Abstract
The rut pathway of pyrimidine catabolism is a novel pathway that allows pyrimidine bases to serve as the sole nitrogen source in suboptimal temperatures. The rut operon in E. coli evaded detection until 2006, yet consists of seven proteins named RutA, RutB, etc. through RutG. The operon is comprised of a pyrimidine transporter and six enzymes that cleave and further process the uracil ring. Herein, we report the structure of RutD, a member of the α/β hydrolase superfamily, which is proposed to enhance the rate of hydrolysis of aminoacrylate, a toxic side product of uracil degradation, to malonic semialdehyde. Although this reaction will occur spontaneously in water, the toxicity of aminoacrylate necessitates catalysis by RutD for efficient growth with uracil as a nitrogen source. RutD has a novel and conserved arrangement of residues corresponding to the α/β hydrolase active site, where the nucleophile’s spatial position occupied by Ser, Cys or Asp of the canonical catalytic triad is replaced by histidine. We have used a combination of crystallographic structure determination, modeling and bioinformatics, to propose a novel mechanism for this enzyme. This approach also revealed that RutD represents a previously undescribed family within the α/β hydrolases. We compare and contrast RutD with PcaD, which is the closest structural homolog to RutD. PcaD is a 3-oxoadipate-enol-lactonase-with a classic arrangement of residues in the active site. We have modeled a substrate in the PcaD active site and proposed a reaction mechanism.
Keywords: α/β hydrolases, rut pathway of pyrimidine degradation, RutD, PcaD, aminoacrylate, 3-oxoadipate-enol-lactonase, 3-oxoadipate enol-lactone, E. coli
INTRODUCTION
While the sequence of the E. coli genome has been known for many years, it is still full of surprises and new discoveries, like the rut operon, the pyrimidine catabolism pathway that remained undiscovered until 20061. This operon encodes six enzymes and a pyrimidine transporter that allow pyrimidines to serve as the sole nitrogen source at room temperature. The operon encodes for seven proteins named RutA, RutB, etc. through RutG, where RutA through RutF are enzymes responsible for cleavage and further processing of the uracil ring. RutG is a pyrimidine base transporter. Expression of the operon is controlled by the TetR family repressor RutR and is under the general control of the nitrogen regulatory protein C2. The pathway is present in many bacterial species, including human pathogens and may contribute to survival upon leaving the gut and entering the environment3.
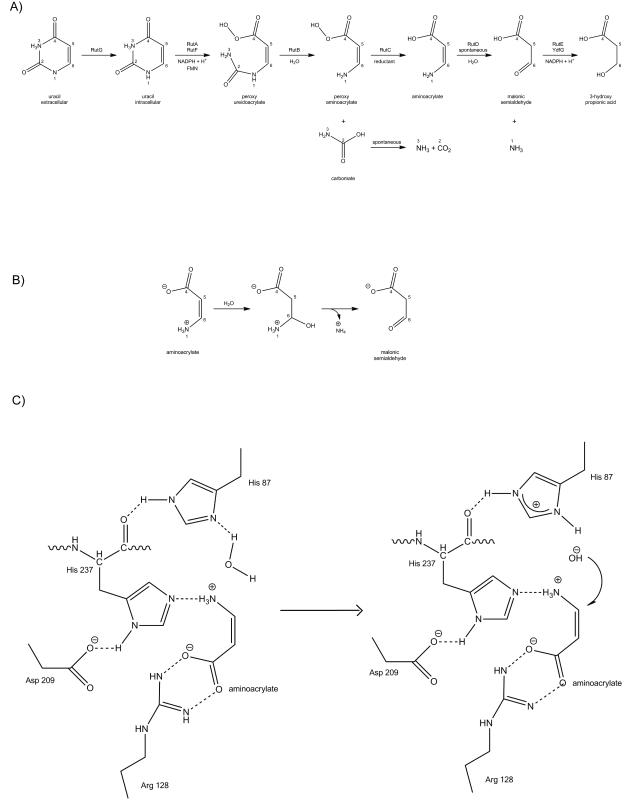
The central enzymes in the pathway are RutA and RutB. RutA works in conjunction with the flavin reductase RutF to catalyze a novel type of reaction where the uracil ring is cleaved between N3 and C4 to yield the intermediate ureidoacrylate4. In vitro, RutB hydrolyzes ureidoacrylate directly to ammonium, malonic semialdehyde and carbon dioxide. In vivo, the rut pathway uses two additional putative enzymes (RutC and RutD) for the neutralization of the toxic effect of the pathway intermediates5 (Fig.2A).
Figure 2.
