Abstract
Bacterial and fungal species produce some of the best-characterized functional amyloids, i.e. extracellular fibres that play key roles in mediating adhesion and biofilm formation. Yet, the molecular details underlying their mechanical strength remain poorly understood. Here, we use single-molecule atomic force microscopy to measure the mechanical properties of amyloids formed by Als cell adhesion proteins from the pathogen Candida albicans. We show that stretching Als proteins through their amyloid sequence yields characteristic force signatures corresponding to the mechanical unzipping of β-sheet interactions formed between surfacearrayed Als proteins. The unzipping probability increases with contact time, reflecting the time necessary for optimal inter β-strand associations. These results demonstrate that amyloid interactions provide cohesive strength to a major adhesion protein from a microbial pathogen, thereby strengthening cell adhesion. We suggest that such functional amyloids may represent a generic mechanism for providing mechanical strength to cell adhesion proteins. In nanotechnology, these single-molecule manipulation experiments provide new opportunities to understand the molecular mechanisms driving the cohesion of functional amyloid-based nanostructures.
Keywords: AFM, amyloids, functional nanomaterials, nanomechanics, pathogens, single-molecule manipulation
Amyloids are partially ordered insoluble fibrous aggregates of proteins that are frequently associated with a range of diseases, including Alzheimer’s, Parkinson’s and prion diseases.1,2 Amyloidogenic proteins assemble into β-sheet-rich fibers where the individual β-strands are oriented perpendicular to the fiber axis and connected through a periodic network of hydrogen bonds that are approximately parallel to the fiber axis.3 The fibers display a unique structural motif, denoted as steric zipper, in which the side chains protruding from two β-sheets are tightly interdigitated and self-associated through van der Waals interactions.4,5 Many polypeptides can form amyloid under modestly denaturing conditions, indicating that amyloid is a widely accessible, low-energy protein quaternary structure.
Unlike pathological amyloids, some amyloid fibrils are used by living cells to perform diverse physiological functions through long-lived protein-protein interactions.6–8 These amyloid-based nanostructures offer promising prospects in nanotechnology for the design of novel self-assembling materials.8 Functional amyloids, which share many features of classical disease-associated amyloids, have been identified in many organisms, from bacteria to humans. Several bacterial and fungal species produce extracellular amyloid compounds that are used to strengthen cell surface interactions such as adhesion, aggregation and biofilm formation. Prominent examples are the proteinaceous fibers (curli) produced by enteric bacteria like Escherichia coli, and known to mediate biofilm formation, host colonization, immune activation and cell invasion.9,10 More recently, Als adhesion proteins from the fungal pathogen Candida albicans have been shown to form functional amyloids.11–15 Cell adhesion mediated by Als proteins depends on conformational change in the molecules, rather than on signal transduction and expression of new surface proteins. The conformational changes result in the formation of nanoscale ordered clusters of Als adhesins on the cell surface, which bind dyes that are used to stain amyloids, like thioflavin T and Congo red.15 These adhesive nanodomains, in turn, mediate high avidity interactions between adhering cells. Consistent with the cellular behavior, Als proteins have conserved amyloid-forming sequences and form amyloid fibers in vitro.12 The V326N single site mutation in the amyloid-forming region of Als5 abrogates amyloid formation and aggregation.13,14,16 The strength of yeast adhesion results from the forceactivated amyloid-like clustering of hundreds of proteins on the cell surface to form arrays of ordered multimeric binding sites.15,16 These amyloid clusters explain why Als proteins often show weak binding to specific ligands, yet mediate remarkably strong adherence.15 Despite the key role that microbial amyloids, like Als amyloids, play in mediating cell adhesion, aggregation and biofilm formation, the molecular details underlying their mechanical strength remain poorly understood.
Recent advances in single-molecule manipulation experiments have offered new opportunities for studying the elasticity of proteins.17,18 Stretching modular proteins, like titin or tenascin, yields characteristic force signatures that reflect the force-induced unfolding of secondary structures (α-helices, β-sheets).19–21 Pulling on amyloid β-peptides associated with Alzheimer’s disease reveals force plateau responses attributed to the unzipping of individual β- sheets from the amyloid fibrils.22,23 To date, however, the mechanical properties of functional microbial amyloids have never been investigated. Here, we use single-molecule atomic force microscopy (AFM) to study the nanomechanics of amyloids made of C. albicans Als adhesion proteins. Our results are consistent with a model in which individual β-sheets formed by the lateral assembly of short β-strands from hundreds of Als proteins can be unzipped with a constant force and under equilibrium conditions. Occurrence of zipper interactions strongly increases with interaction time, which is attributed to the time necessary for conformational changes for optimal β-strand associations. Zipper interactions are detected in Als monolayers displaying an orientation mimicking the yeast cell surface, but not in mature amyloid fibers, suggesting these structures are too stable to be unzipped. Collectively, our results suggest that the Als amyloid functions as a strongly cohesive molecular zipper that mediates long-lived homophilic protein interactions, thereby strengthening cell adhesion.
