Abstract
Over the past decade Hippo kinase signalling has been established as an essential tumour suppressor pathway controlling tissue growth in flies and mammals. All members of the Hippo core signalling cassette are conserved from yeast to humans, whereby the yeast analogues of Hippo, Mats and Lats are central components of the mitotic exit network and septation initiation network in budding and fission yeast, respectively. Here, we discuss how far core Hippo signalling components in Drosophila melanogaster and mammals have reported similar mitotic functions as already established for their highly conserved yeast counterparts.
Keywords: Cell cycle; Kinase signalling; MST, NDR/STK38 and LATS kinases; MOB proteins; Phosphorylation
1. Introduction
One prominent feature of cancer cells is their abnormal composition of chromosomes, which frequently results in the amplification of proto-oncogenes and the loss of tumour suppressor genes. Combined with changes on the epigenetic level, these chromosome alterations play an important role in the long-term survival of cancerous material [1]. Basically, cancer cells need to acquire various changes on the genomic and epigenetic level to ensure resistance to cell death, sustained proliferative potential and evasion of growth suppression [1]. Therefore, Hanahan and Weinberg (2011) included genomic instability as an enabling characteristic in their updated Hallmarks of Cancer review [1]. Current evidence strongly suggests that chromosomal alterations are not only a feature of cancer cells but are also part of the driving force during malignant cellular transformation [2,3]. Significantly, errors in mitosis are prone to result in abnormal chromosome composition. Thus, for decades intensive research efforts have been ongoing to understand the signal transduction machineries that control mitosis and cytokinesis. Not surprisingly, these studies revealed that eukaryotic cells have developed several mechanisms to ensure faithful chromosome segregation [4,5].
Most likely the best studied mechanism is the spindle assembly checkpoint (SAC) [6], which ensures in normal mitotic cells that the meta-to-anaphase transition only occurs once all chromosomes have been attached correctly to the mitotic spindle apparatus. Given that the SAC can play a role in the progression of cancer, compounds targeting the mitotic spindle assembly are now established anti-cancer drugs that are used regularly in the clinic to treat a variety of human tumours [7,8]. However, the SAC is not the only safety mechanism that should be considered when designing anti-cancer drugs targeting mitosis and cytokinesis of cancer cells. Evasion of the SAC can even be a driving force for chromosomal abnormalities [2,3,9]. Therefore, researchers have been studying other mechanisms that play a role in mitosis. In particular, signalling cascades regulating the events after meta-to-anaphase transition were studied intensively one decade ago. Taking advantage of the genetic power of Saccharomyces cerevisiae and Schizosaccharomyces pombe, yeast geneticists found that exit of mitosis is regulated by a conserved signalling pathway, termed the mitotic exit network (MEN) in budding yeast and septation initiation network (SIN) in fission yeast [10,11]. The MEN/SIN achieve the equal distribution of genetic material between mother and daughter cells through a kinase cascade that triggers the dephosphorylation and consequent inactivation of mitotic CDK1 (cyclin dependent kinase 1) by the serine-threonine phosphatase Cdc14. Since mitotic CDK1 inhibits cytokinesis and abscission (cell cleavage) in fungi and metazoans, these studies also suggested that the MEN/SIN controls not only mitotic exit (progression from anaphase to late telophase), but can also play a role in the regulation of cytokinesis and abscission. Intriguingly, the core signalling module of the MEN/SIN is composed of highly conserved members of the Ste20-like kinase family [12-14], the MOB co-activator protein family [15], the LATS/NDR kinase family [16,17], and the Cdc14 protein phosphatase family [18]. The MEN/SIN pathway is very highly conserved from yeast to humans, since human Cdc14 can even compensate for the loss of Cdc14 in budding and fission yeast [19,20].
Over the past decade Ste20-like kinases, MOB proteins, and LATS/NDR kinases have gained even more attention, since Hippo (a GCK-II Ste20-like kinase), Mats/dMOB1, and Warts/Lats (one of two LATS/NDR kinases in flies) have been established as the central signalling module of the Hippo tumour suppressor signalling cascade in Drosophila melanogaster [21-23]. Loss of Hippo, Mats or Warts could be functionally compensated by their mammalian counterparts MST2, LATS1 and MOB1A, respectively [24-26], strongly suggesting that, similar to the MEN/SIN, Hippo signalling is very highly conserved from flies to humans. Over the past years a series of studies using human cells and transgenic mice established that the core components of mammalian Hippo signalling represent tumour suppressor proteins [27-31]. However, in spite of the strong conservation of MEN/SIN and Hippo components, the involvement of the Hippo core cassette in mitotic exit in higher eukaryotes is not yet fully understood. Nevertheless, some studies in flies and human cells suggest that Hippo signalling could play a role in the regulation of mitotic exit and possibly also cytokinesis.
In this review, we briefly give an updated version of the MEN and SIN pathways in budding and fission yeast (see Sections 2 and 3), before providing an overview of our current understanding of the roles of Hippo signalling components in the regulation of mitotic exit and cytokinesis. To make it easier for the reader to appreciate the findings in Drosophila and human cell systems, we have separated the fly and mammalian part into two independent sections (see Sections 4 and 5). We conclude with a discussion of important yet unanswered questions regarding Hippo signalling in the G2/M cell cycle phase. Throughout this review, we aim to emphasise selected key issues that should be addressed in more detail, so that the scientific community may fully appreciate which role(s) Hippo signalling can play in the coordination of two essential mitotic processes: (1) spatial and temporal timing of mitotic chromosome segregation and (2) cleavage furrow formation in cytokinesis.
2. Mitotic exit network (MEN) in S. cerevisiae
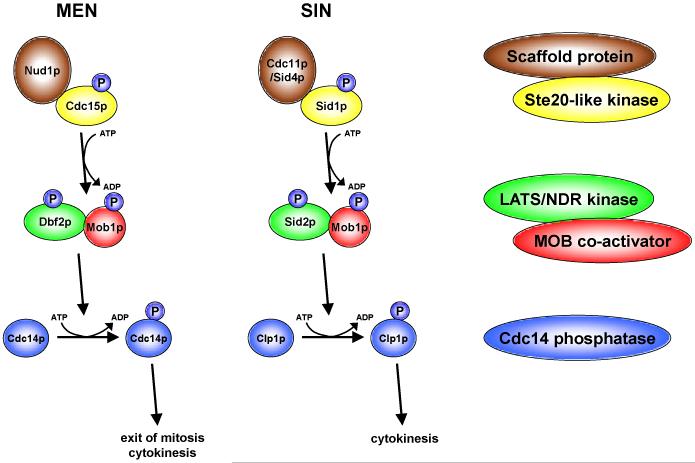
To understand how eukaryotic cells can co-ordinate mitotic chromosome segregation with cytokinesis, researchers turned to the power of yeast genetics. In budding yeast, these efforts revealed a so-called mitotic exit network (MEN) kinase cascade [10,11,32,33]. MEN signalling triggers the transition from the mitotic anaphase through to the G1-phase of the cell cycle by inactivating mitotic CDK1, one key hallmark of the end of mitosis [5]. More specifically, after the initial activation of the GTPase Tem1p at the spindle pole body (SPB; the equivalent of centrosomes in animals), the Cdc15p protein kinase is stimulated by GTP-bound Tem1p. Activation of Cdc15p is followed by increased kinase activity of the Mob1p/Dbf2p (Dbf20p) complex, which in turn results in the release of the serine-threonine phosphatase Cdc14p from the nucleolus into the nucleus and cytoplasm. Released Cdc14p then drives inactivation of mitotic CDK1 and dephosphorylates mitotic CDK1 and other kinase substrates (Fig. 1). This dephosphorylation wave allows spindle breakdown, nuclear division and cytokinesis in order for cells to exit from mitosis in a co-ordinated fashion. Initially, it was unclear how the Mob1p/Dbf2p complex transmitted the signal to Cdc14p. However, Mohl et al. (2009) recently showed that the Mob1p/Dbf2p complex can directly phosphorylate Cdc14p, an event that is sufficient to trigger the relocalisation of Cdc14p at the end of mitosis [34]. To sum up, activation of the Tem1p/Cdc15/Mob1p/Dbf2p/Cdc14p cascade results in CDK1 inactivation, which enables mitotic exit.