RutD catalyzed reaction: A) rut pathway; figure adopted from Kim et al. 5 B) A reaction diagram of the hydrolysis of aminoacrylate to malonic semialdehyde. C) A possible mechanism of the proposed hydrolysis of aminoacrylate by RutD.
The RutD protein belongs to the superfamily of α/β hydrolases and is proposed to increase the rate of spontaneous hydrolysis of aminoacrylate to malonic semialdehyde5. The α/β hydrolase fold is highly conserved and characteristic for many hydrolytic enzymes6-8. The core of each enzyme is an α/β sheet, containing 8 β-strands connected by helices. The structures of some α/β hydrolase enzymes contain an inserted lid domain or are fused with other domains of different functions. The classical feature of α/β hydrolases is a well-conserved catalytic triad which comprises a nucleophile (a serine, cysteine or aspartic acid), an acidic residue (glutamate or aspartate), and an invariant histidine6-8.
Herein we show that RutD is the first structure representing a previously undescribed family of α/β hydrolase that utilizes a unique catalytic triad. The X-ray structure, determined to 2.1 Å resolution, reveals that RutD is a homodimer with a well-defined putative substrate binding site. Although co-crystallization or biochemical characterization with the natural substrate is technically prohibitive due the self-hydrolyzing nature of the substrate, we have used the structure in conjunction with sophisticated bioinformatics and molecular docking to propose a chemical mechanism for RutD. We show that RutD is unusual in that it uses a histidine instead of canonical serine in the catalytic triad. Additionally, this combinatorial approach reveals other features that distinguish RutD-like hydrolases from other members of the superfamily.
MATERIALS AND METHODS
Protein cloning expression and purification
Genomic DNA used for cloning was isolated from XL-1 Blue E. coli according to the protocol from Current Protocols of Molecular Biology9. The gene of interest was amplified using following primers: Forward: 5′-GAA TTC CAT ATG AAA CTA TCA CTC TCA CCT CC-3′and Reverse: 5′-AT CTC GAG TTA CAG GGC GGC TTC GCG GTG-3′, then cloned into a modified pET15b vector, altered by substituting the thrombin cleavage site with a TEV cleavage site. The clones were verified by sequence analysis and validated ones were transformed into E. coli strain BL21 (DE3) Codon Plus RIL (Stratagene). The RutD protein was overexpressed in M9 minimal media with selenomethionine (SeMet) at 37°C until the medium OD600 reached 1.2. The cells were induced with 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) plus a cocktail of inhibitory amino acids and SeMet at 16°C overnight, then harvested and stored at -80°C. Protein was purified as described previously10, with Ni-NTA affinity column (Qiagen) and a HiLoad16/60 Superdex 200 prep grade size-exclusion column attached to the ÅKTA FPLC gel filtration system (GE Healthcare). The protein was then concentrated to a final concentration of 13 mg/ml, and used for further crystallization experiments.
Protein crystallization
RutD protein was crystallized using vapor diffusion in hanging drops at 4°C at 6.5 mg/mL protein concentration. The only diffraction quality crystal of RutD was obtained from the Qiagen JCSG+ initial screen, where the precipitant mother liquor contained 0.15 M malic acid pH=7.0, and 20% w/v PEG 3350. Surprisingly, further optimization gave better looking, but poorly diffracting crystals. The crystal was flash frozen in liquid nitrogen using 1:1 mixture of mother liquor and 50% glycerol for cryoprotection.
Data collection and processing
Diffraction data for selenomethionine substituted RutD were collected on the 21ID-F beam line of the Advanced Photon Source (APS) at Argonne National Laboratory using the SAD protocol at 100K, using a MAR 300 CCD detector. RutD crystals are in the P41212space group with two protein chains in asymmetric unit. Data reduction was performed with HKL-200011. During data collection it became apparent that what visually appeared to be a single crystal was actually a cluster composed of at least 3 crystals. In order to process the data we needed to manually pick peaks corresponding to the main lattice. Structure solution and initial model was obtained by HKL-3000 package12, which interacts with SHELXC/D/E, 13,14, MLPHARE15, DM16, ARP/wARP17, CCP418,19, SOLVE20, RESOLVE21 and COOT 22. Initial sites of the anomalous scattering atoms were found by SHELXD and refined in MLPHARE. Subsequently, the initial phases were improved using solvent flattering followed by autotracing using ARP/wARP. The resulting electron density map was of much better quality than expected from diffraction images. Refinement of the initial model was done in COOT 22 by alternating rounds of manual fitting of the model to electron density maps and maximum likelihood refinement with REFMAC 23,24.MOLPROBITY 25 and ADIT26 were used to validate the structure. Data solution and refinement statistics are presented in Supplementary Table I. The structure was deposited in PDB databank under the code 3V48.
