Abstract
Background
5,10-methylenetetrahydrofolate reductase (MTHFR) variants, C677T and A1298C, have been reported to be associated with decreased risk of acute lymphoblastic leukemia (ALL). However, results derived from individually underpowered studies are conflicting. We carried out an updated meta-analysis on the association between MTHFR polymorphisms and ALL risk.
Methods
Relevant publications were searched through PUBMED and EMBASE databases. The associations between MTHFR C677T and A1298C polymorphisms and the risk of ALL were evaluated by odds ratios (ORs). The heterogeneity and publication bias were estimated. Meta-regression analysis was performed to evaluate the potential sources of heterogeneity.
Results
C677T polymorphism was associated with a reduced risk of ALL (allele contrast: ORRE = 0.91, 95% CI: 0.83-0.99). Subgroup analysis showed MTHFR C677T variant was associated with decreased susceptibility to ALL in children and Caucasians. Meta-regression showed the logOR for the association between T allele and ALL increased as sex ratio (M/F) in the case group increased (P = 0.01). Regarding A1298C polymorphism, no significant association was observed (allele contrast: ORRE = 1.01, 95% CI: 0.91-1.11). There was no publication bias for C677T or A1298C polymorphism.
Conclusions
The present meta-analysis suggests that the C677T polymorphism, not A1298C, in MTHFR gene is associated with a decreased risk of ALL, particularly among children and Caucasians subjects. Our findings suggest that the influence of the C677T polymorphism on ALL susceptibility is modified by sex ratio in cases (M/F). Since folate intake may be a possible confounding factor, including this factor in future prospective studies is warranted. Further meta-analysis studies should be at least stratified for folate levels and gender to give more powerful and informative results.
Keywords: Acute lymphoblastic leukemia; Polymorphism; Meta-analysis; 5,10-methylenetetrahydrofolate reductase; Update
Background
Acute lymphoblastic leukemia (ALL) is a malignant neoplasm of the lymphocyte precursor cells or lymphoblasts. This hematologic malignancy accounts for 75% of pediatric leukemias and 20% of adult leukemias, with an early peak incidence at 2 to 5 years of age followed by a second peak after age 50 years[1-3]. To date, the cause of ALL remains largely unknown and is likely to involve a complex interaction between genetic susceptibility and environmental exposure [4].
Folate metabolism plays an essential role in both DNA synthesis and cellular methylation reactions (e.g., DNA methylation). The enzyme 5,10-methylenetetrahydrofolate reductase (MTHFR) is a key player in folate metabolism, which irreversibly catalyzes the reduction of 5,10-methylenetetrahydrofolate (5,10-methylene THF) into 5-methyltetrahydrofolate (5-methyl THF), the predominant circulatory form of folate (Additional file 1: Figure S1) [5]. The MTHFR gene, containing 11 exons and 10 introns, is located on the short arm of chromosome 1 (1p36.3) [6,7]. Two common polymorphisms in the MTHFR gene, C677T (rs1801133) and A1298C (rs1801131) contribute to reduced enzyme activity and disturbance in folate metabolism. Severe enzymatic activity deficiency results in hyperhomocysteinaemia and is linked to increased risk of neural tube defects and vascular diseases[8-11].
MTHFR variants have been reported to be associated with reduced risk of ALL. However, results derived from individually underpowered studies are conflicting. Here, we reevaluate the association between MTHFR polymorphisms and ALL in a more comprehensive meta-analysis, providing better power to detect small effect size and performing more detailed analysis on the effects of MTHFR C677T and A1298C variants on ALL risk.
Methods
Selection criteria and identification of studies
We conducted a comprehensive search of PUBMED and EMBASE databases for publications on the association between MTHFR C677T and/or A1298C variant(s) and ALL, using the following search terms: methylenetetrahydrofolate reductase or MTHFR; leuk(a)emia, acute lymphocytic or acute lymphoblastic; and gene, polymorphism or genetic variant. The latest searches were undertaken on Oct 3, 2011. All relevant articles identified through the search were scanned on the basis of title and abstract. The articles that clearly did not meet the inclusion criteria were rejected in the initial screening. The appropriateness of remaining articles for inclusion in the meta-analysis was assessed by reading the full text. All references cited in the studies were reviewed to identify additional publications. We also evaluated previous meta-analysis articles [12-17] and manually searched bibliographies to ensure that any relevant but previously omitted articles were included in the present study.
The meta-analysis included case–control, cross-sectional and cohort studies that met all of the inclusion criteria as follows: (i) provided cases of ALL and control subjects without hematologic or other malignancies; (ii) provided relevant data to calculate the odds ratio (OR); (iii) published in English language journals. Case reports, editorials and review articles were excluded. Family-based association studies and genome-wide linkage scans were also excluded for different design considerations. When multiple studies reported on the same population, we used the most recent one only.
Data collection
For each eligible study, the following data were extracted independently by 2 investigators, using a piloted data extraction form: first author, year of publication, demographics (age, sex, and ethnicity), study design, genotyping method, leukemia characteristics, number of case and control subjects, source of controls and blinding of laboratory workers to participant status. The frequencies of the allele and the genotypic distributions were extracted (if not available, the allele frequencies were calculated from genotypes), for both the cases and the controls. When articles presented data for different ethnic groups, results for the subgroups were considered as separate studies. If raw data could not be extracted for the meta-analysis, we attempted to obtain this information by corresponding with the authors. Discrepancies were resolved by discussion, when necessary, adjudicated by a third reviewer.
Data synthesis and analysis
The risk of ALL associated with the MTHFR C677T and A1298C polymorphisms was evaluated by OR with corresponding 95% confidence intervals (CIs) under allele contrast, dominant model, recessive model and additive model[18]. In addition, subgroup analysis was carried out by ethnicity (Caucasians or East Asians) and study population (adults or children).
The interstudy heterogeneity in terms of degree of association was tested using the Cochran’s Q-statistic [19]. If P < 0.10, the heterogeneity was considered significant, which was further explored by I2 statistic. I2 is expressed as the percentage of between-study variability that is attributable to genuine variation rather than sample error[20]. If there was heterogeneity among studies, we used a random-effect (RE) model to pool the ORs; otherwise, a fixed-effect (FE) model was selected [21].
Given that immunophenotypic subtypes of ALL (B or T-lineage ALL), sex ratio (males vs females, M/F) might modulate the effects of MTHFR polymorphisms on ALL risk, and year of publication, journal impact factor (according to the Journal of Citation Report 2010) might lead to publication bias and heterogeneity, we included these factors as covariates in meta-regression. The cumulative and recursive cumulative meta-analysis were performed to demonstrate how evidence concerning the genetic association has evolved over time [18]. The Egger regression test and Begg–Mazumdar test were used to estimate the potential publication bias [22]. Pearson’s χ2 test was used to evaluate Hardy-Weinberg equilibrium (HWE) in the control group for all studies. Studies with controls not in HWE or studies not reporting enough information to evaluate HWE were subjected to a sensitivity analysis. Furthermore, we omitted 1 study at a time to assess the stability of results in the sensitivity analysis. Analyses were performed with Stata software (version 10.0; Stata Corporation, College Station, Texas, USA), using two-side P-values.
Results
Characteristics of the included studies
The literature search identified 155 articles. After abstract examination, 111 articles were excluded, and 44 articles were retrieved and evaluated against the inclusion criteria. Data from 33 articles[5,23-54] that investigated the association between MTHFR polymorphisms and ALL met the inclusion criteria. Figure 1 presents a flowchart for the process of articles inclusion/exclusion, with specification of reasons. All of the articles were in full length except one in letter [32]. Two articles provided separate data for 2 ethnic groups each [33,48]. Thus data were obtained from 35 studies.
