. doi: 10.1111/j.1365-2222.2007.02799.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu SF, Wang CC, Fang WF, Chen YC, Lin MC. MCP1-2518 polymorphism and chronic obstructive pulmonary disease in Taiwanese men. Exp Lung Res. 2010;36(5):277–283. doi: 10.3109/01902140903575989. [DOI] [PubMed] [Google Scholar]
- 6.Harazin M, Parwez Q, Petrasch-Parwez E, et al. Variation in the COL29A1 gene in German patients with atopic dermatitis, asthma and chronic obstructive pulmonary disease. J Dermatol. 2010;37(8):740–742. doi: 10.1111/j.1346-8138.2010.00923.x. [DOI] [PubMed] [Google Scholar]
- 7.Kasuga I, Hogg JC, Pare PD, et al. Role of genetic susceptibility to latent adenoviral infection and decreased lung function. Respir Med. 2009;103(11):1672–1680. doi: 10.1016/j.rmed.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur-Knudsen D, Nordestgaard BG, Tybjaerg-Hansen A, Bojesen SE. CYP1B1 genotype and risk of cardiovascular disease, pulmonary disease, and cancer in 50,000 individuals. Pharmacogenet Genomics. 2009;19(9):685–694. doi: 10.1097/FPC.0b013e32833042cb. [DOI] [PubMed] [Google Scholar]
- 9.Korytina GF, Akhmadishina LZ, Viktorova TV. The CYP1B1 and CYP2F1 genes polymorphisms frequency in three ethnic groups of Bashkortostan and chronic obstructive pulmonary disease patients. Mol Biol (Mosk) 2010;44(1):33–41. Russian. [PubMed] [Google Scholar]
- 10.Arif E, Vibhuti A, Alam P, et al. Association of CYP2E1 and NAT2 gene polymorphisms with chronic obstructive pulmonary disease. Clin Chim Acta. 2007;382(1–2):37–42. doi: 10.1016/j.cca.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 11.van Diemen CC, Postma DS, Vonk JM, Bruinenberg M, Nolte IM, Boezen HM. Decorin and TGF-beta1 polymorphisms and development of COPD in a general population. Respir Res. 2006;7:89. doi: 10.1186/1465-9921-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins GA, Lazarus R, Smith RS, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123(6):1376–1383. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taille C, Guenegou A, Almolki A, et al. ETB receptor polymorphism is associated with airway obstruction. BMC Pulm Med. 2007;7:5. doi: 10.1186/1471-2466-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sunyer J, Pistelli R, Plana E, et al. Systemic inflammation, genetic susceptibility and lung function. Eur Respir J. 2008;32(1):92–97. doi: 10.1183/09031936.00052507. [DOI] [PubMed] [Google Scholar]
- 15.Yanbaeva DG, Dentener MA, Spruit MA, et al. IL6 and CRP haplotypes are associated with COPD risk and systemic inflammation: a case-control study. BMC Med Genet. 2009;10:23. doi: 10.1186/1471-2350-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho MH, Klanderman BJ, Litonjua AA, Sparrow D, Silverman EK, Raby BA. Folliculin mutations are not associated with severe COPD. BMC Med Genet. 2008;9:120. doi: 10.1186/1471-2350-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto T, Yoh K, Kobayashi A, et al. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem Biophys Res Commun. 2004;321(1):72–79. doi: 10.1016/j.bbrc.2004.06.112. [DOI] [PubMed] [Google Scholar]
- 18.Korytina GF, Akhmadishina LZ, Tselousova OS, Zagidullin Sh Z, Viktorova TV. Polymorphism of the genes for antioxidant defense enzymes and their association with the development of chronic obstructive pulmonary disease in the population of Bashkortostan. Genetika. 2009;45(7):967–976. Russian. [PubMed] [Google Scholar]
- 19.Yang IA, Holz O, Jorres RA, et al. Association of tumor necrosis factor-alpha polymorphisms and ozone-induced change in lung function. Am J Respir Crit Care Med. 2005;171(2):171–176. doi: 10.1164/rccm.200402-194OC. [DOI] [PubMed] [Google Scholar]
- 20.Shukla RK, Kant S, Bhattacharya S, Mittal B. Association of genetic polymorphism of GSTT1, GSTM1 and GSTM3 in COPD patients in a north Indian population. COPD. 2011;8(3):167–172. doi: 10.3109/15412555.2011.560128. [DOI] [PubMed] [Google Scholar]
- 21.Xie J, Xu Y, Zhang Z, Ni W, Chen S. Heat shock protein 70 gene polymorphisms in Han nationality of China with chronic obstructive pulmonary diseases. J Huazhong Univ Sci Technolog Med Sci. 2004;24(1):28–31. doi: 10.1007/BF02830699. [DOI] [PubMed] [Google Scholar]
- 22.Beghe B, Hall IP, Parker SG, et al. Polymorphisms in IL13 pathway genes in asthma and chronic obstructive pulmonary disease. Allergy. 2010;65(4):474–481. doi: 10.1111/j.1398-9995.2009.02167.x. [DOI] [PubMed] [Google Scholar]
- 23.Li LJ, Gao LB, Lv ML, et al. Association between SNPs in pre-miRNA and risk of chronic obstructive pulmonary disease. Clin Biochem. 2011;44(10–11):813–816. doi: 10.1016/j.clinbiochem.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 24.Zhu G, Warren L, Aponte J, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176(2):167–173. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 25.Galanter J, Choudhry S, Eng C, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177(11):1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Li B, Zhou Y, Zhong N, Ran P. Genetic analysis of CC16, OGG1 and GCLC polymorphisms and susceptibility to COPD. Respirology. 2007;12(1):29–33. doi: 10.1111/j.1440-1843.2006.00958.x. [DOI] [PubMed] [Google Scholar]
- 27.Young RP, Hopkins R, Black PN, et al. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax. 2006;61(5):394–399. doi: 10.1136/thx.2005.048512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raby BA, Hwang ES, Van Steen K, et al. T-bet polymorphisms are associated with asthma and airway hyperresponsiveness. Am J Respir Crit Care Med. 2006;173(1):64–70. doi: 10.1164/rccm.200503-505OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pabst S, Yenice V, Lennarz M, et al. Toll-like receptor 2 gene polymorphisms Arg677Trp and Arg753Gln in chronic obstructive pulmonary disease. Lung. 2009;187(3):173–178. doi: 10.1007/s00408-009-9144-8. [DOI] [PubMed] [Google Scholar]
- 30.Speletas M, Merentiti V, Kostikas K, et al. Association of TLR4-T399I polymorphism with chronic obstructive pulmonary disease in smokers. Clin Dev Immunol. 2009;2009:260286. doi: 10.1155/2009/260286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffjan S, Stemmler S, Parwez Q, et al. Evaluation of the toll-like receptor 6 Ser249Pro polymorphism in patients with asthma, atopic dermatitis and chronic obstructive pulmonary disease. BMC Med Genet. 2005;6:34. doi: 10.1186/1471-2350-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrarotti I, Zorzetto M, Beccaria M, et al. Tumour necrosis factor family genes in a phenotype of COPD associated with emphysema. Eur Respir J. 2003 Mar;21(3):444–449. doi: 10.1183/09031936.03.00051303. [DOI] [PubMed] [Google Scholar]
Acknowledgment
Yohan Bossé is a research scholar from the Heart and Stroke Foundation of Canada.
Footnotes
Disclosure
The author reports no conflict of interest in this work.
References
- 1.Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990–2020. Global Burden of Disease Study. Lancet. 1997;349(9064):1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 2.Rabe KF, Hurd S, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176(6):532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 3.Halbert RJ, Natoli JL, Gano A, Badamgarav E, Buist AS, Mannino DM. Global burden of COPD: systematic review and meta-analysis. Eur Respir J. 2006;28(3):523–532. doi: 10.1183/09031936.06.00124605. [DOI] [PubMed] [Google Scholar]
- 4.Bossé Y. Genetics of chronic obstructive pulmonary disease: a succinct review, future avenues and prospective clinical applications. Pharmacogenomics. 2009;10(4):655–667. doi: 10.2217/pgs.09.10. [DOI] [PubMed] [Google Scholar]
- 5.Castaldi PJ, Cho MH, Cohn M, et al. The COPD genetic association compendium: a comprehensive online database of COPD genetic associations. Hum Mol Genet. 2010;19(3):526–534. doi: 10.1093/hmg/ddp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanock SJ, Manolio T, Boehnke M, et al. Replicating genotype-phenotype associations. Nature. 2007;447(7145):655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 7.Hu G, Yao W, Zhou Y, et al. Meta- and pooled analyses of the effect of glutathione S-transferase M1 and T1 deficiency on chronic obstructive pulmonary disease. Int J Tuberc Lung Dis. 2008;12(12):1474–1481. [PubMed] [Google Scholar]
- 8.Smolonska J, Wijmenga C, Postma DS, Boezen HM. Meta-analyses on suspected chronic obstructive pulmonary disease genes: a summary of 20 years’ research. Am J Respir Crit Care Med. 2009;180(7):618–631. doi: 10.1164/rccm.200905-0722OC. [DOI] [PubMed] [Google Scholar]
- 9.Zhan P, Wang J, Wei SZ, et al. TNF-308 gene polymorphism is associated with COPD risk among Asians: meta-analysis of data for 6,118 subjects. Mol Biol Rep. 2011;38(1):219–227. doi: 10.1007/s11033-010-0098-y. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, Wang C, Xi B, Li X. Association between the tumour necrosis factor-alpha-308G/A polymorphism and chronic obstructive pulmonary disease: an update. Respirology. 2011;16(1):107–115. doi: 10.1111/j.1440-1843.2010.01879.x. [DOI] [PubMed] [Google Scholar]
- 11.Gingo MR, Silveira LJ, Miller YE, et al. Tumour necrosis factor gene polymorphisms are associated with COPD. Eur Respir J. 2008;31(5):1005–1012. doi: 10.1183/09031936.00100307. [DOI] [PubMed] [Google Scholar]
- 12.Obeidat M, Wain LV, Shrine N, et al. A comprehensive evaluation of potential lung function associated genes in the SpiroMeta general population sample. PLoS One. 2011;6(5):e19382. doi: 10.1371/journal.pone.0019382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong X, Zhou H, Tsai HJ, et al. Cysteinyl leukotriene receptor 1 gene variation and risk of asthma. Eur Respir J. 2009;33(1):42–48. doi: 10.1183/09031936.00057708. [DOI] [PubMed] [Google Scholar]
- 14.Hosgood HD, 3rd, Menashe I, He X, Chanock S, Lan Q. PTEN identified as important risk factor of chronic obstructive pulmonary disease. Respir Med. 2009;103(12):1866–1870. doi: 10.1016/j.rmed.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5(3):e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 19.Cho MH, Boutaoui N, Klanderman BJ, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42(3):200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho MH, Castaldi PJ, Wan ES, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21(4):947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timofeeva MN, McKay JD, Smith GD, et al. Genetic polymorphisms in 15q25 and 19q13 loci, cotinine levels, and risk of lung cancer in EPIC. Cancer Epidemiol Biomarkers Prev. 2011;20(10):2250–2261. doi: 10.1158/1055-9965.EPI-11-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keskitalo K, Broms U, Heliovaara M, et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet. 2009;18(20):4007–4012. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JZ, Tozzi F, Waterworth DM, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillai SG, Kong X, Edwards LD, et al. Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(12):1498–1505. doi: 10.1164/rccm.201002-0151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Broderick P, Matakidou A, Eisen T, Houlston RS. Role of 5p15.33 (TERT-CLPTM1L), 6p21.33 and 15q25.1 (CHRNA5- CHRNA3) variation and lung cancer risk in never-smokers. Carcinogenesis. 2010;31(2):234–238. doi: 10.1093/carcin/bgp287. [DOI] [PubMed] [Google Scholar]
- 28.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100(21):1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85(5):679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chappell SL, Daly L, Lotya J, et al. The role of IREB2 and transforming growth factor beta-1 genetic variants in COPD: a replication case-control study. BMC Med Genet. 2011;12:24. doi: 10.1186/1471-2350-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saccone NL, Culverhouse RC, Schwantes-An TH, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010 Aug;6(8):e1001053. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69(16):6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JC, Cruchaga C, Saccone NL, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18(16):3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang JC, Grucza R, Cruchaga C, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14(5):501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falvella FS, Galvan A, Frullanti E, et al. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin Cancer Res. 2009;15(5):1837–1842. doi: 10.1158/1078-0432.CCR-08-2107. [DOI] [PubMed] [Google Scholar]
- 36.Qiu W, Cho MH, Riley JH, et al. Genetics of sputum gene expression in chronic obstructive pulmonary disease. PLoS One. 2011;6(9):e24395. doi: 10.1371/journal.pone.0024395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilk JB, Walter RE, Laramie JM, Gottlieb DJ, O’Connor GT. Framingham Heart Study genome-wide association: results for pulmonary function measures. BMC Med Genet. 2007;8(Suppl 1):S8. doi: 10.1186/1471-2350-8-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilk JB, Chen TH, Gottlieb DJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5(3):e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42(1):45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Repapi E, Sayers I, Wain LV, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42(1):36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soler Artigas M, Loth DW, Wain LV, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43(11):1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imboden M, Bouzigon E, Curjuric I, et al. Genome-wide association study of lung function decline in adults with and without asthma. J Allergy Clin Immunol. 2012;129(5):1218–1228. doi: 10.1016/j.jaci.2012.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong X, Cho MH, Anderson W, et al. Genome-wide association study identifies BICD1 as a susceptibility gene for emphysema. Am J Respir Crit Care Med. 2011;183(1):43–49. doi: 10.1164/rccm.201004-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan ES, Cho MH, Boutaoui N, et al. Genome-wide association analysis of body mass in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2011;45(2):304–310. doi: 10.1165/rcmb.2010-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverman EK, Vestbo J, Agusti A, et al. Opportunities and challenges in the genetics of COPD 2010: an International COPD Genetics Conference report. COPD. 2011;8(2):121–135. doi: 10.3109/15412555.2011.558864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bossé Y, Sin D, Laviolette M, et al. Hypothesis-driven research on genomic data derived from a large scale lung eQTL mapping study. Webmed Central Lung. 2010;1(9):WMC00724. [Google Scholar]
