Abstract
Background
Human dental pulp stem cells (DPSCs) have potential applications in tissue regeneration because of their convenient cell harvesting procedures and multipotent capacity. However, the tissue regenerative potential of DPSCs is known to be negatively regulated by aging in long-term culture and under oxidative stress. With an aim of reducing cellular senescence and oxidative stress in DPSCs, an intracellular delivery system for superoxide dismutase 1 (SOD1) was developed. We conjugated SOD1 with a cell-penetrating peptide known as low-molecular weight protamine (LMWP), and investigated the effect of LMWP-SOD1 conjugates on hydrogen peroxide-induced cellular senescence and osteoblastic differentiation.
Results
LMWP-SOD1 significantly attenuated enlarged and flattened cell morphology and increased senescence-associated β-galactosidase activity. Under the same conditions, LMWP-SOD1 abolished activation of the cell cycle regulator proteins, p53 and p21Cip1, induced by hydrogen peroxide. In addition, LMWP-SOD1 reversed the inhibition of osteoblastic differentiation and downregulation of osteogenic gene markers induced by hydrogen peroxide. However, LMWP-SOD1 could not reverse the decrease in odontogenesis caused by hydrogen peroxide.
Conclusion
Overall, cell-penetrating LMWP-SOD1 conjugates are effective for attenuation of cellular senescence and reversal of osteoblastic differentiation of DPSCs caused by oxidative stress inhibition. This result suggests potential application in the field of antiaging and tissue engineering to overcome the limitations of senescent stem cells.
Keywords: superoxide dismutase, senescence, oxidative stress, cell-penetrating peptide, dental pulp stem cell, osteoblastic differentiation
Introduction
Because of their unlimited capacity for proliferation, differentiation, and plasticity, mesenchymal stem cells derived from bone marrow, muscle, and adipose tissue are a promising tool for tissue regeneration.1–5 Dental pulp stem cells (DPSCs) are a source of progenitor cells that have the capacity for self-renewal and can undergo differentiation into odontoblastic, osteoblastic, neuronal, and adipocytic cell types in vitro, similar to mesenchymal stem cells.6–8 DPSCs have been studied in odontogenesis, and have been described as an alternative source of mesenchymal stem cells for osteoblastic differentiation.9 We selected DPSCs for use in this study because of their advantages, including their osteogenic potential and the convenient process by which they can be obtained. However, the tissue regenerative potential of DPSCs, as with other mesenchymal stem cells, is known to be negatively affected by aging.10–12 Although little is known about age-related changes in DPSCs, the regenerative capacity of aged DPSCs can be restored by a change in the microenvironment using extrinsic factors.13–15 Cellular aging is associated with damage to tissues by free radicals. An imbalance between generation of free radicals and radical scavenging in antioxidant systems resulting in oxidative stress is considered to be a major factor in the process of senescence. At the cellular level, overproduction of reactive oxygen species can induce cellular apoptosis/necrosis and cellular senescence, which results in cellular damage.
For protection against constant oxidative stress, cells have developed defense mechanisms, such as the superoxide dismutase (SOD), catalase, and glutathione peroxidase enzymes.16 SOD is a key enzyme in the dismutation of superoxide radicals resulting from oxidative cellular metabolism into hydrogen peroxide (H2O2).17 Catalase converts H2O2 into water and oxygen, thus preventing the formation of carbon dioxide bubbles in the blood.17 There is no definitive answer as to which enzyme, SOD or catalase, is more important. Both enzymes have an important role as antioxidants protecting the body. SOD is comprised of three isozymes, ie, SOD1, SOD2, and SOD3. Of these, SOD1 is the most widely distributed in the tissues and represents 90% of total SOD activity. In this study, we selected SOD1 as a conversion factor for senescent DPSCs for the reason that SOD1 has been reported to prevent several severe diseases caused by oxidative stress.18–22
However, direct use of SOD1 for protection against oxidative stress has some limitations, including a short half-life in vivo and an inability to cross the cell membrane to detoxify intracellular reactive oxygen species. Recently, cell-penetrating peptides have been used for cellular delivery of several agents in vitro and in vivo. Cell-penetrating peptides, including peptides from the human immunodeficiency virus transacting activator of transcription (TAT) protein, antennapedia homeodomain transcription factor, and synthetic oligoarginines, have received significant and widespread attention within the pharmaceutical and medical fields, although the mechanism of action of cell-penetrating peptide transduction remains controversial.23–29 In a previous study, the TAT protein was used to deliver SOD for the purpose of overcoming certain diseases, such as inflammation or osteoarthritis, and protecting against ischemic brain injury.30–33 We developed an arginine-rich peptide, termed low molecular weight protamine (LMWP), which is derived from natural protamine. We demonstrated that LMWP achieved protein transduction in various cell types with activity equivalent to that of unconjugated TAT for molecular imaging and therapeutic purposes.34,35 In this study, we investigated transduction of LMWP-SOD1 conjugates into DPSCs and the effect of LMWP-SOD1 conjugates on H2O2-induced senescence by testing changes in cellular morphology, senescence-associated beta-galactosidase (SA-β-gal) activity, mRNA levels of related genes, cell cycle distribution, and levels of p53 and p21Cip1/WAF1 proteins. Furthermore, we determined the effect on odontoblast and osteoblast differentiation after exposure of the cells to sublethal concentrations of H2O2 with or without LMWP-SOD1 conjugates.
Materials and methods
Reagents and antibodies
A protease inhibitor cocktail was obtained from Roche (Penzberg, Germany). Phosphatase inhibitor cocktail 2, H2O2, propidium iodide, dimethyl sulfoxide, phenylmethylsulfonyl fluoride, a SA-β-gal detection kit, bovine SOD1, rabbit polyclonal antibody for SOD1, a SOD1 assay kit, and bovine serum albumin were obtained from Sigma-Aldrich (St Louis, MO). Goat polyclonal antibodies against β-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibodies specific for p53 and Ser15 phosphorylated p53 were obtained from Cell Signaling Technology (Beverly, MA). Mouse anti-human p21Cip1/WAF1 antibody was obtained from BD PharMingen (San Diego, CA).
Cell culture and H2O2 treatment
The human DPSCs used in this study were obtained from Dr Gene Lee.36 Deciduous teeth and adult molars were obtained from the Seoul National University Dental Hospital after securing the approval of the institutional review board at Seoul National University Dental Hospital. The deciduous teeth and adult molars were put into Hank’s balanced salt solution (Welgene, Dae-gu, Korea) supplemented with 3% antibiotic- antimycotic solution (Life Technologies, Carlsbad, CA) at 4°C. Adult dental pulps were extracted after the crown of the tooth was removed using a dental high-speed unit. Deciduous dental pulps were gently extracted with tweezers, and periodontal ligaments were gently separated from the surface of the root. Dental pulps and periodontal ligaments were minced in 1 mg/mL of collagenase type I and 2.4 mg/mL of dispase (Gibco) at 37°C for one hour. Single-cell suspensions were plated and maintained in Minimum Essential Medium alpha (Hyclone, Logan, UT) supplemented with 15% fetal bovine serum (Hyclone), 2 mM L-glutamine, 100 μM ascorbic acid 2-phosphate (Sigma-Aldrich), and 1% antibiotic-antimycotic solution (Gibco). The medium was changed every 3 days, and the cells were subcultured at 70% confluency. DPSCs were cultured in complete medium (Modified Eagle’s Medium [MEM] supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic solution) in a humidified incubator at 37°C and in 5% carbon dioxide. The medium was changed twice weekly, and the cells were detached using trypsin-ethylenediamine tetra-acetic acid (EDTA) and seeded into fresh culture flasks at a ratio of 1 to 4 upon reaching confluence. For induction of premature senescence, DPSCs at approximately 75% confluence were briefly exposed to 200 μM H2O2 (diluted in distilled water) for 2 hours. The cells were washed twice with MEM to remove the H2O2, recultured in fresh complete medium for 24 hours, and then subcultured for various durations as specified in the individual experiments.
For odontogenic and osteogenic differentiation, the cells were washed to remove any remaining H2O2 after exposure to H2O2. The cells were recultured with fresh medium for 28 days in differentiation medium containing 10% fetal bovine serum, 50 μg/mL ascorbic acid, 10 mM β-glycerophosphate, and 5 μM dexamethasone. The medium was changed every 3 days with or without SOD1 or LMWP-SOD1.
Cell viability assay
Cell viability was determined using the 3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay. DPSCs were seeded at a density of 1 × 105 cells/well in a 24-well plate (Nunc, Rochester, NY) with MEM containing 10% fetal bovine serum and 1% antibiotics. The next day, the cells were washed with phosphate-buffered solution and incubated for 24 hours in MEM containing 0.5% fetal bovine serum. One day later, the cells were exposed to 200 μM H2O2 in the presence or absence of SOD1 or LMWP-SOD1. After a specified number of days, the cells were incubated with 500 mg/mL MTT for 2 hours at 37°C. The intensity of the MTT product was measured at 550 nm using a microplate reader. The relative percentage of cell survival was calculated by dividing the absorbance of the treated cells by that of the control in each experiment.