RESULTS AND DISCUSSION
Unzipping individual β-sheets
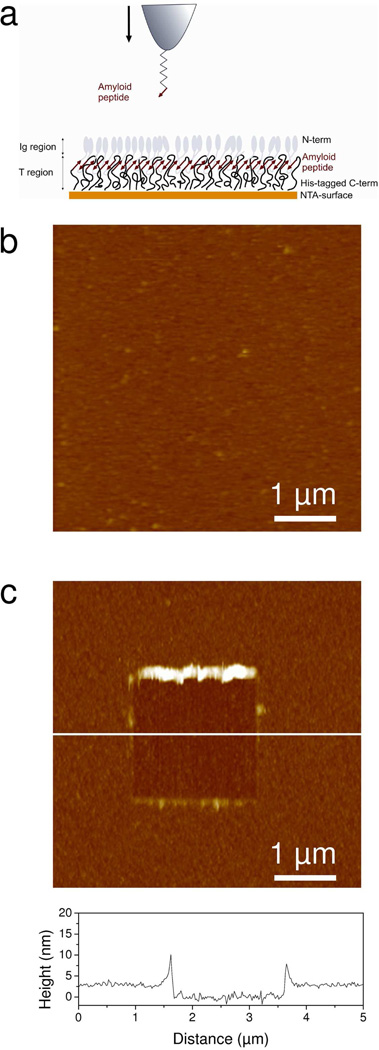
To study the mechanical properties of Als amyloids, oriented monolayers of Als proteins were probed using biologically-modified AFM tips. To this end, 414 amino acid residue fragments of Als5p containing the amyloid region (hereafter denoted as “Ig-T fragments”) were attached through their His-tagged C-terminal regions on an Ni2+-NTA surface in order to mimic the orientation occurring on the yeast cell surface (Figure 1a). Als proteins were probed through their amyloid regions using an AFM tip modified with a short amyloid-forming peptide homologous to the amyloid forming region of Als5p, SNGIVIVATTRTV (hereafter “amyloid-tip”) (Figure 1a).12,14 Thus, the derivatized tips should probe the homologous amyloid-like interactions, rather than the ligand-receptor interactions that we have previously documented.24,25
Figure 1.
Strategy for measuring the nanomechanical response of Als amyloids. (a) Principle of the single-molecule manipulation experiments. Each Als protein protruding from the yeast surface contains a ligand-binding Ig-like region (Ig) made of two β-sheet-rich domains followed by an amyloidogenic threonine-rich region (T). To mimic this orientation, Ig-T fragments terminated with a His-tag at the C terminal were assembled on a gold surface terminated with Ni2+-NTA (10 %) and EG3 (90 %) groups. Note that the cartoon is idealized and that the surface may be less structured than depicted. The surface-displayed Ig-T molecules were stretched via their amyloid region using an AFM tip functionalized with the amyloid-forming peptide SNGIVIVATTRTV. (b) AFM deflection image (5 µm × 5 µm) in PBS confirming the presence of a homogeneous, featureless Ig-T layer. (c) To determine the layer thickness, 2 µm × 2 µm square areas were first scanned at large forces (10 nN), followed by imaging 5 µm × 5 µm images of the same areas under smaller forces. A cross-section taken in the corresponding height image along the white line is shown under the image.
To validate the quality of the functionalized surfaces, Ig-T layers were imaged with a silicon nitride tip in buffer. The morphology of the protein monolayer (Figure 1b) was smooth, devoid of any fibrillar structures, and stable upon repeated scanning, indicating strong attachment of the proteins to the surface. Scanning a small area (2 µm × 2 µm) at large forces (10 nN) resulted in the removal of the protein layer and allowed us to directly determine its thickness, 3.1 ± 0.6 nm (mean ± standard deviation σ, Figure 1c). Applying similar imaging forces on an NTA-surface did not alter its morphology, confirming that only the protein film had been removed. The 3 nm value is much smaller than the length of fully stretched 414 amino acid residue fragments (~150 nm), suggesting that the proteins were folded in their native state.
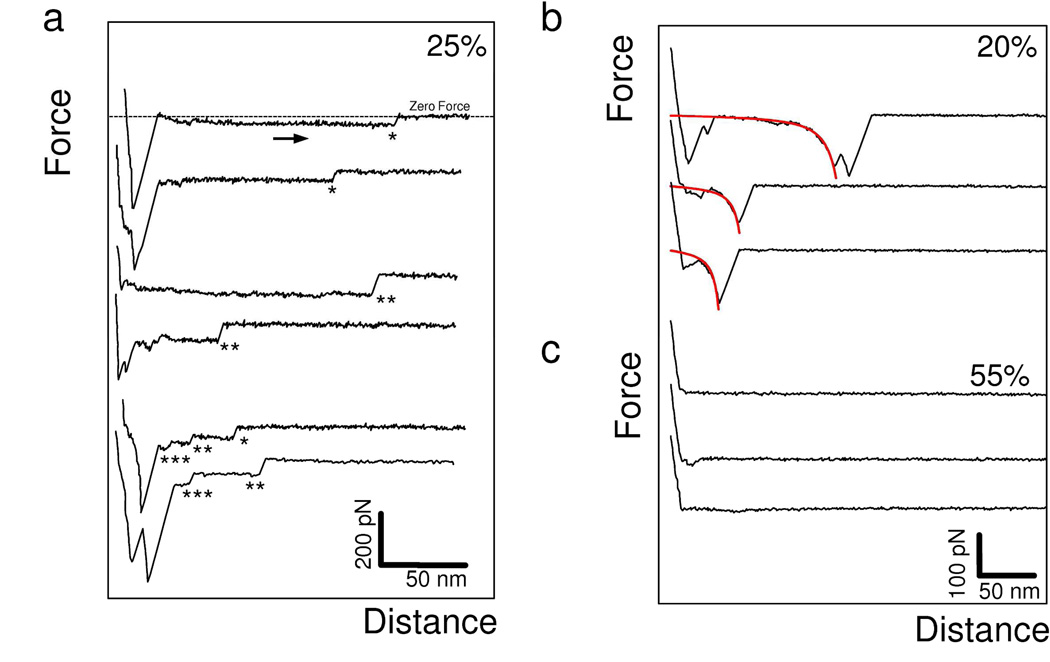
Figure 2 shows representative force-distance curves recorded between an amyloid-tip and an Ig-T monolayer, using a pulling speed of 200 nm/s. Adhesion signatures were frequently detected (45 %) and exhibited two types of behaviors, i.e. pronounced constant force plateaus (25 %, Figure 2a) and single (or double) non-linear force peaks (20 %, Figure 2b). These two features were often sequential (Figure 2a), force plateau being frequently preceded with nonlinear force peaks. Figure 2a shows that the plateau signatures displayed sharp ruptures or “plateau forces” of 31 ± 5 pN, 61 ± 3 pN, and 91 ± 5 pN (each value represents the mean ± σ obtained from n = 150 plateau curves recorded using three independent tips and samples), with a broad distribution of rupture lengths (see Supporting Information Figure 1). Hence, each mean plateau force was a multiple of a ~30 pN unit force.