- 47.DeMeo DL, Mariani T, Bhattacharya S, et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am J Hum Genet. 2009;85(4):493–502. doi: 10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brehm JM, Hagiwara K, Tesfaigzi Y, et al. Identification of FGF7 as a novel susceptibility locus for chronic obstructive pulmonary disease. Thorax. 2011;66(12):1085–1090. doi: 10.1136/thoraxjnl-2011-200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bossé Y. Highlights from the latest articles in chronic obstructive pulmonary disease genetics. Per Med. 2012;9(2):181–184. doi: 10.2217/pme.13.3. [DOI] [PubMed] [Google Scholar]
- 50.Rao DC. Genetic dissection of complex traits: an overview. Adv Genet. 2001;42:13–34. doi: 10.1016/s0065-2660(01)42012-8. [DOI] [PubMed] [Google Scholar]
- 51.Poller W, Barth J, Voss B. Detection of an alteration of the alpha 2-macroglobulin gene in a patient with chronic lung disease and serum alpha 2-macroglobulin deficiency. Hum Genet. 1989;83(1):93–96. doi: 10.1007/BF00274157. [DOI] [PubMed] [Google Scholar]
- 52.Siedlinski M, Boezen HM, Boer JM, Smit HA, Postma DS. ABCC1 polymorphisms contribute to level and decline of lung function in two population-based cohorts. Pharmacogenet Genomics. 2009;19(9):675–684. doi: 10.1097/FPC.0b013e32832f5eff. [DOI] [PubMed] [Google Scholar]
- 53.Budulac SE, Postma DS, Hiemstra PS, et al. Multidrug resistance-associated protein-1 (MRP1) genetic variants, MRP1 protein levels and severity of COPD. Respir Res. 2010;11:60. doi: 10.1186/1465-9921-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dogan OT, Katrancioglu N, Karahan O, Sanli GC, Zorlu A, Manduz S. Frequency of the mdr-1 C > T gene polymorphism in patients with COPD. Clinics (Sao Paulo) 2010;65(11):1115–1117. doi: 10.1590/S1807-59322010001100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Busquets X, MacFarlane NG, Heine-Suner D, et al. Angiotensin-converting- enzyme gene polymorphisms, smoking and chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2007;2(3):329–334. [PMC free article] [PubMed] [Google Scholar]
- 56.Pabst S, Theis B, Gillissen A, et al. Angiotensin-converting enzyme I/D polymorphism in chronic obstructive pulmonary disease. Eur J Med Res. 2009;14(Suppl 4):177–181. doi: 10.1186/2047-783X-14-S4-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahsan A, Ram R, Baig MA, Pasha MA. ACE I allele and eNOS G allele crosstalk may have a role in chronic obstructive pulmonary disease. Clin Biochem. 2004;37(11):1037–1040. doi: 10.1016/j.clinbiochem.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 58.Hopkinson NS, Nickol AH, Payne J, et al. Angiotensin converting enzyme genotype and strength in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170(4):395–399. doi: 10.1164/rccm.200304-578OC. [DOI] [PubMed] [Google Scholar]
- 59.Tkacova R, Joppa P, Stancak B, Salagovic J, Misikova S, Kalina I. The link between angiotensin-converting enzyme genotype and pulmonary artery pressure in patients with COPD. Wien Klin Wochenschr. 2005;117(5–6):210–214. doi: 10.1007/s00508-005-0333-z. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Wang C, Dai H, Lin Y, Zhang J. Association between angiotensin-converting enzyme gene polymorphisms and exercise performance in patients with COPD. Respirology. 2008;13(5):683–688. doi: 10.1111/j.1440-1843.2008.01325.x. [DOI] [PubMed] [Google Scholar]
- 61.Lee J, Nordestgaard BG, Dahl M. Elevated ACE activity is not associated with asthma, COPD, and COPD co-morbidity. Respir Med. 2009;103(9):1286–1292. doi: 10.1016/j.rmed.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Yildiz P, Oflaz H, Cine N, Erginel-Unaltuna N, Erzengin F, Yilmaz V. Gene polymorphisms of endothelial nitric oxide synthase enzyme associated with pulmonary hypertension in patients with COPD. Respir Med. 2003;97(12):1282–1288. doi: 10.1016/j.rmed.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 63.Jongepier H, Boezen HM, Dijkstra A, et al. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy. 2004;34(5):757–760. doi: 10.1111/j.1365-2222.2004.1938.x. [DOI] [PubMed] [Google Scholar]
- 64.Simpson A, Maniatis N, Jury F, et al. Polymorphisms in a disintegrin and metalloprotease 33 (ADAM33) predict impaired early-life lung function. Am J Respir Crit Care Med. 2005;172(1):55–60. doi: 10.1164/rccm.200412-1708OC. [DOI] [PubMed] [Google Scholar]
- 65.van Diemen CC, Postma DS, Vonk JM, Bruinenberg M, Schouten JP, Boezen HM. A disintegrin and metalloprotease 33 polymorphisms and lung function decline in the general population. Am J Respir Crit Care Med. 2005;172(3):329–333. doi: 10.1164/rccm.200411-1486OC. [DOI] [PubMed] [Google Scholar]
- 66.Sadeghnejad A, Ohar JA, Zheng SL, et al. Adam33 polymorphisms are associated with COPD and lung function in long-term tobacco smokers. Respir Res. 2009;10:21. doi: 10.1186/1465-9921-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Li L, Xiao J, et al. Association of ADAM33 gene polymorphisms with COPD in a northeastern Chinese population. BMC Med Genet. 2009;10:132. doi: 10.1186/1471-2350-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schedel M, Depner M, Schoen C, et al. The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respir Res. 2006;7:91. doi: 10.1186/1465-9921-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Diemen CC, Postma DS, Aulchenko YS, et al. Novel strategy to identify genetic risk factors for COPD severity: a genetic isolate. Eur Respir J. 2010;35(4):768–775. doi: 10.1183/09031936.00054408. [DOI] [PubMed] [Google Scholar]
- 70.Pabst S, Pizarro Touron C, Gillissen A, et al. ADAM33 gene polymorphisms in chronic obstructive pulmonary disease. Eur J Med Res. 2009;14(Suppl 4):182–186. doi: 10.1186/2047-783X-14-S4-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hegab AE, Sakamoto T, Saitoh W, et al. Polymorphisms of IL4, IL13, and ADRB2 genes in COPD. Chest. 2004;126(6):1832–1839. doi: 10.1378/chest.126.6.1832. [DOI] [PubMed] [Google Scholar]
- 72.Ho LI, Harn HJ, Chen CJ, Tsai NM. Polymorphism of the beta(2)-adrenoceptor in COPD in Chinese subjects. Chest. 2001;120(5):1493–1499. doi: 10.1378/chest.120.5.1493. [DOI] [PubMed] [Google Scholar]
- 73.Joos L, Weir TD, Connett JE, et al. Polymorphisms in the beta2 adrenergic receptor and bronchodilator response, bronchial hyperresponsiveness, and rate of decline in lung function in smokers. Thorax. 2003;58(8):703–707. doi: 10.1136/thorax.58.8.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim WJ, Oh YM, Sung J, et al. CT scanning-based phenotypes vary with ADRB2 polymorphisms in chronic obstructive pulmonary disease. Respir Med. 2009;103(1):98–103. doi: 10.1016/j.rmed.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 75.Vacca G, Schwabe K, Duck R, et al. Polymorphisms of the beta2 adrenoreceptor gene in chronic obstructive pulmonary disease. Ther Adv Respir Dis. 2009;3(1):3–10. doi: 10.1177/1753465809102553. [DOI] [PubMed] [Google Scholar]
- 76.Papatheodorou A, Makrythanasis P, Kaliakatsos M, et al. Development of novel microarray methodology for the study of mutations in the SERPINA1 and ADRB2 genes – their association with obstructive pulmonary disease and disseminated bronchiectasis in Greek patients. Clin Biochem. 2010;43(1–2):43–50. doi: 10.1016/j.clinbiochem.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 77.Kim WJ, Hoffman E, Reilly J, et al. Association of COPD candidate genes with computed tomography emphysema and airway phenotypes in severe COPD. Eur Respir J. 2011;37(1):39–43. doi: 10.1183/09031936.00173009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matheson MC, Ellis JA, Raven J, Johns DP, Walters EH, Abramson MJ. Beta2-adrenergic receptor polymorphisms are associated with asthma and COPD in adults. J Hum Genet. 2006;51(11):943–951. doi: 10.1007/s10038-006-0043-z. [DOI] [PubMed] [Google Scholar]
- 79.Leung TF, Chan IH, Wong GW, et al. Association between candidate genes and lung function growth in Chinese asthmatic children. Clin Exp Allergy. 2007;37(10):1480–1486. doi: 10.1111/j.1365-2222.2007.02799.x. [DOI] [PubMed] [Google Scholar]
- 80.Turner SW, Khoo SK, Laing IA, et al. Beta2 adrenoceptor Arg16Gly polymorphism, airway responsiveness, lung function and asthma in infants and children. Clin Exp Allergy. 2004;34(7):1043–1048. doi: 10.1111/j.1365-2222.2004.02001.x. [DOI] [PubMed] [Google Scholar]
- 81.Summerhill E, Leavitt SA, Gidley H, Parry R, Solway J, Ober C. Beta(2)-adrenergic receptor Arg16/Arg16 genotype is associated with reduced lung function, but not with asthma, in the Hutterites. Am J Respir Crit Care Med. 2000;162(2 Pt 1):599–602. doi: 10.1164/ajrccm.162.2.9910108. [DOI] [PubMed] [Google Scholar]
- 82.Silverman EK, Kwiatkowski DJ, Sylvia JS, et al. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J Allergy Clin Immunol. 2003;112(5):870–876. doi: 10.1016/s0091-6749(03)02023-2. [DOI] [PubMed] [Google Scholar]
- 83.Brogger J, Steen VM, Eiken HG, Gulsvik A, Bakke P. Genetic association between COPD and polymorphisms in TNF, ADRB2 and EPHX1. Eur Respir J. 2006;27(4):682–688. doi: 10.1183/09031936.06.00057005. [DOI] [PubMed] [Google Scholar]
- 84.Holloway JW, Barton SJ, Holgate ST, Rose-Zerilli MJ, Sayers I. The role of LTA4H and ALOX5AP polymorphism in asthma and allergy susceptibility. Allergy. 2008;63(8):1046–1053. doi: 10.1111/j.1398-9995.2008.01667.x. [DOI] [PubMed] [Google Scholar]
- 85.Ning Y, Ying B, Han S, Wang B, Wang X, Wen F. Polymorphisms of aquaporin5 gene in chronic obstructive pulmonary disease in a Chinese population. Swiss Med Wkly. 2008;138(39–40):573–578. doi: 10.4414/smw.2008.12240. [DOI] [PubMed] [Google Scholar]
- 86.Hansel NN, Sidhaye V, Rafaels NM, et al. Aquaporin 5 polymorphisms and rate of lung function decline in chronic obstructive pulmonary disease. PLoS One. 2010;5(12):e14226. doi: 10.1371/journal.pone.0014226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sata M, Takabatake N, Inoue S, et al. Intronic single-nucleotide polymorphisms in Bcl-2 are associated with chronic obstructive pulmonary disease severity. Respirology. 2007;12(1):34–41. doi: 10.1111/j.1440-1843.2006.00959.x. [DOI] [PubMed] [Google Scholar]
- 88.Hopkinson NS, Eleftheriou KI, Payne J, et al. +9/+9 Homozygosity of the bradykinin receptor gene polymorphism is associated with reduced fat-free mass in chronic obstructive pulmonary disease. Am J Clin Nutr. 2006;83(4):912–917. doi: 10.1093/ajcn/83.4.912. [DOI] [PubMed] [Google Scholar]
- 89.Smith AK, Lange LA, Ampleford EJ, Meyers DA, Bleecker ER, Howard TD. Association of polymorphisms in CASP10 and CASP8 with FEV(1)/FVC and bronchial hyperresponsiveness in ethnically diverse asthmatics. Clin Exp Allergy. 2008;38(11):1738–1744. doi: 10.1111/j.1365-2222.2008.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Korytina GF, Akhmadishina LZ, Tselousova OS, Zagidullin SZ, Viktorova TV. Polymorphism of the genes for antioxidant defense enzymes and their association with the development of chronic obstructive pulmonary disease in the population of Bashkortostan. Genetika. 2009;45(7):967–976. Russian. [PubMed] [Google Scholar]
- 91.Young RP, Hopkins R, Black PN, et al. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax. 2006;61(5):394–399. doi: 10.1136/thx.2005.048512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mak JC, Ho SP, Yu WC, et al. Polymorphisms and functional activity in superoxide dismutase and catalase genes in smokers with COPD. Eur Respir J. 2007;30(4):684–690. doi: 10.1183/09031936.00015207. [DOI] [PubMed] [Google Scholar]
- 93.Hizawa N, Makita H, Nasuhara Y, et al. Functional single nucleotide polymorphisms of the CCL5 gene and nonemphysematous phenotype in COPD patients. Eur Respir J. 2008;32(2):372–378. doi: 10.1183/09031936.00115307. [DOI] [PubMed] [Google Scholar]
- 94.Al-Abdulhadi SA, Al-Rabia MW. Association and preferential transmission of the CCR2V64I polymorphism with absence of asthma in high-risk families. Saudi Med J. 2008;29(12):1711–1718. [PubMed] [Google Scholar]
- 95.Zhou H, Alexis NE, Almond M, et al. Influence of C-159T SNP of the CD14 gene promoter on lung function in smokers. Respir Med. 2009;103(9):1358–1365. doi: 10.1016/j.rmed.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.LeVan TD, Von Essen S, Romberger DJ, et al. Polymorphisms in the CD14 gene associated with pulmonary function in farmers. Am J Respir Crit Care Med. 2005;171(7):773–779. doi: 10.1164/rccm.200404-530OC. [DOI] [PubMed] [Google Scholar]
- 97.Liu Y, Liang WB, Gao LB, Wang YY, Zhang L. Association of CD40-1C/T polymorphism in the 5′-untranslated region and chronic obstructive pulmonary disease. Clin Chim Acta. 2009;408(1–2):56–59. doi: 10.1016/j.cca.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 98.Zhang XZ, Pare PD, Sandford AJ. PMN degranulation in relation to CD63 expression and genetic polymorphisms in healthy individuals and COPD patients. Int J Mol Med. 2007;19(5):817–822. doi: 10.3892/ijmm.19.5.817. [DOI] [PubMed] [Google Scholar]