Figure 1. The MEN and SIN in yeast.
In budding and fission yeast, MEN and SIN core signalling encompasses members of five conserved protein groups. Scaffolding proteins (brown) assist Ste20-like kinases (yellow) in their activation of LATS/NDR kinases (green) in complex with a MOB protein (red). This kinase/co-activator complex then phosphorylates and thereby activates a Cdc14 phosphatase (blue), which finally results in CDK1 inhibition and subsequent exit from mitosis and cytokinesis. Please note that the indicated SIN-phosphorylation of Mob1p is predicted by the conservation between MEN and SIN signalling and not based on scientific evidence. Phosphorylations are indicated by “P” in blue.
However, recent evidence suggests that the role of MEN signalling goes beyond mitotic exit. By activating Cdc14p and subsequently inactivating mitotic CDK1, the MEN indirectly promotes cytokinesis. Nevertheless, recent evidence suggests that MEN signalling components have more direct roles in orchestrating the process of cytokinesis. On one hand, Dbf2p and Mob1p have been found at the actin-myosin ring (AMR) structure that is essential for abscission [35-38], and Cdc15p, Dbf2p and Mob1p are required for AMR contraction [39,40]. Moreover, current evidence suggests that the MEN components Dbf2p and Mob1p can regulate the cytokinetic components Chs2p, Hof1p, and Inn1p (summarised in [33,41]). However, Hof1p (one of at least three F-BAR domain proteins in S. cerevisiae) is so far the only established target of Mob1p/Dbf2p in cytokinesis [41].
On the other hand, by dephosphorylating specific Cdk1 targets in telophase, the Cdc14p phosphatase can promote cytokinesis. By changing the phosphorylation status of the formins Bnr1 and Bni1, Cdc14p can affect the repolarisation of the cytoskeleton at the end of mitosis [42]. Cdc14p may also directly regulate Boi1, Boi2, Chs1, Shs1 and other mitotic factors to facilitate cytokinesis [33], however, the direct regulation of these proteins by Cdc14p is yet to be established. Intriguingly, Cdc14p is further required for the functionality of the LATS/NDR kinase Cbk1p (the second LATS/NDR kinase in budding yeast) [43]. Weiss and colleagues showed recently that Cdc14 is essential for Cbk1 function in vivo, even though Cdc14p does not influence Cbk1p in vitro [43]. Since Cbk1 mainly functions in the G1-phase of the cell cycle [44-46], these findings suggest that the MEN can even play a role beyond mitotic exit and cytokinesis.
Taken together, the mitotic exit of budding yeast is controlled by the MEN signalling machinery (Fig. 1). At the core of this network is a finely tuned cascade of phosphorylations and dephosphorylations events, which co-ordinate the completion of mitosis with cytokinesis and abscission. This spatial and temporal co-ordination is essential for the survival of healthy budding yeast in order to maintain genome integrity over multiple generations. As briefly outlined here, several aspects of the MEN have already been studied in detail with respect to exit of mitosis, however, the importance of the MEN in the regulation of cytokinesis has only been appreciated rather recently. Therefore, we believe the budding yeast system will still be a major contributor to our understanding of MEN in the future.
3. Septation initiation network (SIN) in Schizosaccharomyces pombe
The machinery regulating the controlled exit of mitosis is highly conserved in other yeast species [33]. In particular in Schizosaccharomyces pombe, mitotic exit signalling has been extensively studied in addition to budding yeast [10,11]. Given the differences between budding and fission yeast in mitosis, the budding yeast MEN was termed septation initiation network (SIN) in S. pombe. Although the SIN is analogous to the MEN in budding yeast, the SIN mainly co-ordinates cytokinesis by playing a role in AMR contraction and septum formation rather than controlling mitotic exit in general [10,11,47,48]. Nevertheless, the SIN signalling cascade is very similar to the MEN pathway (Fig. 1). After activation of the GTPase Spg1p, GTP-bound Spg1p activates the Sid1p kinase (the Ste20-like protein kinase orthologue of Cdc15p). Active Sid1p then phosphorylates the Mob1p/Sid2p kinase complex (the fission yeast counterpart of the Mob1p/Dbf2p complex), which in turn activates the Clp1p/Flp1 phosphatase (the counterpart of the budding yeast Cdc14p phosphatase). Using a similar mechanism as reported for the MEN, the Mob1p/Sid2p complex directly phosphorylates Clp1p, thereby affecting the subcellular localisation of Clp1p [49]. Significantly, Clp1p might also regulate similar cytokinetic factors as reported for budding yeast Cdc14p [33,50]. In particular, the regulation of budding yeast Hof1p appears to be conserved between SIN and MEN. Fission yeast Cdc15p is one of two functional homologues of Hof1p [51-53], which is the only established target of budding yeast Mob1p/Dbf2p in cytokinesis [37]. Furthermore, Cdc15p is phosphorylated on a conserved RXXS motif that can be targeted by Mob1p/Sid2p [53]. Therefore it is tempting to speculate that the SIN components Mob1p/Sid2p and Clp1p regulate cytokinesis in a similar fashion as reported for the MEN components Mob1p/Dbf2p and Cdc14p [33]. Last but not least, one should also note that Clp1p has been reported to function together with Aurora kinase at kinetochores in early mitotic stages [54], suggesting that Cdc14 phosphatase family members can have MEN/SIN independent functions.