Cell cycle analysis
DPSCs were seeded at a density of 1 × 106 cells/well in a 60 mm dish (Nunc) in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and 1% antibiotics. The next day, the cells were washed with phosphate- buffered solution and incubated for 24 hours in DMEM containing 0.5% fetal bovine serum. One day later, the cells were exposed to 200 μM H2O2 in the presence or absence of SOD1 or LMWP-SOD1 in DMEM containing 0.5% fetal bovine serum. The cells were collected by centrifugation, washed with phosphate-buffered solution, and fixed with 70% ethanol. The fixed cells were harvested and resuspended in 100 μL of phosphate-buffered solution containing 50.0 μg/mL RNase (Amersco, Solon, OH) and 200 μg/mL propidium iodide (Sigma-Aldrich) at 4°C for an hour. The fluorescence of the cells was measured using a fluorescence-assisted cell sorting (FACS) Calibur flow cytometer (Becton Dickinson, Lincoln Park, NJ).
SA-β-gal assay
SA-β-gal activity was determined using an SA-β-gal staining kit from Cell Signaling Technology according to the manufacturer’s instructions. Briefly, the cells were fixed in 2% (v/v) formaldehyde and 0.2% glutaraldehyde and then incubated in an SA-β-gal staining solution (1 mg/mL 5-bromo-4-chloro-3-indolyl h-D-galactosidase, 40 mM citric acid, pH 6.0, 40 mM sodium phosphate, pH 6.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM sodium chloride, and 2 mM magnesium chloride) at 37°C for 16 hours. The senescent cells were identified as blue-staining by standard light microscopy, and a total of 1000 cells were counted in 20 random fields on a slide to determine the percentage of SA-β-gal-positive cells.
Preparation of LMWP-SOD1 conjugates
SOD1 from bovine erythrocytes (Sigma-Aldrich; 1 mg/mL in phosphate-buffered solution) was mixed with succinimidyl- 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC; Pierce, Rockford, IL) at a molar ratio of 1 to 10 and stirred at room temperature for an hour. The hydrolytic product of unreacted SMCC was removed from the solution by ultra-filtration (molecular weight cutoff 500). The SOD1 solution was then mixed under reducing conditions with sulfhydryl LMWP (20-fold molar excess of sulfhydryl LMWP to SOD1) at pH 9. The solution was stirred overnight at 4°C. Conjugated LMWP-SOD1 was separated from unconjugated SOD1 using heparin-agarose beads (Sigma-Aldrich). A total of 1 mg of the SOD1 and LMWP mixture was applied to a small (0.5–1.0 mL bed volume) column of heparin-agarose in 10 mM Tris-HCl buffer (pH 7.4). The column was washed with this buffer to remove unbound protein and then eluted with a gradient of 0–2.0 M NaCl. The fractions were collected for protein assays. The LMWP-SOD1 conjugate was concentrated, and its concentration was determined. The conjugated LMWP-SOD1 was examined using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and matrix-assisted laser desorption ionization-mass spectrometry to determine the number of LMWP moieties per protein. Molar incorporation of LMWP:SOD1 was calculated to be at a ratio of approximately 1:10.
Fluorescence labeling
The LMWP-SOD1 conjugates were labeled with fluorescein isothiocyanate (FITC) at their N-terminals. Briefly, the conjugate solution (pH 9.3, carbonate buffer) was reacted in a 1.0 to 1.5 molar ratio with an FITC solution (in dimethyl sulfoxide) overnight in the dark at room temperature. The reaction was monitored by high-pressure liquid chromatography for the absorbance change at 215 nm (the peptide peak). The labeled peptides were purified by high-pressure liquid chromatography (purity > 95%), lyophilized, and stored at −20°C in the dark until further use. Only SOD1 was labeled with FITC and used as a conjugate control for FACS and confocal observation.
Flow cytometric analyses
The cells were seeded at a density of 1 × 105 cells/well in six-well plates in 2 mL of culture medium. One day later, the cells were washed and incubated with FITC-labeled LMWP-SOD1 for 30 minutes at 37°C in humidified 5% carbon dioxide. After incubation, the cells were washed with phosphate-buffered solution, treated extensively with trypsin-EDTA to remove any surface-bound FITC-labeled peptides, and washed again. The cells were then washed with phosphate-buffered solution twice. An analysis was conducted on a FACS Calibur flow cytometer (Becton Dickinson) equipped with a 488 nm air-cooled argon laser. The filter settings for emission were 530/30 nm bandpass for FITC. Fluorescence of 5000 vital cells was acquired, and the data were visualized in logarithmic mode.
Live cell uptake experiments
DPSCs were seeded at a density of 1 × 103 cells/well on an observation glass slide (Nunc) in DMEM containing 10% fetal bovine serum and 1% antibiotics. FITC-labeled LMWP-SOD1 and FITC-labeled SOD1 were diluted in DMEM without phenol red and used at the indicated concentrations. FITC-labeled LMWP-SOD1 and FITC-labeled SOD1 were incubated for 30 minutes at 37°C in humidified 5% carbon dioxide. Immediately after addition of the FITC-labeled fusion proteins to the cell, time lapse images were recorded over 60 minutes (with time intervals of one image per 30 seconds). The images were obtained using confocal laser scanning microscopy.
Western blot analysis and enzyme assays
For Western blot analysis, the cells were lysed for 30 minutes on ice in cold lysis buffer (20 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, and 1% Triton X-100) containing a protease inhibitor cocktail and phosphatase inhibitor cocktail 2. The protein concentration was measured using a Bradford protein assay kit (Bio-Rad, Hercules, CA). Equal aliquots of protein (40 μg) were boiled for 5 minutes in 5 × sample buffer (250 mM Tris-HCl [pH 6.8], 50% glycerol, 10% sodium dodecyl sulfate, 500 mM DTT, and 0.5% bromophenol blue) and separated on 10% sodium dodecyl sulfate-polyacrylamide electrophoresis gel. The proteins were transferred to nitrocellulose membranes, washed with 1 × TBS-T (20 mM Tris-HCl, pH 7.6, 137 mM NaCl, and 0.1% Tween-20), and blocked for an hour at room temperature in 1 × TBS-T with 5% nonfat skim milk. The membranes were washed three times and incubated with primary antibodies in TBS-T containing 5% nonfat skim milk for 4 hours at 4°C. After three washes, the membranes were incubated with the secondary antibody (horseradish peroxidase-conjugated, 1:2000 in TBS-T) for 60 minutes, followed by an additional three washes. The protein bands were visualized with chemiluminescence reagents (West-Zol, Intron, Seoul, Korea). The SOD1 activity was assayed using a SOD assay kit from Fluka Chemicals (St Louis, MO) according to the manufacturer’s instructions. The SOD1 activity was indirectly measured using nitroblue tetrazolium according to the standard procedure. Briefly, enzyme activity was defined as the inhibition activity (%) correlating with reduced xanthine oxidase activity which is inhibited by SOD1. The experiments were performed in triplicate.
Estimation of mRNA levels by semiquantitative reverse transcriptase PCR analysis
Human DPSCs were exposed to 200 μM H2O2 in the presence or absence of SOD1 or LMWP-SOD1. At the end of the indicated times, total RNA was isolated using TRIzol RNA isolation (Invitrogen; Life Technologies) and quantified by ultraviolet spectroscopy. cDNA synthesis was performed on DNase I-treated total RNA (1 μg) with oligo (dT) priming using the SuperScript II preamplification system (Invitrogen). Reverse transcriptase polymerase chain reaction (PCR) was performed using a GeneAmp PCR 9700 thermocycler. PCR was performed with primers specific for p53, p21, alkaline phosphatase, type I collagen, osteopontin, runx2, msx1, msx2, dspp, and GAPDH. The density of the PCR products was detected using a Java-based image processing program (Image J, NIH) and was normalized against the density of GAPDH from the same cDNA sample.
Matrix mineralization analysis
Mineralization of the extracellular matrix was determined using alizarin red S staining. For the detection of calcium deposits, the cells were washed three times with cold phosphate- buffered solution and fixed in ice-cold 95% ethanol for 15 minutes at 4°C. After an additional wash in water, the cells were incubated in 2% alizarin red S (Sigma-Aldrich) in water for 30 seconds at room temperature, followed by two washes with water and then air-drying. Matrix mineralization was quantified by extracting the alizarin red S stain with 10% cetylpyridinium chloride (Sigma-Aldrich) at room temperature for 2 hours. The absorbance of the extracted alizarin red S stain was measured at 570 nm.
Statistical analysis
All experiments were independently repeated at least three times, and the data are presented as the mean ± standard error. The data were analyzed by one-way analysis of variance followed by Fisher’s protected least significant difference post hoc test (StatView; SAS Institute, Cary, NC). P < 0.05 was considered to be statistically significant.