Ligand docking procedure
The ICM-PRO software (Internal Coordinate Mechanics Professional; Molsoft LLC) 27 was used for docking of the model substrate to RutD. A rigid ICM model of the protein was prepared from the PDB style coordinates using the ICM conversion procedure, which includes the addition and local minimization of hydrogen atoms in the internal coordinate space and the selection of energetically favorable side chains for the His, Asn, and Gln residues. All water molecules were removed from the structure. Only one protein molecule (chain A) from the asymmetric unit was used for the calculations. ICM-PRO was then used to identify possible binding sites (pockets) in the receptor structure from the maps calculated with the grid size of 0.5 Å. The aminoacrylate substrate model was generated using ICM-PRO. An optimized ligand in cis conformation was placed in the vicinity of the identified pocket and the docking was performed applying the ICM-PRO standard docking approach.
CLANS clustering
Clustering of over 26 000 sequences based on their pair-wise BLAST similarity scores was performed using CLANS (CLuster ANalysis of Sequences)28. As a query we used the following collection of sequences: PFAM family Abhydrolase_6 (PF12697)29 containing only α/β hydrolase domain, and results of a PSI-BLAST 30 search of the NCBI Genbank protein non-redundant database to convergence using the RutD and PcaD31 sequences as queries. After initial clustering with a P-value of 10−5 we removed all sequences that made no connection with the central clusters containing the proteins of interest, and clustered the remaining set of over 14000 sequences with the P-value of 10−22. More permissive values caused over compaction of the whole dataset, while more stringent values caused its dispersion and disengagement of the most diverged clusters. This P-value resulted in obtaining consistent and separate clusters of the RutD and PcaD groups. Since analysis of the whole α/β hydrolase superfamily is beyond the scope of this paper we analyzed only groups containing RutD and PcaD homologues. Sequences from those groups were extracted, saved as FASTA32 files and MSAs (multiple sequence alignments) were constructed using ClustalX233 and then edited by hand in BioEdit34 by adjusting the positions of the gaps. In addition, we used the NCBI CDD Batch35 tool to check family and superfamily annotation of the query.
RESULTS AND DISCUSSION
Overall Structure of RutD monomer
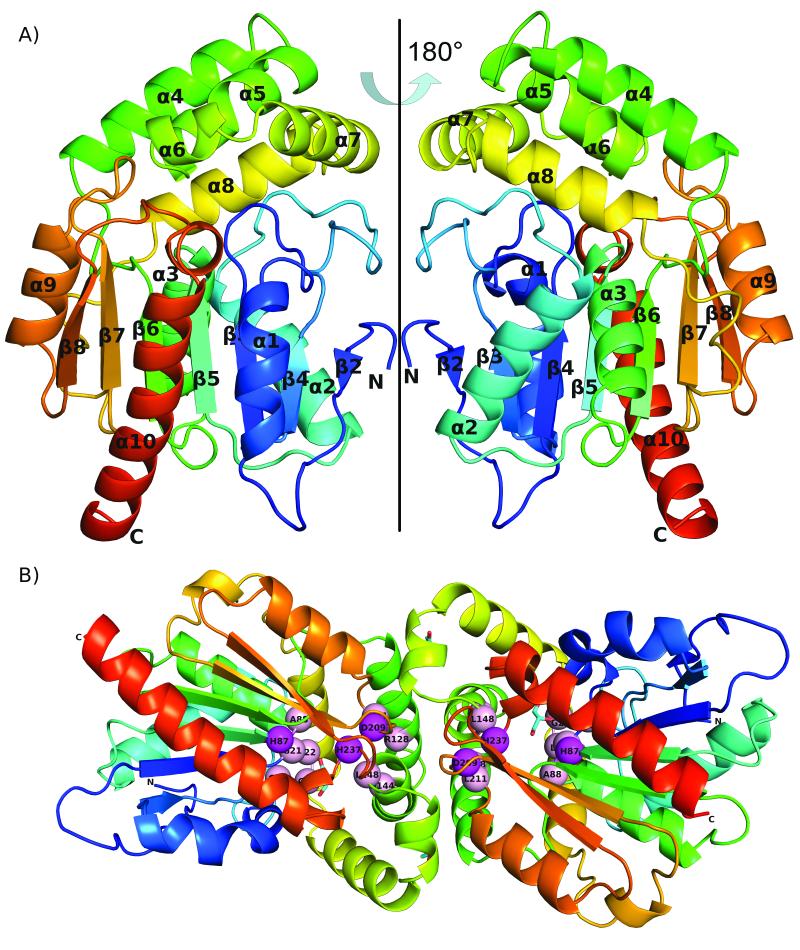
The RutD protein has an α/β hydrolase fold and forms a dimer. The overall structure is presented in Fig.1.A single polypeptide chain of RutD is composed of 266 amino acids, and all residues are visible in the electron density map. The architecture of the RutD monomer is different from the canonical α/β hydrolase fold, as there is a deletion of the first β strand. (To be consistent with previous descriptions7 of α/β hydrolase structures, we start numbering β strands from β2). Thus, the monomer is formed by a 7-stranded (rather than 8-stranded) β-sheet, in which strands β3-β7 are parallel, and β2 is anti-parallel. The β-sheet is significantly twisted, such that strands β2 and β8 are oriented approximately 90° to one another. The β-sheet is surrounded on both sides by five α-helices (α1, α2, α3, α9, and α10), which together with the sheet compose the core of the enzyme. The other five α-helices (α4-α8) form a separate lid domain inserted between β6 and α8. The lid domain is observed in other α/β hydrolases8,31 where it plays a role in active site accessibility, substrate recognition, and oligomerization.