In summary, budding and fission yeast orchestrate mitotic exit and cytokinesis through a conserved kinase signalling cascade (Fig. 1), termed MEN and SIN, respectively. The conserved signalling core cassettes function as follows: Cdc15p and Sid1p (the budding and fission yeast Ste20-like kinase counterparts of Drosophila Hippo) activate Mob1p (the yeast counterpart of Mats/dMOB1) in complex with Dbf2p or Sid2p (the LATS/NDR kinase counterparts of Warts/Lats and Trc), which in turn leads to the activation of protein phosphatase Cdc14p and Clp1p, respectively. Although in yeast additional Ste20-like kinases, LATS/NDR kinases, and MOB proteins are expressed [15,16], the functionality of MEN and SIN is fully dependent on one single signalling core module composed of specific members of the Ste20-like kinase, LATS/NDR, and MOB protein families, namely Cdc15p/Mob1p/Dbf2p and Sid1p/Mob1p/Sid2p, respectively.
4. Hippo signalling in G2/M in Drosophila melanogaster
In flies, members of the Ste20-like kinase, LATS/NDR, and MOB protein families have essential tumour suppressive roles by co-ordination cell growth, proliferation and death [16,21-23]. Considering the impact of initial studies on the Ste20-like Hippo kinase [26,55-58], Hippo/Mats/Lats signalling has been referred to as Hippo signalling ever since. Intriguingly, the Hippo pathway is very similar to the MEN/SIN cascades in budding and fission yeast [15,16,33]. As reported for the MEN/SIN, a GCK-II Ste20-like kinase activates a LATS/NDR kinase in complex with a MOB1 protein. Based on these striking similarities one is very tempted to draw the conclusion that Hippo signalling in flies is controlling mitotic exit in a similar fashion as reported for the MEN/SIN in yeast (Fig. 2). However, in spite of these similarities, scientific reports supporting a role for Hippo signalling in mitotic exit and cytokinesis are limited in number. Nevertheless, there is some evidence suggesting that Hippo signalling (or at least some key components of Hippo signalling) can play a role in mitosis.
Figure 2. Comparison of MEN and Hippo signalling components in mitotic exit and cytokinesis.
S. cerevisiae, D. melanogaster and mammals utilise a highly conserved signalling module composed of scaffolding proteins (brown), Ste20-like kinases (yellow), LATS/NDR kinases (green), MOB proteins (red), and possibly Cdc14 phosphatases (blue). While this signalling cassette is essential for mitotic exit in yeast, the mitotic role of Hippo signalling is debateable. Currently, we do not know whether Hippo signalling as illustrated here is playing any role in mitotic exit and cytokinesis. Nevertheless, some factors (RASSF1A, LATS1/2 and Cdc14) have been reported to play a role in mitotic exit and cytokinesis. Otherwise, question marks highlight proteins that so far have no reported role in mitotic exit and/or cytokinesis. Phosphorylations are indicated by “P” in blue.
In 2004, Bettencourt-Dias and colleagues screened the Drosophila kinome for cell cycle functions by RNA-mediated interference (RNAi), thereby revealing that Hippo and Lats are required for normal cell cycle progression [59]. Depletion of Hippo caused mitotic spindle abnormalities, suggesting that Hippo plays a role in mitotic progression. Knock-down of Lats, on the other hand, resulted in an increase of cells in the G1 phase of the cell cycle. The authors speculated that Lats might accelerate progression through G2/M, thereby shifting more cells into G1 [59]. In light of the finding that Lats can inhibit CDK1 during mitosis in human cells and flies [25], this interpretation might be correct. However, we currently cannot exclude the other possible interpretation of this finding, namely that loss of Lats function results in a G1 cell cycle arrest. Whatever the case, this report [59] suggests that Hippo and Lats do not operate together in cell cycle functions, since their depletion results in very different cell cycle phenotypes. This publication [59] further suggests that Hippo kinase might function in a similar fashion as reported for the Hippo counterparts Cdc15p and Sid1p in MEN/SIN (Fig. 2), but how Hippo could control mitotic exit in Drosophila cells has yet to be determined. Possibly Hippo signals through the Tricornered kinase (Trc; the second LATS/NDR kinase in flies) as already reported for Hippo signalling in sensory neurons [60]. Another possibility is that Hippo can function independent of LATS/NDR kinases during the cell cycle. In this context, one should also note that Hippo mutant cells from dissected developing wings or eyes do not display obvious cell cycle phasing defects [26,55], suggesting that Hippo might not have a role in mitosis in every cell type. However, RasV12 immortalised cells derived from warts null fly embryos display polyploidy, which suggests a defective tetraploidy checkpoint [61]. Given all these indications, future research is warranted to address how Hippo/Lats (and possibly also Trc) signalling plays a role in mitosis.
Given that Hippo can also target Mats [62], the question arises whether Mats could play a role in mitotic Hippo signalling. Intriguingly, Mats (also termed dMOB1) is the counterpart of yeast Mob1p [15], which is an essential component of the MEN/SIN in yeast [10,11]. Considering this high similarity and that Mats is an established core component of Hippo signalling in the control of cell proliferation and death [24,62], it is very tempting to assume that Mats must play a role in mitotic exit. Indeed, one report by the Lai laboratory shows that Mats is required for proper chromosome segregation in developing fly embryos [63]. This suggests that Mats has a role in mitosis, but we do not know which specific function(s) Mats might have in this important cell cycle phase. One possibility is that Mats functions in complex with Warts in mitosis, as already shown for the control of cell proliferation and death [24,62]. Potentially, Mats could function in mitosis independent of Warts, since Mats can also interact with Trc, the second LATS/NDR kinase in flies [64]. A third possibility is that Mats signals downstream of Hippo independent of LATS/NDR kinases. Whatever the case, future research is needed to clarify whether Mats plays a role in mitotic exit and whether this function requires other Hippo signalling components.
A strong argument for pursuing this line of research is the recent report from Gregory et al. (2007) showing that Mats is potentially a direct regulator of cytokinesis [65]. By screening for modulators of Rho signalling during cytokinesis they identified Mats among several other cell cycle regulators [65]. Significantly, this screen [65] also showed that overexpression of the Cdc14 phosphatase modulates the G2/M transition of Drosophila cells. This report [65] proposes that Mats might regulate Cdc14 as already reported for the yeast counterparts Mob1p and Cdc14p/Clp1p (Fig. 1). Similar to budding yeast Cdc14p [18], the fly Cdc14 appears also to be essential, since depletion of Cdc14 (CG7134) by RNAi results in pupal lethality [66]. However, we currently do not understand how Cdc14 functions in Drosophila. Perhaps Cdc14 can counteract CDK1 activity in mitosis, thereby regulating mitotic exit and cytokinesis. On the basis of the functional conservation between yeast and human Cdc14 [19,20] it seems obvious to assume that fly Cdc14 plays a key role in mitosis and cytokinesis, but as already mentioned there is currently no scientific evidence available that strongly supports this assumption. Hopefully future research will unveil whether and how the fly Cdc14 phosphatase is regulating exit of mitosis.