Figure 1.
Flowchart describing the process of articles inclusion/exclusion.
The studies were published from 1999 through 2010 (Additional file 2: Table S1). All the studies were described as case–control in design. Six studies [23,25,28,39,44,46] involved adult ALL patients, 26 [5,24,26,27,30-35,37,38,40,42,43,45,47-54] involved pediatric patients and 3 [29,36,41] involved a mixed population of adult and childhood patients. The identified studies were undertaken in a wide range of ethnicities: 15 [23,27-33,37,38,43,45,47,52,54] providing data on Caucasians, 7 [35,36,39,44,46,48,49] on East Asians (Chinese and Korean), and 13 [5,24-26,33,34,40,42,43,48,50,51,53] on other ethnic origins. Twenty-one studies [5,26,28-31,33,38-41,43-45,49-54] involved general population-based controls and 5 studies [23,27,34,42,45] involved hospital-based controls, 9 studies [24,25,32,35-37,47,48] did not describe the source of their controls. Different genotyping methods were used: polymerase chain reaction (PCR) followed by restriction fragment length polymorphism (RFLP) analysis was used in 26 studies [5,23,24,26,28,29,31,33,34,36-40,44-51,53,54], real-time PCR was used in 8 studies [25,31,32,41-44,52] and allele-specific oligonucleotide hybridization (ASO) was used in 2 studies [27,35]. In most studies, authors reported that the diagnosis of ALL was based on morphologic and immunophenotypic criteria. Eight studies [23,24,28,34,39,47,49,51] stated that the controls were age and gender matched. Only one study [23] mentioned genotyping was performed under blind conditions. Twelve studies provided data for combined genotype distribution of C677T and A1298C variants [23,27,31,32,34,35,38,42,46,50,51] and 4 studies provided analysis of haplotypes for these two variants [39,44,46,53]. In three studies [26,36,39] on C677T and two studies [38,50] on A1298C, the distribution of the genotypes in control group were found to deviate from HWE according to P value (P < 0.05). Thirty-four studies dealt with C677T, 29 studies dealt with A1298C and 28 studies investigated the two polymorphisms together.
There were 5710 cases and 10798 controls included for the association between C677T polymorphism and the risk of ALL. The frequency (%) of T allele/TT genotype in controls in Caucasians was 35.4/12.9, in East Asians was 40.8/16.0, respectively. The studies provided 5356 cases and 9906 controls for A1298C polymorphism. The frequency (%) of C allele/CC genotype in controls in Caucasians was 31.8/10.1, in East Asians was 18.5/3.3, respectively. Detailed information regarding genotype distribution and allele frequency for cases and controls is available in Additional file 3: Table S2 and Additional file 4: Table S3.
C677T Polymorphism Associated with A Decreased Risk of ALL
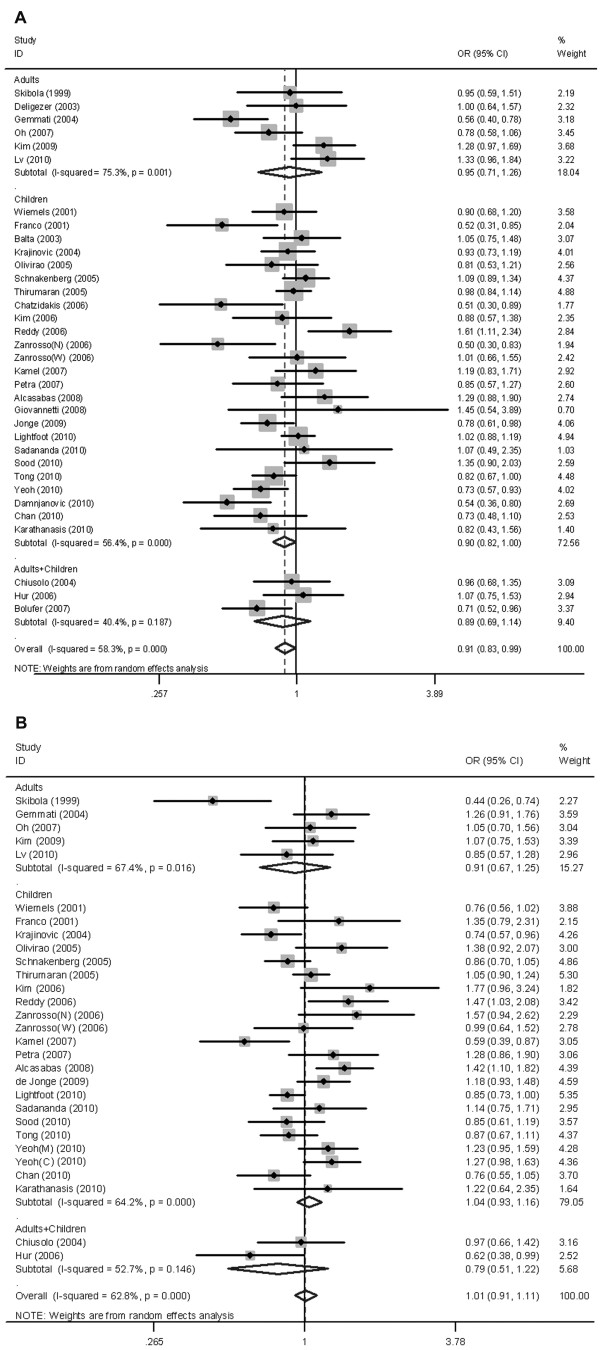
Table 1 shows the meta-analysis results for C677T polymorphism. Overall, significant heterogeneity between studies was found in all genetic contrasts except recessive model (recessive model: PQ-Test = 0.13, I2 = 22%). Marginally significant inverse association was observed in allele contrast, recessive model and additive model (allele contrast: ORRE =0.91, 95% CI: 0.83-0.99; recessive model: ORFE = 0.86, 95% CI: 0.77-0.96; additive model: ORRE =0.80, 95% CI: 0.68-0.95; Figure 2A). After exclusion of the studies lack of agreement of controls with the HWE, there was no significant alteration in the pattern of the results (Table 1). Removal of any one study did not result in movement of the point estimate outside the 95% CIs, suggesting no single study exhibited excessive influence (Additional file 5: Figure S2).
Table 1.