Figure 2.
Unzipping individual β-sheets. (a-c) Representative retraction force curves recorded in buffer between an amyloid-tip and an Ig-T surface, using a pulling speed of 200 nm/s and an interaction time of 250 ms. (a) A substantial fraction (25 %) of the curves showed constant force plateaus, sometimes superimposed onto one another. The dashed line on the top curve represents the zero force line. The number of force plateaus superimposed is highlighted by a corresponding number of asterisks (*). (b) Another fraction of the curves (20 %) recorded in the same conditions showed elastic responses that were well-described by the worm-like-chain model using a persistence length of 0.4 nm (see fits in red). (c) The remaining curves (55 %) did not show any adhesion events. The data shown are from a total of 256 force curves recorded on different locations of a sample. Similar data were obtained using more than 3 different tips and 3 independent samples (> 3,072 force curves).
Non-linear force peaks were well-described by the worm-like-chain model using a persistence length of 0.4 nm: F(x) = kbT/lp [0.25(1− x/Lc)−2 + x/Lc - 0.25], where Lc and lp are the contour length and persistence length of the molecule, kb is the Boltzmann constant and T the absolute temperature. The mean peak magnitude, 198 ± 23 pN (mean ± σ; 250 curves), is in the range of unfolding forces reported for β-folds domains at similar loading rates, such as Ig and fibronectin domains of modular proteins,19,20 and larger than the forces needed to unfold α-helical domains.26 The observed force peaks may reflect the force-induced extension of the Ig and T secondary structures.27,28
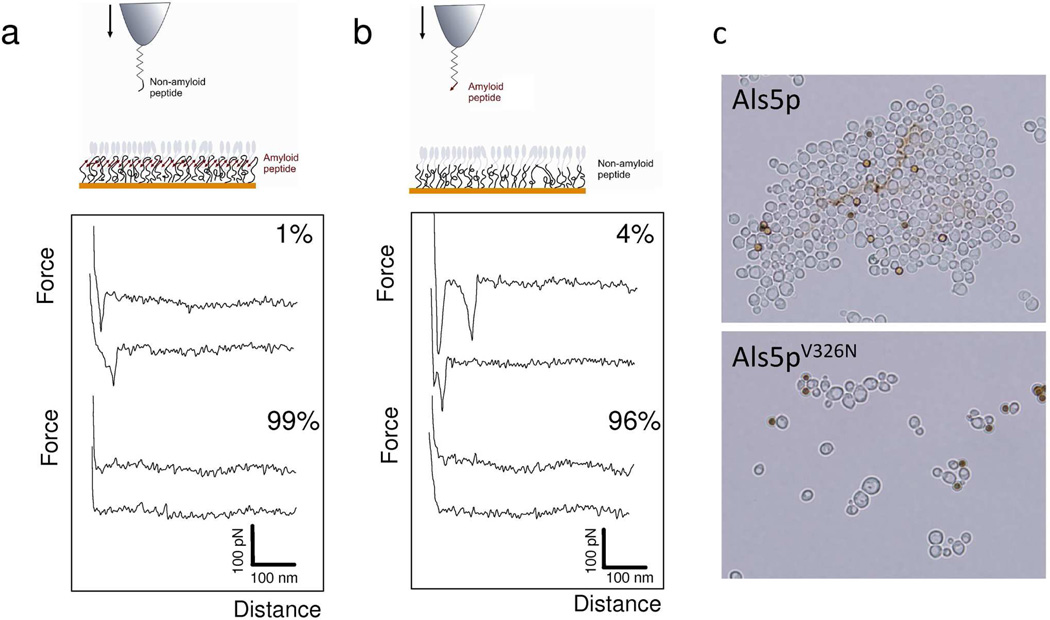
Two series of control experiments suggest that the observed force plateaus represent the characteristic mechanical response of an amlyoid structure. First, force plateaus were never observed when using non-amyloid peptide tips or non-amyloid protein monolayers. Figure 3 shows representative force-distance curves obtained when introducing a single site mutation that prevents amyloid formation. The mutation is V5N in the probing peptide, and corresponds to V326N in the Ig-T fragment of Als5p. When this mutation was present in either the probe sequence or the Ig-T monolayer, force plateaus were never detected, supporting the notion that they originate from amyloid-like interactions (Figure 3a, SNGINIVATTRTV non-amyloid peptide; Figure 3b, V326N Ig-T protein). This mutation in the amyloid-forming region of Als abrogates yeast aggregation (Figure 3c) and amyloid formation.13,14,16 Note that few non-linear force peaks were sometimes observed in these experiments, reflecting non-specific interactions between tip and sample.
Figure 3.
Unzipping interactions are abolished with a single-site mutation in the amyloid sequence. (a, b) Representatives force-distance curves recorded in buffer between a tip bearing a non-amyloid peptide with a V5N substitution (SNGINIVATTRTV) and Ig-T proteins (a), and between a tip bearing the amyloid peptide (SNGIVIVATTRTV) and non-amyloid Ig-T mutant proteins with a V326N substitution in the amyloid sequence (b). The pulling rate was 200 nm/s and the interaction time 250 ms. Both conditions lead to a dramatic reduction in the number of curves with adhesion events and to the disappearance of force plateau signatures, indicating that molecular zippers are formed through amyloid bonds. (c) Effect of the V326N substitution on adhesion and aggregation of S. cerevisiae yeast cells expressing Als5p. The brown spheres are BSA-coated ferromagnetic beads, 2 µm in diameter.38 This substitution in the 1419-residue protein greatly reduce cellular aggregation and amyloid-formation.13,14
Second, control experiments were performed to rule out the possibility that the plateau signatures would correspond to the simple desorption of individual I-g proteins from the substratum (Supporting Information Figure 2). Pulling on Als proteins randomly adsorbed on hydrophilic (EG3) and hydrophobic (CH3) surfaces yielded force curves that did not show force plateaus, confirming that these are only observed with oriented monolayers of Als molecules in which amyloid bonds between parallel proteins are favored. Taken together, the above findings strongly suggest that force plateaus captured the mechanical unzipping of amyloid-like Als5p β- sheet structures.
