Abstract
Barrett’s Esophagus is an increasingly common disease that is strongly associated with reflux of stomach acid and usually a hiatus hernia. Barrett’s Esophagus strongly predisposes to esophageal adenocarcinoma (EAC), a tumour with a very poor prognosis. We have undertaken the first genome-wide association study on Barrett’s Esophagus, comprising 1,852 UK cases and 5,172 UK controls in discovery and 5,986 cases and 12,825 controls in the replication. Two regions were associated with disease risk: chromosome 6p21, rs9257809 (Pcombined=4.09×10−9, OR(95%CI) =1.21(1.13-1.28)) and chromosome 16q24, rs9936833 (Pcombined=2.74×10−10, OR(95%CI) =1.14(1.10-1.19)). The top SNP on chromosome 6p21 is within the major histocompatibility complex, and the closest protein-coding gene to rs9936833 on chromosome 16q24 is FOXF1, which is implicated in esophageal development and structure. We found evidence that the genetic component of Barrett’s Esophagus is mediated by many common variants of small effect and that SNP alleles predisposing to obesity also increase risk for Barrett’s Esophagus.
Keywords: Barrett’s Esophagus, Esophageal adenocarcinoma (EAC), Gastro-esophageal reflux disease (GERD), Genome-wide association (GWA) study, Single nucleotide polymorphisms (SNPs)
Barrett’s Esophagus is one of the most common pre-malignant lesions in the Western world. It affects over 2% of the adult population and, unlike bowel polyps, lacks any proven effective therapy1. In the majority of cases, Barrett’s Esophagus is associated with chronic gastro-esophageal reflux disease (GERD), including esophagitis 2,3. In addition there are structural changes, mainly hiatus hernia, in the lower esophagus in over 80% of patients4. This allows both acid and bile to remain immediately adjacent to the esophageal epithelium. The measured annual risk of esophageal adenocarcinoma (EAC) in Barrett’s Esophagus patient’s varies widely but is approximately 0.4-1% 5-7. Notably the incidence of EAC has been rising by 3% each year for the last 30 years; it is now the fifth commonest cancer in the UK 8. Despite modern multimodality therapy, the prognosis of EAC remains poor, with a 9-15% 5-year survival 9,10.
The etiology of Barrett’s Esophagus is not well characterised. Environmental factors, such as diet, are weakly associated with GERD, Barrett’s Esophagus and EAC, and obesity is a known risk factor for all three conditions11. There is also evidence implicating genetic factors: the relative risks are increased 2-4 fold for GERD, Barrett’s Esophagus and EAC when one first-degree relative is affected12-17. A segregation analysis of 881 pedigrees of familial Barrett’s Esophagus supports an incompletely dominant inheritance model with a polygenic component18. Extensive candidate gene and linkage searches have, to date, failed to identify genetic variants that are associated with risk of Barrett’s Esophagus19.
As part of the Wellcome Trust Case Control Consortium 2 (WTCCC2) study of 15 common disorders and traits, we present the results of the first genome-wide association study of Barrett’s Esophagus susceptibility. Using a discovery cohort from the UK (with case samples from Aspirin and Esomeprazole Chemoprevention Trial of Cancer in Barrett’s Oesophagus (AspECT)20), and five replication cohorts (including case samples from CHemoprevention Of Premalignant Intestinal Neoplasia (ChOPIN) and Esophageal Adenocarcinoma GenEtics Consortium (EAGLE) studies9,20), we identified two variants associated with Barrett’s Esophagus, each with combined evidence at P<5×10−8. The analysis workflow is outlined in Supplementary Figure 1 and characteristics of the case and control samples used can be found in Supplementary Table 1 and Online Methods.
For the discovery analysis, cases with histologically confirmed Barrett’s Esophagus (see methods) were recruited from sites across the UK (Supplementary Table 2). Population controls were taken from the WTCCC2 common set of 1958 Birth Cohort (58C) and National Blood Service (UKBS) samples as previously described21. The case individuals were genotyped on the Illumina 660W-Quad array and controls were genotyped on the Illumina custom Human 1.2M-Duo array, with the analysis performed on the overlapping set of SNPs. Following quality control (see Online Methods, Supplementary Note, Supplementary Figure 2 and Supplementary Table 3), a total of 521,744 SNPs typed in 1,852 cases and 5,172 controls (2,499 UKBS and 2,673 58C) were included in the discovery analysis.