- 99.Liu Y, Liang WB, Gao LB, et al. CTLA4 and CD86 gene polymorphisms and susceptibility to chronic obstructive pulmonary disease. Hum Immunol. 2010;71(11):1141–1146. doi: 10.1016/j.humimm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 100.Takabatake N, Toriyama S, Igarashi A, et al. A novel polymorphism in CDC6 is associated with the decline in lung function of ex-smokers in COPD. Biochem Biophys Res Commun. 2009;381(4):554–559. doi: 10.1016/j.bbrc.2009.02.080. [DOI] [PubMed] [Google Scholar]
- 101.Lee YL, Chen W, Tsai WK, et al. Polymorphisms of p53 and p21 genes in chronic obstructive pulmonary disease. J Lab Clin Med. 2006;147(5):228–233. doi: 10.1016/j.lab.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 102.Bombieri C, Benetazzo M, Saccomani A, et al. Complete mutational screening of the CFTR gene in 120 patients with pulmonary disease. Hum Genet. 1998;103(6):718–722. doi: 10.1007/s004390050897. [DOI] [PubMed] [Google Scholar]
- 103.Dumur V, Lafitte JJ, Gervais R, et al. Abnormal distribution of cystic fibrosis delta F508 allele in adults with chronic bronchial hypersecretion. Lancet. 1990;335(8701):1340. doi: 10.1016/0140-6736(90)91216-w. [DOI] [PubMed] [Google Scholar]
- 104.Gervais R, Lafitte JJ, Dumur V, et al. Sweat chloride and delta F508 mutation in chronic bronchitis or bronchiectasis. Lancet. 1993;342(8877):997. doi: 10.1016/0140-6736(93)92046-v. [DOI] [PubMed] [Google Scholar]
- 105.Pignatti PF, Bombieri C, Benetazzo M, et al. CFTR gene variant IVS8-5T in disseminated bronchiectasis. Am J Hum Genet. 1996;58(4):889–892. [PMC free article] [PubMed] [Google Scholar]
- 106.Poller W, Faber JP, Scholz S, Olek K, Muller KM. Sequence analysis of the cystic fibrosis gene in patients with disseminated bronchiectatic lung disease. Application in the identification of a cystic fibrosis patient with atypical clinical course. Klin Wochenschr. 1991;69(14):657–663. doi: 10.1007/BF01649427. [DOI] [PubMed] [Google Scholar]
- 107.Tzetis M, Efthymiadou A, Strofalis S, et al. CFTR gene mutations – including three novel nucleotide substitutions – and haplotype background in patients with asthma, disseminated bronchiectasis and chronic obstructive pulmonary disease. Hum Genet. 2001;108(3):216–221. doi: 10.1007/s004390100467. [DOI] [PubMed] [Google Scholar]
- 108.Stankovic M, Nikolic A, Divac A, et al. The CFTR M470V gene variant as a potential modifier of COPD severity: study of Serbian population. Genet Test. 2008;12(3):357–362. doi: 10.1089/gte.2007.0069. [DOI] [PubMed] [Google Scholar]
- 109.Artlich A, Boysen A, Bunge S, Entzian P, Schlaak M, Schwinger E. Common CFTR mutations are not likely to predispose to chronic bronchitis in northern Germany. Hum Genet. 1995;95(2):226–228. doi: 10.1007/BF00209408. [DOI] [PubMed] [Google Scholar]
- 110.Gasparini P, Savoia A, Luisetti M, Peona V, Pignatti PF. The cystic fibrosis gene is not likely to be involved in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 1990;2(3):297–299. doi: 10.1165/ajrcmb/2.3.297. [DOI] [PubMed] [Google Scholar]
- 111.Ober C, Tan Z, Sun Y, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358(16):1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lambrechts D, Buysschaert I, Zanen P, et al. The 15q24/25 susceptibility variant for lung cancer and chronic obstructive pulmonary disease is associated with emphysema. Am J Respir Crit Care Med. 2010;181(5):486–493. doi: 10.1164/rccm.200909-1364OC. [DOI] [PubMed] [Google Scholar]
- 113.Young RP, Hopkins RJ, Hay BA, Epton MJ, Black PN, Gamble GD. Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur Respir J. 2008;32(5):1158–1164. doi: 10.1183/09031936.00093908. [DOI] [PubMed] [Google Scholar]
- 114.Hegab AE, Sakamoto T, Uchida Y, et al. CLCA1 gene polymorphisms in chronic obstructive pulmonary disease. J Med Genet. 2004;41(3):e27. doi: 10.1136/jmg.2003.012484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim KM, Park SH, Kim JS, et al. Polymorphisms in the type IV collagen alpha3 gene and the risk of COPD. Eur Respir J. 2008;32(1):35–41. doi: 10.1183/09031936.00076207. [DOI] [PubMed] [Google Scholar]
- 116.Sunyer J, Pistelli R, Plana E, et al. Systemic inflammation, genetic susceptibility and lung function. Eur Respir J. 2008;32(1):92–97. doi: 10.1183/09031936.00052507. [DOI] [PubMed] [Google Scholar]
- 117.van Durme YM, Verhamme KM, Aarnoudse AJ, et al. C-reactive protein levels, haplotypes, and the risk of incident chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;179(5):375–382. doi: 10.1164/rccm.200810-1540OC. [DOI] [PubMed] [Google Scholar]
- 118.Yanbaeva DG, Dentener MA, Spruit MA, et al. IL6 and CRP haplotypes are associated with COPD risk and systemic inflammation: a case-control study. BMC Med Genet. 2009;10:23. doi: 10.1186/1471-2350-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dahl M, Vestbo J, Zacho J, Lange P, Tybjaerg-Hansen A, Nordestgaard BG. C reactive protein and chronic obstructive pulmonary disease: a Mendelian randomisation approach. Thorax. 2011;66(3):197–204. doi: 10.1136/thx.2009.131193. [DOI] [PubMed] [Google Scholar]
- 120.Shen M, Vermeulen R, Chapman RS, et al. A report of cytokine polymorphisms and COPD risk in Xuan Wei, China. Int J Hyg Environ Health. 2008;211(3–4):352–356. doi: 10.1016/j.ijheh.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He JQ, Shumansky K, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Association of genetic variations in the CSF2 and CSF3 genes with lung function in smoking-induced COPD. Eur Respir J. 2008;32(1):25–34. doi: 10.1183/09031936.00040307. [DOI] [PubMed] [Google Scholar]
- 122.Zhu G, Agusti A, Gulsvik A, et al. CTLA4 gene polymorphisms are associated with chronic bronchitis. Eur Respir J. 2009;34(3):598–604. doi: 10.1183/09031936.00141808. [DOI] [PubMed] [Google Scholar]
- 123.Wood AM, Newby PR, Gough SC, Stockley RA. CTLA4 polymorphisms and COPD. Eur Respir J. 2010;35(2):457–458. doi: 10.1183/09031936.00134709. [DOI] [PubMed] [Google Scholar]
- 124.Minematsu N, Nakamura H, Furuuchi M, et al. Common functional polymorphisms in the cathepsin S promoter in Japanese subjects: possible contribution to pulmonary emphysema. Respirology. 2008;13(4):498–504. doi: 10.1111/j.1440-1843.2008.01280.x. [DOI] [PubMed] [Google Scholar]
- 125.Vibhuti A, Arif E, Mishra A, et al. CYP1A1, CYP1A2 and CYBA gene polymorphisms associated with oxidative stress in COPD. Clin Chim Acta. 2010;411(7–8):474–480. doi: 10.1016/j.cca.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 126.Cantlay AM, Lamb D, Gillooly M, et al. Association between the CYP1A1 gene polymorphism and susceptibility to emphysema and lung cancer. Clin Mol Pathol. 1995;48(4):M210–M214. doi: 10.1136/mp.48.4.m210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Korytina GF, Yanbaeva DG, Babenkova LI, Etkina EI, Victorova TV. Genetic polymorphisms in the cytochromes P-450 (1A1, 2E1), microsomal epoxide hydrolase and glutathione S-transferase M1, T1, and P1 genes, and their relationship with chronic bronchitis and relapsing pneumonia in children. J Mol Med. 2005;83(9):700–710. doi: 10.1007/s00109-005-0660-6. [DOI] [PubMed] [Google Scholar]
- 128.Cheng SL, Yu CJ, Yang PC. Genetic polymorphisms of cytochrome p450 and matrix metalloproteinase in chronic obstructive pulmonary disease. Biochem Genet. 2009;47(7–8):591–601. doi: 10.1007/s10528-009-9252-4. [DOI] [PubMed] [Google Scholar]
- 129.Korytina GF, Akhmadishina LZ, Kochetova OV, Zagidullin SZ, Viktorova TV. Association of cytochrome P450 genes polymorphisms (CYP1A1 and CYP1A2) with the development of chronic obstructive pulmonary disease in Bashkortostan. Mol Biol (Mosk) 2008;42(1):32–41. Russian. [PubMed] [Google Scholar]
- 130.Budhi A, Hiyama K, Isobe T, et al. Genetic susceptibility for emphysematous changes of the lung in Japanese. Int J Mol Med. 2003;11(3):321–329. [PubMed] [Google Scholar]
- 131.Uslu A, Ogus C, Ozdemir T, Bilgen T, Tosun O, Keser I. The effect of CYP1A2 gene polymorphisms on theophylline metabolism and chronic obstructive pulmonary disease in Turkish patients. BMB Rep. 2010;43(8):530–534. doi: 10.5483/bmbrep.2010.43.8.530. [DOI] [PubMed] [Google Scholar]
- 132.Arif E, Vibhuti A, Alam P, et al. Association of CYP2E1 and NAT2 gene polymorphisms with chronic obstructive pulmonary disease. Clin Chim Acta. 2007;382(1–2):37–42. doi: 10.1016/j.cca.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 133.Korytina GF, Akhmadishina LZ, Viktorova TV. The CYP1B1 and CYP2F1 genes polymorphisms frequency in three ethnic groups of Bashkortostan and chronic obstructive pulmonary disease patients. Mol Biol (Mosk) 2010;44(1):33–41. Russian. [PubMed] [Google Scholar]
- 134.Seo T, Pahwa P, McDuffie HH, et al. Association between cytochrome P450 3A5 polymorphism and the lung function in Saskatchewan grain workers. Pharmacogenet Genomics. 2008;18(6):487–493. doi: 10.1097/FPC.0b013e3282fb02ba. [DOI] [PubMed] [Google Scholar]
- 135.Hu RC, Xu YJ, Zhang ZX, Ni W, Chen SX. Correlation of HDEFB1 polymorphism and susceptibility to chronic obstructive pulmonary disease in Chinese Han population. Chin Med J (Engl) 2004;117(11):1637–1641. [PubMed] [Google Scholar]
- 136.Matsushita I, Hasegawa K, Nakata K, Yasuda K, Tokunaga K, Keicho N. Genetic variants of human beta-defensin-1 and chronic obstructive pulmonary disease. Biochem Biophys Res Commun. 2002;291(1):17–22. doi: 10.1006/bbrc.2002.6395. [DOI] [PubMed] [Google Scholar]
- 137.Hersh CP, DeMeo DL, Raby BA, et al. Genetic linkage and association analysis of COPD-related traits on chromosome 8p. COPD. 2006;3(4):189–194. doi: 10.1080/15412550601009321. [DOI] [PubMed] [Google Scholar]
- 138.Janssens W, Nuytten H, Dupont LJ, et al. Genomic copy number determines functional expression of {beta}-defensin 2 in airway epithelial cells and associates with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(2):163–169. doi: 10.1164/rccm.200905-0767OC. [DOI] [PubMed] [Google Scholar]
- 139.Sampsonas F, Antonacopoulou A, Spathas D, et al. Positive association between two polymorphic sites (+134 insA/delA and G198T) of the endothelin-1 gene and chronic obstructive pulmonary disease. A case-control study. Respir Med. 2010;104(1):114–120. doi: 10.1016/j.rmed.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 140.Kaparianos A, Argyropoulou E, Efremidis G, Flordellis C, Spiropoulos K. Decline in FEV1 related to genetic polymorphisms (+138 insA/delA and Lys198Asn) of the endothelin-1 gene in COPD. A pilot study. Eur Rev Med Pharmacol Sci. 2010;14(8):705–719. [PubMed] [Google Scholar]
- 141.Kaparianos A, Sampsonas F, Lykouras D, et al. Association of ET-1 gene polymorphisms with COPD phenotypes in a Caucasian population. Monaldi Arch Chest Dis. 2011;75(2):126–131. doi: 10.4081/monaldi.2011.225. [DOI] [PubMed] [Google Scholar]
- 142.Sampsonas E, Lykouras D, Drakatos P, Moschopoulou A, Spiropoulos K, Karkoulias K. Endothelin-1 polymorphisms involved in impaired exercise tolerance in COPD patients. A pilot study. Eur Rev Med Pharmacol Sci. 2011;15(2):123–128. [PubMed] [Google Scholar]
- 143.Taille C, Guenegou A, Almolki A, et al. ETB receptor polymorphism is associated with airway obstruction. BMC Pulm Med. 2007;7:5. doi: 10.1186/1471-2466-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kelleher CM, Silverman EK, Broekelmann T, et al. A functional mutation in the terminal exon of elastin in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2005;33(4):355–362. doi: 10.1165/rcmb.2005-0206OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cho MH, Ciulla DM, Klanderman BJ, et al. Analysis of exonic elastin variants in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2009;40(6):751–755. doi: 10.1165/rcmb.2008-0340OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.DeMeo DL, Hersh CP, Hoffman EA, et al. Genetic determinants of emphysema distribution in the national emphysema treatment trial. Am J Respir Crit Care Med. 2007;176(1):42–48. doi: 10.1164/rccm.200612-1797OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hersh CP, Demeo DL, Lazarus R, et al. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(9):977–984. doi: 10.1164/rccm.200509-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cheng SL, Yu CJ, Chen CJ, Yang PC. Genetic polymorphism of epoxide hydrolase and glutathione S-transferase in COPD. Eur Respir J. 2004;23(6):818–824. doi: 10.1183/09031936.04.00104904. [DOI] [PubMed] [Google Scholar]
- 149.Demeo DL, Campbell EJ, Barker AF, et al. IL10 polymorphisms are associated with airflow obstruction in severe alpha1-antitrypsin deficiency. Am J Respir Cell Mol Biol. 2008;38(1):114–120. doi: 10.1165/rcmb.2007-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hang J, Zhou W, Wang X, et al. Microsomal epoxide hydrolase, endotoxin, and lung function decline in cotton textile workers. Am J Respir Crit Care Med. 2005;171(2):165–170. doi: 10.1164/rccm.200407-888OC. [DOI] [PubMed] [Google Scholar]
- 151.Hersh CP, Demeo DL, Lange C, et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am J Respir Cell Mol Biol. 2005;33(1):71–78. doi: 10.1165/rcmb.2005-0073OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hersh CP, DeMeo DL, Reilly JJ, Silverman EK. Xenobiotic metabolizing enzyme gene polymorphisms predict response to lung volume reduction surgery. Respir Res. 2007;8:59. doi: 10.1186/1465-9921-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Koyama H, Geddes DM. Genes, oxidative stress, and the risk of chronic obstructive pulmonary disease. Thorax. 1998;53(Suppl 2):S10–S14. doi: 10.1136/thx.53.2008.s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Park JY, Chen L, Wadhwa N, Tockman MS. Polymorphisms for microsomal epoxide hydrolase and genetic susceptibility to COPD. Int J Mol Med. 2005;15(3):443–448. [PMC free article] [PubMed] [Google Scholar]