Of note, in addition to the Cdc14 phosphatase, other phosphatases have roles in mitotic exit [67]. In particular, members of the conserved PP2A family of phosphatases are of interest when comparing Hippo signalling with the MEN/SIN. Ribeiro et al. (2010) reported that Hippo signalling is negatively regulated by the STRIPAK complex, which contains a functional PP2A unit [68]. Intriguingly, in fission yeast the SIN is regulated by an SIN-inhibitory PP2A phosphatase (SIP) complex that is very similar to the STRIPAK complex [69]. Five of six SIP components are even conserved from yeast to man [69]. On one hand, this suggests that the STRIPAK complex perhaps regulates Hippo in mitosis as observed for the regulation of the Hippo analogue Sid1p by SIP [69]. On the other hand, these reports emphasise that in animal cells it will not be sufficient to focus on Cdc14 when addressing the regulation of mitotic exit by phosphatases. Most likely a network of phosphatases is to be studied in order to understand how phosphatases regulate the passage through mitosis [67].
When comparing Hippo signalling with the MEN/SIN, one should also mention the conservation of scaffolding proteins between these pathways [16]. In budding and fission yeast, the scaffolding proteins Nud1p and Cdc11p/Sid4p support MEN and SIN signalling, respectively [10,11,16]. In flies, the scaffolding protein Salvador (Sav; the fly counterpart of Nud1p and Cdc11p/Sid4p) functions as a central tumour suppressor in Hippo signalling [26,55-58,70]. However, the involvement of Sav in mitotic processes is yet to be tested. Perhaps the SARAH protein-protein interaction domain shared between Sav, Hippo and Rassf proteins [71] will help with this analysis. The Tapon laboratory reported that dRASSF competes with Sav, and thereby antagonises pro-apoptotic Hippo signalling [72]. Given that RASSF1A (the human counterpart of fly dRASSF) is an established regulator of mitotic progression (see Section 5), one is tempted to speculate that dRASSF might also play a role in mitosis together with Hippo. Nevertheless, as it is the case for most Hippo signalling components, it is yet to be determined whether Sav and dRASSF play any role in mitosis and/or cytokinesis. In this context, one should further note that Sav mutant cells derived from imaginal discs did not display obvious cell cycle defects [70], indicating that Sav might be dispensable for mitosis of certain cell types.
In summary, in D. melanogaster all core components of the yeast MEN/SIN are conserved in the Hippo/Mats/Lats cascade (better known as Hippo tumour suppressor signalling) (Fig. 2). Current reports suggest that Hippo and Mats can function in mitosis, but it is so far unclear whether they play a role in mitotic exit and cytokinesis as reported for their yeast counterparts. Possibly Cdc14 functions downstream of Hippo/Mats. However, the involvement of Hippo signalling in the regulation of mitotic exit is currently not established. Very likely, additional factors such as Trc, PP2A, Sav and dRASSF need to be considered when addressing this research aspect. Taken together, the MEN/SIN core components are conserved from yeast to flies, but animal cells seem to co-ordinate mitotic exit by additional networks, which so far has slowed down research progress with respect to the role of Hippo signalling in mitosis.
5. Mammalian Hippo signalling in the G2/M phase of the cell cycle
Considering that all core components of MEN/SIN and Hippo signalling are conserved from yeast to humans [15,16,73], one might expect that research has already established that a signalling cascade composed of MST1/2 kinases (the human counterpart of Hippo), LATS/NDR kinases (the human analogues of Lats/Trc), and hMOB1 (the human version of Mats) is essential for mitotic exit and cytokinesis. However, somewhat similar to our current picture of mitotic Hippo signalling in Drosophila (see Section 4), our understanding of mammalian Hippo signalling in mitosis is limited (Fig. 2). A large effort has been invested in elucidating the roles of Hippo signalling in human disease, establishing mammalian Hippo signalling as a novel tumour suppressor pathway that is essential for the regulation of tissue growth [27-31]. Unfortunately, these focused approaches also meant that possible mitotic functions were neglected to a certain extent. Nevertheless, there is scientific evidence suggesting that mammalian Hippo signalling (or at least components of this pathway) can play a role in mitosis.
For more than a decade MST1/2 kinases have been known as pro-apoptotic Ste20-like kinases [13,14], and since 2003, their role as tumour suppressor proteins has been established by several studies [27-31]. However, a mitotic role for MST1/2 was described only recently [74,75]. Oh et al. (2010) provided evidence suggesting that Aurora B kinase activity is regulated by MST1 signalling, thereby linking MST1 to the molecular processes of chromosome segregation in mitosis [75]. In line with this finding, a second publication reported that MST2 signalling plays a role in chromosome alignment in metaphase [74]. Whether MST2 can also influence Aurora B activity is currently not known, but MST1 and MST2 both appear to signal through NDR1 kinase (the human counterpart of Trc in flies) [74,75]. Furthermore, they reported that NDR1 kinase activity is elevated in early mitosis [74]. However, the Hemmings laboratory did not detect this increased activity in mitosis [76]. By analysing the cell cycle activation of NDR1/2 via kinase assays and phosphorylation specific antibodies, Cornils et al. (2011) defined human NDR1/2 as cell cycle regulated kinases that are mainly activated upon the G1/S cell cycle transition [76]. They further showed that NDR1/2 are essential for the G1/S cell cycle transition by regulating p21 and c-myc levels [76-78]. Significantly, NDR1/2 kinases require only MST3 kinase activity for this cell cycle role, but not MST1/2 [76,79]. In contrast, MST1 signalling is indispensable for NDR’s regulation of centrosome duplication in S-phase [80,81]. Taken together, these findings indicate that human MST1/2 kinases have diverse roles throughout the cell cycle. In the context of this review, the role of MST1/2 in mitosis appears to be the most interesting [74,75]. Nevertheless, in spite of these studies [74,75] and the described spindle midzone localisation of MST2 in late mitosis [82], MST1/2 signalling in mitotic exit and cytokinesis remains to be studied.
As mentioned above, NDR1/2 kinases can function downstream of MST1/2 signalling although NDR1/2 have not been implicated in the control of mitotic exit. In contrast, LATS1/2 kinases, which can also signal downstream of MST1/2 [13,14,83], have been shown to play a role in later stages of mitosis. A decade ago, it was reported that LATS1 can inhibit CDK1 activity in mitosis, thereby influencing G2/M cell cycle progression [25,84]. Analysis of cells overexpressing LATS1 supported a role for LATS1 in mitosis, since LATS1 overexpression resulted in a G2/M cell cycle arrest [84-86]. Subsequent studies further supported the view that LATS1 is a regulator of mitosis by showing that LATS1 associates with the AMR and midbody in cytokinesis [87], that LATS1 co-localises with Zyxin at the mitotic spindle apparatus [88], that LATS1 can interact with LIMK1 in mitosis, a key regulator of the actin polymerisation machinery [87], and that endogenous human LATS1 plays a role in mitosis [89]. These findings suggest that LATS1 has one major role in the mammalian cell cycle, namely the regulation of mitosis. In contrast, LATS2 (the second human counterpart of Drosophila Lats) seems to function in more than one cell cycle stage. On one hand, it was reported that LATS2 overexpression inhibits G1/S transition [90]. On the other hand, LATS2 overexpression can also lead to a G2/M cell cycle arrest [91]. Most importantly, LATS2 −/− cells display abnormal mitosis and cytokinesis defects [92,93], supporting the view that LATS2 is mainly required for mitotic progression. Taken together, these reports indicate that mammalian LATS1/2 kinases have important roles in mitosis. However, these publications also suggest LATS1/2 are likely to function on more than one level in mitosis. In contrast to LATS2, LATS1 appears to function solely in mitosis, but also this view has been challenged recently by the observation that LATS1 can play a role in the intra-S-phase and G2/M checkpoints upon DNA damage [94]. Therefore, more detailed studies of LATS1/2 functions in the cell cycle are warranted. The mitotic regulation of LATS1/2 by upstream regulators, such as the MST1/2 kinases, also remains to be studied.