Results
Synthesis of LMWP-SOD1 conjugates
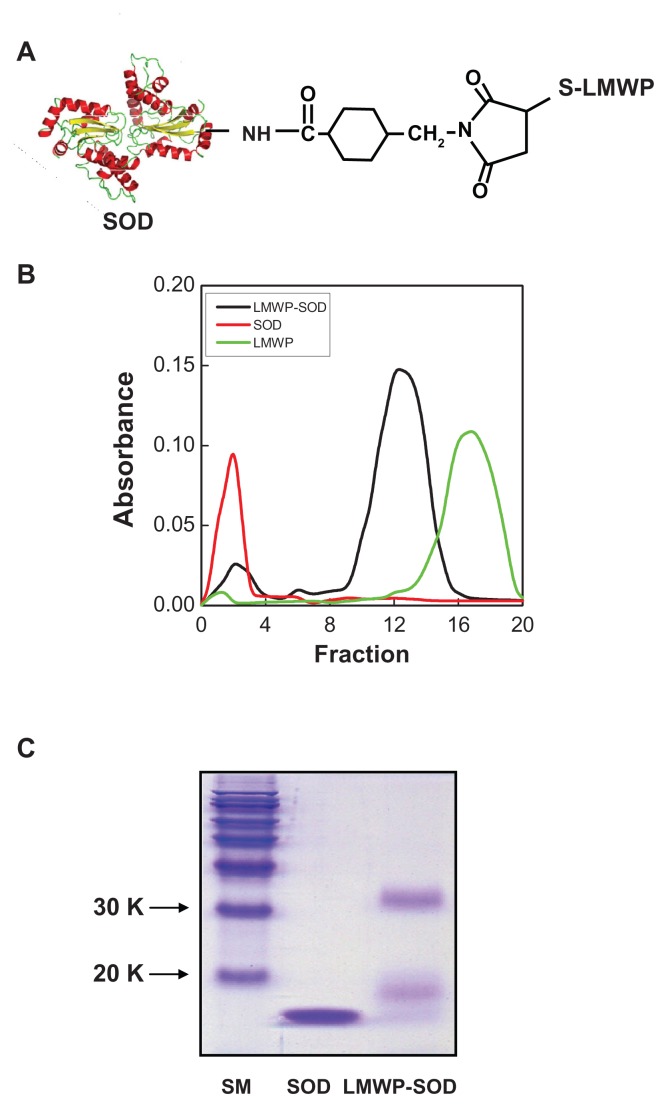
Reactive amino groups in SOD1 enable coupling to various bioactive molecules, including peptides, drugs, and antibodies.25–28 LMWP-SOD1 conjugate synthesis is illustrated in Figure 1A. The amino groups of SOD1 were reacted with SMCC to create SMCC-activated SOD1, followed by coupling with the sulfhydryl group of cysteine in LMWP. The LMWP-SOD1 conjugates were formed via a stable disulfide bond between SMCC and cysteine. Because SOD1 and LMWP were linked via non-cleavable covalent bonds using the crosslinking agent SMCC, the conjugates were stable throughout the entire cell translocation process.
Figure 1.
(A) Diagram of LMWP-SOD1 conjugates. LMWP was chemically conjugated to the amine group of SOD1 via SMCC. (B) Analysis, detection, and purification of LMWP-SOD1 using a heparin affinity column. (C) SOD1 and LMWP-SOD1 were confirmed by Coomassie staining. Lane 1 is the size marker, lane 2 represents SOD1, and lane 3 represents LMWP-SOD1.
Abbreviations: LMWP, low molecular weight protamine; SOD1, superoxide dismutase; SMCC, succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate.
Unreacted SOD1, SMCC, and LMWP were eluted from the heparin column via linear elution using 2.0 M NaCl according to published methods.13 Fraction 17 in Figure 1B had the same retention time as pure LMWP, and thus was identified as unreacted LMWP. Fractions 2 and 12 were collected and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Figure 1C). A size marker was loaded in lane 1, and fractions 2 and 12 were loaded into lanes 2 and 3, respectively. The molecular weights of SOD1 and LMWP are 15,600 Da and 1880 Da, respectively. The molecular weight of the band in lane 3 (LMWP-SOD1 conjugates) was approximately 17,480 Da. The presence of a 33,000 Da band was due to dimer formation of SOD1. Figure 1C demonstrates that the LMWP-SOD1 conjugates were successfully linked.
Transduction of LMWP-SOD1 into human DPSCs
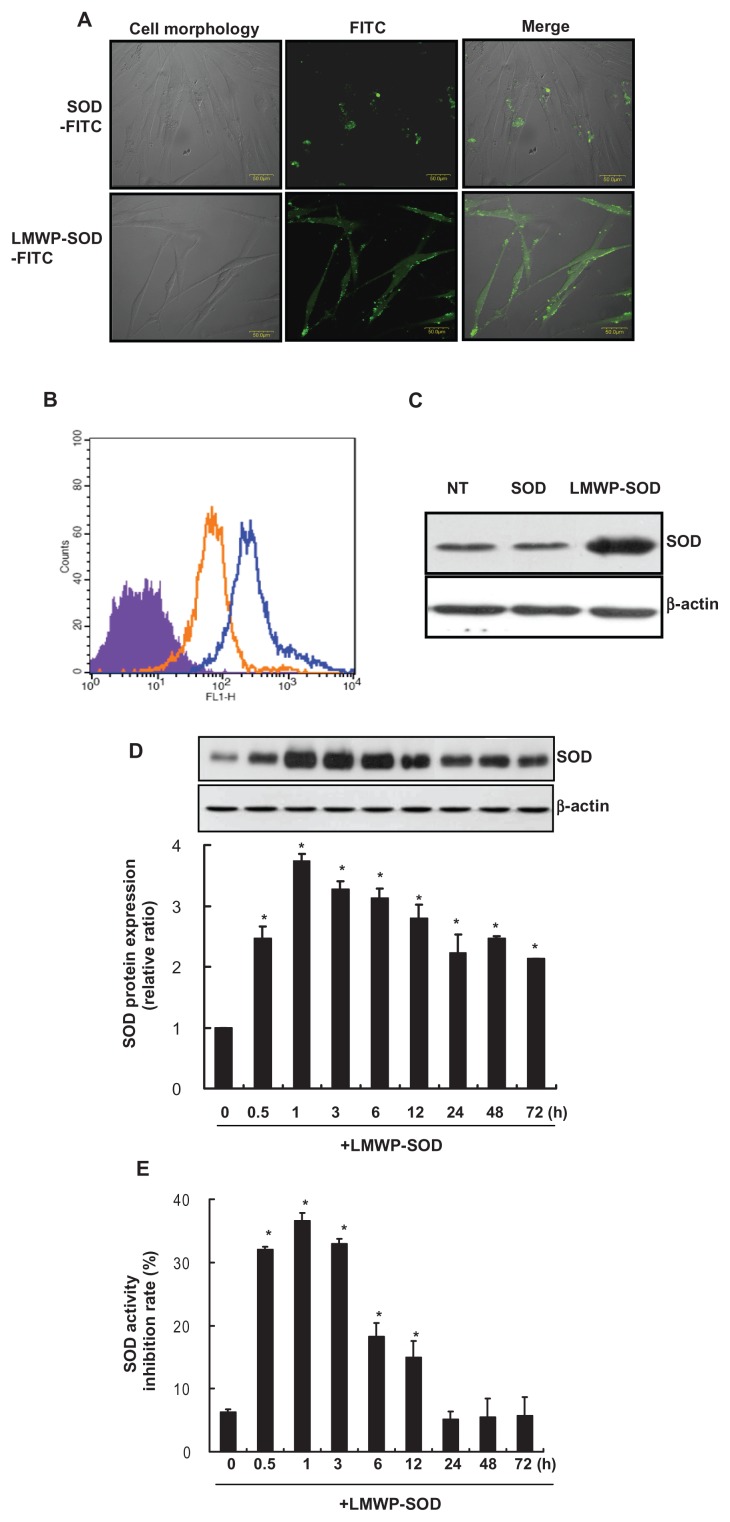
SOD1 is an important enzyme that can detoxify free radicals and protect the cell from oxidative damage. SOD1 is difficult to use in cells because it cannot penetrate the cell due to its large size. We conferred cell-penetrating activity on SOD1 by conjugating it with LMWP. To investigate the ability of LMWP-SOD1 conjugates to transduce into DPSCs, SOD1, and LMWP-SOD1 were labeled with FITC and added to the culture medium of DPSCs at a final concentration of 2 μM for 30 minutes, and analyzed by live cell confocal laser scanning microscopy (Figure 2A). Major fluorescence was observed in the cytosol, indicating that LMWP-SOD1 internalized into the cells within 30 minutes. In contrast with LMWP-SOD1, SOD1 did not internalize into the cell and was instead attached to the cell surface. To verify more precisely that this cell internalization event was mediated by LMWP, we quantified the internalized LMWP-SOD1 conjugates. Cellular uptake of unconjugated SOD1 or LMWP-SOD1 was measured using the mean fluorescent signal of 10,000 collected cells. FACS results were in accordance with confocal microscopic studies (Figure 2B). The reason for the increased intensity in SOD1-treated cells in FACS analysis is that SOD1 protein remained attached to the cell surface despite the harsh washing step and treatment with trypsin-EDTA. This phenomenon might explain the contradiction between Figure 2B and C. The viability of DPSCs treated with SOD1 or LMWP-SOD1 was elucidated by cell cycle analysis using propidium iodide (Figure S1). Before the propidium iodide staining procedure, the cells were washed under the same conditions as shown in Figure 2C. The cell viability of untreated, SOD1-treated, and LMWP-SOD1-treated cells was similar, and treatment with trypsin-EDTA did not affect viability.