- 155.Sandford AJ, Chagani T, Weir TD, Connett JE, Anthonisen NR, Pare PD. Susceptibility genes for rapid decline of lung function in the lung health study. Am J Respir Crit Care Med. 2001;163(2):469–473. doi: 10.1164/ajrccm.163.2.2006158. [DOI] [PubMed] [Google Scholar]
- 156.Smith CA, Harrison DJ. Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema. Lancet. 1997;350(9078):630–633. doi: 10.1016/S0140-6736(96)08061-0. [DOI] [PubMed] [Google Scholar]
- 157.Vibhuti A, Arif E, Deepak D, Singh B, Qadar Pasha MA. Genetic polymorphisms of GSTP1 and mEPHX correlate with oxidative stress markers and lung function in COPD. Biochem Biophys Res Commun. 2007;359(1):136–142. doi: 10.1016/j.bbrc.2007.05.076. [DOI] [PubMed] [Google Scholar]
- 158.Xiao D, Wang C, Du MJ, et al. Association between polymorphisms in the microsomal epoxide hydrolase (mEH) gene and chronic obstructive pulmonary disease. Zhonghua Yi Xue Za Zhi. 2003;83(20):1782–1786. Chinese. [PubMed] [Google Scholar]
- 159.Xiao D, Wang C, Du MJ, et al. Relationship between polymorphisms of genes encoding microsomal epoxide hydrolase and glutathione S-transferase P1 and chronic obstructive pulmonary disease. Chin Med J (Engl) 2004;117(5):661–667. [PubMed] [Google Scholar]
- 160.Yoshikawa M, Hiyama K, Ishioka S, Maeda H, Maeda A, Yamakido M. Microsomal epoxide hydrolase genotypes and chronic obstructive pulmonary disease in Japanese. Int J Mol Med. 2000;5(1):49–53. doi: 10.3892/ijmm.5.1.49. [DOI] [PubMed] [Google Scholar]
- 161.Zidzik J, Slaba E, Joppa P, et al. Glutathione S-transferase and microsomal epoxide hydrolase gene polymorphisms and risk of chronic obstructive pulmonary disease in Slovak population. Croat Med J. 2008;49(2):182–191. doi: 10.3325/cmj.2008.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lakhdar R, Denden S, Knani J, et al. Microsomal epoxide hydrolase gene polymorphisms and susceptibility to chronic obstructive pulmonary disease in the Tunisian population. Genet Test Mol Biomarkers. 2010;14(6):857–863. doi: 10.1089/gtmb.2009.0168. [DOI] [PubMed] [Google Scholar]
- 163.Penyige A, Poliska S, Csanky E, et al. Analyses of association between PPAR gamma and EPHX1 polymorphisms and susceptibility to COPD in a Hungarian cohort, a case-control study. BMC Med Genet. 2010;11:152. doi: 10.1186/1471-2350-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lakhdar R, Denden S, Mouhamed MH, et al. Correlation of EPHX1, GSTP1, GSTM1, and GSTT1 genetic polymorphisms with antioxidative stress markers in chronic obstructive pulmonary disease. Exp Lung Res. 2011;37(4):195–204. doi: 10.3109/01902148.2010.535093. [DOI] [PubMed] [Google Scholar]
- 165.Lakhdar R, Denden S, Knani J, et al. Combined analysis of EPHX1, GSTP1, GSTM1 and GSTT1 gene polymorphisms in relation to chronic obstructive pulmonary disease risk and lung function impairment. Dis Markers. 2011;30(5):253–263. doi: 10.3233/DMA-2011-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fu WP, Sun C, Dai LM, Yang LF, Zhang YP. Relationship between COPD and polymorphisms of HOX-1 and mEPH in a Chinese population. Oncol Rep. 2007;17(2):483–488. [PubMed] [Google Scholar]
- 167.Rodriguez F, Jardi R, Costa X, et al. Detection of polymorphisms at exons 3 (Tyr113-->His) and 4 (His139-->Arg) of the microsomal epoxide hydrolase gene using fluorescence PCR method combined with melting curves analysis. Anal Biochem. 2002;308(1):120–126. doi: 10.1016/s0003-2697(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 168.Matheson MC, Raven J, Walters EH, Abramson MJ, Ellis JA. Microsomal epoxide hydrolase is not associated with COPD in a community-based sample. Hum Biol. 2006;78(6):705–717. doi: 10.1353/hub.2007.0015. [DOI] [PubMed] [Google Scholar]
- 169.Yim JJ, Park GY, Lee CT, et al. Genetic susceptibility to chronic obstructive pulmonary disease in Koreans: combined analysis of polymorphic genotypes for microsomal epoxide hydrolase and glutathione S-transferase M1 and T1. Thorax. 2000;55(2):121–125. doi: 10.1136/thorax.55.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang R, Zhang A, He Q, Lu B. Microsomal epoxide hydrolase gene polymorphism and susceptibility to chronic obstructive pulmonary disease in Han nationality of North China. Zhonghua Nei Ke Za Zhi. 2002;41(1):11–14. Chinese. [PubMed] [Google Scholar]
- 171.Foreman MG, DeMeo DL, Hersh CP, et al. Polymorphic variation in surfactant protein B is associated with COPD exacerbations. Eur Respir J. 2008;32(4):938–944. doi: 10.1183/09031936.00040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chappell S, Daly L, Morgan K, et al. Genetic variants of microsomal epoxide hydrolase and glutamate-cysteine ligase in COPD. Eur Respir J. 2008;32(4):931–937. doi: 10.1183/09031936.00065308. [DOI] [PubMed] [Google Scholar]
- 173.Siedlinski M, Postma DS, Smit HA, Boezen HM. No effects of EPHX1 polymorphisms on the level or change of FEV1 in the general population. Eur Respir J. 2009;33(2):446–449. doi: 10.1183/09031936.00144108. [DOI] [PubMed] [Google Scholar]
- 174.Takeyabu K, Yamaguchi E, Suzuki I, Nishimura M, Hizawa N, Kamakami Y. Gene polymorphism for microsomal epoxide hydrolase and susceptibility to emphysema in a Japanese population. Eur Respir J. 2000;15(5):891–894. doi: 10.1034/j.1399-3003.2000.15e13.x. [DOI] [PubMed] [Google Scholar]
- 175.Hu G, Shi Z, Hu J, Zou G, Peng G, Ran P. Association between polymorphisms of microsomal epoxide hydrolase and COPD: results from meta-analyses. Respirology. 2008;13(6):837–850. doi: 10.1111/j.1440-1843.2008.01356.x. [DOI] [PubMed] [Google Scholar]
- 176.Lee J, Nordestgaard BG, Dahl M. EPHX1 polymorphisms, COPD and asthma in 47,000 individuals and in meta-analysis. Eur Respir J. 2011;37(1):18–25. doi: 10.1183/09031936.00012110. [DOI] [PubMed] [Google Scholar]
- 177.Dijkstra A, Howard TD, Vonk JM, et al. Estrogen receptor 1 polymorphisms are associated with airway hyperresponsiveness and lung function decline, particularly in female subjects with asthma. J Allergy Clin Immunol. 2006;117(3):604–611. doi: 10.1016/j.jaci.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 178.Klar J, Blomstrand P, Brunmark C, et al. Fibroblast growth factor 10 haploinsufficiency causes chronic obstructive pulmonary disease. J Med Genet. 2011;48(10):705–709. doi: 10.1136/jmedgenet-2011-100166. [DOI] [PubMed] [Google Scholar]
- 179.Horne SL, Cockcroft DW, Dosman JA. Possible protective effect against chronic obstructive airways disease by the GC2 allele. Hum Hered. 1990;40(3):173–176. doi: 10.1159/000153926. [DOI] [PubMed] [Google Scholar]
- 180.Ishii T, Keicho N, Teramoto S, et al. Association of Gc-globulin variation with susceptibility to COPD and diffuse panbronchiolitis. Eur Respir J. 2001;18(5):753–757. doi: 10.1183/09031936.01.00094401. [DOI] [PubMed] [Google Scholar]
- 181.Ito I, Nagai S, Hoshino Y, et al. Risk and severity of COPD is associated with the group-specific component of serum globulin 1F allele. Chest. 2004;125(1):63–70. doi: 10.1378/chest.125.1.63. [DOI] [PubMed] [Google Scholar]
- 182.Laufs J, Andrason H, Sigvaldason A, et al. Association of vitamin D binding protein variants with chronic mucus hypersecretion in Iceland. Am J Pharmacogenomics. 2004;4(1):63–68. doi: 10.2165/00129785-200404010-00007. [DOI] [PubMed] [Google Scholar]
- 183.Lu M, Yang B, Cai YY. The relationship between vitamin D binding protein gene polymorphism and chronic obstructive pulmonary disease. Zhonghua Nei Ke Za Zhi. 2004;43(2):117–120. Chinese. [PubMed] [Google Scholar]
- 184.Schellenberg D, Pare PD, Weir TD, Spinelli JJ, Walker BA, Sandford AJ. Vitamin D binding protein variants and the risk of COPD. Am J Respir Crit Care Med. 1998;157(3 Pt 1):957–961. doi: 10.1164/ajrccm.157.3.9706106. [DOI] [PubMed] [Google Scholar]
- 185.Bakke PS, Zhu G, Gulsvik A, et al. Candidate genes for COPD in two large data sets. Eur Respir J. 2011;37(2):255–263. doi: 10.1183/09031936.00091709. [DOI] [PubMed] [Google Scholar]
- 186.Shen LH, Zhang XM, Su DJ, et al. Association of vitamin D binding protein variants with susceptibility to chronic obstructive pulmonary disease. J Int Med Res. 2010;38(3):1093–1098. doi: 10.1177/147323001003800337. [DOI] [PubMed] [Google Scholar]
- 187.Korytina GF, Akhmadishina LZ, Ianbaeva DG, Viktorova TV. Genotypes of vitamin-D-binding protein (DBP) in patients with chronic obstructive pulmonary disease and healthy population of Republic Bashkortostan. Mol Biol (Mosk) 2006;40(2):231–238. Russian. [PubMed] [Google Scholar]
- 188.Siedlinski M, Postma DS, van Diemen CC, Blokstra A, Smit HA, Boezen HM. Lung function loss, smoking, vitamin C intake, and polymorphisms of the glutamate-cysteine ligase genes. Am J Respir Crit Care Med. 2008;178(1):13–19. doi: 10.1164/rccm.200711-1749OC. [DOI] [PubMed] [Google Scholar]
- 189.Liu S, Li B, Zhou Y, Zhong N, Ran P. Genetic analysis of CC16, OGG1 and GCLC polymorphisms and susceptibility to COPD. Respirology. 2007;12(1):29–33. doi: 10.1111/j.1440-1843.2006.00958.x. [DOI] [PubMed] [Google Scholar]
- 190.Hersh CP, Hansel NN, Barnes KC, et al. Transforming growth factor-beta receptor-3 is associated with pulmonary emphysema. Am J Respir Cell Mol Biol. 2009;41(3):324–331. doi: 10.1165/rcmb.2008-0427OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Soler Artigas M, Wain LV, Repapi E, et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med. 2011;184(7):786–795. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Baranova H, Perriot J, Albuisson E, et al. Peculiarities of the GSTM1 0/0 genotype in French heavy smokers with various types of chronic bronchitis. Hum Genet. 1997;99(6):822–826. doi: 10.1007/s004390050455. [DOI] [PubMed] [Google Scholar]
- 193.Calikoglu M, Tamer L, Ates Aras N, Karakas S, Ercan B. The association between polymorphic genotypes of glutathione S-transferases and COPD in the Turkish population. Biochem Genet. 2006;44(7–8):307–319. doi: 10.1007/s10528-006-9031-4. [DOI] [PubMed] [Google Scholar]
- 194.Gilliland FD, Gauderman WJ, Vora H, Rappaport E, Dubeau L. Effects of glutathione-S-transferase M1, T1, and P1 on childhood lung function growth. Am J Respir Crit Care Med. 2002;166(5):710–716. doi: 10.1164/rccm.2112065. [DOI] [PubMed] [Google Scholar]
- 195.Harrison DJ, Cantlay AM, Rae F, Lamb D, Smith CA. Frequency of glutathione S-transferase M1 deletion in smokers with emphysema and lung cancer. Hum Exp Toxicol. 1997;16(7):356–360. doi: 10.1177/096032719701600703. [DOI] [PubMed] [Google Scholar]
- 196.He JQ, Ruan J, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Antioxidant gene polymorphisms and susceptibility to a rapid decline in lung function in smokers. Am J Respir Crit Care Med. 2002;166(3):323–328. doi: 10.1164/rccm.2111059. [DOI] [PubMed] [Google Scholar]
- 197.Imboden M, Downs SH, Senn O, et al. Glutathione S-transferase genotypes modify lung function decline in the general population: SAPALDIA cohort study. Respir Res. 2007;8:2. doi: 10.1186/1465-9921-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Faramawy MM, Mohammed TO, Hossaini AM, Kashem RA, Abu Rahma RM. Genetic polymorphism of GSTT1 and GSTM1 and susceptibility to chronic obstructive pulmonary disease (COPD) J Crit Care. 2009;24(3):e7–e10. doi: 10.1016/j.jcrc.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 199.Lakhdar R, Denden S, Knani J, et al. Association of GSTM1 and GSTT1 polymorphisms with chronic obstructive pulmonary disease in a Tunisian population. Biochem Genet. 2010;48(7–8):647–657. doi: 10.1007/s10528-010-9346-z. [DOI] [PubMed] [Google Scholar]
- 200.Shukla RK, Kant S, Bhattacharya S, Mittal B. Association of genetic polymorphism of GSTT1, GSTM1 and GSTM3 in COPD patients in a north Indian population. COPD. 2011;8(3):167–172. doi: 10.3109/15412555.2011.560128. [DOI] [PubMed] [Google Scholar]
- 201.Tkacova R, Salagovic J, Ceripkova M, Tkac I, Stubna J, Kalina I. Glutathione S-transferase M1 gene polymorphism is related to COPD in patients with non-small-cell lung cancer. Wien Klin Wochenschr. 2004;116(4):131–134. doi: 10.1007/BF03040750. [DOI] [PubMed] [Google Scholar]
- 202.Yanchina ED, Ivchik TV, Shvarts EI, Kokosov AN, Khodzhayantz NE. Gene-gene interactions between glutathione-s transferase M1 and matrix metalloproteinase 9 in the formation of hereditary predisposition to chronic obstructive pulmonary disease. Bull Exp Biol Med. 2004;137(1):64–66. doi: 10.1023/b:bebm.0000024389.16247.0a. [DOI] [PubMed] [Google Scholar]
- 203.Chan-Yeung M, Ho SP, Cheung AH, et al. Polymorphisms of glutathione S-transferase genes and functional activity in smokers with or without COPD. Int J Tuberc Lung Dis. 2007;11(5):508–514. [PubMed] [Google Scholar]
- 204.He JQ, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Glutathione S-transferase variants and their interaction with smoking on lung function. Am J Respir Crit Care Med. 2004;170(4):388–394. doi: 10.1164/rccm.200312-1763OC. [DOI] [PubMed] [Google Scholar]
- 205.Mehrotra S, Sharma A, Kumar S, Kar P, Sardana S, Sharma JK. Polymorphism of glutathione S-transferase M1 and T1 gene loci in COPD. Int J Immunogenet. 2010;37(4):263–267. doi: 10.1111/j.1744-313X.2010.00918.x. [DOI] [PubMed] [Google Scholar]
- 206.Thakur H, Gupta L, Sobti RC, Janmeja AK, Seth A, Singh SK. Association of GSTM1T1 genes with COPD and prostate cancer in north Indian population. Mol Biol Rep. 2011;38(3):1733–1739. doi: 10.1007/s11033-010-0287-8. [DOI] [PubMed] [Google Scholar]
- 207.Yanbaeva DG, Wouters EF, Dentener MA, Spruit MA, Reynaert NL. Association of glutathione-S-transferase omega haplotypes with susceptibility to chronic obstructive pulmonary disease. Free Radic Res. 2009;43(8):738–743. doi: 10.1080/10715760903038440. [DOI] [PubMed] [Google Scholar]