ORs and heterogeneity results for the genetic contrasts of MTHFR C677T polymorphism for ALL risk
| Genetic Model | Studies | I2, (%) | P, Q Testa |
Fixed-effect |
Random-effect |
|
|---|---|---|---|---|---|---|
| OR | OR | |||||
| All |
Allele contrast |
34 |
58 |
0.000 |
0.92(0.88-0.97) |
0.91(0.83-0.99) |
| |
Dominant model |
34 |
58 |
0.000 |
0.92(0.86-0.98) |
0.90(0.80-1.02) |
| |
Recessive model |
33 |
22 |
0.129 |
0.86(0.77-0.96) |
0.85(0.74-0.98) |
| |
Additive mode |
33 |
39 |
0.013 |
0.82(0.73-0.93) |
0.80(0.68-0.95) |
| All in HWE |
Allele contrast |
31 |
61 |
0.000 |
0.92(0.87-0.97) |
0.90(0.82-0.99) |
| |
Dominant model |
31 |
60 |
0.000 |
0.92(0.86-0.99) |
0.90(0.80-1.02) |
| |
Recessive model |
30 |
25 |
0.109 |
0.85(0.76-0.96) |
0.84(0.72-0.97) |
| |
Additive model |
30 |
41 |
0.011 |
0.82(0.72-0.92) |
0.78(0.65-0.94) |
| Children |
Allele contrast |
25 |
56 |
0.000 |
0.92(0.87-0.98) |
0.90(0.82-0.997) |
| |
Dominant model |
25 |
60 |
0.000 |
0.92(0.85-0.99) |
0.90(0.79-1.03) |
| |
Recessive model |
24 |
0 |
0.479 |
0.85(0.74-0.96) |
0.86(0.75-0.98) |
| |
Additive model |
24 |
23 |
0.158 |
0.82(0.71-0.94) |
0.80(0.67-0.95) |
| Adults |
Allele contrast |
6 |
75 |
0.001 |
0.95(0.83-1.09) |
0.95(0.71-1.26) |
| |
Dominant model |
6 |
67 |
0.010 |
0.94(0.77-1.14) |
0.96(0.68-1.35) |
| |
Recessive model |
6 |
66 |
0.012 |
0.93(0.72-1.22) |
0.85(0.52-1.40) |
| |
Additive model |
6 |
74 |
0.002 |
0.89(0.67-1.19) |
0.82(0.44-1.52) |
| Adults+children |
Allele contrast |
3 |
40 |
0.187 |
0.88(0.72-1.06) |
0.89(0.69-1.14) |
| |
Dominant model |
3 |
52 |
0.126 |
0.85(0.64-1.12) |
0.86(0.57-1.29) |
| |
Recessive model |
3 |
31 |
0.236 |
0.85(0.60-1.20) |
0.85(0.55-1.39) |
| |
Additive model |
3 |
30 |
0.241 |
0.77(0.52-1.14) |
0.79(0.49-1.26) |
| East Asians |
Allele contrast |
7 |
65 |
0.009 |
0.92(0.83-1.02) |
0.95(0.79-1.15) |
| |
Dominant model |
7 |
60 |
0.020 |
0.90(0.78-1.04) |
0.95(0.74-1.22) |
| |
Recessive model |
7 |
53 |
0.047 |
0.89(0.72-1.09) |
0.91(0.66-1.24) |
| |
Additive model |
7 |
60 |
0.020 |
0.87(0.69-1.09) |
0.90(0.62-1.32) |
| Caucasians |
Allele contrast |
15 |
54 |
0.006 |
0.89(0.83-0.96) |
0.85(0.76-0.95) |
| |
Dominant model |
15 |
54 |
0.007 |
0.88(0.80-0.96) |
0.82(0.70-0.95) |
| |
Recessive model |
15 |
27 |
0.155 |
0.85(0.73-0.98) |
0.82(0.68-0.99) |
| Additive model | 15 | 40 | 0.053 | 0.80(0.69-0.93) | 0.74(0.59-0.92) |
All = all of the studies meet the inclusion criteria; All in HWE = all of the studies meet the inclusion criteria except ones with genotype distribution of the controls deviating from HWE; HWE = Hardy-Weinberg Equilibrium.
a Q test is to estimate heterogeneity between studies. P < 0.10 indicates significant heterogeneity.
Figure 2.
Random-effect OR estimates with the corresponding 95% CIs for the allele contrast of MTHFR C677T and A1298C polymorphisms and the risk of ALL. (A) C677T polymorphism and ALL; (B) A1298C polymorphism and ALL. Studies are displayed by ascending order of publication year. The size of the square represents the weight of the corresponding study. C, Chinese; M, Malays; N, non-Caucasians, admixture of Amerindians, Europeans and Africans; W, mainly Brazilians of Caucasian descent.
When the analysis was carried out in different age subgroups, the MTHFR 677 T variant was associated with a decreased susceptibility to pediatric ALL (n = 25, allele contrast: ORRE = 0.90, 95%CI: 0.82-0.997; recessive model: ORFE = 0.85, 95% CI: 0.74-0.96; additive model: ORFE =0.82, 95% CI: 0.71-0.94; Figure 2A), whereas it did not show reduced risk for ALL in adults (n = 6). Higher degree of heterogeneity was observed in adults in comparison with children subgroup (Table 1). Importantly, recessive model showed the absence of heterogeneity in children (I2 = 0%). Analysis stratified by ethnicity showed 677 T variant was associated with a significantly decreased risk of ALL in Caucasians under all genetic contrasts (n = 15, allele contrast: ORRE = 0.85, 95% CI: 0.76-0.95). However, no significant association was observed in East Asians (Table 1).
No association between A1298C polymorphism and risk of ALL observed
Regarding the MTHFR A1298C polymorphism, no significant association was observed in any genetic model test when all the 29 studies pooled together (allele contrast: ORRE = 1.01, 95% CI: 0.91-1.11; dominant model: ORRE = 1.02, 95% CI: 0.90-1.16; recessive model: ORFE = 0.99, 95% CI: 0.88-1.12; additive model: ORRE = 1.01, 95% CI: 0.83-1.22; Figure 2B). Heterogeneity between studies was significant in all genetic contrasts except recessive model. Exclusion of studies deviating from HWE did not alter the pattern of results (Table 2). None of the single study exhibited excessive influence on the pooled results (Additional file 6: Figure S3). Subgroup analysis showed evidence for a relationship between A1298C polymorphism and a decreased risk of ALL neither in pediatric (n = 22) nor in adult subjects (n = 5). Stratification taking into account ethnicity also produced no significant results, with a magnitude of effects similar to that found in the main analysis (Table 2).
Table 2.
ORs and heterogeneity results for the genetic contrasts ofMTHFRA1298C polymorphism for ALL risk
| Genetic model | Studies |
I2, |
P, Q Testa |
Fixed-effect |
Random-effect |
|
|---|---|---|---|---|---|---|
| (%) | OR | OR | ||||
| All |
Allele contrast |
29 |
63 |
0.000 |
0.99(0.94-1.05) |
1.01(0.91-1.11) |
| |
Dominant model |
29 |
65 |
0.000 |
0.99 (0.92-1.06) |
1.02(0.90-1.16) |
| |
Recessive model |
29 |
20 |
0.171 |
0.99(0.88-1.12) |
1.00(0.86-1.17) |
| |
Additive model |
29 |
41 |
0.012 |
0.99(0.87-1.13) |
1.01(0.83-1.22) |
| All in HWE |
Allele contrast |
27 |
64 |
0.000 |
0.99(0.94-1.05) |
1.01(0.91-1.11) |
| |
Dominant model |
27 |
66 |
0.000 |
0.99(0.92-1.07) |
1.02(0.89-1.17) |
| |
Recessive model |
27 |
26 |
0.113 |
0.99(0.87-1.22) |
1.00(0.84-1.18) |
| |
Additive model |
27 |
44 |
0.008 |
0.99(0.87-1.13) |
1.01(0.82-1.24) |
| Children |
Allele contrast |
22 |
64 |
0.000 |
1.01(0.95-1.07) |
1.04(0.93-1.16) |
| |
Dominant model |
22 |
67 |
0.000 |
1.01(0.93-1.09) |
1.07(0.92-1.24) |
| |
Recessive model |
22 |
26 |
0.129 |
1.00(0.88-1.14) |
1.00(0.85-1.18) |
| |
Additive model |
22 |
47 |
0.008 |
1.00(0.87-1.15) |
1.02(0.83-1.25) |
| Adults |
Allele contrast |
5 |
67 |
0.016 |
0.96(0.81-1.14) |
0.92(0.67-1.25) |
| |
Dominant model |
5 |
68 |
0.014 |
0.97(0.79-1.19) |
0.92(0.63-1.33) |
| |
Recessive model |
5 |
41 |
0.150 |
0.84(0.50-1.42) |
0.85(0.37-1.93) |
| |
Additive model |
5 |
51 |
0.086 |
0.84(0.49-1.42) |
0.80(0.32-2.01) |
| East Asians |
Allele contrast |
7 |
53 |
0.046 |
1.01(0.89-1.15) |
1.01(0.82-1.23) |
| |
Dominant model |
7 |
63 |
0.013 |
1.01(0.87-1.17) |
1.01(0.78-1.31) |
| |
Recessive model |
7 |
0 |
0.730 |
1.02(0.70-1.50) |
1.07(0.72-1.58) |
| |
Additive model |
7 |
0 |
0.671 |
1.03(0.70-1.51) |
1.07(0.72-1.59) |
| Caucasians |
Allele contrast |
12 |
62 |
0.003 |
0.96(0.89-1.04) |
0.98(0.86-1.12) |
| |
Dominant model |
12 |
55 |
0.011 |
0.95(0.86-1.04) |
0.98(0.83-1.15) |
| |
Recessive model |
12 |
42 |
0.062 |
0.97(0.82-1.14) |
0.97(0.76-1.25) |
| Additive model | 12 | 52 | 0.019 | 0.95(0.80-1.12) | 0.98(0.73-1.30) |
All = all of the studies meet the inclusion criteria; All in HWE = all of the studies meet the inclusion criteria except ones with genotype distribution of the controls deviating from HWE; HWE = Hardy-Weinberg Equilibrium.