Association analysis was carried out under a logistic regression model as implemented in SNPTEST. The genomic over-dispersion factor22 λ was 1.10 and this was reduced to 1.05 when incorporating the first principal component as a covariate, suggesting that population structure was not a major problem in the discovery analyses (Supplementary Figure 3). For all of the following results presented, unless otherwise stated, the first principal component was used as a covariate.
Following analysis of the genome-wide association results (Figure 1), we adopted a staged approach to replication, outlined below and in Supplementary Figure 1.
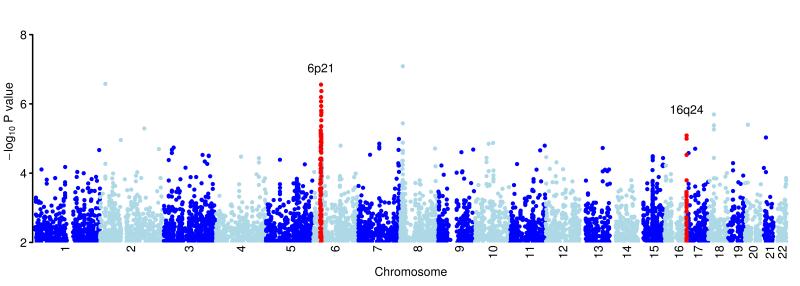
Figure 1.
Plot of the genome-wide association results after fitting the multiplicative model in SNPTEST. Results shown for the 521,744 SNPs passing quality control filters. Chromosomes are coloured dark blue and light blue alternatively, as labelled on the x-axis. The y-axis shows the −log10 P values. Regions in red show the loci newly identified as associated with BE, as described in Table 1.
Stage 1
100 SNPs that showed evidence of association in the discovery data (at P<5×10−4) were analysed in another UK sample set. This comprised 1,105 cases from ChOPIN and EAGLE and 4,421 controls from the 58C control dataset, all genotyped on the Illumina Immunochip23 (WTCCC2 contributed SNPs to the Immunochip design to allow for its replication studies, and the set of 100 SNPs followed up in our Stage 1 replication were all on the Immunochip), and a further set of 2,578 UK controls (the People of the British Isles (PoBI) collection 24) genotyped on the Illumina custom Human 1.2M-Duo array. Results of this first stage of replication are shown in Supplementary Table 4.
Stage 2
The 16 top SNPs (Pcombined<10−5) from meta-analysis of the discovery and Stage 1 replication were replicated in silico in a Dutch collection of 473 cases and 1,780 controls genotyped on the Immunochip23. Results from Stage 2 replication are shown in Supplementary Table 5.
Stage 3
Two SNPs with Pcombined<5×10−8 after Stage 2 replication (rs9257809 on chromosome 6p21 and rs9936833 on chromosome 16q24) were studied in three additional replication sample sets. They were directly genotyped in an Irish cohort of 245 cases and 473 controls and a UK cohort of 1,765 cases and 1,586 controls, and data from these SNPs was retrieved from the BEACON consortium for 2,398 cases and 2,167 controls from European, Australian and American individuals with European ancestry.
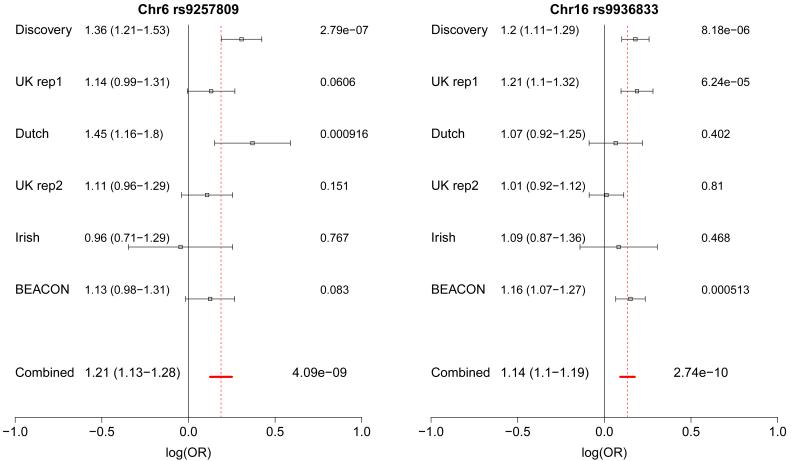
After these three stages of replication, the two SNPs on chromosome 6p21 and 16q24 showed compelling evidence for association, with combined P values of 4.09×10−9 for rs9257809, OR(95%CI)=1.21(1.13-1.28) and 2.74×10−10 for rs9936833, OR(95%CI) =1.14(1.10-1.19) (Table 1, Figures 2, 3).