- 208.Ishii T, Matsuse T, Teramoto S, et al. Glutathione S-transferase P1 (GSTP1) polymorphism in patients with chronic obstructive pulmonary disease. Thorax. 1999;54(8):693–696. doi: 10.1136/thx.54.8.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Rodriguez F, de la Roza C, Jardi R, Schaper M, Vidal R, Miravitlles M. Glutathione S-transferase P1 and lung function in patients with alpha1- antitrypsin deficiency and COPD. Chest. 2005;127(5):1537–1543. doi: 10.1378/chest.127.5.1537. [DOI] [PubMed] [Google Scholar]
- 210.Lakhdar R, Denden S, Knani J, et al. Relationship between glutathione S-transferase P1 polymorphisms and chronic obstructive pulmonary disease in a Tunisian population. Genet Mol Res. 2010;9(2):897–907. doi: 10.4238/vol9-2gmr770. [DOI] [PubMed] [Google Scholar]
- 211.Lu B, He Q. Correlation between exon5 polymorphism of glutathione S-transferase P1 gene and susceptibility to chronic obstructive pulmonary disease in northern Chinese population of Han nationality living in Beijing, China. Zhonghua Nei Ke Za Zhi. 2002;41(10):678–681. Chinese. [PubMed] [Google Scholar]
- 212.Yim JJ, Yoo CG, Lee CT, Kim YW, Han SK, Shim YS. Lack of association between glutathione S-transferase P1 polymorphism and COPD in Koreans. Lung. 2002;180(2):119–125. doi: 10.1007/s004080000086. [DOI] [PubMed] [Google Scholar]
- 213.Yan F, Chen C, Jing J, Li W, Shen H, Wang X. Association between polymorphism of glutathione S-transferase P1 and chronic obstructive pulmonary disease: a meta-analysis. Respir Med. 2010;104(4):473–480. doi: 10.1016/j.rmed.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 214.Zhong L, Zhang YP, Fu WP, Dai LM, Sun C, Wang YQ. The relationship between GSTP1 I105V polymorphism and COPD: a reappraisal. Am J Respir Crit Care Med. 2010;181(7):763–765. doi: 10.1164/ajrccm.181.7.763. [DOI] [PubMed] [Google Scholar]
- 215.Zhang X, Mahmudi-Azer S, Connett JE, et al. Association of Hck genetic polymorphisms with gene expression and COPD. Hum Genet. 2007;120(5):681–690. doi: 10.1007/s00439-006-0253-7. [DOI] [PubMed] [Google Scholar]
- 216.Van Durme YM, Eijgelsheim M, Joos GF, et al. Hedgehog-interacting protein is a COPD susceptibility gene: the Rotterdam Study. Eur Respir J. 2010;36(1):89–95. doi: 10.1183/09031936.00129509. [DOI] [PubMed] [Google Scholar]
- 217.Keicho N, Tokunaga K, Nakata K, et al. Contribution of HLA genes to genetic predisposition in diffuse panbronchiolitis. Am J Respir Crit Care Med. 1998;158(3):846–850. doi: 10.1164/ajrccm.158.3.9712125. [DOI] [PubMed] [Google Scholar]
- 218.Sugiyama Y, Kudoh S, Maeda H, Suzaki H, Takaku F. Analysis of HLA antigens in patients with diffuse panbronchiolitis. Am Rev Respir Dis. 1990;141(6):1459–1462. doi: 10.1164/ajrccm/141.6.1459. [DOI] [PubMed] [Google Scholar]
- 219.Kasuga I, Hogg JC, Pare PD, et al. Role of genetic susceptibility to latent adenoviral infection and decreased lung function. Respir Med. 2009;103(11):1672–1680. doi: 10.1016/j.rmed.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kasuga I, Ruan J, Connett JE, Anthonisen NR, Sandford AJ. Lack of association of human leukocyte antigen-B7 with COPD and rate of decline in lung function. Respir Med. 2005;99(12):1528–1533. doi: 10.1016/j.rmed.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 221.Aubier M, Guenegou A, Benessiano J, Leynaert B, Boczkowski J, Neukirch F. Association of lung function decline with the microsatellite polymorphism in the heme oxygenase-1 gene promoter, in a general population sample. Results from the Longitudinal European Community Respiratory Health Survey (ECRHS-France) Bull Acad Natl Med. 2006;190(4–5):877–891. French. [PubMed] [Google Scholar]
- 222.Guenegou A, Boczkowski J, Aubier M, Neukirch F, Leynaert B. Interaction between a heme oxygenase-1 gene promoter polymorphism and serum beta-carotene levels on 8-year lung function decline in a general population: the European Community Respiratory Health Survey (France) Am J Epidemiol. 2008;167(2):139–144. doi: 10.1093/aje/kwm282. [DOI] [PubMed] [Google Scholar]
- 223.Guenegou A, Leynaert B, Benessiano J, et al. Association of lung function decline with the heme oxygenase-1 gene promoter microsatellite polymorphism in a general population sample. Results from the European Community Respiratory Health Survey (ECRHS), France. J Med Genet. 2006;43(8):e43. doi: 10.1136/jmg.2005.039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yamada N, Yamaya M, Okinaga S, et al. Microsatellite polymorphism in the heme oxygenase-1 gene promoter is associated with susceptibility to emphysema. Am J Hum Genet. 2000;66(1):187–195. doi: 10.1086/302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tanaka G, Aminuddin F, Akhabir L, et al. Effect of heme oxygenase-1 polymorphisms on lung function and gene expression. BMC Med Genet. 2011;12:117. doi: 10.1186/1471-2350-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.He JQ, Burkett K, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Interferon gamma polymorphisms and their interaction with smoking are associated with lung function. Hum Genet. 2006;119(4):365–375. doi: 10.1007/s00439-006-0143-z. [DOI] [PubMed] [Google Scholar]
- 227.Trajkov D, Mirkovska-Stojkovikj J, Petlichkovski A, et al. Association of cytokine gene polymorphisms with chronic obstructive pulmonary disease in Macedonians. Iran J Allergy Asthma Immunol. 2009;8(1):31–42. [PubMed] [Google Scholar]
- 228.Hackett TL, Stefanowicz D, Aminuddin F, et al. Effect of gene environment interactions on lung function and cardiovascular disease in COPD. Int J Chron Obstruct Pulmon Dis. 2011;6:277–287. doi: 10.2147/COPD.S18279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Asada M, Yamaya M, Ebihara S, et al. Interleukin-1beta gene polymorphisms associated with COPD. Chest. 2005;128(2):1072–1073. doi: 10.1378/chest.128.2.1072. author reply 1073. [DOI] [PubMed] [Google Scholar]
- 230.Hegab AE, Sakamoto T, Saitoh W, et al. Polymorphisms of TNFalpha, IL1beta, and IL1RN genes in chronic obstructive pulmonary disease. Biochem Biophys Res Commun. 2005;329(4):1246–1252. doi: 10.1016/j.bbrc.2005.02.099. [DOI] [PubMed] [Google Scholar]
- 231.Joos L, McIntyre L, Ruan J, et al. Association of IL-1beta and IL-1 receptor antagonist haplotypes with rate of decline in lung function in smokers. Thorax. 2001;56(11):863–866. doi: 10.1136/thorax.56.11.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lee JM, Kang YR, Park SH, et al. Polymorphisms in interleukin-1B and its receptor antagonist genes and the risk of chronic obstructive pulmonary disease in a Korean population: a case-control study. Respir Med. 2008;102(9):1311–1320. doi: 10.1016/j.rmed.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 233.Broekhuizen R, Grimble RF, Howell WM, et al. Pulmonary cachexia, systemic inflammatory profile, and the interleukin 1beta- 511 single nucleotide polymorphism. Am J Clin Nutr. 2005;82(5):1059–1064. doi: 10.1093/ajcn/82.5.1059. [DOI] [PubMed] [Google Scholar]
- 234.Danilko KV, Korytina GF, Akhmidishina LZ, Ianbaeva DG, Zagidullin SZ, Victorova TV. Association of cytokines genes (ILL, IL1RN, TNF, LTA, IL6, IL8, IL0) polymorphic markers with chronic obstructive pulmonary disease. Mol Biol (Mosk) 2007;41(1):26–36. Russian. [PubMed] [Google Scholar]
- 235.Josyula AB, Kurzius-Spencer M, Littau SR, Yucesoy B, Fleming J, Burgess JL. Cytokine genotype and phenotype effects on lung function decline in firefighters. J Occup Environ Med. 2007;49(3):282–288. doi: 10.1097/JOM.0b013e3180322584. [DOI] [PubMed] [Google Scholar]
- 236.Hsieh MH, Chong IW, Hwang JJ, et al. Lack of associations between several polymorphisms in cytokine genes and the risk of chronic obstructive pulmonary diseases in Taiwan. Kaohsiung J Med Sci. 2008;24(3):126–137. doi: 10.1016/S1607-551X(08)70140-2. [DOI] [PubMed] [Google Scholar]
- 237.Ishii T, Matsuse T, Teramoto S, et al. Neither IL-1beta, IL-1 receptor antagonist, nor TNF-alpha polymorphisms are associated with susceptibility to COPD. Respir Med. 2000;94(9):847–851. doi: 10.1053/rmed.2000.0808. [DOI] [PubMed] [Google Scholar]
- 238.Yucesoy B, Kurzius-Spencer M, Johnson VJ, et al. Association of cytokine gene polymorphisms with rate of decline in lung function. J Occup Environ Med. 2008;50(6):642–648. doi: 10.1097/JOM.0b013e31816515e1. [DOI] [PubMed] [Google Scholar]
- 239.Huang N, Liu L, Wang XZ, Liu D, Yin SY, Yang XD. Association of interleukin (IL)-12 and IL-27 gene polymorphisms with chronic obstructive pulmonary disease in a Chinese population. DNA Cell Biol. 2008;27(9):527–531. doi: 10.1089/dna.2007.0715. [DOI] [PubMed] [Google Scholar]
- 240.Burchard EG, Silverman EK, Rosenwasser LJ, et al. Association between a sequence variant in the IL-4 gene promoter and FEV(1) in asthma. Am J Respir Crit Care Med. 1999;160(3):919–922. doi: 10.1164/ajrccm.160.3.9812024. [DOI] [PubMed] [Google Scholar]
- 241.Beghe B, Hall IP, Parker SG, et al. Polymorphisms in IL13 pathway genes in asthma and chronic obstructive pulmonary disease. Allergy. 2010;65(4):474–481. doi: 10.1111/j.1398-9995.2009.02167.x. [DOI] [PubMed] [Google Scholar]
- 242.van der Pouw Kraan TC, Kucukaycan M, Bakker AM, et al. Chronic obstructive pulmonary disease is associated with the -1055 IL-13 promoter polymorphism. Genes Immun. 2002;3(7):436–439. doi: 10.1038/sj.gene.6363896. [DOI] [PubMed] [Google Scholar]
- 243.He JQ, Connett JE, Anthonisen NR, Sandford AJ. Polymorphisms in the IL13, IL13RA1, and IL4RA genes and rate of decline in lung function in smokers. Am J Respir Cell Mol Biol. 2003;28(3):379–385. doi: 10.1165/rcmb.4885. [DOI] [PubMed] [Google Scholar]
- 244.Hong SJ, Lee SY, Kim HB, et al. IL-5 and thromboxane A2 receptor gene polymorphisms are associated with decreased pulmonary function in Korean children with atopic asthma. J Allergy Clin Immunol. 2005;115(4):758–763. doi: 10.1016/j.jaci.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 245.Cordoba-Lanus E, de-Torres JP, Lopez-Aguilar C, et al. Association of IL-6 gene polymorphisms and COPD in a Spanish population. Respir Med. 2008;102(12):1805–1811. doi: 10.1016/j.rmed.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 246.He JQ, Foreman MG, Shumansky K, et al. Associations of IL6 polymorphisms with lung function decline and COPD. Thorax. 2009;64(8):698–704. doi: 10.1136/thx.2008.111278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.van den Borst B, Souren NY, Gielen M, et al. Association between the IL6-174G/C SNP and maximally attained lung function. Thorax. 2011;66(2):179. doi: 10.1136/thx.2009.128108. author reply 179–180. [DOI] [PubMed] [Google Scholar]
- 248.Seifart C, Dempfle A, Plagens A, et al. TNF-alpha-, TNF-beta-, IL-6-, and IL-10-promoter polymorphisms in patients with chronic obstructive pulmonary disease. Tissue Antigens. 2005;65(1):93–100. doi: 10.1111/j.1399-0039.2005.00343.x. [DOI] [PubMed] [Google Scholar]
- 249.Arinir U, Klein W, Rohde G, Stemmler S, Epplen JT, Schultze-Werninghaus G. Polymorphisms in the interleukin-8 gene in patients with chronic obstructive pulmonary disease. Electrophoresis. 2005;26(15):2888–2891. doi: 10.1002/elps.200500095. [DOI] [PubMed] [Google Scholar]
- 250.Matheson MC, Ellis JA, Raven J, Walters EH, Abramson MJ. Association of IL8, CXCR2 and TNF-alpha polymorphisms and airway disease. J Hum Genet. 2006;51(3):196–203. doi: 10.1007/s10038-005-0344-7. [DOI] [PubMed] [Google Scholar]
- 251.Stemmler S, Arinir U, Klein W, et al. Association of interleukin-8 receptor alpha polymorphisms with chronic obstructive pulmonary disease and asthma. Genes Immun. 2005;6(3):225–230. doi: 10.1038/sj.gene.6364181. [DOI] [PubMed] [Google Scholar]
- 252.Burgess JL, Fierro MA, Lantz RC, et al. Longitudinal decline in lung function: evaluation of interleukin-10 genetic polymorphisms in firefighters. J Occup Environ Med. 2004;46(10):1013–1022. doi: 10.1097/01.jom.0000141668.70006.52. [DOI] [PubMed] [Google Scholar]
- 253.Hu RC, Xu YJ, Zhang ZX, Xie JG. Polymorphism of interleukin-10 gene promoter and its association with susceptibility to chronic obstructive pulmonary disease in Chinese Han people. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20(6):504–507. Chinese. [PubMed] [Google Scholar]
- 254.Lyon H, Lange C, Lake S, et al. IL10 gene polymorphisms are associated with asthma phenotypes in children. Genet Epidemiol. 2004;26(2):155–165. doi: 10.1002/gepi.10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.He JQ, Shumansky K, Zhang X, Connett JE, Anthonisen NR, Sandford AJ. Polymorphisms of interleukin-10 and its receptor and lung function in COPD. Eur Respir J. 2007;29(6):1120–1126. doi: 10.1183/09031936.00002907. [DOI] [PubMed] [Google Scholar]
- 256.He B, Zhao MW, Ning LD, Jiang L, Yao WZ. The study of interleukin- 13 gene promoter polymorphism in patients with chronic obstructive pulmonary disease. Zhonghua Jie He He Hu Xi Za Zhi. 2004;27(8):529–532. Chinese. [PubMed] [Google Scholar]
- 257.Hu RC, Xu YJ, Zhang ZX. Study on the correlation of interleukin-13 polymorphism and susceptibility to chronic obstructive pulmonary disease in Chinese Han population. Zhonghua Liu Xing Bing Xue Za Zhi. 2004;25(7):607–611. Chinese. [PubMed] [Google Scholar]
- 258.Sadeghnejad A, Meyers DA, Bottai M, Sterling DA, Bleecker ER, Ohar JA. IL13 promoter polymorphism 1112C/T modulates the adverse effect of tobacco smoking on lung function. Am J Respir Crit Care Med. 2007;176(8):748–752. doi: 10.1164/rccm.200704-543OC. [DOI] [PubMed] [Google Scholar]
- 259.Liu SF, Chen YC, Wang CC, et al. Il13 promoter (-1055) polymorphisms associated with chronic obstructive pulmonary disease in Taiwanese. Exp Lung Res. 2009;35(10):807–816. doi: 10.3109/01902140902893644. [DOI] [PubMed] [Google Scholar]
- 260.Kang MJ, Lee SY, Kim HB, et al. Association of IL-13 polymorphisms with leukotriene receptor antagonist drug responsiveness in Korean children with exercise-induced bronchoconstriction. Pharmacogenet Genomics. 2008;18(7):551–558. doi: 10.1097/FPC.0b013e3282fe94c5. [DOI] [PubMed] [Google Scholar]