a Q test is to estimate heterogeneity between studies. P < 0.10 indicates significant heterogeneity.
For combined genotypes analysis, we did not observe significant pooled ORs when using 677CC/1298AA combination as a baseline (Additional file 7: Table S4). We did not pool the ORs for haplotypes in the meta-analysis because of the limited data.
Potential Bias
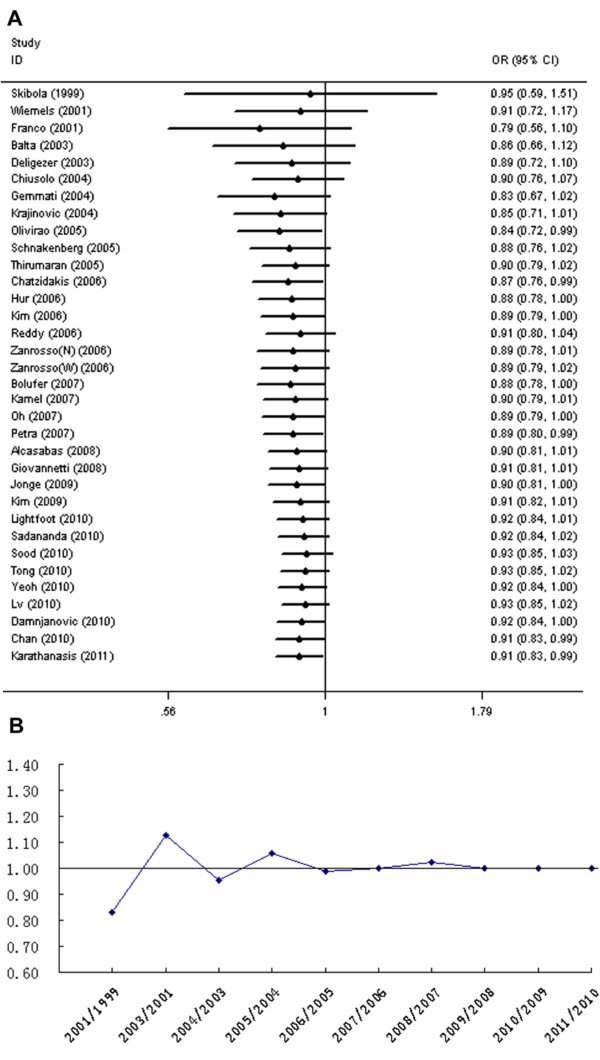
Meta-regression results indicated a significant correlation between sex ratio (M/F) in ALL cases and genetic effect (n = 25, P =0.01), which could explain 28% of the variance (Figure 3), whereas year of publication, journal impact factor, immunophenotypic subtypes and sex ratio in controls did not contribute significantly to between-study heterogeneity (P > 0.05). The logOR for the association between T allele and ALL increased as M/F in the case group increased. No covariates modulating the effect of A1298C polymorphism on ALL risk was found (data not shown). Cumulative meta-analysis showed a trend of inverse association between C677T variant and ALL risk as evidence accumulated. Recursive cumulative meta-analysis showed that the relative change in ORs for the C677T polymorphism fluctuated in the beginning years (from 1999 to 2006) and then stabilized at around 1.0 (Figure 4).
Figure 3.
Meta-regression of sex ratio (M/F) in ALL cases and MTHFR C677T genetic effect using allele contrast. The size of the circles represents the weight of the corresponding study in the meta-regression.
Figure 4.
Cumulative and recursive cumulative meta-analysis for the association betweenMTHFRC677T polymorphism and the risk of ALL. (A) Cumulative meta-analysis; (B) Recursive cumulative meta-analysis. Allele contrast was used in cumulative and recursive cumulative meta-analysis. N, non-Caucasians, admixture of Amerindians, Europeans and Africans; W, mainly Brazilians of Caucasian descent.
No significant publication bias was detected for C677T or A1298C polymorphism by formal statistics (C677T: Egger's test, P = 0.42, Begg's test, P = 0.51; A1298C: Egger's test, P = 0.61, Begg's test, P = 0.81; respectively), indicating there is no differential magnitude of effect in large vs small studies.
Discussion
Two meta-analyses published in 2006, focused on the associations between MTHFR C677T and A1298C polymorphisms and ALL risk [12,13]. Nevertheless, inconsistent results were obtained between the two studies for C677T and A1298C variants, which might mainly be due to limited number of the included studies and/or different selection criteria[18,55]. In recent two years, another 4 meta-analyses also investigated the associations between MTHFR polymorphisms and ALL risk [14-17]. None of the authors carried out analysis in detail which could be done as more studies were available. Additionally, fewer studies were collected in these meta-analyses compared with ours (Additional file 8: Table S5), probably due to insufficient attention in the search strategy or screening process, which might introduce selection bias. Thus, it is not surprising these meta-analyses also failed to give consistent results. Here, the strength of the present analysis is that our study is based on a larger amount of published data and gives comprehensive and intensive information to evaluate the effects of MTHFR C677T and A1298C polymorphisms on ALL risk.
It is possible to draw several conclusions from the current data. The results of main analysis support a protective role of the C677T variant in the development of ALL, but not A1298C polymorphism. The stability in sensitive analysis and recursive cumulative meta-analysis indicates that there is sufficient evidence to confirm the beneficial effect of C677T variant in ALL. It has been well documented that MTHFR 677 T variant encodes a thermolabile enzyme with reduced catalytic activity and increases plasma homocysteine levels [1,2]. MTHFR enzyme function influences cancer risk in two pathways. Polymorphisms that affect MTHFR enzyme activity decreases the methylation of homocysteine to methionine and in turn the level of S-adenosylmethionine (SAM), resulting in DNA hypomethylation (Additional file 1: Figure S1). This phenomenon can increase the risk of some cancers (e.g. esophageal[56] and gastric cancers[57]). On the other hand, the mechanism proposed to explain the reduced risk of leukemia[23], colorectal carcinoma[58] and other neoplasias is that impaired MTHFR activity, because of polymorphic variation, leads to an accumulation of cytosolic 5,10-methylene THF available for purine and pyrimidine synthesis, thus a lower incorporation of uracil into DNA and a lower cancer risk (Additional file 1: Figure S1). The A1298C polymorphism contributes to less effect on enzyme activity. Decrease in enzyme activity in individuals homozygous for the A1298C polymorphism (30-40% of the wild type) is less than that of C677T homozygotes (60-70% of the wild type). A1298C polymorphism does not seem to be powerful enough to affect plasma homocysteine level, except when accompanied by C677T variant [3,4]. This may partly explain why MTHFR C677T variant but not A1298C was found to be protective for ALL.