Figure 2.
Transduction ability of LMWP-SOD1 in DPSCs. FITC-labeled SOD1 and LMWP-SOD1 were incubated with DPSCs at a concentration of 2 μM for 30 minutes (A) The intracellular distribution of SOD1 and LMWP-SOD1 without fixation was observed by live cell confocal laser scanning microscopy. After FITC-labeled SOD1 and LMWP-SOD1 (2 μM) were incubated with the cells, they were washed twice with phosphate-buffered solution and immediately observed by confocal microscopy. (B) FITC-labeled SOD1 and LMWP-SOD1 (2 μM) were incubated with cells for 30 minutes, and the cells were washed twice with phosphate-buffered solution and incubated with 1% trypsin-ethylenediamine tetra-acetic acid for 10 minutes. After incubation, the cells were washed twice with phosphate-buffered solution and immediately observed by flow cytometry. Purple, untreated cells; orange, SOD1-FITC; blue, LMWP-SOD1-FITC. (C) Transduction of LMWP-SOD1 was analyzed by Western blotting using an anti-SOD1 antibody. β-actin was detected as a loading control. Stability of the transduced LMWP-SOD1 protein in cultures of human dental pulp stem cells is shown. A total of 2 μM LMWP-SOD1 was added to the culture medium and incubated for various time periods. As a control, 2 μM SOD1 protein without the transduction domain was added to the culture medium for 3 hours. (D) Transduction of the proteins into the cells was analyzed by Western blotting. β-actin was detected as a loading control. (E) Transduced proteins in the cells were analyzed using a SOD1 activity assay.
Notes: Each bar represents the mean ± standard error of the mean obtained from four experiments. Four independent experiments were performed in duplicate, and significant differences are denoted by symbols. *P < 0.05 versus the 0-hour incubation.
Abbreviations: LMWP, low molecular weight protamine; SOD1, superoxide dismutase; FITC, fluorescein isothiocyanate; DPSCs, human dental pulp stem cells.
Overall, the cell uptake studies clearly demonstrate that LMWP can deliver SOD1 effectively to the DPSC cytosol. To confirm penetration of LMWP-SOD1 and SOD1, each enzyme was added to DPSCs at a 2 μM concentration for 30 minutes, followed by Western blot analysis (Figure 2C). The cells treated with the LMWP-SOD1 conjugate were detected only in comparison with the control and the SOD1- treated cell cultures. To detect the stability of LMWP in cells, the investigators treated DPSCs with LMWP (2 μM) in a time-dependent manner and observed the remaining LMWP by FACS analysis. LMWP itself penetrated into the cell within 30 minutes and remained unchanged until 6 hours. After 12 hours of treatment, the remaining LMWP in the DPSCs had reduced to nearly half the level in the control group (Figure S2A and B). Next, to clarify that LMWP alone has no effect on cells, we determined cell viability by MTT assay (Figure S3A) and cell cycle distribution by FACS analysis (Figure S3B). The cell viability curve for the LMWP-treated group was similar to that of the untreated cells. The LMWP did not affect cell cycle distribution when compared with the untreated cells. Previous studies have demonstrated that LMWP itself has no effect on cells.34,35,37,38
Next, we determined the transduction kinetics for the LMWP-SOD1 conjugate. The SOD1 and LMWP-SOD1 conjugate was added to the culture medium of DPSCs at a concentration of 2 μM for various time periods. The resulting levels of transduced proteins were analyzed by Western blotting, and SOD1 enzyme activity was measured using a SOD1 determination assay kit. The intracellular level of the LMWP-SOD1 conjugate was initially detected after 30 minutes, peaked at one hour, and persisted in the cells until 72 hours ( Figure 2D). However, the enzyme activity of the LMWP-SOD1 that penetrated the cells peaked at 3 hours and then declined gradually over 72 hours (Figure 2E). SOD1 activity includes that for endogenous SOD1. The SOD1-treated cells were also subjected to Western blot analysis and a SOD1 activity assay (Figure S4A and B). Treatment of non-conjugated SOD1 did not affect intracellular SOD1 levels or its activity. These data suggest that the LMWP-SOD1 conjugate is capable of being internalized and demonstrates that protein transduction can be applied generally to DPSCs with high efficiency.
LMWP-SOD1 suppresses cellular senescence induced by H2O2 in DPSCs
Internalization of SOD1 by LWMP suppressed H2O2-induced enlargement of cell size and flattened cell morphology, and restored the natural morphology of DPSCs compared with SOD1-treated cells (Figure 3A and B). Control DPSCs (+H2O2) and SOD1-treated DPSCs (+H2O2) were 28.5% and 26.8% positive, respectively, for SA-β-gal staining ( Figure 3B and C, arrow head); however, LMWP-SOD1-treated DPSCs (+H2O2) were 12.3% positive for SA-β-gal staining ( Figure 3B and C, arrow head). Furthermore, we investigated the effect of LMWP-SOD1 on cell viability and the cell cycle. LMWP-SOD1 slightly restored cell viability which had been inhibited by H2O2 (Figure 3D). Similarly, for the control DPSCs (+H2O2) and the SOD1-treated DPSCs (+H2O2), the number of cells in G1 phase was increased, but the number of LMWP-SOD1-treated DPSCs (+H2O2) in G1 phase slightly and significantly decreased compared with control DPSCs (+H2O2) and SOD1-treated DPSCs ( Figure 3E). These results suggest that LMWP-SOD1 conjugates have an ability to suppress H2O-induced cellular senescence-associated morphological changes and cell proliferation.
Figure 3.
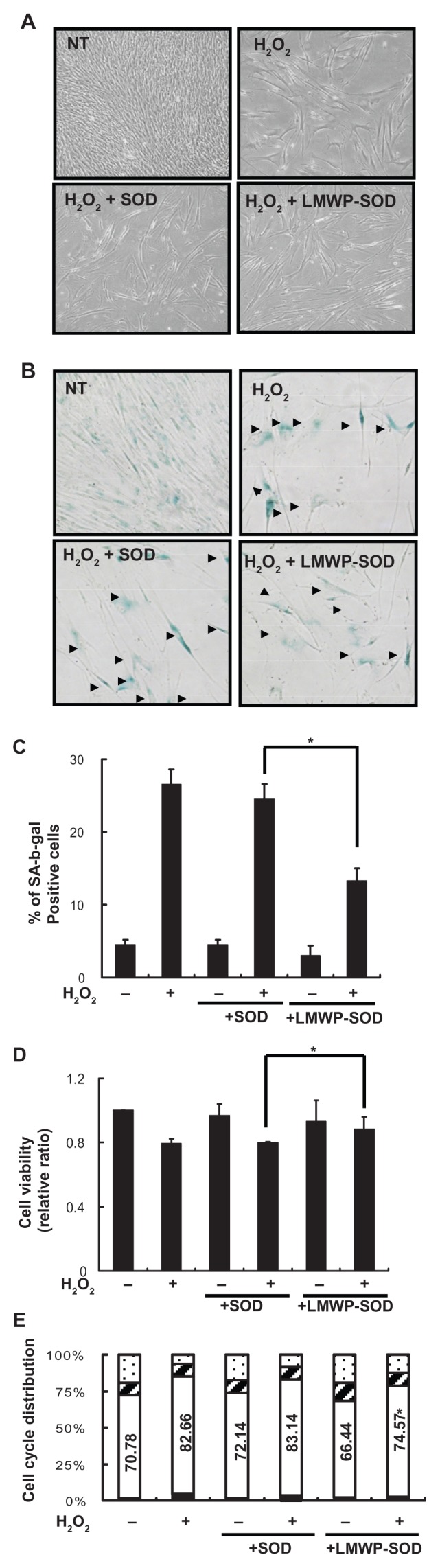
Effect of LWMP-SOD1 on H2O2-induced senescence in human DPSCs. DPSCs were pretreated with SOD1 (2 μM) or LMWP-SOD1 (2 μM) for 3 hours before 2 hours of incubation with H2O2. After removal of H2O2, the medium was refreshed, and the cells were continuously cultured for an additional 3 days. (A) The altered morphology of the DPSCs was observed by light microscopy (50×). (B) SA-β-gal-positive DPSCs observed by light microscopy (100×). (C) The percentage of SA-β-gal-positive cells treated with SOD1 or LMWP-SOD1. (D) Growth curves were monitored via MTT assay. Cell viability was determined at day 3 after brief H2O2 exposure, and LMWP-SOD1 slightly reversed the H2O2- induced attenuation of cell viability. (E) FACS analysis of the cell cycle distribution in controls or 3 days after brief H2O2 exposure.