- 261.Park HW, Lee JE, Kim SH, et al. Genetic variation of IL13 as a risk factor of reduced lung function in children and adolescents: a cross-sectional population-based study in Korea. Respir Med. 2009;103(2):284–288. doi: 10.1016/j.rmed.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 262.Jiang L, He B, Zhao MW, Ning LD, Li XY, Yao WZ. Association of gene polymorphisms of tumour necrosis factor-alpha and interleukin-13 with chronic obstructive pulmonary disease in Han nationality in Beijing. Chin Med J (Engl) 2005;118(7):541–547. [PubMed] [Google Scholar]
- 263.Hizawa N, Kawaguchi M, Huang SK, Nishimura M. Role of interleukin-17F in chronic inflammatory and allergic lung disease. Clin Exp Allergy. 2006;36(9):1109–1114. doi: 10.1111/j.1365-2222.2006.02550.x. [DOI] [PubMed] [Google Scholar]
- 264.Seibold MA, Wang B, Eng C, et al. An African-specific functional polymorphism in KCNMB1 shows sex-specific association with asthma severity. Hum Mol Genet. 2008;17(17):2681–2690. doi: 10.1093/hmg/ddn168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Siedlinski M, Postma DS, Boer JM, et al. Level and course of FEV1 in relation to polymorphisms in NFE2L2 and KEAP1 in the general population. Respir Res. 2009;10:73. doi: 10.1186/1465-9921-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ye XW, Xiao M, Ye J, et al. The polymorphism -2548G/A in leptin and severity of chronic obstructive pulmonary disease. Int J Immunogenet. 2011;38(1):45–50. doi: 10.1111/j.1744-313X.2010.00968.x. [DOI] [PubMed] [Google Scholar]
- 267.Hansel NN, Gao L, Rafaels NM, et al. Leptin receptor polymorphisms and lung function decline in COPD. Eur Respir J. 2009;34(1):103–110. doi: 10.1183/09031936.00120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ianbaeva DG, Korytina GF, Viktorova TV. Allelic variants of the tumor necrosis factor superfamily as markers of the severity of the course of chronic obstructive lung disease and bronchiectatic disease. Genetika. 2004;40(4):545–551. Russian. [PubMed] [Google Scholar]
- 269.Ruse CE, Hill MC, Tobin M, et al. Tumour necrosis factor gene complex polymorphisms in chronic obstructive pulmonary disease. Respir Med. 2007;101(2):340–344. doi: 10.1016/j.rmed.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 270.Zhang H, Hang J, Wang X, et al. TNF polymorphisms modify endotoxin exposure-associated longitudinal lung function decline. Occup Environ Med. 2007;64(6):409–413. doi: 10.1136/oem.2006.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Yang IA, Holz O, Jorres RA, et al. Association of tumor necrosis factor-alpha polymorphisms and ozone-induced change in lung function. Am J Respir Crit Care Med. 2005;171(2):171–176. doi: 10.1164/rccm.200402-194OC. [DOI] [PubMed] [Google Scholar]
- 272.Randolph AG, Lange C, Silverman EK, Lazarus R, Weiss ST. Extended haplotype in the tumor necrosis factor gene cluster is associated with asthma and asthma-related phenotypes. Am J Respir Crit Care Med. 2005;172(6):687–692. doi: 10.1164/rccm.200501-122OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ferrarotti I, Zorzetto M, Beccaria M, et al. Tumour necrosis factor family genes in a phenotype of COPD associated with emphysema. Eur Respir J. 2003;21(3):444–449. doi: 10.1183/09031936.03.00051303. [DOI] [PubMed] [Google Scholar]
- 274.Patuzzo C, Gile LS, Zorzetto M, et al. Tumor necrosis factor gene complex in COPD and disseminated bronchiectasis. Chest. 2000;117(5):1353–1358. doi: 10.1378/chest.117.5.1353. [DOI] [PubMed] [Google Scholar]
- 275.Tanaka G, Sandford AJ, Burkett K, et al. Tumour necrosis factor and lymphotoxin A polymorphisms and lung function in smokers. Eur Respir J. 2007;29(1):34–41. doi: 10.1183/09031936.00045206. [DOI] [PubMed] [Google Scholar]
- 276.Lin CL, Siu LK, Lin JC, et al. Mannose-binding lectin gene polymorphism contributes to recurrence of infective exacerbation in patients with COPD. Chest. 2011;139(1):43–51. doi: 10.1378/chest.10-0375. [DOI] [PubMed] [Google Scholar]
- 277.Yang IA, Seeney SL, Wolter JM, et al. Mannose-binding lectin gene polymorphism predicts hospital admissions for COPD infections. Genes Immun. 2003;4(4):269–274. doi: 10.1038/sj.gene.6363961. [DOI] [PubMed] [Google Scholar]
- 278.Hui J, Palmer LJ, James AL, Musk AW, Beilby JP. AluyMICB dimorphism within the class I region of the major histocompatibility complex is associated with asthma and airflow obstruction in the Busselton population. Clin Exp Allergy. 2006;36(6):728–734. doi: 10.1111/j.1365-2222.2006.02495.x. [DOI] [PubMed] [Google Scholar]
- 279.Li LJ, Gao LB, Lv ML, et al. Association between SNPs in pre-miRNA and risk of chronic obstructive pulmonary disease. Clin Biochem. 2011;44(10–11):813–816. doi: 10.1016/j.clinbiochem.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 280.Joos L, He JQ, Shepherdson MB, et al. The role of matrix metalloproteinase polymorphisms in the rate of decline in lung function. Hum Mol Genet. 2002;11(5):569–576. doi: 10.1093/hmg/11.5.569. [DOI] [PubMed] [Google Scholar]
- 281.Tesfaigzi Y, Myers OB, Stidley CA, et al. Genotypes in matrix metalloproteinase 9 are a risk factor for COPD. Int J Chron Obstruct Pulmon Dis. 2006;1(3):267–278. doi: 10.2147/copd.2006.1.3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Korytina GF, Akhmadishina LZ, Ianbaeva DG, Viktorova TV. Polymorphism in promoter regions of matrix metalloproteinases (MMP1, MMP9, and MMP12) in chronic obstructive pulmonary disease patients. Genetika. 2008;44(2):242–249. [PubMed] [Google Scholar]
- 283.Haq I, Chappell S, Johnson SR, et al. Association of MMP-2 polymorphisms with severe and very severe COPD: a case control study of MMPs-1, 9 and 12 in a European population. BMC Med Genet. 2010;11:7. doi: 10.1186/1471-2350-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Lee SY, Kim MJ, Kang HG, et al. Polymorphisms in matrix metalloproteinase-1, -9 and -12 genes and the risk of chronic obstructive pulmonary disease in a Korean population. Respiration. 2010;80(2):133–138. doi: 10.1159/000284926. [DOI] [PubMed] [Google Scholar]
- 285.van Diemen CC, Postma DS, Siedlinski M, Blokstra A, Smit HA, Boezen HM. Genetic variation in TIMP1 but not MMPs predict excess FEV1 decline in two general population-based cohorts. Respir Res. 2011;12:57. doi: 10.1186/1465-9921-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Santus P, Casanova F, Biondi ML, Blasi F, Di Marco F, Centanni S. Stromelysin-1 polymorphism as a new potential risk factor in progression of chronic obstructive pulmonary disease. Monaldi Arch Chest Dis. 2009;71(1):15–20. doi: 10.4081/monaldi.2009.371. [DOI] [PubMed] [Google Scholar]
- 287.Schirmer H, Basso da Silva L, Teixeira PJ, Moreira JS, Moreira AL, Simon D. Matrix metalloproteinase gene polymorphisms: lack of association with chronic obstructive pulmonary disease in a Brazilian population. Genet Mol Res. 2009;8(3):1028–1034. doi: 10.4238/vol8-3gmr596. [DOI] [PubMed] [Google Scholar]
- 288.Ito I, Nagai S, Handa T, et al. Matrix metalloproteinase-9 promoter polymorphism associated with upper lung dominant emphysema. Am J Respir Crit Care Med. 2005;172(11):1378–1382. doi: 10.1164/rccm.200506-953OC. [DOI] [PubMed] [Google Scholar]
- 289.Minematsu N, Nakamura H, Tateno H, Nakajima T, Yamaguchi K. Genetic polymorphism in matrix metalloproteinase-9 and pulmonary emphysema. Biochem Biophys Res Commun. 2001;289(1):116–119. doi: 10.1006/bbrc.2001.5936. [DOI] [PubMed] [Google Scholar]
- 290.Zhou M, Huang SG, Wan HY, Li B, Deng WW, Li M. Genetic polymorphism in matrix metalloproteinase-9 and the susceptibility to chronic obstructive pulmonary disease in Han population of south China. Chin Med J (Engl) 2004;117(10):1481–1484. [PubMed] [Google Scholar]
- 291.Hunninghake GM, Cho MH, Tesfaigzi Y, et al. MMP12, lung function, and COPD in high-risk populations. N Engl J Med. 2009;361(27):2599–2608. doi: 10.1056/NEJMoa0904006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Haq I, Lowrey GE, Kalsheker N, Johnson SR. Matrix metalloproteinase- 12 (MMP-12) SNP affects MMP activity, lung macrophage infiltration and protects against emphysema in COPD. Thorax. 2011;66(11):970–976. doi: 10.1136/thx.2011.159087. [DOI] [PubMed] [Google Scholar]
- 293.Saitoh W, Sakamoto T, Hegab AE, et al. MMP14 gene polymorphisms in chronic obstructive pulmonary disease. Int J Mol Med. 2006;17(4):621–626. [PubMed] [Google Scholar]
- 294.Ohar JA, Hamilton RF, Jr, Zheng S, et al. COPD is associated with a macrophage scavenger receptor-1 gene sequence variation. Chest. 2010;137(5):1098–1107. doi: 10.1378/chest.09-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Hua CC, Chang LC, Tseng JC, Chu CM, Liu YC, Shieh WB. Functional haplotypes in the promoter region of transcription factor Nrf2 in chronic obstructive pulmonary disease. Dis Markers. 2010;28(3):185–193. doi: 10.3233/DMA-2010-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Arif E, Ahsan A, Vibhuti A, et al. Endothelial nitric oxide synthase gene variants contribute to oxidative stress in COPD. Biochem Biophys Res Commun. 2007;361(1):182–188. doi: 10.1016/j.bbrc.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 297.Novoradovsky A, Brantly ML, Waclawiw MA, et al. Endothelial nitric oxide synthase as a potential susceptibility gene in the pathogenesis of emphysema in alpha1-antitrypsin deficiency. Am J Respir Cell Mol Biol. 1999;20(3):441–447. doi: 10.1165/ajrcmb.20.3.3144. [DOI] [PubMed] [Google Scholar]
- 298.Corvol H, Nathan N, Charlier C, et al. Glucocorticoid receptor gene polymorphisms associated with progression of lung disease in young patients with cystic fibrosis. Respir Res. 2007;8:88. doi: 10.1186/1465-9921-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Schwabe K, Vacca G, Duck R, Gillissen A. Glucocorticoid receptor gene polymorphisms and potential association to chronic obstructive pulmonary disease susceptibility and severity. Eur J Med Res. 2009;14(Suppl 4):210–215. doi: 10.1186/2047-783X-14-S4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Yang SF, Xu YJ, Xie JG, Zhang ZX. hOGG1 Ser326Cys and XRCC1 Arg399Gln polymorphisms associated with chronic obstructive pulmonary disease. Chin Med J (Engl) 2009;122(8):960–966. [PubMed] [Google Scholar]
- 301.Takabatake N, Toriyama S, Takeishi Y, et al. A nonfunctioning single nucleotide polymorphism in olfactory receptor gene family is associated with the forced expiratory volume in the first second/ the forced vital capacity values of pulmonary function test in a Japanese population. Biochem Biophys Res Commun. 2007;364(3):662–667. doi: 10.1016/j.bbrc.2007.10.068. [DOI] [PubMed] [Google Scholar]
- 302.Homma S, Sakamoto T, Hegab AE, et al. Association of phosphodiesterase 4D gene polymorphisms with chronic obstructive pulmonary disease: relationship to interleukin 13 gene polymorphism. Int J Mol Med. 2006;18(5):933–939. [PubMed] [Google Scholar]
- 303.Stewart CE, Hall IP, Parker SG, et al. PLAUR polymorphisms and lung function in UK smokers. BMC Med Genet. 2009;10:112. doi: 10.1186/1471-2350-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Barton SJ, Koppelman GH, Vonk JM, et al. PLAUR polymorphisms are associated with asthma, PLAUR levels, and lung function decline. J Allergy Clin Immunol. 2009;123(6):1391–1400. doi: 10.1016/j.jaci.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 305.Tsai YJ, Choudhry S, Kho J, et al. The PTGDR gene is not associated with asthma in 3 ethnically diverse populations. J Allergy Clin Immunol. 2006;118(6):1242–1248. doi: 10.1016/j.jaci.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 306.Wu L, Merrilees M, Young RP, Black PN. The cyclooxygenase-2- 765C promoter polymorphism protects against the development of chronic obstructive pulmonary disease. Respir Med. 2011;105(3):506–510. doi: 10.1016/j.rmed.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 307.Arif E, Vibhuti A, Deepak D, Singh B, Siddiqui MS, Pasha MA. COX2 and p53 risk-alleles coexist in COPD. Clin Chim Acta. 2008;397(1–2):48–50. doi: 10.1016/j.cca.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 308.Kwok JS, Lawton JW, Yew WW, Chau CH, Lee J, Wong PC. Protease inhibitor phenotypes and serum alpha-1-antitrypsin levels in patients with COPD: a study from Hong Kong. Respirology. 2004;9(2):265–270. doi: 10.1111/j.1440-1843.2004.00560.x. [DOI] [PubMed] [Google Scholar]
- 309.Hodgson I, Kalsheker N. DNA polymorphisms of the human alpha 1 antitrypsin gene in normal subjects and in patients with pulmonary emphysema. J Med Genet. 1987;24(1):47–51. doi: 10.1136/jmg.24.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Chappell S, Daly L, Morgan K, et al. Cryptic haplotypes of SERPINA1 confer susceptibility to chronic obstructive pulmonary disease. Hum Mutat. 2006;27(1):103–109. doi: 10.1002/humu.20275. [DOI] [PubMed] [Google Scholar]
- 311.Abboud RT, Rushton JM, Grzybowski S. Interrelationships between neutrophil elastase, serum alpha, -antitrypsin, lung function and chest radiography in patients with chronic airflow obstruction. Am Rev Respir Dis. 1979;120(1):31–40. doi: 10.1164/arrd.1979.120.1.31. [DOI] [PubMed] [Google Scholar]
- 312.Barnett TB, Gottovi D, Johnson AM. Protease inhibitors in chronic obstructive pulmonary disease. Am Rev Respir Dis. 1975;111(5):587–593. doi: 10.1164/arrd.1975.111.5.587. [DOI] [PubMed] [Google Scholar]
- 313.Bartmann K, Fooke-Achterrath M, Koch G, et al. Heterozygosity in the Pi-system as a pathogenetic cofactor in chronic obstructive pulmonary disease (COPD) Eur J Respir Dis. 1985;66(4):284–296. [PubMed] [Google Scholar]