Our data show that C677T variant plays a protective role in a group of pediatric patients, but has no significantly beneficial effect in adult subjects. There is significant difference between adult and childhood acute leukemia. The most common form of acute leukemia in adults is acute myeloid leukemia (AML), whereas in childhood is ALL[59]. The effect of C677T polymorphism in disease susceptibility may vary depending on folate status. Individuals during the periods of rapid cell division and growth, such as infants and children, have higher folate requirement and are more susceptible to DNA damage as a result of folate insufficiency than adults. In addition, children are not exposed to many environmentally carcinogenic factors, so a diet- and genetic-related etiology of leukaemia is more likely [14].
We observed MTHFR C677T polymorphism was associated with a significant reduction of ALL risk in Caucasian subjects, whereas it failed to show any protective effect in East Asians. As the role of the MTHFR genotypes in the development of ALL may differ among population origin due to the different dietary customs and racial backgrounds [60], we retrieved the T allele frequency in controls and folate intake in the included studies. There was no obvious difference in the average T allele frequency in controls between Caucasians and East Asians (35.4% and 40.8%, respectively). No study provided data on folate intake. Only one study provided data on the serum folate level in ALL [49]. There may be lower folate status or even folate deficiency in the Asian populations, especially in China where women taking insufficient folate during pregnancy result in a high incidence of infant neural tube defects [61]. Therefore, it is unexpected that the protective effect of C677T polymorphism is not observed in East Asian populations. Importantly, we calculated the pooled results in Caucasian children and East Asian children, separately, and found significant results in both groups (allele contrast: Caucasian children, ORRE = 0.89, 95% CI: 0.79-0.997, n = 11; East Asian children, ORRE = 0.79, 95% CI: 0.69-0.92, n = 3). As we included English studies only in this meta-analysis, the results might be influenced by language bias, especially in non-English ethnicity. We pooled the studies on Chinese children without any restriction on language in another article and found a significant protective effect of MTHFR C677T variant on ALL risk (n = 7, data not shown). We cannot exclude the possibility that the difference between Caucasians and East Asians is attributable to chance factors or age composition. Thus, the conclusion derived from such subgroup should be interpreted with caution.
We carried out the meta-regression study to evaluate the potential sources of heterogeneity. For C677T polymorphism, the heterogeneity could be partly attributed to variation in M/F in ALL cases of the original studies. The OR increased as M/F in the case group increased. It was surprising to find OR > 1 in studies with M/F ≥ 2 in case group, indicating the protective effect of T allele disappeared or even turned to the opposite in these studies [25,26,36,50,51]. We limited our analysis to the pediatric patients and observed a similar trend (data not shown). This interesting result motivated us to check three studies providing data on the separate sex group [26,34,46]. Belta et al.[26] and Reddy et al.[34] showed the CT/TT genotypes were more frequent in male than female cases, but Lv et al.[46] did not show any difference on the CT/TT genotypes distribution between male and female cases. The ORs in the male group were also not consistent, with OR < 1 in Reddy’s study and OR > 1 in the Lv’s study. Because of the discrepant results, we could not give the conclusion whether the genetic effects are different or not in separate gender here. It has been reported gender difference exists in ALL susceptibility and ALL is more common in males of all age group, despite the underlying mechanisms for sex difference remain unknown[62]. Our study showed sex ratio in case group modified the C677T variant effects with regard to ALL risk. However, this result is preliminary and deserved further investigation stratified for gender.
Gene-environment interaction between the MTHFR genotypes and dietary folate intake has been documented in previous studies concerning colorectal cancer, which might alter the effects of the polymorphic variants [5,6]. With respect to the risk of ALL, epidemiological studies proved a protective effect of maternal folate supplementation during pregnancy against childhood ALL[63,64]. As mentioned above, however, no studies to date described the effect of folate status on the association between MTHFR polymorphic variants and ALL susceptibility. Conflicting results among studies may be due to the lack of information on folate status. Other enzymes involved in folate metabolism, including methionine synthase (MS), thymidylate synthase (TS) and serine hydroxymethyltransferase (SHMT), may regulate intracellular folate metabolism. The variant forms of these enzymes may be associated with the risk of ALL and interactions between these candidate genes may exist. Three included studies described the interactions between MTHFR polymorphisms and other gene variants. Jonge et al. [45] found the MTHFR C677T and NNMT C-151 T variants interacted to decrease the risk of pediatric ALL. Petra et al. [38] reported the MTHFR C677T, MS A2756G and MTRR A66G interaction was associated with a reduced risk of pediatric ALL, whereas Gemmati et al. [28] did not detect a significant interaction between MTHFR C677T and MS A2756G polymorphisms. No further analysis on these interactions in our study was performed due to limited number of reports on each item.
Conclusion
In conclusion, the present meta-analysis suggests that the C677T polymorphism in MTHFR gene is associated with decreased susceptibility to ALL, and indicates a lack of positive relationship between A1298C polymorphism and ALL. The C677T variant plays a protective role in pediatric patients and Caucasian subjects. M/F in cases could modulate the influence of the C677T polymorphism on ALL susceptibility. Although more than 30 genetic association studies are included in this meta-analysis to draw relative safe conclusions, it is also worth mentioning that several interesting but unsolved issues are raised from current meta-analysis and thus further studies are needed which should be at least stratified for folate levels and gender.
Competing interest
The authors declare that they have no competing interests.
Authors' contributions
HGW drafted the manuscript and performed data analysis. JLW edited the manuscript. HGW, JLW and LXZ participated in data collection. XCL and WJM conceived of the study and participated in designing the manuscript. All authors participated in revising the manuscript. All authors have read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Figure S1.Folate metabolism and the role of MTHFR. Modified from Wiemels et al. (5).
Table S1.General characteristics of studies included in the meta-analysis on MTHFR polymorphisms and ALL risk.
Table S2.MTHFRC677T genotype distribution and allele frequency in ALL cases and controls.
Table S3.MTHFRA1298C genotype distribution and allele frequency in ALL cases and controls.
Figure S2.Sensitive analysis to assess the influence of a single study in the meta-analysis on MTHFR C677T polymorphism and the risk of ALL. Allele contrast was used in sensitive analysis. N, non-Caucasians, admixture of Amerindians, Europeans and Africans; W, mainly Brazilians of Caucasian descent.
Figure S3.Sensitive analysis to assess the influence of a single study in the meta-analysis on MTHFR A1298C polymorphism and the risk of ALL. Allele contrast was used in sensitive analysis. C, Chinese; M, Malays; N, non-Caucasians, admixture of Amerindians, Europeans and Africans; W, mainly Brazilians of Caucasian descent.
Table S4.Results of the pooled OR and the corresponding 95% CIs for each combined genotype for MTHFR C677T and A1298C polymorphisms.
Table S5.Characteristics of meta-analyses regarding MTHFR polymorphisms and ALL risk. (DOC 42 kb)
Contributor Information
Haigang Wang, Email: sdwfbf@qq.com.
Jiali Wang, Email: wangjiali_2000@126.com.
Lixia Zhao, Email: qlyypharmacy@126.com.
Xinchun Liu, Email: pivaslxc@126.com.
Wenjie Mi, Email: pivasmwj@126.com.