Recent research progress on hMOB1, the human counterpart of fly Mats and yeast Mob1p [15], should also be mentioned when discussing Hippo signalling in mitosis. hMOB1, a known co-activator of LATS/NDR kinases [15], has been detected in the spindle midzone in late mitotic stages [89,95]. Moreover, Bothos et al. (2005) found that endogenous hMOB1 can play a role in mitosis [89]. This suggests that also the third core component of mammalian Hippo signalling might have a role in mitotic exit and cytokinesis. However, it is currently not known whether hMOB1 functions in mitosis together with LATS1/2 and MST1/2 kinases. Since hMOB1 further plays a role in S-phase controlled centrosome duplication by regulating NDR signalling [80,81], the mitotic analysis of hMOB1 is likely to be more complicated. This further suggests that hMOB1 can function in different cell cycle phases, which is very similar to observations made for budding yeast Mob1p. In yeast, Mob1p plays a role in spindle pole body duplication (the yeast equivalent of centrosome duplication in multicellular organisms) [96] in addition to being essential for mitotic exit [10,11,32,33]. Nevertheless, despite this recent research progress, a role for hMOB1 in mitotic exit and cytokinesis has as yet to be established.
In yeast, activation of the Cdc14 phosphatase by the MEN/SIN is a key event to trigger mitotic exit (see Sections 2 and 3). Is this also the case in mammalian cells? Considering that human Cdc14 can compensate for the loss of Cdc14 in budding and fission yeast [19,20], the initial answer is a clear yes. However, as is the case for most cellular processes in mammals, the picture is more complicated. Several studies showed that Cdc14 is dispensable for exiting mitosis in different model systems other than yeast [18]. However, this does not mean that Cdc14 has no role to play in mitosis, since overexpression of Cdc14 leads to cytokinesis defects in mammalian cells [18]. In addition, Cdc14 can be detected at the spindle midzone and midbody in animal cells [97-101]. These reports suggest that mammalian Cdc14 phosphatases are not essential for mitotic progression, although they can have negative effects on mitotic processes upon overexpression. Of note, Cdc14A (one of two Cdc14 phosphatases expressed in human cells) can further play a role in the G2/M transition by preventing premature activation of CDK1 through regulating the phosphatases Cdc25A/B [102], suggesting that mammalian Cdc14 has function beyond mitotic exit and cytokinesis. This recent report [102] is interesting, since Clp1p, the fission yeast Cdc14, has already been connected with the yeast Cdc25p phosphatase [103]. Furthermore, Clp1p has been reported to function in meta-/anaphase on the yeast kinetochore in addition to its role in the SIN [54]. Therefore, given that Cdc14 has more than one function in yeast [54,103], it should not be surprising that mammalian Cdc14 phosphatases have different roles in mitosis [18]. Furthermore, one needs to keep in mind that other phosphatases, in particular members of the PP1 and PP2A families, have been reported to play a role in mitotic exit of mammalian cells [67]. Therefore, we believe that it will not be sufficient to focus on Cdc14 when addressing the regulation of mitotic exit by mammalian Hippo signalling.
In spite of all these reports linking MST1/2, LATS1/2, hMOB1, and Cdc14 to mitotic functions, we have to date not understood the real nature of the mitotic signalling cascades involving core Hippo components (Fig. 2). Possibly by understanding the trigger for mitotic activation of MST1/2 we can unveil a more detailed picture of mammalian Hippo signalling in mitosis. Interestingly, the MST1/2 binding partners RASSF1A and mammalian Salvador/WW45 (the analogues of Drosophila dRASSF and Sav, respectively) are potentially part of this trigger. MST1/2 kinases can be activated by the tumour suppressor proteins WW45 and RASSF1A [13,14]. The interaction of WW45 with MST2 can regulate mitotic centrosome disjunction [104], but any further roles for WW45 in mitosis have yet to be reported. In contrast, the regulation of MST1/2 by RASSF1A has been studied more extensively. In human cells, RASSF1A signals through MST/LATS1 to induce apoptosis [105-107], and can also trigger apoptosis by MST1/NDR signalling [108]. Part of the mode of action appears to be that RASSF1A protects MST1/2 from dephosphorylation by PP2A to induce apoptosis [109]. Whatever the case, RASSF1A has been repeatedly reported as an activator of MST1/2 signalling [105-108,110]. RASSF1A can form complexes with MST1/2 and WW45 through conserved SARAH domains [71,82,83,111], which seems to play a role in Hippo dependent and independent apoptotic signalling [112]. Significantly, RASSF1A null cells display cytokinesis failure [82], which is a similar phenotype as observed in LATS2 null cells [92,93]. Furthermore, it has been reported that RASSF1A overexpression induces a G2/M cell cycle arrest [113], most likely by RASSF1A regulating APC-Cdc20 E3 ligase activity which affects mitotic progression [114]. In addition, it was found that Aurora A regulates RASSF1A during prometaphase progression [115,116], while Aurora B phosphorylation of RASSF1A plays a role in the regulation of cytokinesis [117]. These reports strongly suggest that RASSF1A has diverse roles in mitosis. However, according to our knowledge, any direct link between RASSF1A and mitotic Hippo signalling is yet to be established. Furthermore, the contradiction between Drosophila and human data regarding RASSF signalling should be clarified. In flies, the Tapon laboratory reported that dRASSF antagonises Hippo signalling [72], while in human cells RASSF1A activates MST/LATS/NDR signalling [105-108,110]. Possibly these differences can be explained by considering other RASSF family members [118,119]. In this context it is noteworthy that Ikeda et al. (2009) published that RASSF6 can regulate MST2 dependent and independent apoptosis pathways [120]. In other words, at least two RASSF family members play a role in mammalian Hippo signalling.