Notes:
 G2, ▨ S, □ G1, ■ sub-G1. Each bar represents the mean ± standard error of the mean obtained from four experiments. Four independent experiments were performed in duplicate, and significant differences are denoted by symbols: *P < 0.05 versus H2O2 and SOD1-treated group (C) P = 0.018, (D) P = 0.04, (E) P = 0.015.
G2, ▨ S, □ G1, ■ sub-G1. Each bar represents the mean ± standard error of the mean obtained from four experiments. Four independent experiments were performed in duplicate, and significant differences are denoted by symbols: *P < 0.05 versus H2O2 and SOD1-treated group (C) P = 0.018, (D) P = 0.04, (E) P = 0.015.
Abbreviations: LMWP, low molecular weight protamine; SOD1, superoxide dismutase; DPSCs, human dental pulp stem cells; SA-β-gal, senescence-associated β-galactosidase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; FACS, fluorescence-assisted cell sorting.
LMWP-SOD1 influences H2O2 induction of p53-p21cip/WAF1 pathway in DPSCs
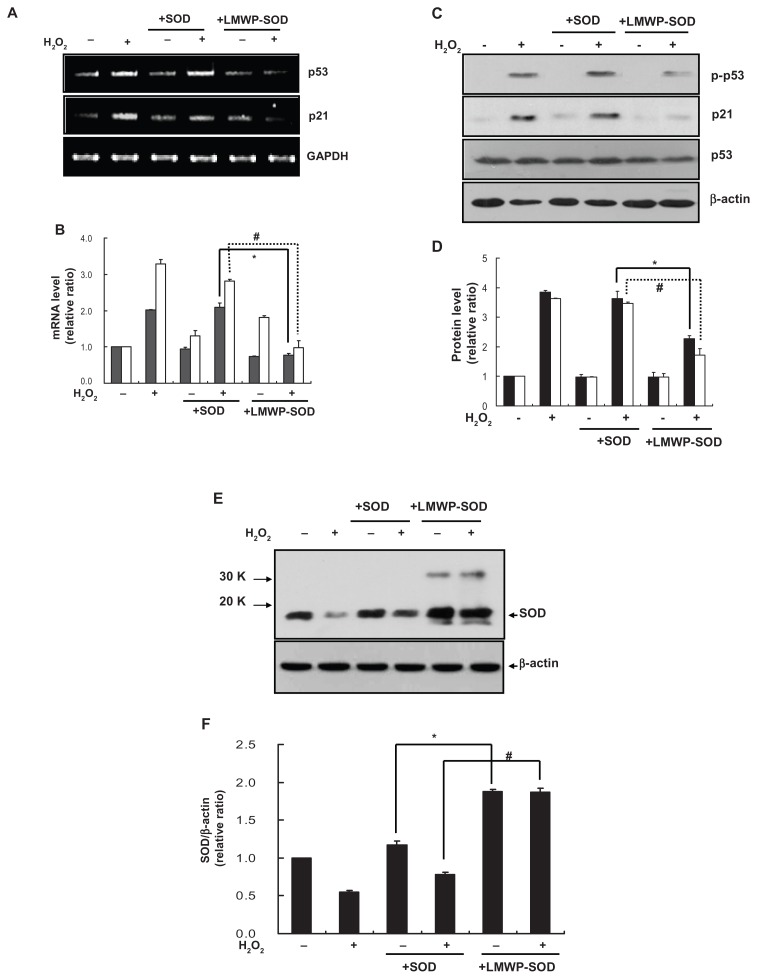
To investigate the effect of LMWP-SOD1 on the H2O2- activated p53–p21Cip1/WAF1 pathway, p53 and p21Cip1 mRNA and protein expression levels were measured. The p53 and p21Cip1/WAF1 mRNA levels were increased by H2O2 2.0- fold and 3.15-fold, respectively (Figure 4A and B). The control DPSCs (+H2O2) and SOD1-treated DPSCs (+H2O2) had no influence on mRNA expression, but LMWP-SOD1- treated DPSCs (+H2O2) significantly abolished the H2O-induced increase in p53 and p21Cip1/WAF1 mRNA expression levels to the basal state. The p53 and p21Cip1/WAF1 protein levels were also quantified by Western blot analysis. As with the mRNA levels, treatment with LMWP-SOD1 partially abolished H2O2-induced p53 and p21Cip1/WAF1 protein expression in human DPSCs (Figure 4C and D). Next, levels of the remaining intracellular SOD1 proteins were assessed using a specific antibody. H2O2 exposure decreased SOD1 levels, but treatment of DPSCs with the LMWP-SOD1 conjugate maintained SOD1 levels despite H2O2 exposure. SOD1 expression was 2.3-fold higher than that of the control (Figure 4E and F). These results suggest that pretreatment with LMWP-SOD1 not only enables SOD1 to remain in the cell for at least 3 days, but also suppresses H2O2-induced cellular senescence via regulation of the p53-p21Cip1/WAF1 signaling pathway.
Figure 4.
Effect of LMWP-SOD1 on H2O2-induced activation of the p53-p21cip/WAF1 pathway in human DPSCs. DPSCs were pretreated with SOD1 (2 μM) or LMWP-SOD1 (2 μM) for 3 hours before 2 hours of incubation with H2O2. After removal of H2O2, the cells were continuously cultured for an additional 3 days. (A) The p53 and p21Cip1/WAF1 genes were detected by reverse transcriptase polymerase chain reaction. (B) Transcription levels of p53 and p21Cip1/WAF1 were quantified. GAPDH was detected as a loading control. ■ p53, □ p21 Cip1/WAF1. (C) Phosphorylated p53, p53, and p21Cip1/WAF1 proteins were detected by Western blotting. (D) Protein levels of phosphorylated p53/p53 and p21Cip1/WAF1 were quantified. β-actin was detected as a loading control. ■ p-p53/p53, □ p21 Cip1/WAF1. DPSCs were pretreated with SOD1 (2 μM) or LMWP-SOD1 (2 μM) for 3 hours before 2 hours of incubation with H2O2. After removal of H2O2, the cells were continuously cultured for an additional 3 days. (E) Levels of SOD1 protein were detected by Western blotting. (F) The SOD1 expression level was quantified and normalized with β-actin.
Notes: Each bar represents the mean ± standard error of the mean obtained from four experiments. Four independent experiments were performed in duplicate, and significant differences are denoted by symbols: (B, D) *P < 0.05 versus the H2O2- and SOD1-treated group; #P < 0.05 versus the H2O2- and SOD1-treated group. (B) *P = 0.012; #P = 0.04; (D) *P = 0.023; #P = 0.02; (F) *P < 0.05 versus the SODI-treated group; #P < 0.05 versus the H2O2- and SODI-treated group. (F) *P = 0.018; #P = 0.03.
Abbreviations: LMWP, low molecular weight protamine; SOD1, superoxide dismutase; DPSCs, human dental pulp stem cells.