Figure 1.
Overall structure of RutD protein: A) monomer shown in two orientations; the protein chain is colored according to residue number with N-terminus being blue and C-terminus red B) dimer; Putative active site residues (listed in Table 1) are shown as spheres, the catalytic triad in magenta, structural residues in pink. Glycerol molecules in the active site and on the dimer interface are shown in cyan
Quaternary structure of RutD
The asymmetric unit contains two monomers, with a decrease in solvent-accessible surface area of 910 Å2 for the dimer assembly. The oligomeric state is consistent with gel filtration results observed during the purification process (data not shown). The dimer interface is formed by helices α4 - α7. The majority of interactions on the dimerization interface are hydrophobic, but there are salt bridges formed by two arginine residues (R123 and R130 from the first chain) to one glutamic acid residue (E142 from the second chain), as well as a π-π stacking interaction between the Y234 side chains of each monomer. Despite their possible importance in the formation of monomer-monomer interface they are not conserved in the α/β hydrolase family.
In the crystal structure of the RutD protein we observed four ordered glycerol molecules, two of them occupying the putative active site, and two within the dimer interface. Glycerol molecules that are bound between two monomers form contacts with the following residues involved in the dimerization process: M154, H120, R123 and A158.
Putative active site of RutD
The α/β hydrolases have a well conserved and characteristic active site, which is composed of a catalytic triad: a nucleophile, an acidic residue and a histidine. The nucleophile can be serine, cysteine or aspartic acid and is located on the tight turn between β5 and α3: the so-called “nucleophilic elbow”. The nucleophilic elbow is defined by the motif Sm-X-Nu-X-Sm, where Sm is a small side chain amino acid, X any amino acid, and Nu nucleophile. The acidic residue can be an aspartic or glutamic acid, and the histidine is invariant6-8. In RutD, the residues corresponding to the α/β hydrolase catalytic triad are of unusual composition – the first position of the triad (the nucleophile position in other α/β hydrolases) is substituted by histidine (H87). This substitution is 100% conserved among all known RutD proteins (Table I).
Table I.
RutD putative active site composition. Residues in the 3.5 Å sphere in vicinity of glycerol molecule are listed. To obtain percentage value of conservancy for the table, MSA of RutD and RutD-like sequences obtained from CLANS analysis were used, and the percentage was calculated by BioEdit 34. Residues involved in ligand binding and reaction shown in bold.