We attribute these force responses to the rupture of bonds formed between amyloid strands from hundreds of parallel Als molecules. The ~30 pN unit force would correspond to the unzipping of the β strands from a single β-sheet, specifically to the rupture of single interstrand H-bonds, known to be stronger and shorter than H-bonds in α-helices.29 The unzipping of two (or three) β-sheets in parallel would lead to a two (or three)-fold increase of the unit force value (Figure 2a). The force plateaus in our experiments are different from those observed in amyloid β fibrils associated with Alzheimer’s disease in that the latter were attributed to the unzipping of individual β-sheets from the Aβ fibrils.22,23 In our experiments, fibrils were not present but the similarity in force-extension characteristics of the plateaus implies that similar amyloid-like bonds are ruptured in each case.
Lastly, we note that the observed force plateaus clearly differ from the multiple unfolding force peaks observed earlier by pulling on Ig-T-TR6 proteins.14,16,24 This behavior is attributed to differences in probe specificity: in previous studies, Als proteins were stretched through their Ig terminal domains with an antibody or full Als5p protein bound to the tip, whereas here the tip was functionalized with a short amyloid peptide expected to be able to reach and specifically bind the amyloid region.
Dynamics of unzipping
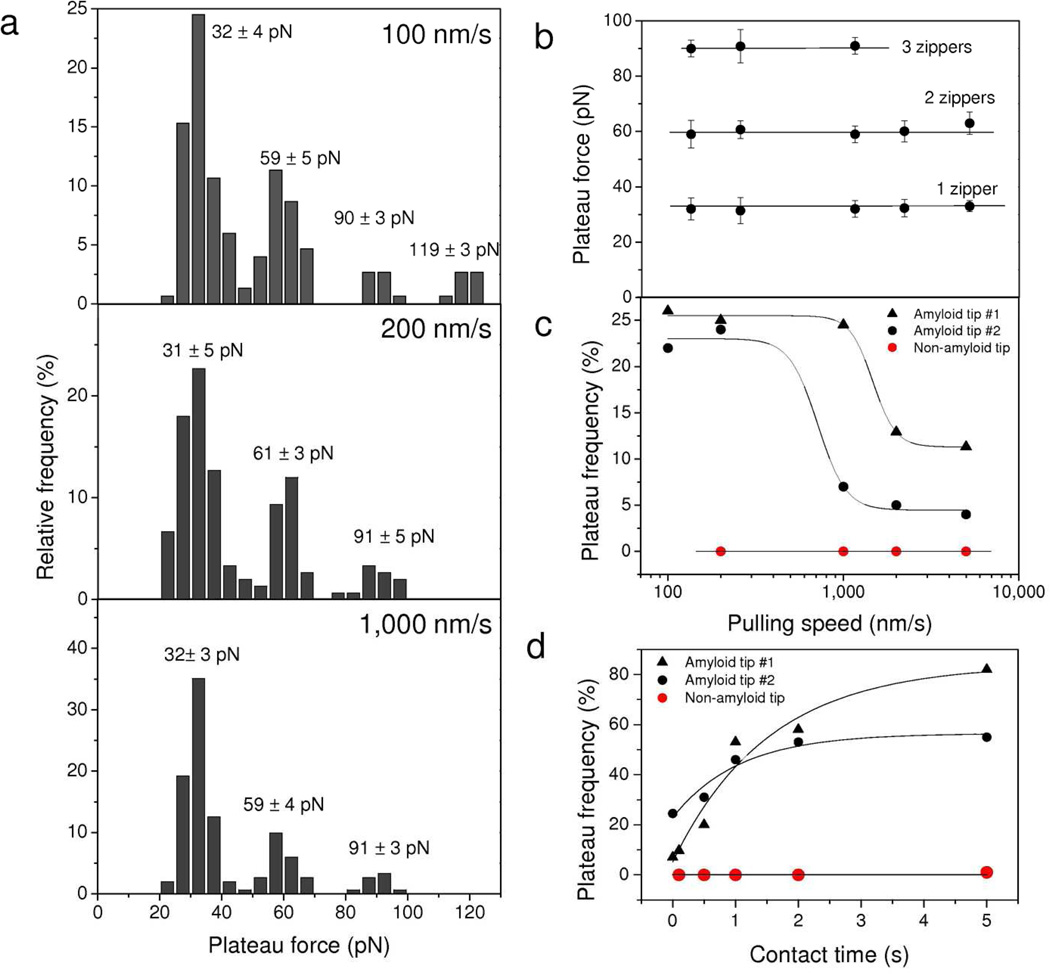
The dynamics of the unzipping interaction was investigated by recording force curves between an amyloid-tip and an Ig-T-surface at various pulling speeds. Figure 4a,b shows that the mean plateau forces corresponding to the unzipping of single or multiple β-sheets did not substantially depend on the stretching speed, over the range experimentally accessible. This finding is in sharp contrast with the behavior observed for the rupture of receptor-ligand complexes 30,31 and for the unfolding force of water soluble proteins 19,32 which usually increase with the logarithm of the stretching speed. Rather, our data are reminiscent of the behavior of pathological amyloids,22,23 and indicate that the mechanical measurements were made near thermodynamic equilibrium, thus that the time for establishing and maintaining β-sheet associations is smaller than the time scale of our experiment (i.e. the 0.1 – 1.0 s range).