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56(2):106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- Karathanasis NV, Choumerianou DM, Kalmanti M. Gene polymorphisms in childhood ALL. Pediatr Blood Cancer. 2009;52(3):318–323. doi: 10.1002/pbc.21825. [DOI] [PubMed] [Google Scholar]
- Faderl S, O'Brien S, Pui CH, Stock W, Wetzler M, Hoelzer D, Kantarjian HM. Adult acute lymphoblastic leukemia: concepts and strategies. Cancer. 2010;116(5):1165–1176. doi: 10.1002/cncr.24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onciu M. Acute lymphoblastic leukemia. Hematol Oncol Clin North Am. 2009;23(4):655–674. doi: 10.1016/j.hoc.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Wiemels JL, Smith RN, Taylor GM, Eden OB, Alexander FE, Greaves MF. Methylenetetrahydrofolate reductase (MTHFR) polymorphisms and risk of molecularly defined subtypes of childhood acute leukemia. Proc Natl Acad Sci U S A. 2001;98(7):4004–4009. doi: 10.1073/pnas.061408298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, Rozen R. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat Genet. 1994;7(2):195–200. doi: 10.1038/ng0694-195. [DOI] [PubMed] [Google Scholar]
- Gaughan DJ, Barbaux S, Kluijtmans LA, Whitehead AS. The human and mouse methylenetetrahydrofolate reductase (MTHFR) genes: genomic organization, mRNA structure and linkage to the CLCN6 gene. Gene. 2000;257(2):279–289. doi: 10.1016/S0378-1119(00)00392-9. [DOI] [PubMed] [Google Scholar]
- Ueland PM, Hustad S, Schneede J, Refsum H, Vollset SE. Biological and clinical implications of the MTHFR C677T polymorphism. Trends Pharmacol Sci. 2001;22(4):195–201. doi: 10.1016/S0165-6147(00)01675-8. [DOI] [PubMed] [Google Scholar]
- Kluijtmans LA, Kastelein JJ, Lindemans J, Boers GH, Heil SG, Bruschke AV, Jukema JW, van den Heuvel LP, Trijbels FJ, Boerma GJ, Verheugt FW, Willems F, Blom HJ. Thermolabile methylenetetrahydrofolate reductase in coronary artery disease. Circulation. 1997;96(8):2573–2577. doi: 10.1161/01.CIR.96.8.2573. [DOI] [PubMed] [Google Scholar]
- van der Put NM, Gabreels F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel LP, Blom HJ. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects. Am J Hum Genet. 1998;62(5):1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CY, Stevenson RE, Brown VK, Schwartz CE, Allen WP, Khoury MJ, Rozen R, Oakley GP, Adams MJ. 5,10 Methylenetetrahydrofolate reductase genetic polymorphism as a risk factor for neural tube defects. Am J Med Genet. 1996;63(4):610–614. doi: 10.1002/(SICI)1096-8628(19960628)63:4<610::AID-AJMG15>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Pereira TV, Rudnicki M, Pereira AC, Pombo-de-Oliveira MS, Franco RF. 5,10-Methylenetetrahydrofolate reductase polymorphisms and acute lymphoblastic leukemia risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1956–1963. doi: 10.1158/1055-9965.EPI-06-0334. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Koufakis T, Ziakas PD, Rodopoulou P, Giannouli S, Voulgarelis M. A meta-analysis of genotypes and haplotypes of methylenetetrahydrofolate reductase gene polymorphisms in acute lymphoblastic leukemia. Eur J Epidemiol. 2006;21(7):501–510. doi: 10.1007/s10654-006-9027-8. [DOI] [PubMed] [Google Scholar]
- Koppen IJ, Hermans FJ, Kaspers GJ. Folate related gene polymorphisms and susceptibility to develop childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;148(1):3–14. doi: 10.1111/j.1365-2141.2009.07898.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhan P, Chen B, Zhou R, Yang Y, Ouyang J. MTHFR C677T polymorphisms and childhood acute lymphoblastic leukemia: a meta-analysis. Leuk Res. 2010;34(12):1596–1600. doi: 10.1016/j.leukres.2010.03.034. [DOI] [PubMed] [Google Scholar]
- Zacho J, Yazdanyar S, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Hyperhomocysteinemia, methylenetetrahydrofolate reductase c.677C>T polymorphism and risk of cancer: cross-sectional and prospective studies and meta-analyses of 75,000 cases and 93,000 controls. Int J Cancer. 2011;128(3):644–652. doi: 10.1002/ijc.25375. [DOI] [PubMed] [Google Scholar]
- Yan J, Yin M, Dreyer ZE, Scheurer ME, Kamdar K, Wei Q, Okcu MF. A meta-analysis of MTHFR C677T and A1298C polymorphisms and risk of acute lymphoblastic leukemia in children. Pediatr Blood Cancer. 2012;58(4):513–518. doi: 10.1002/pbc.23137. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol. 2008;61(7):634–645. doi: 10.1016/j.jclinepi.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2005;28(2):123–137. doi: 10.1002/gepi.20048. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira TV, Rudnicki M, Cheung BM, Baum L, Yamada Y, Oliveira PS, Pereira AC, Krieger JE. Three endothelial nitric oxide (NOS3) gene polymorphisms in hypertensive and normotensive individuals: meta-analysis of 53 studies reveals evidence of publication bias. J Hypertens. 2007;25(9):1763–1774. doi: 10.1097/HJH.0b013e3281de740d. [DOI] [PubMed] [Google Scholar]
- Macaskill P, Walter SD, Irwig L. A comparison of methods to detect publication bias in meta-analysis. Stat Med. 2001;20(4):641–654. doi: 10.1002/sim.698. [DOI] [PubMed] [Google Scholar]
- Skibola CF, Smith MT, Kane E, Roman E, Rollinson S, Cartwright RA, Morgan G. Polymorphisms in the methylenetetrahydrofolate reductase gene are associated with susceptibility to acute leukemia in adults. Proc Natl Acad Sci U S A. 1999;96(22):12810–12815. doi: 10.1073/pnas.96.22.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco RF, Simoes BP, Tone LG, Gabellini SM, Zago MA, Falcao RP. The methylenetetrahydrofolate reductase C677T gene polymorphism decreases the risk of childhood acute lymphocytic leukaemia. Br J Haematol. 2001;115(3):616–618. doi: 10.1046/j.1365-2141.2001.03140.x. [DOI] [PubMed] [Google Scholar]
- Deligezer U, Akisik E, Dalay N. Genotyping of the MTHFR gene polymorphism, C677T in patients with leukemia by melting curve analysis. Mol Diagn. 2003;7(3–4):181–185. doi: 10.1007/BF03260036. [DOI] [PubMed] [Google Scholar]
- Balta G, Yuksek N, Ozyurek E, Ertem U, Hicsonmez G, Altay C, Gurgey A. Characterization of MTHFR, GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes in childhood acute leukemia. Am J Hematol. 2003;73(3):154–160. doi: 10.1002/ajh.10339. [DOI] [PubMed] [Google Scholar]
- Krajinovic M, Lamothe S, Labuda D, Lemieux-Blanchard E, Theoret Y, Moghrabi A, Sinnett D. Role of MTHFR genetic polymorphisms in the susceptibility to childhood acute lymphoblastic leukemia. Blood. 2004;103(1):252–257. doi: 10.1182/blood-2003-06-1794. [DOI] [PubMed] [Google Scholar]