Table 1.
Loci associated with risk of Barrett’s Esophagus
| Chr Position* |
rsID | Risk allele |
Discovery (1852/5172) |
Stage 1 | Stage 2 | Stage 3 | Combined P value OR (95%CI) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UK replication 1 (1105/6819) |
Dutch Replication (473/1780) |
UK Replication 2 (1765/1586) |
Irish Replication (245/473) |
BEACON (2398/2167) |
|||||||||||
| RAF Case /Con |
P value OR (95%CI) |
RAF Case /Con |
P value OR (95%CI) |
RAF Case /Con |
P value OR (95%CI) |
RAF Case /Con |
P value OR (95%CI) |
RAF Case /Con |
P value OR (95%CI) |
RAF Case /Con |
P value OR (95%CI) |
||||
| 6p21 29464310 |
rs9257809 | A | 0.90 /0.87 |
2.78×10−7 1.36 (1.21-1.53) |
0.89 /0.87 |
0.0606 1.14 (0.99-1.31) |
0.91 /0.87 |
9.16×10−4 1.45 (1.16-1.80) |
0.88 /0.87 |
0.151 1.11 (0.96-1.29) |
0.85 /0.86 |
0.767 0.96 (0.71-1.29) |
0.91 /0.90 |
0.083 1.13 (0.98-1.31) |
4.09×10−9 1.21 (1.13-1.28) |
| 16q24 84960619 |
rs9936833 | C | 0.42 /0.38 |
8.18×10−6 1.20 (1.11-1.29) |
0.42 /0.37 |
6.24×10−5 1.21 (1.10-1.32) |
0.35 /0.34 |
0.402 1.07 (0.92-1.25) |
0.39 /0.39 |
0.810 1.01 (0.92-1.12) |
0.41 /0.39 |
0.468 1.09 (1.06-1.11) |
0.40 /0.36 |
5.13 ×10−4 1.16 (1.07-1.27) |
2.74×10−10 1.14 (1.10-1.19) |
Discovery and replication results at the lead SNPs at the two loci for which there is combined evidence of P< 5×10−8. P values are two-sided. ‘RAF’- Risk allele frequency, *NCBI Build 36. The number of cases and controls, respectively, in each cohort is shown under the title of the cohort.
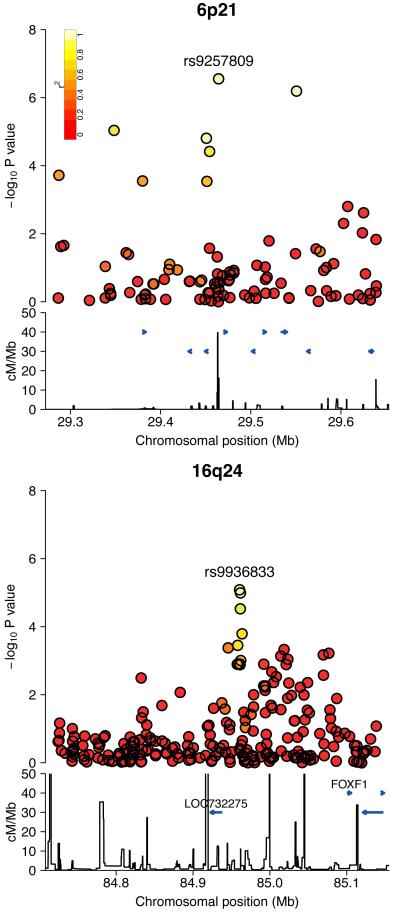
Figure 2.
Regional association plot of the associated loci as detailed in Table 1, showing the signal at the lead SNP. The −log10 P values for the SNPs are shown on the upper part of each plot. SNPs are coloured based on their r2 with the labelled hit SNP which has the smallest P value in the region. r2 is calculated from the 58C data. The bottom section of each plot shows the fine scale recombination rates estimated from individuals in the HapMap population, and genes are marked by horizontal blue lines.
Figure 3.