Figure 4.
Dynamics of the zipper interaction. (a) Histograms showing the distribution of plateau forces measured between an amyloid-tip and an Ig-T surface at different pulling speeds (100 nm/s, 200 nm/s and 1,000 nm/s; surface delay = 0 ms). ). For each loading rate, the data correspond to 150 plateau curves from a total of 3,072 force curves recorded using three different tips and samples. (b, c) Dependence of the mean plateau forces (b) and plateau force frequency (c) on the pulling speed. Each data point in (b) represents the mean ± σ calculated from 150 plateau curves from a total of 3,072 force curves obtained using three different tips and samples. (d) Dependence of the plateau force frequency on contact time measured at a constant pulling speed of 1,000 nm/s. The plateau force probability showed a strong dependence both on the pulling speed and the interaction time. Each value in (c) and (d) was obtained from a total of 1,024 forces curves. The two sets of black symbols represent two independent experiments with amyloid-tips, while the red symbols correspond to control experiments with tips bearing non-amyloid peptides.
Figure 4c shows the influence of pulling speed on the force plateau frequency, i.e. the number of curves with plateau events. Lowering the pulling rate (from 1,000 nm/s to 200 nm/s) dramatically increased the frequency of force plateaus, although some variations were found from one independent experiment to another (see duplicates experiments). To assess whether this could be related to a time dependence in the zipper interaction, the contact time was varied while keeping the pulling rate constant (1,000 nm/s). Under these conditions, the force plateau frequency increased exponentially with time to reach a plateau after 5 s (Figure 4d). The dependence of the force plateau frequency on pulling rate and contact time supports the notion that zipper-like interactions are established between the β-strand on the tip and anti-parallel β-strands from self-associated adhesion proteins. The prolonged contact time required to form a zipper is suggested to reflect the time necessary for conformational changes and optimal fitting of neighbored β-strands.
In the light of these results and of currently available X-ray diffraction data on Aβ fibrils,4,5 we suggest that parallel Als proteins interact through their β-strands to form a two-dimensional protein array stabilized by amyloid-like hydrogen bond networks. The protein orientation in the Als monolayers mimics that in vivo: on yeast cell walls, Als molecules are anchored to insoluble wall polysaccharides through their C-terminals, and form two-dimensional amyloid-like nanodomains.15,16
Als proteins form amyloid fibrils that cannot be unzipped
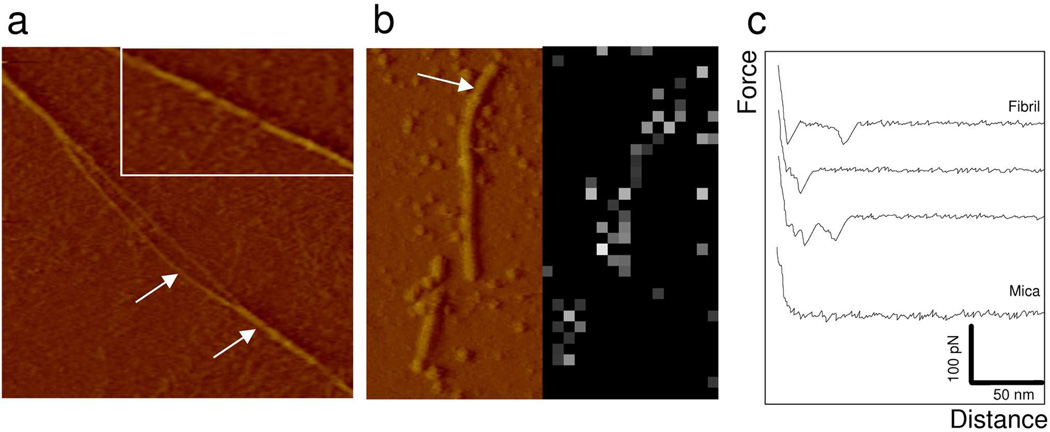
To assess whether the force plateau pattern is also observed with mature Als fibrils, we probed the force response of fibrils formed from Ig-T proteins. Proteins were suspended in deionized water (0.2 mg/ml), stirred for 2 days at 5°C, left unstirred for 2 days at 5°C and then incubated with a mica substrate.12 Inspection of the protein preparation using AFM imaging in PBS revealed the presence of unbranched straight linear fibrils with diameters of 3.5 ± 0.8 nm (mean ± σ, n = 25 from 5 independent samples) (Figure 5a). Some of these were found to locally separate into thinner filaments of 1.5 nm in diameter, which is about the size of protofilaments making the structural unit of fibrils formed from Aβ 1–40 peptides.23 This feature is consistent with the notion that amyloids are highly hierarchical structures assembled from individual subunit structures. No fibrils were formed from Ig-T proteins bearing the V326N single-site mutation, indicating that amyloid interactions are critical for fibril formation.15
Figure 5.
Morphology and mechanical response of mature amyloid fibrils. (a) AFM deflection image (1 µm × 1 µm) recorded with a silicon nitride tip in buffer for a preparation of Ig-T molecules (0.2 mg/ml) incubated during 4 days at 4°C in PBS buffer and adsorbed on mica. Arrows show an amyloid fiber separated into two strands. The inset is an enlarged view of a fiber showing helical markings. (b) Deflection image and adhesion force map (16 × 32 force curves on a 0.5 µm × 1 µm area) recorded in buffer with an amyloid-tip. The arrow emphasizes a fibril. Bright pixels in the map correspond to binding events. (c) Representative force curves recorded on mica and on a fibril, documenting single unbinding forces (32 ± 5 pN) without any evidence for unzipping interactions.