- Gemmati D, Ongaro A, Scapoli GL, Della PM, Tognazzo S, Serino ML, Di BE, Rodeghiero F, Gilli G, Reverberi R, Caruso A, Pasello M, Pellati A, De Mattei M. Common gene polymorphisms in the metabolic folate and methylation pathway and the risk of acute lymphoblastic leukemia and non-Hodgkin's lymphoma in adults. Cancer Epidemiol Biomarkers Prev. 2004;13(5):787–794. [PubMed] [Google Scholar]
- Chiusolo P, Reddiconto G, Cimino G, Sica S, Fiorini A, Farina G, Vitale A, Sora F, Laurenti L, Bartolozzi F, Fazi P, Mandelli F, Leone G. Methylenetetrahydrofolate reductase genotypes do not play a role in acute lymphoblastic leukemia pathogenesis in the Italian population. Haematologica. 2004;89(2):139–144. [PubMed] [Google Scholar]
- Schnakenberg E, Mehles A, Cario G, Rehe K, Seidemann K, Schlegelberger B, Elsner HA, Welte KH, Schrappe M, Stanulla M. Polymorphisms of methylenetetrahydrofolate reductase (MTHFR) and susceptibility to pediatric acute lymphoblastic leukemia in a German study population. BMC Med Genet. 2005;6:23. doi: 10.1186/1471-2350-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E, Alves S, Quental S, Ferreira F, Norton L, Costa V, Amorim A, Prata MJ. The MTHFR C677T and A1298C polymorphisms and susceptibility to childhood acute lymphoblastic leukemia in Portugal. J Pediatr Hematol Oncol. 2005;27(8):425–429. doi: 10.1097/01.mph.0000177513.81465.94. [DOI] [PubMed] [Google Scholar]
- Thirumaran RK, Gast A, Flohr T, Burwinkel B, Bartram C, Hemminki K, Kumar R. MTHFR genetic polymorphisms and susceptibility to childhood acute lymphoblastic leukemia. Blood. 2005;106(7):2590–2591. doi: 10.1182/blood-2005-04-1719. author reply 2591–2592. [DOI] [PubMed] [Google Scholar]
- Zanrosso CW, Hatagima A, Emerenciano M, Ramos F, Figueiredo A, Felix TM, Segal SL, Giugliani R, Muniz MT, Pombo-de-Oliveira MS. The role of methylenetetrahydrofolate reductase in acute lymphoblastic leukemia in a Brazilian mixed population. Leuk Res. 2006;30(4):477–481. doi: 10.1016/j.leukres.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Reddy H, Jamil K. Polymorphisms in the MTHFR gene and their possible association with susceptibility to childhood acute lymphocytic leukemia in an Indian population. Leuk Lymphoma. 2006;47(7):1333–1339. doi: 10.1080/10428190600562773. [DOI] [PubMed] [Google Scholar]
- Kim NK, Chong SY, Jang MJ, Hong SH, Kim HS, Cho EK, Lee JA, Ahn MJ, Kim CS, Oh D. Association of the methylenetetrahydrofolate reductase polymorphism in Korean patients with childhood acute lymphoblastic leukemia. Anticancer Res. 2006;26(4B):2879–2881. [PubMed] [Google Scholar]
- Hur M, Park JY, Cho HC, Lee KM, Shin HY, Cho HI. Methylenetetrahydrofolate reductase A1298C genotypes are associated with the risks of acute lymphoblastic leukaemia and chronic myelogenous leukaemia in the Korean population. Clin Lab Haematol. 2006;28(3):154–159. doi: 10.1111/j.1365-2257.2006.00769.x. [DOI] [PubMed] [Google Scholar]
- Chatzidakis K, Goulas A, Athanassiadou-Piperopoulou F, Fidani L, Koliouskas D, Mirtsou V. Methylenetetrahydrofolate reductase C677T polymorphism: association with risk for childhood acute lymphoblastic leukemia and response during the initial phase of chemotherapy in greek patients. Pediatr Blood Cancer. 2006;47(2):147–151. doi: 10.1002/pbc.20574. [DOI] [PubMed] [Google Scholar]
- Petra BG, Janez J, Vita D. Gene-gene interactions in the folate metabolic pathway influence the risk for acute lymphoblastic leukemia in children. Leuk Lymphoma. 2007;48(4):786–792. doi: 10.1080/10428190601187711. [DOI] [PubMed] [Google Scholar]
- Oh D, Kim NK, Jang MJ, Kim HC, Lee JH, Lee JA, Ahn MJ, Kim CS, Kim HS, Park S, Chio HS, Min YH. Association of the 5,10-methylenetetrahydrofolate reductase (MTHFR C677T and A1298C) polymorphisms in Korean patients with adult acute lymphoblastic leukemia. Anticancer Res. 2007;27(5A):3419–3424. [PubMed] [Google Scholar]
- Kamel AM, Moussa HS, Ebid GT, Bu RR, Bhatia KG. Synergistic effect of methyltetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphism as risk modifiers of pediatric acute lymphoblastic leukemia. J Egypt Natl Canc Inst. 2007;19(2):96–105. [PubMed] [Google Scholar]
- Bolufer P, Collado M, Barragan E, Cervera J, Calasanz MJ, Colomer D, Roman-Gomez J, Sanz MA. The potential effect of gender in combination with common genetic polymorphisms of drug-metabolizing enzymes on the risk of developing acute leukemia. Haematologica. 2007;92(3):308–314. doi: 10.3324/haematol.10752. [DOI] [PubMed] [Google Scholar]
- Giovannetti E, Ugrasena DG, Supriyadi E, Vroling L, Azzarello A, de Lange D, Peters GJ, Veerman AJ, Cloos J. Methylenetetrahydrofolate reductase (MTHFR) C677T and thymidylate synthase promoter (TSER) polymorphisms in Indonesian children with and without leukemia. Leuk Res. 2008;32(1):19–24. doi: 10.1016/j.leukres.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Alcasabas P, Ravindranath Y, Goyette G, Haller A, Del RL, Lesaca-Medina MY, Darga L, Ostrea EM, Taub JW, Everson RB. 5,10-methylenetetrahydrofolate reductase (MTHFR) polymorphisms and the risk of acute lymphoblastic leukemia (ALL) in Filipino children. Pediatr Blood Cancer. 2008;51(2):178–182. doi: 10.1002/pbc.21511. [DOI] [PubMed] [Google Scholar]
- Kim HN, Kim YK, Lee IK, Yang DH, Lee JJ, Shin MH, Park KS, Choi JS, Park MR, Jo DY, Won JH, Kwak JY, Kim HJ. Association between polymorphisms of folate-metabolizing enzymes and hematological malignancies. Leuk Res. 2009;33(1):82–87. doi: 10.1016/j.leukres.2008.07.026. [DOI] [PubMed] [Google Scholar]
- de Jonge R, Tissing WJ, Hooijberg JH, Jansen G, Kaspers GJ, Lindemans J, Peters GJ, Pieters R. Polymorphisms in folate-related genes and risk of pediatric acute lymphoblastic leukemia. Blood. 2009;113(10):2284–2289. doi: 10.1182/blood-2008-07-165928. [DOI] [PubMed] [Google Scholar]
- Lv L, Wu C, Sun H, Zhu S, Yang Y, Chen X, Fu H, Bao L. Combined 677CC/1298 AC genotypes of methylenetetrahydrofolate reductase (MTHFR ) reduce susceptibility to precursor B lymphoblastic leukemia in a Chinese population. Eur J Haematol. 2010;84(6):506–512. doi: 10.1111/j.1600-0609.2010.01430.x. [DOI] [PubMed] [Google Scholar]
- Damnjanovic T, Milicevic R, Novkovic T, Jovicic O, Bunjevacki V, Jekic B, Lukovic L, Novakovic I, Redzic D, Milasin J. Association between the methylenetetrahydrofolate reductase polymorphisms and risk of acute lymphoblastic leukemia in Serbian children. J Pediatr Hematol Oncol. 2010;32(4):e148–150. doi: 10.1097/MPH.0b013e3181cbd252. [DOI] [PubMed] [Google Scholar]