In summary, mammalian Hippo signalling components can play a role in the regulation of mitosis (Fig. 2). In how far the MST/hMOB1/LATS core signalling cassette is functioning together in mitosis is still an open question. Perhaps by taking into account that MST1/2 and hMOB1 signalling are also important for the regulation of NDR1/2 kinases (the second group of LATS/NDR kinases in human cells), the complexity of mammalian Hippo signalling in mitosis could be better understood. Moreover, the downstream targets of mammalian Hippo signalling are yet to be determined in mitosis. Any involvement of YAP and TAZ, the two established Hippo targets [27-31], in mitotic exit and cytokinesis has not been reported so far. Possibly Cdc14 phosphatases can be activated by Hippo signalling, which could be part of the regulation of PSTPIP1, the mammalian homolog of yeast Hof1p [121]. Taken together, by considering all these open questions, we look forward to exciting new discoveries in mammalian Hippo signalling and its role in mitotic progression.
6. Conclusions
Eukaryotic cells have developed a range of mechanisms to ensure the equal distribution of genetic material between daughter cells at the end of the cell cycle. Here, we have summarised our current understanding of a signalling cascade that regulates mitotic exit and cytokinesis after the meta-to-anaphase transition. Intriguingly, the core components of this pathway are highly conserved from yeast to humans. In budding/fission yeast, the Ste20-like kinases Cdc15p/Sid1p function together with the LATS/NDR kinases Dbf2p/Sid2p and the Mob1p co-activator as a central hub in the regulation of mitotic exit and cytokinesis. Therefore, the Cdc15p/Dbf2p/Mob1p and Sid1p/Sid2p/Mob1p cascades were termed the MEN and SIN, respectively. In Drosophila melanogaster, the Ste20-like kinase Hippo functions together with the LATS/NDR kinase Lats and the Mats co-activator as a central module in Hippo tumour suppressor signalling, which is essential for the co-ordination of cell growth, proliferation, and death in tissue growth. Current reports suggest that Hippo and Mats can function in mitosis, but it is so far unclear whether they play any role in mitotic exit and cytokinesis, as reported for their yeast counterparts. In mammalian cells, the Ste20-like kinases MST1/2 function together with the LATS/NDR kinases LATS1/2 and the hMOB1 co-activator as central players in mammalian Hippo tumour suppressor signalling. Nevertheless, in spite of the high conservation between the MEN/SIN factors and MST/hMOB1/LATS it is yet to be established whether the Hippo core signalling cassette is functioning together in mitosis. This lack of knowledge in metazoan model systems is somewhat surprising. Very likely, the complexity of mammalian cell systems (human cells express five Ste20-like kinases that are similar to MST1/2; four different LATS/NDR kinases; and six MOB proteins) will need to be considered when tackling mitotic mammalian Hippo signalling. Currently, we do not know whether the Hippo growth repressor pathway and Hippo signalling in mitosis represent one pathway that has different roles dependent on developmental and cell cycle stages. Possibly, not all components are shared between these two signalling cascades, which would mean that at least two distinct Hippo pathways can operate in multicellular organisms. In this respect we are very confident that exciting times lay ahead of us in translating lessons learned from mitosis in yeast into more complex mitotic events in metazoans.
Acknowledgements
We thank J. Lisztwan and our lab members for their critical input on this manuscript. This work was supported by the Wellcome Trust grant 090090/Z/09/Z. A.H. is a Wellcome Trust Research Career Development fellow at the UCL Cancer Institute. The Friedrich Miescher Institute is part of the Novartis Research Foundation.
References
- [1].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [2].Fang X, Zhang P. Aneuploidy and tumorigenesis. Semin Cell Dev Biol. 2011;22:595–601. doi: 10.1016/j.semcdb.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nat Rev Mol Cell Biol. 2010;11:91–102. doi: 10.1038/nrm2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- [6].Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- [7].Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- [8].Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- [9].Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- [10].Bardin AJ, Amon A. Men and sin: what’s the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- [11].McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- [12].Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- [13].Ling P, Lu TJ, Yuan CJ, Lai MD. Biosignaling of mammalian Ste20-related kinases. Cell Signal. 2008;20:1237–1247. doi: 10.1016/j.cellsig.2007.12.019. [DOI] [PubMed] [Google Scholar]
- [14].Matallanas D, Romano D, Hamilton G, Kolch W, O’Neill E. A Hippo in the ointment: MST signalling beyond the fly. Cell Cycle. 2008;7:879–884. doi: 10.4161/cc.7.7.5630. [DOI] [PubMed] [Google Scholar]
- [15].Hergovich A. MOB control: reviewing a conserved family of kinase regulators. Cell Signal. 2011;23:1433–1440. doi: 10.1016/j.cellsig.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- [17].Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- [18].Mocciaro A, Schiebel E. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci. 2010;123:2867–2876. doi: 10.1242/jcs.074815. [DOI] [PubMed] [Google Scholar]
- [19].Li L, Ernsting BR, Wishart MJ, Lohse DL, Dixon JE. A family of putative tumor suppressors is structurally and functionally conserved in humans and yeast. J Biol Chem. 1997;272:29403–29406. doi: 10.1074/jbc.272.47.29403. [DOI] [PubMed] [Google Scholar]
- [20].Vazquez-Novelle MD, Esteban V, Bueno A, Sacristan MP. Functional homology among human and fission yeast Cdc14 phosphatases. J Biol Chem. 2005;280:29144–29150. doi: 10.1074/jbc.M413328200. [DOI] [PubMed] [Google Scholar]
- [21].Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- [22].Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- [23].Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- [24].Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- [25].Tao W, Zhang S, Turenchalk GS, Stewart RA, St John MA, Chen W, Xu T. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet. 1999;21:177–181. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- [26].Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- [27].Badouel C, McNeill H. SnapShot: The hippo signaling pathway. Cell. 2011;145:484–484. doi: 10.1016/j.cell.2011.04.009. [DOI] [PubMed] [Google Scholar]
- [28].Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bosl WJ, Li R. Mitotic-exit control as an evolved complex system. Cell. 2005;121:325–333. doi: 10.1016/j.cell.2005.04.006. [DOI] [PubMed] [Google Scholar]
- [33].Meitinger F, Palani S, Pereira G. The power of MEN in cytokinesis. Cell Cycle. 2012:11. doi: 10.4161/cc.11.2.18857. [DOI] [PubMed] [Google Scholar]
- [34].Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, Deshaies RJ. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J Cell Biol. 2009;184:527–539. doi: 10.1083/jcb.200812022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Frenz LM, Lee SE, Fesquet D, Johnston LH. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J Cell Sci. 2000;113(Pt 19):3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- [36].Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25:875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yoshida S, Toh-e A. Regulation of the localization of Dbf2 and mob1 during cell division of saccharomyces cerevisiae. Genes Genet Syst. 2001;76:141–147. doi: 10.1266/ggs.76.141. [DOI] [PubMed] [Google Scholar]