Figure 5b,c shows the force data obtained between a single fibril and an amyloid-tip. The force signatures (Figure 5c) were different from those recorded on the Ig-T monolayer (Figure 2), in that force plateaus were never observed. Rather, most curves showed single force peaks of 32 ± 5 pN (mean ± σ; 1024 curves) magnitude that were not observed with a bare tip. Hence, while the amyloid-tip is able to bind the mature fibrils, the latter cannot be unzipped using our methodology. This result is compatible with the formation and rupture of a single 30 pN bond between the tip and the fiber and suggests that β-sheets in Als fibrils are assembled through amyloid interactions that are too strong to be unraveled by AFM. This finding is also consistent with the high stability of amyloid-like fibrils. Nelson et al.4 estimated the free energy associated with the formation of an amyloid-like protofibril made of a seven-residue peptide from the yeast protein Sup35 to be ~24 kcal mol−1. This high stability results from the combination of multiple H-bonds forming individual β-sheets with dry van der Waals forces associated with the tight self-complementation of two sheets.4,33 As this ~24 kcal mol−1 value is 10 times larger than the ~2.5 kcal mol−1 energy associated with an H-bond between two β-strands, we suggest that the 30 pN tip-fibril binding force will be too weak to dissociate a fibril.
Biological implications of the Als molecular zipper
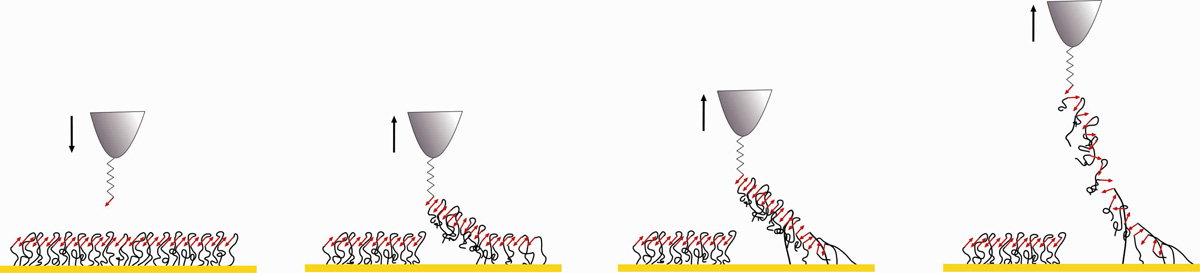
Taken together, our results are consistent with a molecular zipper model in which amyloid-like β-sheets form between parallel surface-displayed Als molecules, and can be unzipped at constant force and under equilibrium conditions, using an amyloid probing peptide (Figure 6). Amyloid interactions are additive and increase with interaction time, supporting a zipper model in which amyloid bonds are established between many lateral Als proteins. On yeast cells, such amyloid bonds form upon mechanical stimulation by cell-cell contact14 or an AFM tip,16 and are facilitated by mobility of the amyloid regions, which are tethered to the surface through flexible glycopeptide “stalks” up to 140 nm long.15 Amyloid clustering activates yeast cells for strong adhesion by increasing the avidity of binding through hundreds of parallel binding events or more.14–16,33,34 The proposed model (Figure 6) is also consistent with the observed rupture lengths, which have mean values in multiples of ~30–35 nm (see Supporting Information Figure 1), the predicted length for the T domain sequence when fully extended. The multiple lengths may be caused by the tip remaining attached to the surface through two, three, or more T domains.
Figure 6.
Nanomechanics of Als amyloid zippers. Model showing homotypic binding of lateral Als adhesins through antiparallel β-strands (red arrows) leading to the formation of a surface amyloid zipper (for sake of clarity, Ig regions are not shown). Note that the surface may be less structured than depicted, and we do not know whether the amyloid-like interactions result from association of parallel or anti-parallel β-strands. From left to right: approaching an amyloid peptide towards the Ig-T β-strands leads to the formation of an amyloid bond; retracting the probing peptide stretches and lifts amyloid regions from the surface, breaks H bonds sequentially between two strands (30 pN) or more (60 pN or 90 pN) at a time. In nature, this molecular zipper provides a powerful mechanism to strengthen cell-cell adhesion.
Our finding that homologous interactions take place between a single Als amyloid peptide and a monolayer of oriented Als proteins indicates that zipper-like amyloid bonds can form between opposing adhesion molecules and is consistent with in vivo data showing that amyloid bonds can form between cells.15 Amyloid bonds between opposite cells would be highly stable, because the dissociation rates would be lowered to near zero as a result of the summed energies of H bonding29 and van der Waals forces33 in amyloids. Considering that in our unzipping experiments the lifetime of the bond between two opposing amyloids is given by the plateau rupture length divided by the tip velocity, a zipper interaction between two cells will substantially increase the lifetime – thus lower the dissociation constant - of the cell-cell bond. Amyloid bonds between cells are consistent with the strong cell-cell binding characteristic of the cohesion of C. albicans colonies and mats.34 Indeed, the physical characteristics of C. albicans colonization of human tissues is such that physicians sampling superficial lesions find the fungus is so strongly bound that the epithelial layer is usually adherent to the colony rather than the host.35 Our proposed cell-cell zipper interaction is reminiscent of the homophilic cadherin-cadherin interaction, in which multiple binding contacts between opposing surfaces have been demonstrated and suggested to allow for greater stability in cell-cell interactions,36 and implies that similar zipper-like interactions can occur in other amyloid-mediated interactions such as platelet activation37 and bacterial biofilms.10
CONCLUSIONS
Adhesion of C. albicans cells is known to be mediated by amyloid-like interactions involving clusters of Als adhesion molecules on the cell surface.13 Yet, how amyloid bonds affect the high mechanical strength of Als proteins was not known. We have shown that stretching Als proteins through their amyloid sequences yields characteristic force responses corresponding to the mechanical unzipping of β-sheet interactions formed between parallel Als proteins, oriented in the same way as they are on the yeast cell surface. We have also found that amyloid-like bonds form between amyloid molecules on two opposing surfaces (the amyloid monolayer and the amyloid probing peptide), an interaction that we believe strengthen the bonds between two opposing cells expressing homologous proteins. Hence, these results suggest that the Als amyloid can function as a strongly cohesive molecular zipper that mediates long-lived homophilic protein interactions between cells. The adhesion zipper interaction unravelled here may represent a general mechanism for providing remarkable mechanical strength to cell adhesion proteins from various cell types, from microbes to plant and animal cells.