| residues forming active site | % of conservancy |
|---|---|
| G21 | 100 |
| L22 | 99 |
| G23 | 100 |
| G24 | 100 |
| H87 | 100 |
| A88 | 88 |
| R128 | 100 |
| Q144 | 88 |
| L148 | 91 |
| D209 | 100 |
| L211 | 91 |
| H237 | 100 |
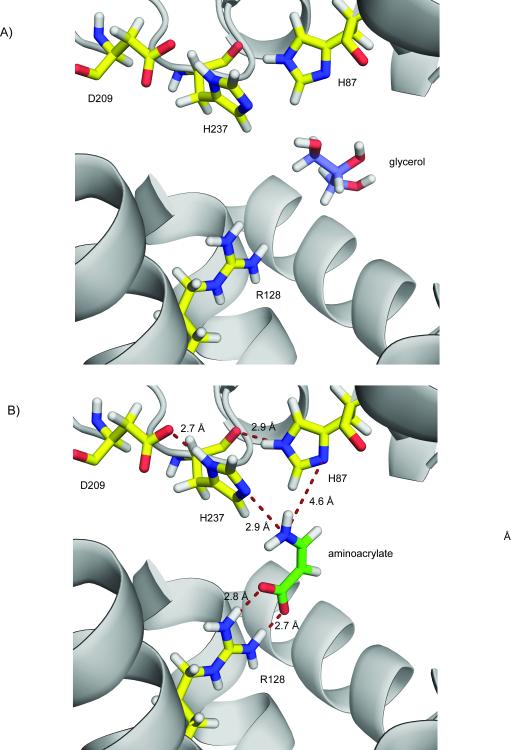
After determining the X-ray structure, we observed a glycerol molecule bound in the cleft created by cap helices 6 and 7 and loops from the core of the enzyme. However, while the glycerol molecule does not make any significant contacts with the protein apart from one hydrogen bond with the main chain nitrogen of G23 bridged through water molecules, it occupies the cleft presumed to be an active site.
Secondly, we predicted and calculated the volumes of the clefts present in the RutD crystal structure. Only one predicted cavity is well defined, which is 320 A3 in volume and corresponding to the cleft occupied by the glycerol molecule. The cavity has the amino acid composition characteristic for the α/β hydrolase active site. A list of residues forming the putative active site of the RutD protein, as well as the degree of conservation of each among members of the RutD-like proteins group, is shown in Table I.
We constructed a MSA of proteins homologous to RutD and observed 100% conservation of the following residues: H87, R128, D209, and H237 (Fig. S1). We also superimposed similar structures predicted by the DALI server36 on the RutD structure using the SSM algorithm37, and observed that the positions of the three residues corresponding to the catalytic triad are also conserved (Table II).
Table II.
Composition of residues of RutD protein corresponding to active site residues in other representatives of α/β hydrolases superfamily. H87 is in the spatial position of the nucleophile in the catalytic triad.
| Protein name |
RutD | PcaD | Bromo- peroxidase |
RP2 Lipase |
Lipase G.candidum |
Halokane Dehalo genase |
Acetyl choline esterase |
Serine carboxy Peptidase |
|---|---|---|---|---|---|---|---|---|
|
PDB code
Position in active site: |
3V48 | 2XUA | 3FOB | 1GPL | 1THG | 2HAD | 2XI4 | 3SC2 |
| Nucleophile | H87 | S100 | S99 | S152 | S217 | D124 | S200 | S146 |
|
Acidic
residue |
D209 | D217 | D229 | D205 | E354 | D260 | E237 | D338 |
| Histidine | H237 | H244 | H258 | H263 | H463 | H289 | H440 | H397 |
H87 is located on the sharp turn between β5 and α3, and the composition of the “nucleophilic elbow” is V85, G86, H87, A88, L89, and G90. The RutD residue corresponding to the acidic residue in the α/β hydrolase active site is D209, which is located on the loop between β7 and α9. H237 is located on the loop between β8 and α10. Another interesting element of the putative RutD active site is R128, which is also conserved. We suggest that the residue has a significant structural role for the positioning of aminoacrylate in the putative active site. It is located on the lid forming α4. This arrangement of active site arginines was previously reported in 3-oxoadipate-enol-lactonase PcaD from Burkholderia xenovorans LB40031 and C-C hydrolase MhpC from E. coli 7,38.
In the active site area there is also a flexible loop composed of the 4 highly conserved (up to 100%) residues G21, L22, G23, G24 (Table 1).
Substrate docking and reaction mechanism
According to Kim et al. the hydrolysis of aminoacrylate to malonic semialdehyde is the fourth step in the Rut pathway (Fig. 2A). Despite the fact that it happens spontaneously in aqueous solvents, the presence of the RutD suggests that the efficiency of this reaction is a key factor for the Rut pathway to function correctly5. It has previously been suggested that the toxic intermediates of this pathway may serve as a means of regulating growth during periods of suboptimal conditions5,3. Indeed the product of the RutD reaction, malonic semialdehyde, is likely more toxic than aminoacrylate. RutE or the short-chain dehydrogenase YdfG is required to reduce the semialdehyde to 3-hydroxypropionic acid5.