- [39].Meitinger F, Petrova B, Lombardi IM, Bertazzi DT, Hub B, Zentgraf H, Pereira G. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J Cell Sci. 2010;123:1851–1861. doi: 10.1242/jcs.063891. [DOI] [PubMed] [Google Scholar]
- [40].Menssen R, Neutzner A, Seufert W. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr Biol. 2001;11:345–350. doi: 10.1016/s0960-9822(01)00095-1. [DOI] [PubMed] [Google Scholar]
- [41].Roberts-Galbraith RH, Gould KL. Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle. 2010;9:4091–4097. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- [42].Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR. Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem. 2011;286:5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Brace J, Hsu J, Weiss EL. Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis. Mol Cell Biol. 2011;31:721–735. doi: 10.1128/MCB.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jansen JM, Barry MF, Yoo CK, Weiss EL. Phosphoregulation of Cbk1 is critical for RAM network control of transcription and morphogenesis. J Cell Biol. 2006;175:755–766. doi: 10.1083/jcb.200604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mazanka E, Weiss EL. Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol Biol Cell. 2010;21:2809–2820. doi: 10.1091/mbc.E10-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nelson B, Kurischko C, Horecka J, Mody M, Nair P, Pratt L, Zougman A, McBroom LD, Hughes TR, Boone C, Luca FC. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Krapp A, Gulli MP, Simanis V. SIN and the art of splitting the fission yeast cell. Curr Biol. 2004;14:R722–730. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- [48].Krapp A, Simanis V. An overview of the fission yeast septation initiation network (SIN) Biochem Soc Trans. 2008;36:411–415. doi: 10.1042/BST0360411. [DOI] [PubMed] [Google Scholar]
- [49].Chen CT, Feoktistova A, Chen JS, Shim YS, Clifford DM, Gould KL, McCollum D. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol. 2008;18:1594–1599. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Roberts-Galbraith RH, Gould KL. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–3088. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- [51].Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1995;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- [52].Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol. 2009;184:113–127. doi: 10.1083/jcb.200806044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, 3rd, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Trautmann S, Rajagopalan S, McCollum D. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev Cell. 2004;7:755–762. doi: 10.1016/j.devcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- [55].Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- [56].Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- [58].Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- [59].Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, Cooper M, Jones D, Frenz L, Glover DM. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- [60].Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- [61].Simcox A, Mitra S, Truesdell S, Paul L, Chen T, Butchar JP, Justiniano S. Efficient genetic method for establishing Drosophila cell lines unlocks the potential to create lines of specific genotypes. PLoS Genet. 2008;4:e1000142. doi: 10.1371/journal.pgen.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. Embo J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shimizu T, Ho LL, Lai ZC. The mob as tumor suppressor gene is essential for early development and regulates tissue growth in Drosophila. Genetics. 2008;178:957–965. doi: 10.1534/genetics.107.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].He Y, Emoto K, Fang X, Ren N, Tian X, Jan YN, Adler PN. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol Biol Cell. 2005;16:4139–4152. doi: 10.1091/mbc.E05-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gregory SL, Shandala T, O’Keefe L, Jones L, Murray MJ, Saint R. A Drosophila overexpression screen for modifiers of Rho signalling in cytokinesis. Fly (Austin) 2007;1:13–22. doi: 10.4161/fly.3806. [DOI] [PubMed] [Google Scholar]
- [66].Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol. 2011;12:469–482. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- [68].Ribeiro PS, Josue F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- [69].Singh NS, Shao N, McLean JR, Sevugan M, Ren L, Chew TG, Bimbo A, Sharma R, Tang X, Gould KL, Balasubramanian MK. SIN-Inhibitory Phosphatase Complex Promotes Cdc11p Dephosphorylation and Propagates SIN Asymmetry in Fission Yeast. Curr Biol. 2011;21:1968–1978. doi: 10.1016/j.cub.2011.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- [71].Scheel H, Hofmann K. A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr Biol. 2003;13:R899–900. doi: 10.1016/j.cub.2003.11.007. [DOI] [PubMed] [Google Scholar]
- [72].Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Curr Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hergovich A, Hemmings BA. Mammalian NDR/LATS protein kinases in hippo tumor suppressor signaling. Biofactors. 2009;35:338–345. doi: 10.1002/biof.47. [DOI] [PubMed] [Google Scholar]
- [74].Chiba S, Ikeda M, Katsunuma K, Ohashi K, Mizuno K. MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr Biol. 2009;19:675–681. doi: 10.1016/j.cub.2009.02.054. [DOI] [PubMed] [Google Scholar]
- [75].Oh HJ, Kim MJ, Song SJ, Kim T, Lee D, Kwon SH, Choi EJ, Lim DS. MST1 limits the kinase activity of aurora B to promote stable kinetochore-microtubule attachment. Curr Biol. 2010;20:416–422. doi: 10.1016/j.cub.2009.12.054. [DOI] [PubMed] [Google Scholar]
- [76].Cornils H, Kohler RS, Hergovich A, Hemmings BA. Human NDR kinases control G(1)/S cell cycle transition by directly regulating p21 stability. Mol Cell Biol. 2011;31:1382–1395. doi: 10.1128/MCB.01216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cornils H, Kohler RS, Hergovich A, Hemmings BA. Downstream of human NDR kinases: impacting on c-myc and p21 protein stability to control cell cycle progression. Cell Cycle. 2011;10:1897–1904. doi: 10.4161/cc.10.12.15826. [DOI] [PubMed] [Google Scholar]
- [78].Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, Rajbhandari P, Shen Q, Nemenman I, Basso K, Margolin AA, Klein U, Dalla-Favera R, Califano A. Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat Biotechnol. 2009;27:829–839. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol Cell Biol. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr Biol. 2009;19:1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- [81].Hergovich A, Lamla S, Nigg EA, Hemmings BA. Centrosome-associated NDR kinase regulates centrosome duplication. Mol Cell. 2007;25:625–634. doi: 10.1016/j.molcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- [82].Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17:700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- [83].Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- [84].Xia H, Qi H, Li Y, Pei J, Barton J, Blackstad M, Xu T, Tao W. LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene. 2002;21:1233–1241. doi: 10.1038/sj.onc.1205174. [DOI] [PubMed] [Google Scholar]