LMWP-SOD1 reverses H2O2 inhibition of osteogenic but not odontoblast differentiation
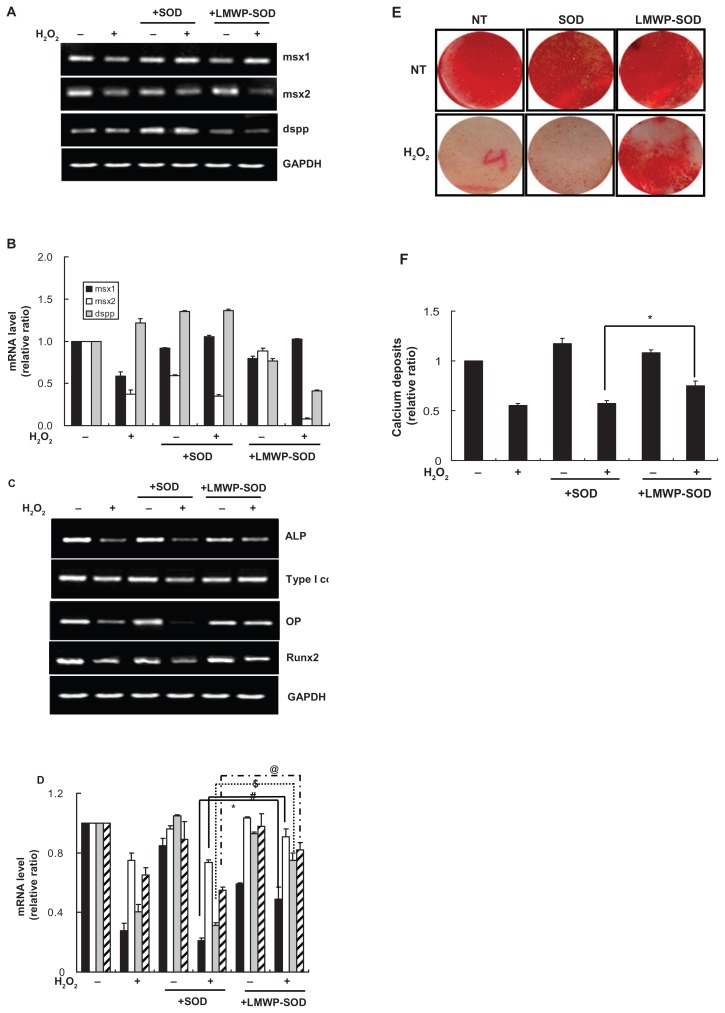
We investigated the effect of LMWP-SOD1 on odontoblastic differentiation by detecting odontogenesis markers (msx1, msx2, dspp). Odontogenesis was inhibited in H2O2-treated cells but pretreatment with SOD1 or LMWP-SOD1 did not induce any change in these gene markers. This result suggests that internalization of SOD1 by LMWP had no significant effect on any H2O2-induced decrease in odontogenesis ( Figure 5A and B). For this reason, the investigators focused on osteoblastic differentiation of DPSCs to clarify the effect of LMWP-SOD1 in H2O2-induced DPSC senescence. To elucidate the osteogenic differentiation, osteogenic genes such as alkaline phosphatase, type I collagen, osteopontin, and runx2 were detected by reverse transcriptase PCR ( Figure 5C) and quantified using the Image J program ( Figure 5D). Treatment with LMWP-SOD1 partially restored the alkaline phosphatase, type I collagen, osteopontin, and runx2 mRNA levels, but there was no significant change in expression of osteocalcin (data not shown), a late gene marker of osteogenesis. Osteogenesis was inhibited in H2O2-treated cells and H2O2-treated cells with SOD1. Cells treated with LMWP-SOD1 prior to H2O2 exposure showed significantly increased formation of calcium phosphate crystals (Figure 5E). This result suggests that inhibition of osteogenesis by H2O2 was reversed by LMWP-SOD1. The calcified areas stained with alizarin red S were quantified (Figure 5F). These data confirm that intracellular penetration of LMWP-SOD1 reversed the H2O2-induced decrease in osteogenic differentiation by restoring the expression of osteogenic genes, such as alkaline phosphatase, type I collagen, osteopontin, and runx2, without affecting odontogenic gene markers.
Figure 5.
Effect of LMWP-SOD1 on H2O2 inhibition of odontogenic or osteogenic differentiation in human DPSCs. DPSCs were pretreated with SOD1 (2 μM) or LMWP-SOD1 (2 μM) for 3 hours before 2 hours of incubation with H2O2. After removal of H2O2, the cells were washed and recultured with fresh differentiation medium for 28 days. (A and B) Odontogenic gene markers were detected by reverse transcriptase polymerase chain reaction (A) and gene expression levels were quantified (B). (C) Osteogenic gene markers were detected by reverse transcriptase polymerase chain reaction. (D) Gene expression levels were quantified. ■ Alkaline phosphatase, □ type I collagen,
 osteopontin, ▨ runx2. Each bar represents the mean ± standard error of the mean obtained from four experiments. (E) Matrix mineralization was detected by alizarin red S staining. (F) The calcified area stained with alizarin red S was quantified.
osteopontin, ▨ runx2. Each bar represents the mean ± standard error of the mean obtained from four experiments. (E) Matrix mineralization was detected by alizarin red S staining. (F) The calcified area stained with alizarin red S was quantified.
Notes: Four independent experiments were performed in duplicate, and significant differences are denoted by symbols: *P < 0.05; #P < 0.05; $P < 0.05; aP < 0.05 versus the H2O2- and SOD1-treated group (D) *P = 0.015; #P: 0.021; $P: 0.025; @P: 0.043; (F) *P = 0.019.
Abbreviations: LMWP, low molecular weight protamine; SOD1, superoxide dismutase; DPSCs, human dental pulp stem cells; ALP, alkaline phosphatase; NT, no treatment; OP, osteopontin.
Discussion
Reactive oxygen species, such as superoxide anion (O2−) and H2O2, induce oxidative stress and mediate pathological events leading to development of several diseases, including cancer, allergies, and Alzheimer’s disease, as well as irreversible senescence.17,39 In addition to the several negative effects of reactive oxygen species, one of the major problems concerning use of stem cells is their decreasing potential for differentiation and self-renewal. As reported previously, cells with premature senescence induced by H2O2 showed increased p53 activation and p21Cip1/WAF1 expression, which is likely due to H2O-induced oxidative DNA damage.40–42 Increased levels of p53 and p21Cip1/WAF1 may mediate initiation of H2O2-induced cell cycle arrest.42–45 H2O2-induced cellular senescence was indicated by increased SA-β-gal activity, a flattened cell morphology, and an enlarged cell size.43–45 Antioxidant enzymes, including SOD1, have been considered to be potential therapeutic agents because of their detoxifying abilities, which can reduce oxidative stress caused by various factors. Unfortunately, SOD1 has the limitation of being unable to penetrate into cells to protect against oxidative stress. We developed an intracellular delivery system for SOD1 by conjugating it with a cell-penetrating peptide to restore osteoblastic differentiation from oxidative stress-induced senescence of DPSCs. Previous studies have shown that cell-penetrating recombinant catalase protein fused with HIV-1 TAT (RKKRRQRRR) or arginine- rich peptides (RRRRRRRRR) are transported into the epidermis of the skin.17 It would be more appropriate to study the effect of conjugated catalase rather than SOD1 which is also involved in generation of H2O2. However, we focused on the delivery of SOD, rather than the kind of antioxidant enzyme, into cells to eliminate the reactive oxygen species and reverse the cellular senescence by penetrated SOD.
In this study, we conjugated LMWP with SOD1 and evaluated its effects on cellular senescence and differentiation capacity induced by H2O2. Treatment with LMWP-SOD1 suppressed H2O2-induced cell enlargement and flattened morphology and significantly attenuated increased SA-β-gal expression, but only slightly affected H2O2-induced cell cycle arrest and cell viability. Further, induction of p53 and p21Cip1/WAF1 mRNA and protein levels by H2O2 was significantly suppressed by LMWP-SOD1. Herein, the authors focused on the regulation of cell cycle proteins, such as p53 and p21Cip1/WAF1,46,47 to determine the possibility of attenuation of cellular senescence, because these are major targeting proteins directly influenced by H2O2. Further study might be needed for other cell cycle proteins, such as p27, retinoblastoma protein, cell-division cycle proteins, and cyclins, to explain the observed drop in number of cells at G1 phase in cells treated with LMWP-SOD1. LMWP-SOD1 did not show any reversal of the decrease in odontogenesis induced by H2O2. In contrast, osteoblastic differentiation ability was partially restored when the cells were pretreated with LMWP-SOD1. This study suggests that LMWP-SOD1 conjugates may be useful for preventing stem cell senescence induced by oxidative stress, and specifically for application in osteoblastic differentiation.
Conclusion
SOD1 conjugated with LMWP can penetrate into DPSCs directly, where they have a protective effect against H2O2. LMWP-SOD1 conjugates can be used not only in protein therapy for various diseases related to this antioxidant enzyme but also to overcome stem cell senescence induced by oxidative stress.
Supplementary figures
The viability of DPSCs treated with SOD1 or LMWP-SOD1 was elucidated by cell cycle analysis.
Notes: SOD1 and LMWP-SOD1 (2 μM) were incubated with cells for 30 minutes, the cells were washed twice with phosphate-buffered solution, and then incubated with 1% trypsin-ethylenediamine tetra-acetic acid for 10 minutes. After incubation, the cells were washed twice with phosphate-buffered solution and fixed with 70% ethyl alcohol for one hour. The cells were then washed twice with phosphate-buffered solution and incubated with propidium iodide and RNase. Cells stained with propidium iodide were observed by flow cytometry.
Abbreviations: LMWP, low molecular weight protamine; SOD1, superoxide dismutase; DPSCs, human dental pulp stem cells, NT, no treatment.
Transduction ability of LMWP into DPSCs. FITC-labeled LMWP (2 μM) was incubated for various time intervals. After incubation of the cells with FITC-labeled LMWP, the cells were washed twice with phosphate-buffered solution and incubated with 1% trypsin-ethylenediamine tetra-acetic acid for 10 minutes. After incubation, the cells were washed twice with phosphate-buffered solution and immediately observed by flow cytometry. (A) FACS analysis of the LMWP which had penetrated into the cells. (B) Penetration level of LMWP.