METHODS
Expression and purification of Als5p fragments
Ig-T fragments made of 414 amino acid residues were expressed and secreted from S. cerevisiae harboring plasmid pRL09 and purified by His-Trap chromatography as previously described.12,38 The SNGIVIVATTRTV amyloid peptide and the SNGINIVATTRTV non-amyloid peptide used to functionalize the tips were synthesized and purified to over 95% by the Rockefeller University Proteomics Facility.12,14
Atomic force microscopy
AFM measurements were performed at room temperature (20°C) in buffered solutions (Phosphate Buffer Saline; pH 7.2), using a Nanoscope V Multimode AFM (Veeco Metrology Group, Santa Barbara, CA) and oxide sharpened microfabricated Si3N4 cantilevers (Microlevers, Veeco Metrology Group). The spring constants of the cantilevers were measured using the thermal noise method (Picoforce, Veeco Metrology Group), yielding values ranging from 0.007 to 0.019 N/m. Unless otherwise specified, all force measurements were recorded with a pulling speed of 200 nm/s and a surface delay of 0 s.
To prepare Ig-T surfaces, silicon wafers (Siltronix) were coated using electron beam thermal evaporation with a 5 nm thick chromium layer followed by a 30 nm thick gold layer. The gold coated surfaces were cleaned for 15 min by UV and ozone treatment, rinsed with ethanol, dried with a gentle nitrogen flow, and immersed overnight in ethanol containing 0.1 mM of nitrilotriacetate (NTA)-terminated (10 %) and triethylene-glycol (EG3)-terminated (90%) alkanethiols (Prochimia, Poland). After rinsing with ethanol, the surfaces were immersed in a 40 mM aqueous solution of NiSO4 for 1 h, rinsed in water, incubated in PBS containing 200 µg/mL of His-tagged Ig-T fragments (wild-type and V326N) for 1h, and finally rinsed in buffer. Control experiments were performed in which His-tagged Ig-T fragments (200 µg/mL in PBS) were simply adsorbed on hydrophilic and hydrophobic substrata, prepared by treating gold coated surfaces with EG3-terminated (100 %) and CH3-terminated (100 %) alkanethiols, respectively.
AFM tips were functionalized with amyloid and non-amyloid peptides through their -NH2 groups using acetal-PEG-NHS linkers as described by Wildling et al.39 Cantilevers were washed with chloroform and ethanol, placed in an UV-ozone-cleaner for 30 min, immersed overnight into an ethanolamine solution (3.3 g ethanolamine into 6 mL of DMSO), then washed 3 times with DMSO and 2 times with ethanol, and dried with N2. The ethanolamine-coated cantilevers were immersed for two hours in a solution prepared by mixing 1 mg Acetal-PEG-NHS dissolved in 0.5 mL chloroform with 10 µL triethylamine, then washed with chloroform and dried with N2. Cantilevers were further immersed for 10 min in a 1% citric acid solution, washed 3 times in acetone, dried under N2, and then covered with a 200 µL droplet of a PBS solution containing peptides (0.2 mg/mL) to which 2 µL of a 1 M NaCNBH3 solution were added. After 50 min, cantilevers were incubated with 5 µL of a 1 M ethanolamine solution in order to passivate unreacted aldehyde groups, and then washed with and stored in PBS 10 min later.
Supplementary Material
Acknowledgements
Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Work at Brooklyn College was supported by NIH grant SC1 GM083756. Y.F.D. and D.A. are Senior Research Associate and Postdoctoral Researcher of the FRS-FNRS. We thank Cho Tan for the micrographs in Figure 3c.
Footnotes
Author Contributions. D.A., C.B.R., P.N.L. and Y.F.D. designed the experiments, analysed the data and wrote the article. C.B.R. produced and purified the proteins. D.A. collected the AFM data.
Supporting Information Available. Rupture lengths of the zipper interactions are given in Supplementary Figure 1. Supplementary Figure 2 shows that zipper interactions do not originate from the random desorption of single Ig-T molecules from the substratum. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dobson CM. Protein Misfolding, Evolution and Disease. Trends Biochem. Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 2.Dobson CM. Structural Biology: Prying into Prions. Nature. 2005;435:747–749. doi: 10.1038/435747a. [DOI] [PubMed] [Google Scholar]
- 3.Shewmaker F, McGlinchey RP, Wickner RB. Structural Insights into Functional and Pathological Amyloid. J. Biol. Chem. 2011;286:16533–16540. doi: 10.1074/jbc.R111.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the Cross-β Spine of Amyloid-Like Fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. Atomic Structures of Amyloid Cross-β Spines Reveal Varied Steric Zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 6.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional Amyloid - from Bacteria to Humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Chiti F, Dobson CM. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 8.Knowles TPJ, Buehler MJ. Nanomechanics of Functional and Pathological Amyloid Materials. Nature Nanotechnology. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 9.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli Curli Operons in Directing Amyloid Fiber Formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Diversity, Biogenesis and Function of Microbial Amyloids. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauceo JM, Gaur NK, Lee KG, Edwards JE, Klotz SA, Lipke PN. Global Cell Surface Conformational Shift Mediated by a Candida albicans Adhesin. Infect. Immun. 