- Yeoh AE, Lu Y, Chan JY, Chan YH, Ariffin H, Kham SK, Quah TC. Genetic susceptibility to childhood acute lymphoblastic leukemia shows protection in Malay boys: results from the Malaysia-Singapore ALL Study Group. Leuk Res. 2010;34(3):276–283. doi: 10.1016/j.leukres.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Tong N, Fang Y, Li J, Wang M, Lu Q, Wang S, Tian Y, Rong L, Sun J, Xu J, Zhang Z. Methylenetetrahydrofolate reductase polymorphisms, serum methylenetetrahydrofolate reductase levels, and risk of childhood acute lymphoblastic leukemia in a Chinese population. Cancer Sci. 2010;101(3):782–786. doi: 10.1111/j.1349-7006.2009.01429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood S, Das R, Trehan A, Ahluwalia J, Sachdeva MU, Varma N, Bansal D, Marwaha RK. Methylenetetrahydrofolate reductase gene polymorphisms: association with risk for pediatric acute lymphoblastic leukemia in north Indians. Leuk Lymphoma. 2010;51(5):928–932. doi: 10.3109/10428191003719023. [DOI] [PubMed] [Google Scholar]
- Sadananda AMN, Chandy S, Ramachandra N, Appaji L, Aruna KBS, Ramaswamy G, Savithri HS, Krishnamoorthy L. Methylenetetrahydrofolate reductase gene polymorphisms and risk of acute lymphoblastic leukemia in children. Indian J Cancer. 2010;47(1):40–45. doi: 10.4103/0019-509X.58858. [DOI] [PubMed] [Google Scholar]
- Lightfoot TJ, Johnston WT, Painter D, Simpson J, Roman E, Skibola CF, Smith MT, Allan JM, Taylor GM. Genetic variation in the folate metabolic pathway and risk of childhood leukemia. Blood. 2010;115(19):3923–3929. doi: 10.1182/blood-2009-10-249722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Ugrasena DG, Lum DW, Lu Y, Yeoh AE. Xenobiotic and folate pathway gene polymorphisms and risk of childhood acute lymphoblastic leukaemia in Javanese children. Hematol Oncol. 2011;29(3):116–123. doi: 10.1002/hon.965. [DOI] [PubMed] [Google Scholar]
- Karathanasis NV, Stiakaki E, Goulielmos GN, Kalmanti M. The role of the methylenetetrahydrofolate reductase 677 and 1298 polymorphisms in Cretan children with acute lymphoblastic leukemia. Genet Test Mol Biomarkers. 2011;15(1–2):5–10. doi: 10.1089/gtmb.2010.0083. [DOI] [PubMed] [Google Scholar]
- Pereira TV, Rudnicki M, Pereira AC, Pombo-de-Oliveira MS, Franco RF. Do polymorphisms of 5,10-methylenetetrahydrofolate reductase (MTHFR) gene affect the risk of childhood acute lymphoblastic leukemia. Eur J Epidemiol. 2006;21(12):885–886. doi: 10.1007/s10654-006-9079-9. [DOI] [PubMed] [Google Scholar]
- Zhao P, Lin F, Li Z, Lin B, Lin J, Luo R. Folate intake, methylenetetrahydrofolate reductase polymorphisms, and risk of esophageal cancer. Asian Pac J Cancer Prev. 2011;12(8):2019–2023. [PubMed] [Google Scholar]
- Boccia S, Hung R, Ricciardi G, Gianfagna F, Ebert MP, Fang JY, Gao CM, Gotze T, Graziano F, Lacasana-Navarro M, Lin D, Lopez-Carrillo L, Qiao YL, Shen H, Stolzenberg-Solomon R, Takezaki T, Weng YR, Zhang FF, van Duijin CM, Boffetta P, Taioli E. Meta- and pooled analyses of the methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and gastric cancer risk: a huge-GSEC review. Am J Epidemiol. 2008;167(5):505–516. doi: 10.1093/aje/kwm344. [DOI] [PubMed] [Google Scholar]
- Taioli E, Garza MA, Ahn YO, Bishop DT, Bost J, Budai B, Chen K, Gemignani F, Keku T, Lima CS, Le ML, Matsuo K, Moreno V, Plaschke J, Pufulete M, Thomas SB, Toffoli G, Wolf CR, Moore CG, Little J. Meta- and pooled analyses of the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and colorectal cancer: a HuGE-GSEC review. Am J Epidemiol. 2009;170(10):1207–1221. doi: 10.1093/aje/kwp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolufer P, Barragan E, Collado M, Cervera J, Lopez JA, Sanz MA. Influence of genetic polymorphisms on the risk of developing leukemia and on disease progression. Leuk Res. 2006;30(12):1471–1491. doi: 10.1016/j.leukres.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Kim YI. Methylenetetrahydrofolate reductase polymorphisms, folate, and cancer risk: a paradigm of gene-nutrient interactions in carcinogenesis. Nutr Rev. 2000;58(7):205–209. doi: 10.1111/j.1753-4887.2000.tb01863.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hao L, Zhang L, Tian Y, Cao Y, Xia H, Deng Y, Wang T, Yu M, Li Z. Plasma folate status and dietary folate intake among Chinese women of childbearing age. Matern Child Nutr. 2009;5(2):104–116. doi: 10.1111/j.1740-8709.2008.00172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassan R, Gatta G, Tondini C, Willemze R. Adult acute lymphoblastic leukaemia. Crit Rev Oncol Hematol. 2004;50(3):223–261. doi: 10.1016/j.critrevonc.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Milne E, Royle JA, Miller M, Bower C, de Klerk NH, Bailey HD, van Bockxmeer F, Attia J, Scott RJ, Norris MD, Haber M, Thompson JR, Fritschi L, Marshall GM, Armstrong BK. Maternal folate and other vitamin supplementation during pregnancy and risk ofacute lymphoblastic leukemia in the offspring. Int J Cancer. 2010;126(11):2690–2699. doi: 10.1002/ijc.24969. [DOI] [PubMed] [Google Scholar]
- Thompson JR, Gerald PF, Willoughby ML, Armstrong BK. Maternal folate supplementation in pregnancy and protection against acute lymphoblastic leukaemia in childhood: a case–control study. Lancet. 2001;358(9297):1935–1940. doi: 10.1016/S0140-6736(01)06959-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.Folate metabolism and the role of MTHFR. Modified from Wiemels et al. (5).
Table S1.General characteristics of studies included in the meta-analysis on MTHFR polymorphisms and ALL risk.
Table S2.MTHFRC677T genotype distribution and allele frequency in ALL cases and controls.
Table S3.MTHFRA1298C genotype distribution and allele frequency in ALL cases and controls.
Figure S2.Sensitive analysis to assess the influence of a single study in the meta-analysis on MTHFR C677T polymorphism and the risk of ALL. Allele contrast was used in sensitive analysis. N, non-Caucasians, admixture of Amerindians, Europeans and Africans; W, mainly Brazilians of Caucasian descent.
Figure S3.Sensitive analysis to assess the influence of a single study in the meta-analysis on MTHFR A1298C polymorphism and the risk of ALL. Allele contrast was used in sensitive analysis. C, Chinese; M, Malays; N, non-Caucasians, admixture of Amerindians, Europeans and Africans; W, mainly Brazilians of Caucasian descent.
Table S4.Results of the pooled OR and the corresponding 95% CIs for each combined genotype for MTHFR C677T and A1298C polymorphisms.
Table S5.Characteristics of meta-analyses regarding MTHFR polymorphisms and ALL risk. (DOC 42 kb)