Notes: Each bar represents the mean ± standard error of the mean obtained from four experiments. Three independent experiments were performed in duplicate.
Abbreviations: FITC, fluorescein isothiocyanate; LMWP, low molecular weight protamine; SOD1, superoxide dismutase; DPSCs, human dental pulp stem cells.
Effect of LWMP on human DPSCs. DPSCs were pretreated with LMWP (2 μM) for various time intervals. (A) Growth curves were monitored by MTT assay. (B) FACS analysis of cell cycle distribution was determined at 0, 24, 48, and 72 hours after incubation with LMWP.
Note: Four independent experiments were performed in duplicate.
Abbreviations: LMWP, low molecular weight protamine; DPSCs, human dental pulp stem cells; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
Transduction ability of SOD1 into DPSCs. SOD1 protein (2 μM) was incubated for various time intervals. (A) Transduction of SOD1 was analyzed by Western blotting with an anti-SOD1 antibody. β-actin was detected as a loading control. (B) The activity of the transduced SOD1 protein was analyzed in cultures of human DPSCs.
Notes: Each bar represents the mean ± standard error of the mean obtained from four experiments. Four independent experiments were performed in duplicate.
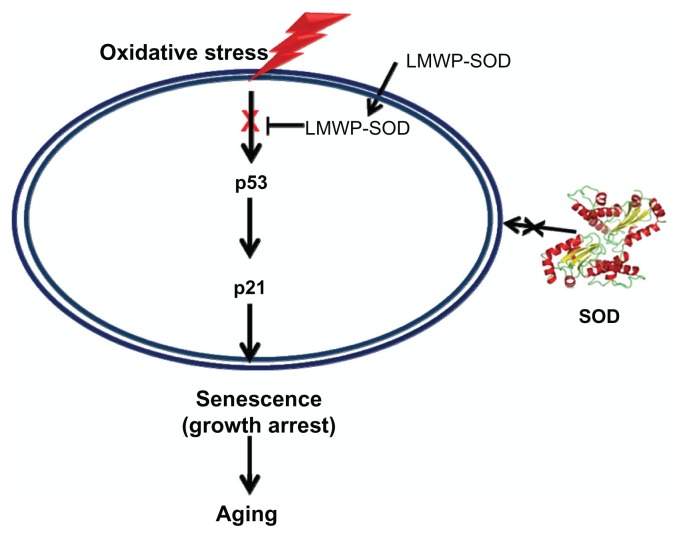
Figure 6.
Attenuation mechanism of the LMWP-SOD1 conjugate for oxidative stress-induced senescence via regulation of the p53-p21 pathway.
Abbreviations: LMWP, low molecular weight protamine; SOD1, superoxide dismutase.
Acknowledgment
This study was supported by the Korean Research Foundation Nanobiotechnology Development Regenomics Program (2008-00889) and the Ministry of Health, Welfare, and Family Affairs (A085056) in Korea.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Carroll SH, Wigner NA, Kulkarni N, Johnston-Cox H, Gerstenfeld LC, Ravid K. A2B adenosine receptor promotes mesenchymal stem cell differentiation to osteoblasts and bone formation in vivo. J Biol Chem. 2012;287:15718–15727. doi: 10.1074/jbc.M112.344994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lund P, Pilgaard L, Duroux M, Fink T, Zachar V. Effect of growth media and serum replacements on the proliferation and differentiation of adipose-derived stem cells. Cytotherapy. 2009;11:189–197. doi: 10.1080/14653240902736266. [DOI] [PubMed] [Google Scholar]
- 3.Meligy FY, Shigemura K, Behnsawy HM, Fujisawa M, Kawabata M, Shirakawa T. The efficiency of in vitro isolation and myogenic differentiation of MSCs derived from adipose connective tissue, bone marrow, and skeletal muscle tissue. In Vitro Cell Dev Biol Anim. 2012;48:203–215. doi: 10.1007/s11626-012-9488-x. [DOI] [PubMed] [Google Scholar]
- 4.Park BW, Kang EJ, Byun JH, et al. In vitro and in vivo osteogenesis of human mesenchymal stem cells derived from skin, bone marrow and dental follicle tissues. Differentiation. 2012;83:249–259. doi: 10.1016/j.diff.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Vishnubalaji R, Al-Nbaheen M, Kadalmani B, Aldahmash A, Ramesh T. Comparative investigation of the differentiation capability of bone-marrow- and adipose-derived mesenchymal stem cells by qualitative and quantitative analysis. Cell Tissue Res. 2012;347:419–427. doi: 10.1007/s00441-011-1306-3. [DOI] [PubMed] [Google Scholar]
- 6.Unda FJ, Martin A, Hilario E, Begue-Kirn C, Ruch JV, Arechaga J. Dissection of the odontoblast differentiation process in vitro by a combination of FGF1, FGF2, and TGFbeta1. Dev Dyn. 2000;218:480–489. doi: 10.1002/1097-0177(200007)218:3<480::AID-DVDY1011>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 7.Yokose S, Kadokura H, Tajima Y, et al. Establishment and characterization of a culture system for enzymatically released rat dental pulp cells. Calcif Tissue Int. 2000;66:139–144. doi: 10.1007/s002230010028. [DOI] [PubMed] [Google Scholar]
- 8.Tsukamoto Y, Fukutani S, Shin-Ike T, et al. Mineralized nodule formation by cultures of human dental pulp-derived fibroblasts. Arch Oral Biol. 1992;37:1045–1055. doi: 10.1016/0003-9969(92)90037-9. [DOI] [PubMed] [Google Scholar]
- 9.Karbanová J, Soukup T, Suchánek J, Mokrý J. Osteogenic differentiation of human dental pulp-derived stem cells under various ex-vivo culture conditions. Acta Medica (Hradec Kralove) 2010;53:79–84. doi: 10.14712/18059694.2016.64. [DOI] [PubMed] [Google Scholar]
- 10.Ma D, Ma Z, Zhang X, et al. Effect of age and extrinsic microenvironment on the proliferation and osteogenic differentiation of rat dental pulp stem cells in vitro. J Endod. 2009;35:1546–1553. doi: 10.1016/j.joen.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 12.Modino SA, Sharpe PT. Tissue engineering of teeth using adult stem cells. Arch Oral Biol. 2005;50:255–258. doi: 10.1016/j.archoralbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Stenderup K, Justesen J, Clausen C, Kassem M. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone. 2003;33:919–926. doi: 10.1016/j.bone.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Roura S, Farré J, Soler-Botija C, et al. Effect of aging on the pluripotential capacity of human CD105+ mesenchymal stem cells. Eur J Heart Fail. 2006;8:555–563. doi: 10.1016/j.ejheart.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Ito T, Sawada R, Fujiwara Y, Seyama Y, Tsuchiya T. FGF-2 suppresses cellular senescence of human mesenchymal stem cells by downregulation of TGF-beta2. Biochem Biophys Res Commun. 2007;359:108–114. doi: 10.1016/j.bbrc.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 16.Forman HJ, Torres M. Redox signaling in macrophages. Mol Aspects Med. 2001;22:189–216. doi: 10.1016/s0098-2997(01)00010-3. [DOI] [PubMed] [Google Scholar]
- 17.Jin LH, Bahn JH, Eum WS, et al. Transduction of human catalase mediated by an HIV-1 TAT protein basic domain and arginine-rich peptides into mammalian cells. Free Radic Biol Med. 2001;31:1509–1519. doi: 10.1016/s0891-5849(01)00734-1. [DOI] [PubMed] [Google Scholar]
- 18.Arteel GE. Oxidants and antioxidants in alcohol-induced liver disease. Gastroenterology. 2003;124:778–790. doi: 10.1053/gast.2003.50087. [DOI] [PubMed] [Google Scholar]
- 19.Fujimura M, Morita-Fujimura Y, Noshita N, Sugawara T, Kawase M, Chan PH. The cytosolic antioxidant copper/zinc-superoxide dismutase prevents the early release of mitochondrial cytochrome c in ischemic brain after transient focal cerebral ischemia in mice. J Neurosci. 2000;20:2817–2824. doi: 10.1523/JNEUROSCI.20-08-02817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura Y, Noda S, Hashizume K, et al. Drusen, choroidal neovascularization, and retinal pigment epithelium dysfunction in SOD1-deficient mice: a model of age-related macular degeneration. Proc Natl Acad Sci U S A. 2006;103:11282–11287. doi: 10.1073/pnas.0602131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo T, Reaume AG, Huang TT, et al. Reduction of CuZn-superoxide dismutase activity exacerbates neuronal cell injury and edema formation after transient focal cerebral ischemia. J Neurosci. 2007;17:4180–4189. doi: 10.1523/JNEUROSCI.17-11-04180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler MD, Kono H, Yin M, et al. Delivery of the Cu/Zn-superoxide dismutase gene with adenovirus reduces early alcohol-induced liver injury in rats. Gastroenterology. 2001;120:1241–1250. doi: 10.1053/gast.2001.23253. [DOI] [PubMed] [Google Scholar]
- 23.Park YJ, Liang JF, Song H, Li YT, Naik S, Yang VC. ATTEMPTS: a heparin/protamine-based triggered release system for the delivery of enzyme drugs without associated side-effects. Adv Drug Deliv Rev. 2003;55:251–265. doi: 10.1016/s0169-409x(02)00181-3. [DOI] [PubMed] [Google Scholar]
- 24.Ziegler A, Nervi P, Durrenberger M, Seelig J. The cationic cell-penetrating peptide CPP (TAT) derived from the HIV-1 protein TAT is rapidly transported into living fibroblasts: optical, biophysical, and metabolic evidence. Biochemistry. 2005;44:138–148. doi: 10.1021/bi0491604. [DOI] [PubMed] [Google Scholar]