2004;72:4948–4955. doi: 10.1128/IAI.72.9.4948-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otoo HN, Lee KG, Qiu WG, Lipke PN. Candida Albicans Als Adhesins Have Conserved Amyloid-Forming Sequences. Eukaryot. Cell. 2008;7:768–782. doi: 10.1128/EC.00309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, Henry R, Litewka A, O'Meally S, Otoo HN, Khalaf RA, et al. Yeast Cell Adhesion Molecules Have Functional Amyloid-Forming Sequences. Eukaryot. Cell. 2010;9:393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrene YF, Lipke PN. A Role for Amyloid in Cell Aggregation and Biofilm Formation. PLoS ONE. 2011;6:e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrene YF. Strengthening Relationships: Amyloids Create Adhesion Nanodomains in Yeasts. Trends Microbiol. 2012;20:59–65. doi: 10.1016/j.tim.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. Force-Induced Formation and Propagation of Adhesion Nanodomains in Living Fungal Cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puchner EM, Gaub HE. Force and Function: Probing Proteins with AFM-Based Force Spectroscopy. Curr. Opin. Struct. Biol. 2009;19:605–614. doi: 10.1016/j.sbi.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Li HB, Cao Y. Protein Mechanics: From Single Molecules to Functional Biomaterials. Acc. Chem. Res. 2010;43:1331–1341. doi: 10.1021/ar100057a. [DOI] [PubMed] [Google Scholar]
- 19.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible Unfolding of Individual Titin Immunoglobulin Domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 20.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The Molecular Elasticity of the Extracellular Matrix Protein Tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 21.Lv S, Dudek DM, Cao Y, Balamurali MM, Gosline J, Li H. Designed Biomaterials to Mimic the Mechanical Properties of Muscles. Nature. 2010;465:69–73. doi: 10.1038/nature09024. [DOI] [PubMed] [Google Scholar]
- 22.Kellermayer MSZ, Grama L, Karsai Á, Nagy A, Kahn A, Datki ZL, Penke B. Reversible Mechanical Unzipping of Amyloid β -Fibrils. J. Biol. Chem. 2005;280:8464–8470. doi: 10.1074/jbc.M411556200. [DOI] [PubMed] [Google Scholar]
- 23.Karsai A, Mártonfalvi Z, Nagy A, Grama L, Penke B, Kellermayer MSZ. Mechanical Manipulation of Alzheimer's Amyloid β 1-42 Fibrils. J. Struct. Biol. 2006;155:316–326. doi: 10.1016/j.jsb.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 24.Alsteens D, Dupres V, Klotz SA, Gaur NK, Lipke PN, Dufrêne YF. Unfolding Individual Als5p Adhesion Proteins on Live Cells. ACS Nano. 2009;3:1677–1682. doi: 10.1021/nn900078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klotz SA, Gaur NK, Lake DF, Chan V, Rauceo J, Lipke PN. Degenerate Peptide Recognition by Candida albicans Adhesins Als5p and Als1p. Infect. Immun. 2004;72:2029–2034. doi: 10.1128/IAI.72.4.2029-2034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rief M, Pascual J, Saraste M, Gaub HE. Single Molecule Force Spectroscopy of Spectrin Repeats: Low Unfolding Forces in Helix Bundles. J. Mol. Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez M, de Groot PWJ, Klis FM, Lipke PN. Glycoconjugate Structure and Function in Fungal Cell Walls. In: AP M, editor. Microbial Glycobiology. San Diego: Academic Press; 2009. pp. 169–183. [Google Scholar]
- 28.Salgado PS, Yan R, Taylor JD, Burchell L, Jones R, Hoyer LL, Matthews SJ, Simpson PJ, Cota E. Structural Basis for the Broad Specificity to Host-Cell Ligands by the Pathogenic Fungus Candida Albicans. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15775–15779. doi: 10.1073/pnas.1103496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viswanathan R, Asensio A, Dannenberg JJ. Cooperative Hydrogen-Bonding in Models of Antiparallel β-Sheets. J. Phys. Chem. A. 2004;108:9205–9212. [Google Scholar]
- 30.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy Landscapes of Receptor-Ligand Bonds Explored with Dynamic Force Spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 31.Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, Drenckhahn D. Cadherin Interaction Probed by Atomic Force Microscopy. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans EA, Calderwood DA. Forces and Bond Dynamics in Cell Adhesion. Science. 2007;316:1148–1153. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- 33.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the Amylome, Proteins Capable of Forming Amyloid-Like Fibrils. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds TB, Jansen A, Peng X, Fink GR. Mat Formation in Saccharomyces Cerevisiae Requires Nutrient and Ph Gradients. Eukaryot. Cell. 2008;7:122–130. doi: 10.1128/EC.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rippon JW. The Pathogneic Fungi and the Pathogenic Actinomycetes. Third Edition. Philadelphia-London-Toronto-Montreal-Sydney- Tokyo: W.B. Saunders Company-Harcourt Brace Jovanovich Inc.; 1988. Vol. [Google Scholar]
- 36.Wu Y, Jin X, Harrison O, Shapiro L, Honig BH, Ben-Shaul A. Cooperativity between Trans and Cis Interactions in Cadherin-Mediated Junction Formation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17592–17597. doi: 10.1073/pnas.1011247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herczenik E, Bouma B, Korporaal SJ, Strangi R, Zeng Q, Gros P, Van Eck M, Van Berkel TJ, Gebbink MF, Akkerman JW. Activation of Human Platelets by Misfolded Proteins. Arterioscler. Thromb. Vasc. Biol. 2007;27:1657–1665. doi: 10.1161/ATVBAHA.107.143479. [DOI] [PubMed] [Google Scholar]
- 38.Rauceo JM, De Armond R, Otoo H, Kahn PC, Klotz SA, Gaur NK, Lipke PN. Threonine-Rich Repeats Increase Fibronectin Binding in the Candida albicans Adhesin Als5p. Eukaryot. Cell. 2006;5:1664–1673. doi: 10.1128/EC.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wildling L, Unterauer B, Zhu R, Rupprecht A, Haselgrübler T, Rankl C, Ebner A, Vater D, Pollheimer P, Pohl EE, et al. Linking of Sensor Molecules with Amino Groups to Amino-Functionalized AFM Tips. Bioconjug. Chem. 2011;22:1239–1248. doi: 10.1021/bc200099t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.