- [85].Iida S, Hirota T, Morisaki T, Marumoto T, Hara T, Kuninaka S, Honda S, Kosai K, Kawasuji M, Pallas DC, Saya H. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 2004;23:5266–5274. doi: 10.1038/sj.onc.1207623. [DOI] [PubMed] [Google Scholar]
- [86].Yang X, Li DM, Chen W, Xu T. Human homologue of Drosophila lats, LATS1, negatively regulate growth by inducing G(2)/M arrest or apoptosis. Oncogene. 2001;20:6516–6523. doi: 10.1038/sj.onc.1204817. [DOI] [PubMed] [Google Scholar]
- [87].Yang X, Yu K, Hao Y, Li DM, Stewart R, Insogna KL, Xu T. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat Cell Biol. 2004;6:609–617. doi: 10.1038/ncb1140. [DOI] [PubMed] [Google Scholar]
- [88].Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000;149:1073–1086. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005;65:6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- [90].Li Y, Pei J, Xia H, Ke H, Wang H, Tao W. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene. 2003;22:4398–4405. doi: 10.1038/sj.onc.1206603. [DOI] [PubMed] [Google Scholar]
- [91].Kamikubo Y, Takaori-Kondo A, Uchiyama T, Hori T. Inhibition of cell growth by conditional expression of kpm, a human homologue of Drosophila warts/lats tumor suppressor. J Biol Chem. 2003;278:17609–17614. doi: 10.1074/jbc.M211974200. [DOI] [PubMed] [Google Scholar]
- [92].McPherson JP, Tamblyn L, Elia A, Migon E, Shehabeldin A, Matysiak-Zablocki E, Lemmers B, Salmena L, Hakem A, Fish J, Kassam F, Squire J, Bruneau BG, Hande MP, Hakem R. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. Embo J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, Okabe M, Nojima H. Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem. 2007;282:19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- [94].Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- [95].Wilmeth LJ, Shrestha S, Montano G, Rashe J, Shuster CB. Mutual dependence of Mob1 and the chromosomal passenger complex for localization during mitosis. Mol Biol Cell. 2010;21:380–392. doi: 10.1091/mbc.E09-06-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Luca FC, Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol Biol Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Cho HP, Liu Y, Gomez M, Dunlap J, Tyers M, Wang Y. The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol Cell Biol. 2005;25:4541–4551. doi: 10.1128/MCB.25.11.4541-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kaiser BK, Zimmerman ZA, Charbonneau H, Jackson PK. Disruption of centrosome structure, chromosome segregation, and cytokinesis by misexpression of human Cdc14A phosphatase. Mol Biol Cell. 2002;13:2289–2300. doi: 10.1091/mbc.01-11-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Krasinska L, de Bettignies G, Fisher D, Abrieu A, Fesquet D, Morin N. Regulation of multiple cell cycle events by Cdc14 homologues in vertebrates. Exp Cell Res. 2007;313:1225–1239. doi: 10.1016/j.yexcr.2006.12.022. [DOI] [PubMed] [Google Scholar]
- [100].Mailand N, Lukas C, Kaiser BK, Jackson PK, Bartek J, Lukas J. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat Cell Biol. 2002;4:317–322. doi: 10.1038/ncb777. [DOI] [PubMed] [Google Scholar]
- [101].Mishima M, Pavicic V, Gruneberg U, Nigg EA, Glotzer M. Cell cycle regulation of central spindle assembly. Nature. 2004;430:908–913. doi: 10.1038/nature02767. [DOI] [PubMed] [Google Scholar]
- [102].Vazquez-Novelle MD, Mailand N, Ovejero S, Bueno A, Sacristan MP. Human Cdc14A phosphatase modulates the G2/M transition through Cdc25A and Cdc25B. J Biol Chem. 2010;285:40544–40553. doi: 10.1074/jbc.M110.133009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wolfe BA, Gould KL. Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation. Embo J. 2004;23:919–929. doi: 10.1038/sj.emboj.7600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Matallanas D, Romano D, Al-Mulla F, O’Neill E, Al-Ali W, Crespo P, Doyle B, Nixon C, Sansom O, Drosten M, Barbacid M, Kolch W. Mutant K-Ras Activation of the Proapoptotic MST2 Pathway Is Antagonized by Wild-Type K-Ras. Mol Cell. 2011;44:893–906. doi: 10.1016/j.molcel.2011.10.016. [DOI] [PubMed] [Google Scholar]
- [106].Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O’Neill E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, Im CR, Lee JO, Yonehara S, Lim DS. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–2569. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- [108].Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008;18:1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- [109].Guo C, Zhang X, Pfeifer GP. The tumor suppressor RASSF1A prevents dephosphorylation of the mammalian STE20-like kinases MST1 and MST2. J Biol Chem. 2011;286:6253–6261. doi: 10.1074/jbc.M110.178210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Hamilton G, Yee KS, Scrace S, O’Neill E. ATM regulates a RASSF1A-dependent DNA damage response. Curr Biol. 2009;19:2020–2025. doi: 10.1016/j.cub.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Callus BA, Verhagen AM, Vaux DL. Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. Febs J. 2006;273:4264–4276. doi: 10.1111/j.1742-4658.2006.05427.x. [DOI] [PubMed] [Google Scholar]
- [112].Donninger H, Allen N, Henson A, Pogue J, Williams A, Gordon L, Kassler S, Dunwell T, Latif F, Clark GJ. Salvador protein is a tumor suppressor effector of RASSF1A with hippo pathway-independent functions. J Biol Chem. 2011;286:18483–18491. doi: 10.1074/jbc.M110.214874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Rong R, Jin W, Zhang J, Sheikh MS, Huang Y. Tumor suppressor RASSF1A is a microtubule-binding protein that stabilizes microtubules and induces G2/M arrest. Oncogene. 2004;23:8216–8230. doi: 10.1038/sj.onc.1207901. [DOI] [PubMed] [Google Scholar]
- [114].Song MS, Song SJ, Ayad NG, Chang JS, Lee JH, Hong HK, Lee H, Choi N, Kim J, Kim H, Kim JW, Choi EJ, Kirschner MW, Lim DS. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat Cell Biol. 2004;6:129–137. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- [115].Rong R, Jiang LY, Sheikh MS, Huang Y. Mitotic kinase Aurora-A phosphorylates RASSF1A and modulates RASSF1A-mediated microtubule interaction and M-phase cell cycle regulation. Oncogene. 2007;26:7700–7708. doi: 10.1038/sj.onc.1210575. [DOI] [PubMed] [Google Scholar]
- [116].Song SJ, Song MS, Kim SJ, Kim SY, Kwon SH, Kim JG, Calvisi DF, Kang D, Lim DS. Aurora A regulates prometaphase progression by inhibiting the ability of RASSF1A to suppress APC-Cdc20 activity. Cancer Res. 2009;69:2314–2323. doi: 10.1158/0008-5472.CAN-08-3984. [DOI] [PubMed] [Google Scholar]
- [117].Song SJ, Kim SJ, Song MS, Lim DS. Aurora B-mediated phosphorylation of RASSF1A maintains proper cytokinesis by recruiting Syntaxin16 to the midzone and midbody. Cancer Res. 2009;69:8540–8544. doi: 10.1158/0008-5472.CAN-09-1554. [DOI] [PubMed] [Google Scholar]
- [118].Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- [119].van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Ikeda M, Kawata A, Nishikawa M, Tateishi Y, Yamaguchi M, Nakagawa K, Hirabayashi S, Bao Y, Hidaka S, Hirata Y, Hata Y. Hippo pathway-dependent and - independent roles of RASSF6. Sci Signal. 2009;2:ra59. doi: 10.1126/scisignal.2000300. [DOI] [PubMed] [Google Scholar]
- [121].Spencer S, Dowbenko D, Cheng J, Li W, Brush J, Utzig S, Simanis V, Lasky LA. PSTPIP: a tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J Cell Biol. 1997;138:845–860. doi: 10.1083/jcb.138.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]