Abstract
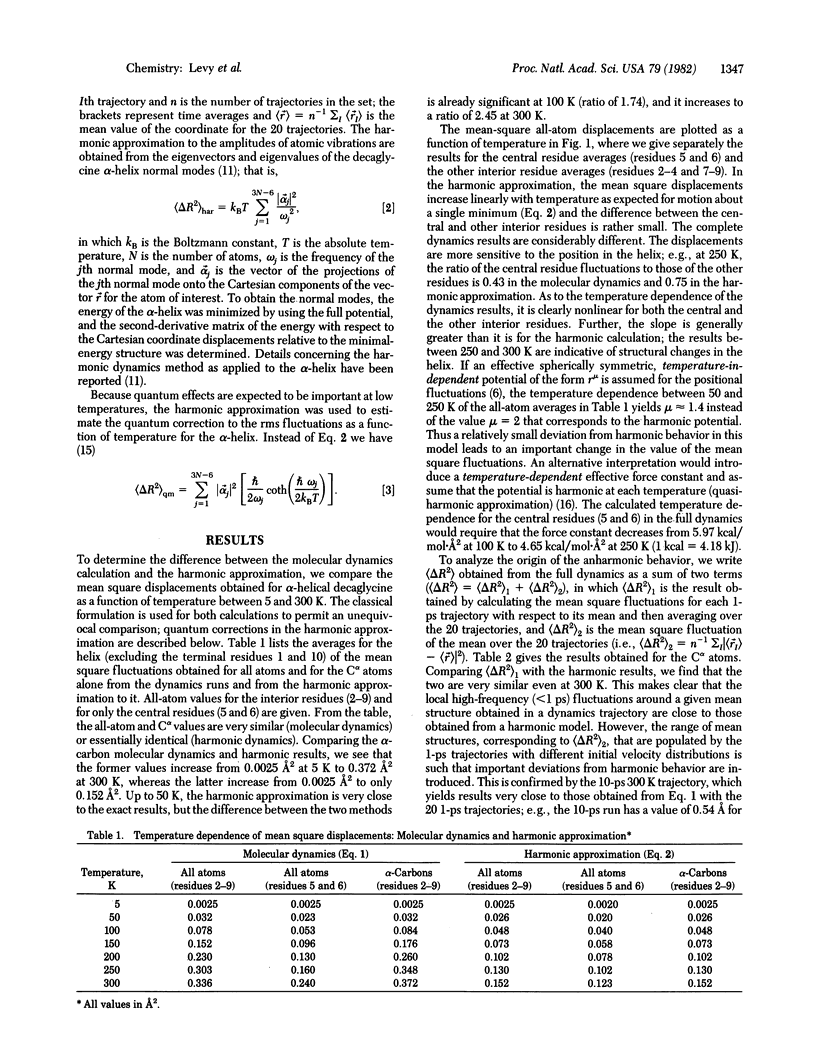
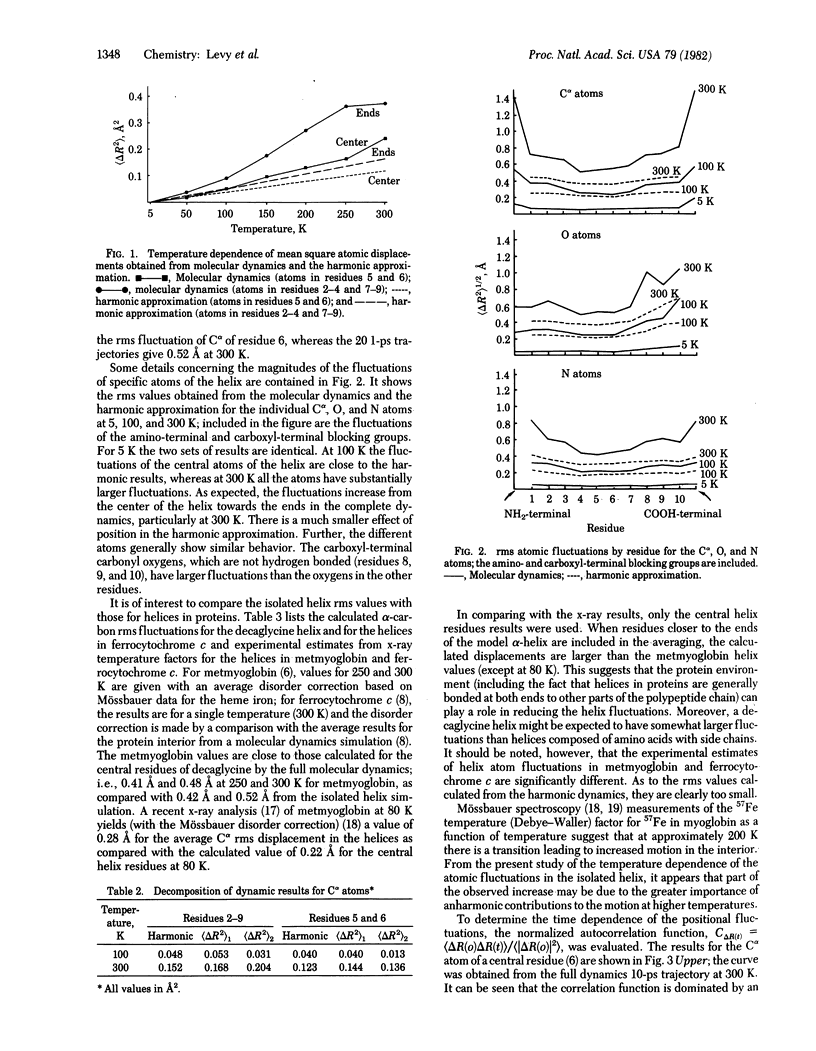
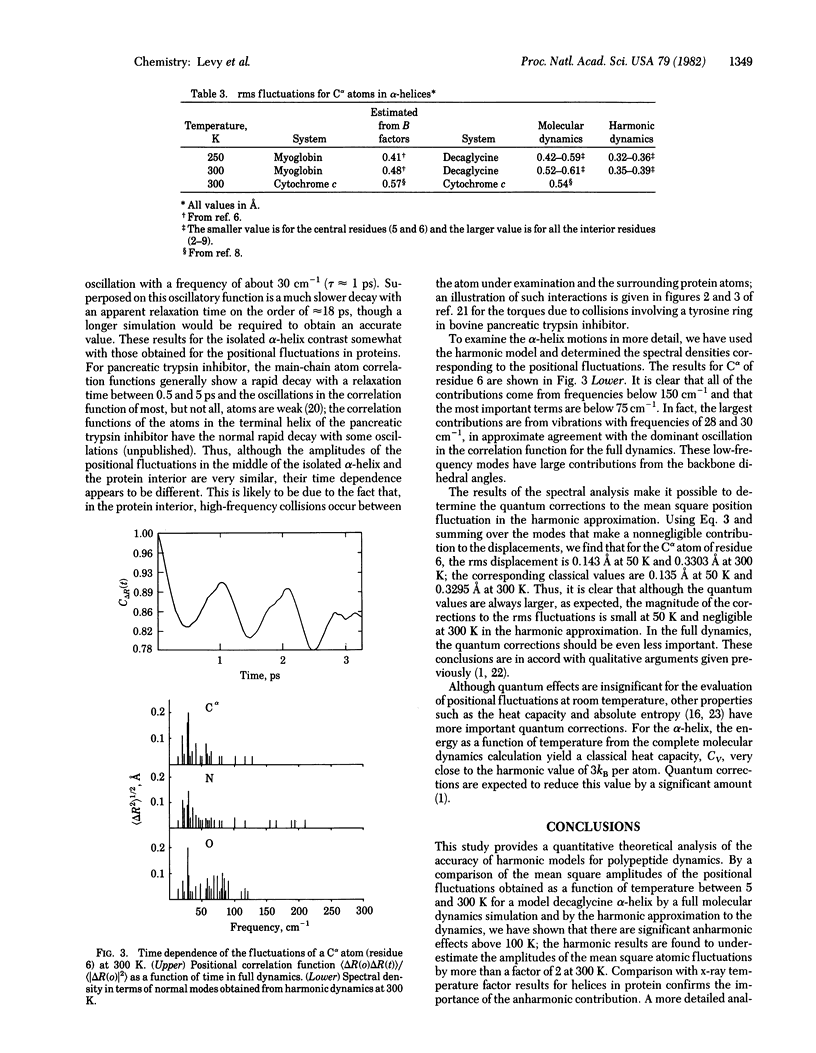
The mean square amplitudes of atomic fluctuations for a polypeptide (decaglycine) α-helix evaluated from molecular dynamics simulations at seven temperatures between 5 and 300 K are compared with analytic harmonic results and with experimental values. Above 100 K the harmonic approximation significantly underestimates the amplitudes of the displacements. Analysis of the time dependence of the fluctuations shows that low-frequency modes (<75 cm-1) dominate the atomic fluctuations and that there is a contribution with a very long relaxation time (>10 ps). Quantum corrections to the amplitude of the fluctuations are found to be small above 50 K. The mean square amplitudes obtained from the molecular dynamics simulations are compared with the values derived from x-ray temperature (Debye-Waller) factors for metmyoglobin (80, 250, and 300 K) and ferrocytochrome c (300 K).
Keywords: protein dynamics, temperature factor, quantum corrections, fluctuations, normal modes