- 314.Cox DW, Hoeppner VH, Levison H. Protease inhibitors in patients with chronic obstructive pulmonary disease: the alpha-antitrypsin heterozygote controversy. Am Rev Respir Dis. 1976;113(5):601–606. doi: 10.1164/arrd.1976.113.5.601. [DOI] [PubMed] [Google Scholar]
- 315.Dahl M, Nordestgaard BG, Lange P, Vestbo J, Tybjaerg-Hansen A. Molecular diagnosis of intermediate and severe alpha(1)-antitrypsin deficiency: MZ individuals with chronic obstructive pulmonary disease may have lower lung function than MM individuals. Clin Chem. 2001;47(1):56–62. [PubMed] [Google Scholar]
- 316.Denden S, Haj Khelil A, Perrin P, et al. Alpha 1 antitrypsin polymorphism in the Tunisian population with special reference to pulmonary disease. Pathol Biol (Paris) 2008;56(3):106–110. doi: 10.1016/j.patbio.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 317.Gupta J, Bhadoria DP, Lal MK, et al. Association of the PIM3 allele of the alpha-1-antitrypsin gene with chronic obstructive pulmonary disease. Clin Biochem. 2005;38(5):489–491. doi: 10.1016/j.clinbiochem.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 318.Lieberman J, Winter B, Sastre A. Alpha 1-antitrypsin Pi-types in 965 COPD patients. Chest. 1986;89(3):370–373. doi: 10.1378/chest.89.3.370. [DOI] [PubMed] [Google Scholar]
- 319.Matzen RN, Bader PI, Block WD. alpha1-Antitrypsin deficiency in clinic patients. Ann Clin Res. 1977;9(2):88–92. [PubMed] [Google Scholar]
- 320.Poller W, Meisen C, Olek K. DNA polymorphisms of the alpha 1-antitrypsin gene region in patients with chronic obstructive pulmonary disease. Eur J Clin Invest. 1990;20(1):1–7. doi: 10.1111/j.1365-2362.1990.tb01769.x. [DOI] [PubMed] [Google Scholar]
- 321.Sandford AJ, Weir TD, Spinelli JJ, Pare PD. Z and S mutations of the alpha1-antitrypsin gene and the risk of chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 1999;20(2):287–291. doi: 10.1165/ajrcmb.20.2.3177. [DOI] [PubMed] [Google Scholar]
- 322.Kalsheker NA, Hodgson IJ, Watkins GL, White JP, Morrison HM, Stockley RA. Deoxyribonucleic acid (DNA) polymorphism of the alpha 1-antitrypsin gene in chronic lung disease. Br Med J (Clin Res Ed) 1987;294(6586):1511–1514. doi: 10.1136/bmj.294.6586.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Dahl M, Tybjaerg-Hansen A, Lange P, Vestbo J, Nordestgaard BG. Change in lung function and morbidity from chronic obstructive pulmonary disease in alpha1-antitrypsin MZ heterozygotes: a longitudinal study of the general population. Ann Intern Med. 2002;136(4):270–279. doi: 10.7326/0003-4819-136-4-200202190-00006. [DOI] [PubMed] [Google Scholar]
- 324.Klayton R, Fallat R, Cohen AB. Determinants of chronic obstructive pulmonary disease in patients with intermediate levels of alpha-antitrypsin. Am Rev Respir Dis. 1975;112(1):71–75. doi: 10.1164/arrd.1975.112.1.71. [DOI] [PubMed] [Google Scholar]
- 325.Shigeoka JW, Hall WJ, Hyde RW, et al. The prevalence of alpha-antitrypsin heterozygotes (Pi MZ) in patients with obstructive pulmonary disease. Am Rev Respir Dis. 1976;114(6):1077–1084. doi: 10.1164/arrd.1976.114.6.1077. [DOI] [PubMed] [Google Scholar]
- 326.Fujimoto K, Ikeda S, Arai T, et al. Polymorphism of SERPINE2 gene is associated with pulmonary emphysema in consecutive autopsy cases. BMC Med Genet. 2010;11:159. doi: 10.1186/1471-2350-11-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Quint JK, Donaldson GC, Kumari M, Talmud PJ, Hurst JR. SERPINA1 11478G→A variant, serum alpha1-antitrypsin, exacerbation frequency and FEV1 decline in COPD. Thorax. 2011;66(5):418–424. doi: 10.1136/thx.2010.152975. [DOI] [PubMed] [Google Scholar]
- 328.Gulsvik A, Fagerhol MK. Alpha 1-antitrypsin phenotypes and obstructive lung disease in the city of Oslo. Scand J Respir Dis. 1979;60(5):267–274. [PubMed] [Google Scholar]
- 329.Roberts A, Kagan A, Rhoads GG, Pierce JA, Bruce RM. Antitrypsin and chronic obstructive pulmonary disease among Japanese-American men. Chest. 1977;72(4):489–491. doi: 10.1378/chest.72.4.489. [DOI] [PubMed] [Google Scholar]
- 330.Lochon B, Vercaigne D, Lochon C, et al. Pan-lobular emphysema: relationship with serum alpha-1-antitrypsin levels, Pi phenotype and the HLA system (author’s transl) Nouv Presse Med. 1978;7(14):1167–1170. French. [PubMed] [Google Scholar]
- 331.Ostrow DN, Manfreda J, Dorman T, Cherniack RM. Alpha1-antitrypsin phenotypes and lung function in a moderately polluted northern Ontario community. Can Med Assoc J. 1978;118(6):669–672. [PMC free article] [PubMed] [Google Scholar]
- 332.Benetazzo MG, Gile LS, Bombieri C, et al. Alpha 1-antitrypsin TAQ I polymorphism and alpha 1-antichymotrypsin mutations in patients with obstructive pulmonary disease. Respir Med. 1999;93(9):648–654. doi: 10.1016/s0954-6111(99)90105-1. [DOI] [PubMed] [Google Scholar]
- 333.Kim CH, Yim JJ, Yoo CG, et al. Alpha-antitrypsin genotypes in Korean patients with chronic obstructive pulmonary disease. Respirology. 2005;10(2):223–228. doi: 10.1111/j.1440-1843.2005.00693.x. [DOI] [PubMed] [Google Scholar]
- 334.Sandford AJ, Spinelli JJ, Weir TD, Pare PD. Mutation in the 3′ region of the alpha-1-antitrypsin gene and chronic obstructive pulmonary disease. J Med Genet. 1997;34(10):874–875. doi: 10.1136/jmg.34.10.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Zhu YJ. Epidemiological survey of chronic obstructive pulmonary disease and alpha-1-deficiency in China. Respirology. 2001;(Suppl 6):S13–S15. doi: 10.1046/j.1440-1843.2001.00306.x. [DOI] [PubMed] [Google Scholar]
- 336.Shim YS. Epidemiological survey of chronic obstructive pulmonary disease and alpha-1 antitrypsin deficiency in Korea. Respirology. 2001;6(Suppl):S9–S11. doi: 10.1046/j.1440-1843.2001.00305.x. [DOI] [PubMed] [Google Scholar]
- 337.Eriksson S, Lindmark B, Lilja H. Familial alpha 1-antichymotrypsin deficiency. Acta Med Scand. 1986;220(5):447–453. [PubMed] [Google Scholar]
- 338.Faber JP, Poller W, Olek K, et al. The molecular basis of alpha 1- antichymotrypsin deficiency in a heterozygote with liver and lung disease. J Hepatol. 1993;18(3):313–321. doi: 10.1016/s0168-8278(05)80275-2. [DOI] [PubMed] [Google Scholar]
- 339.Lindmark BE, Arborelius M, Jr, Eriksson SG. Pulmonary function in middle-aged women with heterozygous deficiency of the serine protease inhibitor alpha 1-antichymotrypsin. Am Rev Respir Dis. 1990;141(4 Pt 1):884–888. doi: 10.1164/ajrccm/141.4_Pt_1.884. [DOI] [PubMed] [Google Scholar]
- 340.Poller W, Faber JP, Scholz S, et al. Mis-sense mutation of alpha 1-antichymotrypsin gene associated with chronic lung disease. Lancet. 1992;339(8808):1538. doi: 10.1016/0140-6736(92)91301-n. [DOI] [PubMed] [Google Scholar]
- 341.Poller W, Faber JP, Weidinger S, et al. A leucine-to-proline substitution causes a defective alpha 1-antichymotrypsin allele associated with familial obstructive lung disease. Genomics. 1993;17(3):740–743. doi: 10.1006/geno.1993.1396. [DOI] [PubMed] [Google Scholar]
- 342.Samilchuk EI, Chuchalin AG. Mis-sense mutation of alpha 1- antichymotrypsin gene and chronic lung disease. Lancet. 1993;342(8871):624. doi: 10.1016/0140-6736(93)91455-u. [DOI] [PubMed] [Google Scholar]
- 343.Ishii T, Matsuse T, Teramoto S, et al. Association between alpha- 1-antichymotrypsin polymorphism and susceptibility to chronic obstructive pulmonary disease. Eur J Clin Invest. 2000;30(6):543–548. doi: 10.1046/j.1365-2362.2000.00655.x. [DOI] [PubMed] [Google Scholar]
- 344.Sandford AJ, Chagani T, Weir TD, Pare PD. Alpha 1-antichymotrypsin mutations in patients with chronic obstructive pulmonary disease. Dis Markers. 1998;13(4):257–260. doi: 10.1155/1998/867620. [DOI] [PubMed] [Google Scholar]
- 345.Demeo DL, Mariani TJ, Lange C, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet. 2006;78(2):253–264. doi: 10.1086/499828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zhu G, Warren L, Aponte J, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176(2):167–173. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 347.Sorheim IC, Gulsvik A. Genetics of chronic obstructive pulmonary disease: a case-control study in Bergen, Norway. Clin Respir J. 2008;2(Suppl 1):129–131. doi: 10.1111/j.1752-699X.2008.00097.x. [DOI] [PubMed] [Google Scholar]
- 348.Cha SI, Kang HG, Choi JE, et al. SERPINE2 polymorphisms and chronic obstructive pulmonary disease. J Korean Med Sci. 2009;24(6):1119–1125. doi: 10.3346/jkms.2009.24.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Chappell S, Daly L, Morgan K, et al. The SERPINE2 gene and chronic obstructive pulmonary disease. Am J Hum Genet. 2006 Jul;79(1):184–186. doi: 10.1086/505268. author reply 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Zhong L, Fu WP, Sun C, Dai LM, Zhang YP. Absence of association between SERPINE2 genetic polymorphisms and chronic obstructive pulmonary disease in Han Chinese: a case-control cohort study. BMC Med Genet. 2009;10:66. doi: 10.1186/1471-2350-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Guo X, Lin HM, Lin Z, et al. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J. 2001;18(3):482–490. doi: 10.1183/09031936.01.00043401. [DOI] [PubMed] [Google Scholar]
- 352.Hu R, Xu Y, Zhang Z. Surfactant protein B 1580 polymorphism is associated with susceptibility to chronic obstructive pulmonary disease in Chinese Han population. J Huazhong Univ Sci Technolog Med Sci. 2004;24(3):216–218. 238. doi: 10.1007/BF02831993. [DOI] [PubMed] [Google Scholar]
- 353.Seifart C, Plagens A, Brodje D, Muller B, von Wichert P, Floros J. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Dis Markers. 2002;18(3):129–136. doi: 10.1155/2002/194075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Baekvad-Hansen M, Dahl M, Tybjaerg-Hansen A, Nordestgaard BG. Surfactant protein-B 121ins2 heterozygosity, reduced pulmonary function, and chronic obstructive pulmonary disease in smokers. Am J Respir Crit Care Med. 2010;181(1):17–20. doi: 10.1164/rccm.200906-0963OC. [DOI] [PubMed] [Google Scholar]
- 355.Baekvad-Hansen M, Nordestgaard BG, Dahl M. Surfactant protein B polymorphisms, pulmonary function and COPD in 10,231 individuals. Eur Respir J. 2011;37(4):791–799. doi: 10.1183/09031936.00026410. [DOI] [PubMed] [Google Scholar]
- 356.Tsvetkova OA, Veselovskaia MV. Polymorphic variants of a surfactant protein C gene in patients with chronic obstructive pulmonary disease. Ter Arkh. 2007;79(9):65–69. Russian. [PubMed] [Google Scholar]
- 357.Baekvad-Hansen M, Nordestgaard BG, Tybjaerg-Hansen A, Dahl M. Two novel mutations in surfactant protein-C, lung function and obstructive lung disease. Respir Med. 2010;104(3):418–425. doi: 10.1016/j.rmed.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 358.Ishii T, Hagiwara K, Kamio K, et al. Involvement of surfactant protein D in emphysema revealed by genetic association study. Eur J Hum Genet. 2012;20(2):230–235. doi: 10.1038/ejhg.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Foreman MG, Kong X, DeMeo DL, et al. Polymorphisms in surfactant protein-D are associated with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2011;44(3):316–322. doi: 10.1165/rcmb.2009-0360OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Ishii T, Wakabayashi R, Kurosaki H, Gemma A, Kida K. Association of serotonin transporter gene variation with smoking, chronic obstructive pulmonary disease, and its depressive symptoms. J Hum Genet. 2011;56(1):41–46. doi: 10.1038/jhg.2010.133. [DOI] [PubMed] [Google Scholar]
- 361.Kim EJ, Kim KM, Park SH, et al. SLC11A1 polymorphisms are associated with the risk of chronic obstructive pulmonary disease in a Korean population. Biochem Genet. 2008;46(7–8):506–519. doi: 10.1007/s10528-008-9166-6. [DOI] [PubMed] [Google Scholar]
- 362.Yang T, Ying B, Song X, et al. Single-nucleotide polymorphisms in SMAD3 are associated with chronic obstructive pulmonary disease. Exp Biol Med (Maywood) 2010;235(5):599–605. doi: 10.1258/ebm.2010.009268. [DOI] [PubMed] [Google Scholar]
- 363.Wilk JB, Herbert A, Shoemaker CM, Gottlieb DJ, Karamohamed S. Secreted modular calcium-binding protein 2 haplotypes are associated with pulmonary function. Am J Respir Crit Care Med. 2007;175(6):554–560. doi: 10.1164/rccm.200601-110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Siedlinski M, van Diemen CC, Postma DS, Vonk JM, Boezen HM. Superoxide dismutases, lung function and bronchial responsiveness in a general population. Eur Respir J. 2009;33(5):986–992. doi: 10.1183/09031936.00171507. [DOI] [PubMed] [Google Scholar]
- 365.Pietras T, Szemraj J, Witusik A, et al. The sequence polymorphism of MnSOD gene in subjects with respiratory insufficiency in COPD. Med Sci Monit. 2010;16(9):CR427–CR432. [PubMed] [Google Scholar]
- 366.Pietras T, Witusik A, Panek M, Galecki P, Szemraj J, Gorski P. Anxiety, depression and polymorphism of the gene encoding superoxide dismutase in patients with chronic obstructive pulmonary disease. Pol Merkur Lekarski. 2010;29(171):165–168. Polish. [PubMed] [Google Scholar]
- 367.Juul K, Tybjaerg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(8):858–864. doi: 10.1164/rccm.200509-1387OC. [DOI] [PubMed] [Google Scholar]