- 25.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J Biol Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 26.Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. A comprehensive model for the cellular uptake of cationic cell- penetrating peptides. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 27.Watkins CL, Schmaljohann D, Futaki S, Jones AT. Low concentration thresholds of plasma membranes for rapid energy-independent translocation of a cell-penetrating peptide. Biochem J. 2009;420:179–189. doi: 10.1042/BJ20090042. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs SM, Raines RT. Pathway for polyarginine entry into mammalian cells. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saalik P, Elmquist A, Hansen M, et al. Protein cargo delivery properties of cell-penetrating peptides. A comparative study. Bioconjug Chem. 2004;15:1246–1253. doi: 10.1021/bc049938y. [DOI] [PubMed] [Google Scholar]
- 30.Marikovsky M, Ziv V, Nevo N, Harris-Cerruti C, Mahler O. Cu/Zn superoxide dismutase plays important role in immune response. J Immunol. 2003;170:2993–3001. doi: 10.4049/jimmunol.170.6.2993. [DOI] [PubMed] [Google Scholar]
- 31.Okumura K, Nishiguchi K, Tanigawara Y, Mori S, Iwakawa S, Komada F. Enhanced anti-inflammatory effects of Cu, Zn-superoxide dismutase delivered by genetically modified skin fibroblasts in vitro and in vivo. Pharm Res. 1997;14:1223–1227. doi: 10.1023/a:1012115109525. [DOI] [PubMed] [Google Scholar]
- 32.Eum WS, Jang SH, Kim DW, et al. Enhanced transduction of Cu, Zn-superoxide dismutase with HIV-1 Tat protein transduction domains at both termini. Mol Cells. 2005;19:191–197. [PubMed] [Google Scholar]
- 33.Kim DW, Eum WS, Jang SH, et al. Transduced Tat-SOD fusion protein protects against ischemic brain injury. Mol Cells. 2005;19:88–96. [PubMed] [Google Scholar]
- 34.Park YJ, Chang LC, Liang JF, Moon C, Chung CP, Yang VC. Nontoxic membrane translocation peptide from protamine, low molecular weight protamine (LMWP), for enhanced intracellular protein delivery: in vitro and in vivo study. FASEB J. 2005;19:1555–1557. doi: 10.1096/fj.04-2322fje. [DOI] [PubMed] [Google Scholar]
- 35.Suh JS, Lee JY, Choi YS, et al. Efficient labeling of mesenchymal stem cells using cell permeable magnetic nanoparticles. Biochem Biophys Res Commun. 2009;379:669–675. doi: 10.1016/j.bbrc.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 36.Lee JY, Nam H, Park YJ, et al. The effects of platelet-rich plasma derived from human umbilical cord blood on the osteogenic differentiation of human dental stem cells. In Vitro Cell Dev Biol Anim. 2011;47:157–164. doi: 10.1007/s11626-010-9364-5. [DOI] [PubMed] [Google Scholar]
- 37.Choi YS, Lee JY, Suh JS, et al. The systemic delivery of siRNAs by a cell penetrating peptide, low molecular weight protamine. Biomaterials. 2010;31:1429–1443. doi: 10.1016/j.biomaterials.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Suh JS, Kim KS, Lee JY, et al. A cell-permeable fusion protein for the mineralization of human dental pulp stem cells. J Dent Res. 2012;91:90–96. doi: 10.1177/0022034511424746. [DOI] [PubMed] [Google Scholar]
- 39.Greenwald RA. Superoxide dismutase and catalase as therapeutic agents for human diseases. A critical review. Free Radic Biol Med. 1990;8:201–209. doi: 10.1016/0891-5849(90)90092-w. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Meng A, Zhou D. Inhibition of phosphatidylinostol 3-kinase uncouples H2O2-induced senescent phenotype and cell cycle arrest in normal human diploid fibroblasts. Exp Cell Res. 2004;298:188–196. doi: 10.1016/j.yexcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 41.Duan J, Zhang Z, Tong T. Irreversible cellular senescence induced by prolonged exposure to H2O2 involves DNA-damage-and-repair genes and telomere shortening. Int J Biochem Cell Biol. 2005;37:1407–1420. doi: 10.1016/j.biocel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 42.Chen QM, Bartholomew JC, Campisi J, Acosta M, Reagan JD, Ames BN. Molecular analysis of H2O2-induced senescent-like growth arrest in normal human fibroblasts: p53 and Rb control G1 arrest but not cell replication. Biochem J. 1998;332(Pt 1):43–50. doi: 10.1042/bj3320043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serrano M, Blasco MA. Putting the stress on senescence. Curr Opin Cell Biol. 2001;13:748–753. doi: 10.1016/s0955-0674(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 44.Chen QM. Replicative senescence and oxidant-induced premature senescence. Beyond the control of cell cycle checkpoints. Ann N Y Acad Sci. 2002;908:111–125. doi: 10.1111/j.1749-6632.2000.tb06640.x. [DOI] [PubMed] [Google Scholar]
- 45.Marcotte R, Wang E. Replicative senescence revisited. J Gerontol A Biol Sci Med Sci. 2002;57:B257–269. doi: 10.1093/gerona/57.7.b257. [DOI] [PubMed] [Google Scholar]
- 46.Itahana K, Dimri G, Campisi J. Regulation of cellular senescence by p53. Eur J Biochem. 2001;268:2784–2791. doi: 10.1046/j.1432-1327.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- 47.Itahana K, Dimri GP, Hara E, et al. A role for p53 in maintaining and establishing the quiescence growth arrest in human cells. J Biol Chem. 2002;277:18206–18214. doi: 10.1074/jbc.M201028200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The viability of DPSCs treated with SOD1 or LMWP-SOD1 was elucidated by cell cycle analysis.
Notes: SOD1 and LMWP-SOD1 (2 μM) were incubated with cells for 30 minutes, the cells were washed twice with phosphate-buffered solution, and then incubated with 1% trypsin-ethylenediamine tetra-acetic acid for 10 minutes. After incubation, the cells were washed twice with phosphate-buffered solution and fixed with 70% ethyl alcohol for one hour. The cells were then washed twice with phosphate-buffered solution and incubated with propidium iodide and RNase. Cells stained with propidium iodide were observed by flow cytometry.
Abbreviations: LMWP, low molecular weight protamine; SOD1, superoxide dismutase; DPSCs, human dental pulp stem cells, NT, no treatment.
Transduction ability of LMWP into DPSCs. FITC-labeled LMWP (2 μM) was incubated for various time intervals. After incubation of the cells with FITC-labeled LMWP, the cells were washed twice with phosphate-buffered solution and incubated with 1% trypsin-ethylenediamine tetra-acetic acid for 10 minutes. After incubation, the cells were washed twice with phosphate-buffered solution and immediately observed by flow cytometry. (A) FACS analysis of the LMWP which had penetrated into the cells. (B) Penetration level of LMWP.
Notes: Each bar represents the mean ± standard error of the mean obtained from four experiments. Three independent experiments were performed in duplicate.
Abbreviations: FITC, fluorescein isothiocyanate; LMWP, low molecular weight protamine; SOD1, superoxide dismutase; DPSCs, human dental pulp stem cells.
Effect of LWMP on human DPSCs. DPSCs were pretreated with LMWP (2 μM) for various time intervals. (A) Growth curves were monitored by MTT assay. (B) FACS analysis of cell cycle distribution was determined at 0, 24, 48, and 72 hours after incubation with LMWP.
Note: Four independent experiments were performed in duplicate.
Abbreviations: LMWP, low molecular weight protamine; DPSCs, human dental pulp stem cells; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
Transduction ability of SOD1 into DPSCs. SOD1 protein (2 μM) was incubated for various time intervals. (A) Transduction of SOD1 was analyzed by Western blotting with an anti-SOD1 antibody. β-actin was detected as a loading control. (B) The activity of the transduced SOD1 protein was analyzed in cultures of human DPSCs.
Notes: Each bar represents the mean ± standard error of the mean obtained from four experiments. Four independent experiments were performed in duplicate.