To address the question of the possible mechanism for the proposed hydrolysis of aminoacrylate to malonic semialdehyde, we used ICM-PRO to dock the substrate in the RutD protein structure. The substrate binding cavity identified by the docking agreed with the glycerol binding site in the crystal structure of RutD. In the docking model, the optimal conformation of the aminoacrylate molecule is well positioned in the active site and is coordinated by the conserved residues identified above: H87, R128, D209, and H237 (bold residues in Table I, aminoacrylate docked in putative active site is presented on Fig.3B). We hypothesize that conserved residues H87, H237, and D209 could serve as catalytic residues. We suggest that D209 is critical for maintaining the correct protonation state of H237 side chain. If RutD hydrolyzes aminoacrylate in the orientation observed in the docking model, the correct protonation state of both H87 and H237 has to be achieved. H237 may be forced into in a deprotonated state on nitrogen 2 of the imidazole ring by hydrogen bond formation between side-chain D209 and the opposite nitrogen atom on the H237 ring. Hydrogen bonding between H87 and the main chain carbonyl of H237 will likewise ensure that the second nitrogen of the H87 imidazole ring is available for water activation. Both histidine sidechains may be in forced protonation states which would allow the formation of hydrogen bonds with the amino group of aminoacrylate molecule. The sidechain of R128 forms an electrostatic interaction with the carboxyl group of aminoacrylate. According to our model the sidechain of arginine 128 is forming hydrogen bond network with carboxyl group of aminoacrylate in similar fashion as reported previously39.
Figure 3.
Close-up of RutD active site: A) glycerol bound in the putative active site cleft (waters are not shown). B) Docking results: aminoacrylate coordinated by presumed active site residues.
Docking results allowed to propose the basis of mechanism for RutD catalyzed reaction. The amino acid composition of RutD putative active site may be determined by the chemical character of aminoacrylate hydrolysis (amino group hydrolysis). We hypothesize that RutD catalyzes the hydrolysis of aminoacrylate by the following mechanism. The aminoacrylate is positioned in the active site by a combination of a strong interaction of the carboxylate with R128, and the amino group is coordinated by a deprotonated nitrogen of the H237 imidazole ring. The nearby H87 hydrogen bonds with a backbone carbonyl, allowing the Nδ to act as a base to activate water. It is important to note that the positions of waters in our structure will most likely not directly correspond to the positions of waters in an aminoacrylate bound form. Substitution of the canonical nucleophile in the first position of the catalytic triad with histidine may permit subsequent nucleophilic attack of the hydroxyl ion (from the activated water) on the carbon atom at the sixth position in the aminoacrylate molecule. If the aminoacrylate molecule is in cis conformation (which would be the natural conformation for the RutC product), then the carbon in the sixth position in the molecule faces H87 and the proposed activated water molecule.
The putative catalytic triad in RutD appears to be rearranged spatially in comparison to other α/β hydrolases active sites. The conserved H237 position in RutD appears to be responsible for substrate amino group coordination in the docking model. In most α/β hydrolases, this spatial position is occupied by a His residue responsible for water activation. However, in the proposed RutD mechanism, water activation is done by H87, which spatially occupies the position of conserved serine nucleophile in other α/β hydrolases. Binding of carboxyl of aminoacrylate by side chain of R128 is likely to neutralize the negative charge of the carboxyl group. The proposed reaction mechanism is shown in Fig. 2C.
New family
The α/β hydrolase fold is widely distributed in nature and the overall structure is highly conserved in evolution despite relatively low similarity on the sequence level. In the SCOP40 classification, there are 41 families within the α/β hydrolase superfamily, but proteins with new hydrolytic functions are being reported. The Uniprot41 and CDD35 databases annotate RutD as a part of the α/β hydrolase superfamily, but do not distinguish it as a separate family.
As reported by Loh et al1, rut genes are clustered and found in α- and γ-proteobacteria, mostly in Enterobacteriaceae. Detailed analysis of the occurrence of the rut operon was presented in the Loh paper reporting existence of the new metabolic pathway in E. coli1. After PSI-BLAST42, 30 analysis using the RutD protein sequence as a query, we obtained sequences of RutD-like proteins, all having conserved characteristic features. We found that the most similar homologs are 3-oxoadipate-enol-lactonases. The features that distinguish RutD-like proteins from other α/β hydrolase superfamily members are: (1) histidine in the position of a nucleophile in the catalytic triad, (2) an absolutely conserved R128, and (3) deletion of the first β-strand. These conserved features led us to speculate that the RutD-like proteins form a new family within the α/β hydrolase superfamily. To investigate this idea further, we analyzed the α/β hydrolase superfamily by clustering the α/βhydrolase_6 PFAM superfamily members based on their pair-wise BLAST similarity scores using CLANS. The results of the clustering analysis (shown in Fig.S2) revealed that RutD creates a single compact cluster with a few satellite (less similar) sequences weakly connected to the main cluster. Further analyses (see Materials and Methods) via construction of MSAs (Fig. S1) and CDD annotation checks showed that the main cluster is composed of sequences showing all the characteristics of the proposed new RutD family listed above. Interestingly, the less similar sequences at the position of the catalytic triad nucleophile have a histidine followed by a serine residue. Since no information is published about any of those proteins we cannot say which residue for sure is the active part of a catalytic triad. We hypothesize that a RutD could have evolved from an enzyme in which mutation of serine to alanine gave new function to histidine placed immediately upstream.