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artymiuk P. J., Blake C. C., Grace D. E., Oatley S. J., Phillips D. C., Sternberg M. J. Crystallographic studies of the dynamic properties of lysozyme. Nature. 1979 Aug 16;280(5723):563–568. doi: 10.1038/280563a0. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Petsko G. A., Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979 Aug 16;280(5723):558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- Go M., Go N. Fluctuations of an alpha-helix. Biopolymers. 1976 Jun;15(6):1119–1127. doi: 10.1002/bip.1976.360150608. [DOI] [PubMed] [Google Scholar]
- Gurd F. R., Rothgeb T. M. Motions in proteins. Adv Protein Chem. 1979;33:73–165. doi: 10.1016/s0065-3233(08)60459-3. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. The internal dynamics of globular proteins. CRC Crit Rev Biochem. 1981;9(4):293–349. doi: 10.3109/10409238109105437. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Gelin B. R., Karplus M. Dynamics of folded proteins. Nature. 1977 Jun 16;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Karplus M. Dynamics of activated processes in globular proteins. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3585–3589. doi: 10.1073/pnas.76.8.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon J. A., Wolynes P. G., Karplus M. Picosecond dynamics of tyrosine side chains in proteins. Biochemistry. 1979 Mar 20;18(6):927–942. doi: 10.1021/bi00573a001. [DOI] [PubMed] [Google Scholar]
- Northrup S. H., Pear M. R., McCammon J. A., Karplus M. Molecular dynamics of ferrocytochrome c. Nature. 1980 Jul 17;286(5770):304–305. doi: 10.1038/286304a0. [DOI] [PubMed] [Google Scholar]
- Parak F., Frolov E. N., Mössbauer R. L., Goldanskii V. I. Dynamics of metmyoglobin crystals investigated by nuclear gamma resonance absorption. J Mol Biol. 1981 Feb 5;145(4):825–833. doi: 10.1016/0022-2836(81)90317-x. [DOI] [PubMed] [Google Scholar]
- Williams R. J. The conformation properties of proteins in solution. Biol Rev Camb Philos Soc. 1979 Nov;54(4):389–437. doi: 10.1111/j.1469-185x.1979.tb00843.x. [DOI] [PubMed] [Google Scholar]