Forest plots showing evidence for association at each of the loci described in Table 1. The effect size and 95% CI are shown to the right of the cohort label for the discovery and replication cohorts and for the fixed effects meta-analysis. The red dashed line marks the effect size calculated from the fixed effects meta-analysis. P values for each cohort are shown at the right of the plot and the meta-analysis P value is also given, all P values are two-sided.
We performed tests for pair-wise interaction (see Supplementary Note) in the discovery data between all pairs of the 16 SNPs taken forward to Stage 2 replication (Supplementary Table 5), but no significant interactions (P<0.01) were found.
Imputation was carried out in the discovery data for the chromosome 6p21 and 16q24 regions, using the 1000 Genomes June 2010 CEU reference panel. In each case, rs9257809 and rs9936833 respectively remained as the strongest signal of association in each region (Supplementary Figure 4).
The lead SNP on 16q24, rs9936833, maps 24kb from the spliced, non-coding transcript LOC732275. The closest coding gene, 141kb towards the telomere, is FOXF1, a forkhead family transcription factor that acts in the hedgehog signaling pathway. FOXF1 is known to have a role in the development of the gastrointestinal tract and has been reported to cause esophageal structural alterations, especially atresia, when inactivated 25. The region around rs9936833 contains multiple binding sites for specific transcription factors, such as FOXP2, that are known to control FOXF1 expression (assessed using ENCODE data, see URLs).
The lead SNP on 6p21, rs9257809, lies on the telomeric edge of the major histocompatibility complex (MHC) region between olfactory receptor genes OR2D12 and OR2D13. It is in strong long-range linkage disequilibrium (r2>0.6 calculated in the control data) with SNPs over 1 Mb away, including two at which Stage 2 replication was attempted, rs13211507 (Pcombined =8.77×10−9) and rs9262143 (Pcombined=2.18×10−8). When conditioning on rs9257809, no other SNP in the MHC was significant at P<10−5.
To further investigate the SNP signal in the MHC region, we took two approaches: GENECLUSTER, which is a Bayesian tree building method26,27; and HLA*IMP, which is a method for imputing classical HLA alleles from SNP data28. Both methods provided evidence of association in the discovery data for reduced risk conferred by three classical HLA alleles that are in strong LD with each other (HLA-C*07:01, HLA-A*01:01 and HLA-B*08:01), see Supplementary Table 6. However, conditional analysis suggested that rs9257809 better captures the association in our discovery data and none of these three classical HLA alleles showed an association signal in the replication data (P>0.1, Supplementary Table 6).
We used standard UK criteria, in accordance with the British Society of Gastoenterology, for diagnosis of Barrett’s Esophagus. However some countries use the American College of Gastroenterology criteria that require the presence of intestinal metaplasia for the diagnosis of Barrett’s Esophagus. To investigate this, we analysed the two replicated loci using only the subset of discovery and replication cases (86%) with histological evidence of intestinal metaplasia. Both signals remained significant, with combined evidence across discovery and all stages of replication of P<5×10−8 (Supplementary Table 7A and 7B).
We also investigated associations with the related quantitative traits of circumferential extent (C) and maximal extent (M) of the length of Barrett’s segment. In the discovery cohort, the C measurement was available for 1,744 cases, and the M measurement for 1,618 cases. In a linear regression analysis of cases, neither SNP showed evidence of association with C or M status (for rs9936833, P=0.63 and P=0.87 respectively; for rs925809, P=0.10 and P=0.09 respectively). We then extended the C and M analysis genome-wide. No SNP reached P<10−6 in the analysis of C. One SNP (rs1023313) reached P<10−6 in the analysis of M, but this association was not confirmed in Stage 1 or Stage 2 replication (see Supplementary Table 8).
There is an established sex bias in BE susceptibility, with men at greater risk than women3,29. The ratio of males to females is 4:1 in our case discovery data. To see whether there might be sex-specific effects of any predisposition SNPs, we performed a sex-stratified analysis for the 16 SNPs analysed in Stage 2 (Supplementary Table 9). The SNP showing the most evidence for a sex-specific effect from the combined discovery and Stage 1 and 2 replication was rs9257809. The association signal was stronger in males than females (uncorrected P=0.01 for difference of effects between sexes), corresponding to a male odds ratio of 1.38 (95%CI 1.25-1.53, Pcombined=1.71×10−10) and a female odds ratio of 1.11 (95%CI 0.95-1.30, Pcombined=0.19), see Supplementary Note for further details. This finding warrants further investigation.