PcaD
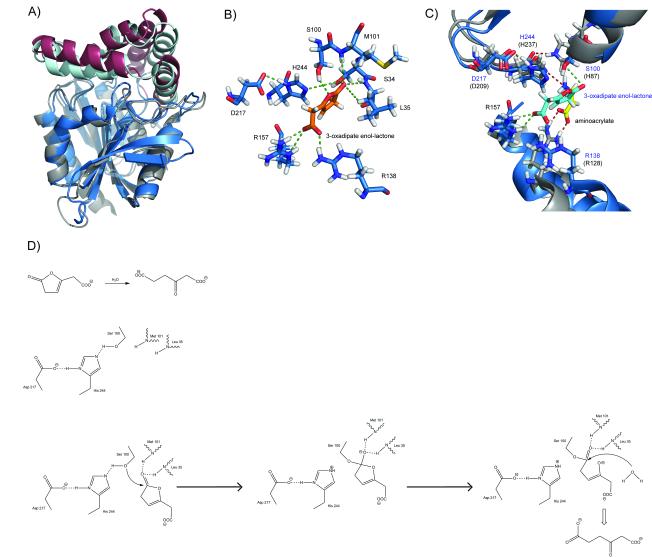
The closest structural homolog of RutD protein, found by DALI36 search is 3-oxoadipate-enol-lactonase (PcaD) from Burkholderia xenovorans LB400 (PDB id:2XUA)31. They share sequence identity of 24%, and superposition of these two structures using the SSM algorithm gives an RMSD (Root Mean Square Deviation) value of 1.6 Å for 233 aligned Cα atoms. This superposition is presented in Fig.4A. Both structures have a similar overall fold, except that PcaD has the classic number of 8 β-strands. Both proteins have a cap domain formed by five α-helices, which are shifted compared to the RutD protein. The active site of PcaD has a classic α/β hydrolase catalytic triad with S100 as the nucleophile, D217 and H244. It also contains two arginine residues (R138 and R157, conserved in 100% and 93% of PcaD sequence homologs, (data not shown) protruding from the α-helices of the cap domain. This enzyme catalyzes the hydrolysis of 3-oxoadipate-enol-lactone to 3-oxoadipate in the protocatechuate pathway31. Bains et al reports the structure of PcaD protein with a product analog (levulinic acid) bound within the active site. We performed docking of the PcaD physiological substrate (3-oxoadipate-enol-lactone) into the PcaD structure and on the basis of those results we propose both a possible binding mode for the substrate and a suggested reaction mechanism (Fig. 4B, 4D). Despite the fact that the spatial composition of the PcaD active site is virtually identical with the putative active site of RutD, the mechanism of hydrolysis for PcaD protein is very different and falls into the category of canonical hydrolysis of carbonyl-group containing substrates (Fig 4C). The clustering analysis also showed that PcaD proteins form a single cluster in the proximity, but with no strong connection to the RutD cluster. MSAs and reciprocal CDD analysis of sequence confirm the PcaD sequence characteristics.
Figure 4.
PcaD protein: A) Superposition of the RutD and PcaD proteins: the RutD core is shown in gray, the RutD lid domain in burgundy, the PcaD core in blue, and the RutD lid in cyan. B) Arrangement of the active site residues with a physiological ligand docked: the catalytic triad S100, D217 and H244; the residues forming the oxyanion hole M101 and S34 /L35 R157 and R138; and 3-oxoadipate-enol-lactone (shown in orange). C) A detailed view of the superposition of the RutD and PcaD active sites: RutD residues shown in grey and residues’ names shown in black, PcaD active site residues in blue and residues; names shown in navy, 3-oxoadipate enol-lactone shown in cyan, and aminoacrylate shown in yellow. D) Proposed reaction mechanism of PcaD-catalyzed hydrolysis of 3-oxoadipate enol-lactone to 3-oxoadipate.