- 368.Dahl M, Bowler RP, Juul K, Crapo JD, Levy S, Nordestgaard BG. Superoxide dismutase 3 polymorphism associated with reduced lung function in two large populations. Am J Respir Crit Care Med. 2008;178(9):906–912. doi: 10.1164/rccm.200804-549OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Ganguly K, Depner M, Fattman C, et al. Superoxide dismutase 3, extracellular (SOD3) variants and lung function. Physiol Genomics. 2009;37(3):260–267. doi: 10.1152/physiolgenomics.90363.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Sorheim IC, DeMeo DL, Washko G, et al. Polymorphisms in the superoxide dismutase-3 gene are associated with emphysema in COPD. COPD. 2010;7(4):262–268. doi: 10.3109/15412555.2010.496821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Hersh CP, Silverman EK, Gascon J, et al. SOX5 is a candidate gene for chronic obstructive pulmonary disease susceptibility and is necessary for lung development. Am J Respir Crit Care Med. 2011;183(11):1482–1489. doi: 10.1164/rccm.201010-1751OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Litonjua AA, Tantisira KG, Lake S, et al. Polymorphisms in signal transducer and activator of transcription 3 and lung function in asthma. Respir Res. 2005;6:52. doi: 10.1186/1465-9921-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Hawkins GA, Lazarus R, Smith RS, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123(6):1376–1383. doi: 10.1016/j.jaci.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Leung TF, Tang NL, Lam CW, Li AM, Chan IH, Ha G. Thromboxane A2 receptor gene polymorphism is associated with the serum concentration of cat-specific immunoglobulin E as well as the development and severity of asthma in Chinese children. Pediatr Allergy Immunol. 2002;13(1):10–17. doi: 10.1034/j.1399-3038.2002.01033.x. [DOI] [PubMed] [Google Scholar]
- 375.Celedon JC, Lange C, Raby BA, et al. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) Hum Mol Genet. 2004;13(15):1649–1656. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- 376.Ogawa E, Ruan J, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Transforming growth factor-beta1 polymorphisms, airway responsiveness and lung function decline in smokers. Respir Med. 2007;101(5):938–943. doi: 10.1016/j.rmed.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 377.Su ZG, Wen FQ, Feng YL, Xiao M, Wu XL. Transforming growth factor-beta1 gene polymorphisms associated with chronic obstructive pulmonary disease in Chinese population. Acta Pharmacol Sin. 2005;26(6):714–720. [PubMed] [Google Scholar]
- 378.van Diemen CC, Postma DS, Vonk JM, Bruinenberg M, Nolte IM, Boezen HM. Decorin and TGF-beta1 polymorphisms and development of COPD in a general population. Respir Res. 2006;7:89. doi: 10.1186/1465-9921-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Wu L, Chau J, Young RP, et al. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax. 2004;59(2):126–129. doi: 10.1136/thorax.2003.005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Ito M, Hanaoka M, Droma Y, et al. The association of transforming growth factor beta 1 gene polymorphisms with the emphysema phenotype of COPD in Japanese. Intern Med. 2008;47(15):1387–1394. doi: 10.2169/internalmedicine.47.1116. [DOI] [PubMed] [Google Scholar]
- 381.Liu DS, Li XO, Ying BW, et al. Effects of single nucleotide polymorphisms 869 T/C and 915 G/C in the exon 1 locus of transforming growth factor-beta1 gene on chronic obstructive pulmonary disease susceptibility in Chinese. Chin Med J (Engl) 2010;123(4):390–394. [PubMed] [Google Scholar]
- 382.Arkwright PD, Laurie S, Super M, et al. TGF-beta(1) genotype and accelerated decline in lung function of patients with cystic fibrosis. Thorax. 2000;55(6):459–462. doi: 10.1136/thorax.55.6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Yoon HI, Silverman EK, Lee HW, et al. Lack of association between COPD and transforming growth factor-beta1 (TGFB1) genetic polymorphisms in Koreans. Int J Tuberc Lung Dis. 2006;10(5):504–509. [PubMed] [Google Scholar]
- 384.Gong Y, Fan L, Wan H, et al. Lack of association between the TGF-beta( 1) gene and development of COPD in Asians: a case-control study and meta-analysis. Lung. 2011;189(3):213–223. doi: 10.1007/s00408-011-9294-3. [DOI] [PubMed] [Google Scholar]
- 385.Hirano K, Sakamoto T, Uchida Y, et al. Tissue inhibitor of metalloproteinases-2 gene polymorphisms in chronic obstructive pulmonary disease. Eur Respir J. 2001;18(5):748–752. doi: 10.1183/09031936.01.00102101. [DOI] [PubMed] [Google Scholar]
- 386.Hegab AE, Sakamoto T, Uchida Y, et al. Promoter activity of human tissue inhibitor of metalloproteinase 2 gene with novel single nucleotide polymorphisms. Respirology. 2005;10(1):27–30. doi: 10.1111/j.1440-1843.2005.00654.x. [DOI] [PubMed] [Google Scholar]
- 387.Hegab AE, Sakamoto T, Uchida Y, et al. Association analysis of tissue inhibitor of metalloproteinase2 gene polymorphisms with COPD in Egyptians. Respir Med. 2005;99(1):107–110. doi: 10.1016/j.rmed.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 388.Speletas M, Merentiti V, Kostikas K, et al. Association of TLR4-T399I polymorphism with chronic obstructive pulmonary disease in smokers. Clin Dev Immunol. 2009;2009:260286. doi: 10.1155/2009/260286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Rohde G, Klein W, Arinir U, et al. Association of the ASP299GLY TLR4 polymorphism with COPD. Respir Med. 2006;100(5):892–896. doi: 10.1016/j.rmed.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 390.Huang SL, Su CH, Chang SC. Tumor necrosis factor-alpha gene polymorphism in chronic bronchitis. Am J Respir Crit Care Med. 1997;156(5):1436–1439. doi: 10.1164/ajrccm.156.5.9609138. [DOI] [PubMed] [Google Scholar]
- 391.Kucukaycan M, Van Krugten M, Pennings HJ, et al. Tumor necrosis factor-alpha +489G/A gene polymorphism is associated with chronic obstructive pulmonary disease. Respir Res. 2002;3:29. doi: 10.1186/rr194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Papatheodorou A, Latsi P, Vrettou C, et al. Development of a novel microarray methodology for the study of SNPs in the promoter region of the TNF-alpha gene: their association with obstructive pulmonary disease in Greek patients. Clin Biochem. 2007;40(12):843–850. doi: 10.1016/j.clinbiochem.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 393.Sakao S, Tatsumi K, Igari H, Shino Y, Shirasawa H, Kuriyama T. Association of tumor necrosis factor alpha gene promoter polymorphism with the presence of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163(2):420–422. doi: 10.1164/ajrccm.163.2.2006031. [DOI] [PubMed] [Google Scholar]
- 394.Stankovic MM, Nestorovic AR, Tomovic AM, et al. TNF-alpha-308 promotor polymorphism in patients with chronic obstructive pulmonary disease and lung cancer. Neoplasma. 2009;56(4):348–352. doi: 10.4149/neo_2009_04_348. [DOI] [PubMed] [Google Scholar]
- 395.Sapey E, Wood AM, Ahmad A, Stockley RA. Tumor necrosis factor-{alpha} rs361525 polymorphism is associated with increased local production and downstream inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(2):192–199. doi: 10.1164/rccm.200912-1846OC. [DOI] [PubMed] [Google Scholar]
- 396.Chen YC, Liu SF, Chin CH, et al. Association of tumor necrosis factor-alpha-863C/A gene polymorphism with chronic obstructive pulmonary disease. Lung. 2010;188(4):339–347. doi: 10.1007/s00408-010-9236-5. [DOI] [PubMed] [Google Scholar]
- 397.Cordoba-Lanus E, Baz-Davila R, de-Torres JP, et al. TNFA-863 polymorphism is associated with a reduced risk of chronic obstructive pulmonary disease: a replication study. BMC Med Genet. 2011;12:132. doi: 10.1186/1471-2350-12-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Keatings VM, Cave SJ, Henry MJ, et al. A polymorphism in the tumor necrosis factor-alpha gene promoter region may predispose to a poor prognosis in COPD. Chest. 2000;118(4):971–975. doi: 10.1378/chest.118.4.971. [DOI] [PubMed] [Google Scholar]
- 399.Chappell S, Daly L, Morgan K, et al. Variation in the tumour necrosis factor gene is not associated with susceptibility to COPD. Eur Respir J. 2007;30(4):810–812. doi: 10.1183/09031936.00057107. [DOI] [PubMed] [Google Scholar]
- 400.Chierakul N, Wongwisutikul P, Vejbaesya S, Chotvilaiwan K. Tumor necrosis factor-alpha gene promoter polymorphism is not associated with smoking-related COPD in Thailand. Respirology. 2005;10(1):36–39. doi: 10.1111/j.1440-1843.2005.00626.x. [DOI] [PubMed] [Google Scholar]
- 401.Higham MA, Pride NB, Alikhan A, Morrell NW. Tumour necrosis factor-alpha gene promoter polymorphism in chronic obstructive pulmonary disease. Eur Respir J. 2000;15(2):281–284. doi: 10.1034/j.1399-3003.2000.15b10.x. [DOI] [PubMed] [Google Scholar]
- 402.Sakao S, Tatsumi K, Igari H, et al. Association of tumor necrosis factor-alpha gene promoter polymorphism with low attenuation areas on high-resolution CT in patients with COPD. Chest. 2002;122(2):416–420. doi: 10.1378/chest.122.2.416. [DOI] [PubMed] [Google Scholar]
- 403.Teramoto S, Ishii T, Ishii M, et al. Variation in the tumour necrosis factor-alpha gene is not associated with susceptibility to Asian COPD. Eur Respir J. 2008;31(3):682–683. doi: 10.1183/09031936.00130607. [DOI] [PubMed] [Google Scholar]
- 404.Zhu G, Gulsvik A, Bakke P, et al. Association of TRPV4 gene polymorphisms with chronic obstructive pulmonary disease. Hum Mol Genet. 2009;18(11):2053–2062. doi: 10.1093/hmg/ddp111. [DOI] [PubMed] [Google Scholar]
- 405.Masuko H, Sakamoto T, Kaneko Y, et al. Lower FEV1 in non-COPD, nonasthmatic subjects: association with smoking, annual decline in FEV1, total IgE levels, and TSLP genotypes. Int J Chron Obstruct Pulmon Dis. 2011;6:181–189. doi: 10.2147/COPD.S16383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Janssens W, Bouillon R, Claes B, et al. Vitamin D deficiency is highly prevalent in COPD and correlates with variants in the vitamin D-binding gene. Thorax. 2010;65(3):215–220. doi: 10.1136/thx.2009.120659. [DOI] [PubMed] [Google Scholar]
- 407.Hopkinson NS, Li KW, Kehoe A, et al. Vitamin D receptor genotypes influence quadriceps strength in chronic obstructive pulmonary disease. Am J Clin Nutr. 2008;87(2):385–390. doi: 10.1093/ajcn/87.2.385. [DOI] [PubMed] [Google Scholar]
- 408.Raby BA, Lazarus R, Silverman EK, et al. Association of vitamin D receptor gene polymorphisms with childhood and adult asthma. Am J Respir Crit Care Med. 2004;170(10):1057–1065. doi: 10.1164/rccm.200404-447OC. [DOI] [PubMed] [Google Scholar]
- 409.Shaheen SO, Jameson KA, Robinson SM, et al. Relationship of vitamin D status to adult lung function and COPD. Thorax. 2011;66(8):692–698. doi: 10.1136/thx.2010.155234. [DOI] [PubMed] [Google Scholar]
- 410.Sharma S, Murphy AJ, Soto-Quiros ME, et al. Association of VEGF polymorphisms with childhood asthma, lung function and airway responsiveness. Eur Respir J. 2009;33(6):1287–1294. doi: 10.1183/09031936.00113008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Sakao S, Tatsumi K, Hashimoto T, et al. Vascular endothelial growth factor and the risk of smoking-related COPD. Chest. 2003;124(1):323–327. doi: 10.1378/chest.124.1.323. [DOI] [PubMed] [Google Scholar]
- 412.Hersh CP, Pillai SG, Zhu G, et al. Multistudy fine mapping of chromosome 2q identifies XRCC5 as a chronic obstructive pulmonary disease susceptibility gene. Am J Respir Crit Care Med. 2010;182(5):605–613. doi: 10.1164/rccm.200910-1586OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Genes tested but showing lack of association with chronic obstructive pulmonary disease
| Symbol | Name | Chromosome | References | |||
|---|---|---|---|---|---|---|
|
|
||||||
| Single studies | Meta-analyses | |||||
|
|
|
|||||
| Positive | Negative | Positive | Negative | |||
| AGER | Advanced glycosylation end product-specific receptor | 6 | 1,2 | |||
| CASP8 | Caspase 8, apoptosis-related cysteine peptidase | 2 | 3 | |||
| CCL17 (TARC) | Chemokine (C-C motif) ligand 17 | 16 | 4 | |||
| CCL2 | Chemokine (C-C motif) ligand 2 | 17 | 5 | |||
| CFLAR | CASP8 and FADD-like apoptosis regulator | 2 | 3 | |||
| COL6A5 | Collagen, type VI, alpha 5 | 3 | 6 | |||
| CXADR | Coxsackie virus and adenovirus receptor | 21 | 7 | |||
| CYP1B1 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 2 | 8,9 | |||
| CYP2D6 | Cytochrome P450, family 2, subfamily D, polypeptide 6 | 22 | 10 | |||
| DCN | Decorin | 12 | 11 | |||
| DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 19 | 12 | |||
| EDNRA | Endothelin receptor type A | 4 | 13 | |||
| FGA | Fibrinogen alpha chain | 4 | 14 | |||
| FGB | Fibrinogen beta chain | 4 | 14,15 | |||
| FGG | Fibrinogen gamma chain | 4 | 14 | |||
| FKBP4 | FK506 binding protein 4, 59 kDa | 12 | 12 | |||
| FKBP5 | FK506 binding protein 5 | 6 | 12 | |||
| FLCN | Folliculin | 17 | 16 | |||
| GABPA | GA binding protein transcription factor, alpha subunit 60 kDa | 21 | 17 | |||
| GPX1 | Glutathione peroxidase 1 | 3 | 18,19 | |||
| GSTM3 | Glutathione S-transferase mu 3 (brain) | 1 | 20 | |||
| HDAC2 | Histone deacetylase 2 | 6 | 1 | |||
| HDAC5 | Histone deacetylase 5 | 17 | 1 | |||
| HSP90AA1 (HSPCA) | Heat shock protein 90 kDa alpha (cytosolic), class A member 1 | 14 | 12 | |||
| HSP90AB1 (HSPCB) | Heat shock protein 90 kDa alpha (cytosolic), class B member 1 | 6 | 12 | |||
| HSPA1A | Heat shock 70 kDa protein 1A | 6 | 21 | |||
| HSPA1B | Heat shock 70 kDa protein 1B | 6 | 21 | |||
| HSPA1L | Heat shock 70 kDa protein 1-like | 6 | 21 | |||
| HSPA8 | Heat shock 70 kDa protein 8 | 11 | 12 | |||
| IL11 | Interleukin 11 | 19 | 1 | |||
| IL13RA2 | Interleukin 13 receptor, alpha 2 | X | 22 | |||
| ITGB5 | Integrin, beta 5 | 3 | 7 | |||
| JAK3 | Janus kinase 3 | 19 | 1 | |||
| KCND2 | Potassium voltage-gated channel, Shal-related subfamily, member 2 | 7 | 1 | |||
| MAP3K5 | Mitogen-activated protein kinase kinase kinase 5 | 6 | 1 | |||
| MIR146a | MicroRNA 146a | 5 | 23 | |||
| MRPL44 | Mitochondrial ribosomal protein L44 | 2 | 24 | |||
| ORMDL3 | ORM1-like 3 (S cerevisiae) | 17 | 25 | |||
| PTGES3 | Prostaglandin E synthase 3 (cytosolic) | 12 | 12 | |||
| RARRES2 | Retinoic acid receptor responder (tazarotene induced) 2 | 7 | 1 | |||
| SCGB1A1 (CC16) | Secretoglobin, family 1A, member 1 (uteroglobin) | 11 | 26 | |||
| SOD1 | Superoxide dismutase 1, soluble | 21 | 18,27 | |||
| TBX21 | T-box 21 | 17 | 28 | |||
| THSD4 | Thrombospondin, type I, domain containing 4 | 15 | 2 | |||
| TLR2 | Toll-like receptor 2 | 4 | 29,30 | |||
| TLR6 | Toll-like receptor 6 | 4 | 31 | |||
| TNFRSF1A | Tumor necrosis factor receptor superfamily, member 1A | 12 | 32 | |||
| TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B | 1 | 32 | |||