CONCLUSIONS
The determination of the RutD structure is an important step towards the full structural characterization and understanding of the rut pathway. While RutD clearly belongs to the α/β hydrolase superfamily, its structural features, like the unusual composition of the residues corresponding to the catalytic triad, suggest that it might be a member of a separate family in the α/β hydrolase superfamily. The Rut pathway is a very recently discovered metabolic pathway and further structural characterization of RutD and other members of Rut operon will be crucial for complete understanding of the enzymology and physiology of pyrimidine degradation in Enterobacteriacae, which in turn may lead to the design of novel antibiotics and bacteriostatic drugs.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like thank Dr. Matthew D. Zimmerman for valuable discussions and Michal Szpak for help with preparation of figure 2 and 4. The work described in this paper was supported by NIH grant GM094662 to New York Structural Genomics Consortium.
Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817)
REFERENCES
- 1.Loh KD, Gyaneshwar P, Papadimitriou E Markenscoff, Fong R, Kim KS, Parales R, Zhou Z, Inwood W, Kustu S. A previously undescribed pathway for pyrimidine catabolism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(13):5114–5119. doi: 10.1073/pnas.0600521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimada T, Hirao K, Kori A, Yamamoto K, Ishihama A. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol Microbiol. 2007;66(3):744–757. doi: 10.1111/j.1365-2958.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- 3.Parales RE, Ingraham JL. The surprising Rut pathway: an unexpected way to derive nitrogen from pyrimidines. J Bacteriol. 2010;192(16):4086–4088. doi: 10.1128/JB.00573-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukherjee T, Zhang Y, Abdelwahed S, Ealick SE, Begley TP. Catalysis of a flavoenzyme-mediated amide hydrolysis. Journal of the American Chemical Society. 2010;132(16):5550–5551. doi: 10.1021/ja9107676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim KS, Pelton JG, Inwood WB, Andersen U, Kustu S, Wemmer DE. The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J Bacteriol. 2010;192(16):4089–4102. doi: 10.1128/JB.00201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. The alpha/beta hydrolase fold. Protein Eng. 1992;5(3):197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 7.Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9(6):732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 8.Holmquist M. Alpha/Beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr Protein Pept Sci. 2000;1(2):209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 9.Current protocols in molecular biology. Greene Pub. Associates; Brooklyn, N.Y. Media, Pa.: 1987. J. Wiley, order fulfillment. [Google Scholar]
- 10.Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure. 2001;9(11):1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallographica Section D-Biological Crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 13.Sheldrick GM. A short history of SHELX. Acta crystallographica Section A, Foundations of crystallography. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 14.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta crystallographica Section D, Biological crystallography. 2010;66(Pt 4):479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Wolf W, Evans PR, Leslie AGW, editors. Isomorphous replacement and anomalous scattering. Daresbury Laboratory; Warrington, UK: 1991. pp. 80–86. [Google Scholar]
- 16.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72(3):245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 17.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6(5):458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 18.The CCP4 suite: programs for protein crystallography. Acta crystallographica Section D, Biological crystallography. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallographica Section D-Biological Crystallography. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov Garib N, Lebedev Andrey A, Pannu Navraj S, Steiner Roberto A, Nicholls Robert A, Winn Martyn D, Long Fei, Vagin Alexei A. REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallographica Section D Biological Crystallography. 2011;67(4):355–367. doi: 10.1107/S0907444911001314. PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson DC, Lovell SC, Davis IW, Adrendall WB, de Bakker PIW, Word JM, Prisant MG, Richardson JS. Structure validation by C alpha geometry: phi,psi and C beta deviation. Proteins-Structure Function and Genetics. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 26.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta crystallographica Section D, Biological crystallography. 2004;60(Pt 10):1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 27.Abagyan R, Totrov M. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins-Structure Function and Genetics. 1997:215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 28.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20(18):3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 29.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32(Database issue):D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bains J, Kaufman L, Farnell B, Boulanger MJ. A product analog bound form of 3-oxoadipate-enol-lactonase (PcaD) reveals a multifunctional role for the divergent cap domain. J Mol Biol. 2011;406(5):649–658. doi: 10.1016/j.jmb.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Pearson WR. Rapid and Sensitive Sequence Comparison with Fastp and Fasta. Methods in Enzymology. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. Clustal-W - Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium. 1999:41. [Google Scholar]
- 35.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta crystallographica Section D, Biological crystallography. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Li JJ, Montgomery MG, Wood SP, Bugg TD. Catalytic role for arginine 188 in the C-C hydrolase catalytic mechanism for Escherichia coli MhpC and Burkholderia xenovorans LB400 BphD. Biochemistry. 2006;45(41):12470–12479. doi: 10.1021/bi061253t. [DOI] [PubMed] [Google Scholar]
- 39.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer ELL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36(Database issue):D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.