Previous genome-wide association studies of common diseases or phenotypes have found evidence for a model where many common variants of small effect influence risk 30,31. We looked for these en masse effects in Barrett’s Esophagus using two methods (see Online Methods). Firstly, taking the top K SNPs (for different values of K) in independent regions in the discovery data, we performed a sign test to see whether there was an excess (over the proportion expected under the null of 50%) of SNPs for which the effect was in the same direction in the Stage 1 replication data. Secondly a disease-score test analysis was undertaken, as described by the International Schizophrenia Consortium 30. Both methods found evidence of an excess of SNPs that have the same risk allele in both cohorts. The strongest evidence in the sign test was for the top 1,100 SNPs, for which the sign test gave Puncorrected=2.30×10−5 (Supplementary Figure 5). From the disease-score analysis, the strongest evidence was for the top 1,710 SNPs, for which Puncorrected=7.07×10−11 (Supplementary Figure 6). Both analyses thus implicate a large number of common SNPs of small effect in susceptibility to Barrett’s Esophagus.
There is a well-established link between Barrett’s Esophagus and obesity32,33. To investigate whether this may in part reflect genetic effects, we repeated the sign test at 40 of the SNPs that have been found to be associated with either Body Mass Index (BMI) or Waist Hip Ratio (WHR), where genotype data or tag SNPs were available in our discovery samples 34-38. In our discovery data, a total of 29 out of 40 BMI/WHR-associated SNPs (14 genotyped, 15 tagging, Supplementary Tables 10A and 10B) shared the same risk alleles in Barrett’s Esophagus as they did for BMI/WHR (P=6.42×10−3).
Our results provide direct evidence that Barrett’s Esophagus aetiology has a genetic component. Inference as to the underlying genes must be cautious, especially for the variant (tagged by rs9257809) in the gene-rich MHC region in which linkage disequilibrium is long-range and complex. However, the location of the other associated SNP, rs9936833, near FOXF1 suggests a role for structural factors in the esophagus and stomach as a disease-predisposing factor, consistent with the evidence that changes such as hiatus hernia are known to be strongly associated with Barrett’s Esophagus. We also found evidence to show that body weight SNPs are more likely than by chance to show effects in the same direction in Barrett’s Esophagus, suggesting that genetic effects may in part underpin the epidemiological observation that BMI is a risk factor for Barrett’s Esophagus 39. Given that Barrett’s Esophagus is an accepted status as a precursor lesion, the SNPs that we have identified could also be de facto risk factors for esophageal adenocarcinoma and may give clues as to the biology of both of these important phenotypes.
Online Methods
Samples
Cases from Discovery, Stages 1 and 2 replication, and Stage 3 UK and Irish
For the discovery, we ascertained cases of histologically-confirmed Barrett’s Esophagus through the United Kingdom-based ASPECT clinical trial of proton pump-inhibitor (esomeprazole) and aspirin as preventive agents for progression of Barrett’s Esophagus to EAC20. UK, Irish and Dutch replication cases were from the Chemoprevention of Premalignant Intestinal Neoplasia (ChOPIN) genetic study and the Esophageal Adenocarcinoma GenE (EAGLE) consortium9. Replication cases were diagnosed with Barrett’s Esophagus with lengths of at least 1cm (CIMI) circumferential Barrett’s Esophagus or at least a 2cm tongue (C0M2) according to the Prague criteria40. Case collection was in accordance with the British Society of Gastoenterology criteria41, the standard practice for collaborating Histopathologists in the UK and much of Europe. We found that 90% of our discovery samples (for which full clinico-pathological data were available) had evidence of intestinal metaplasia and therefore also met the American College of Gastroenterology criteria that are widely used in the USA42 . For full details of the ethnicity, age and sex distributions and Prague criteria measurements of the cases see Supplementary Table 1.
Discovery
The full data set comprised of 1,991 cases and 5,667 controls. After QC, 1,852 cases and 5,172 controls were analysed. Controls were taken from the WTCCC2 set, made up of samples from the 1958 British Birth Cohort (58C) and the National Blood Service collection (UKBS). Samples were genotyped at the Wellcome Trust Sanger Institute (WTSI), cases on the Illumina Human660W-Quad array, and controls on the Illumina custom Human 1.2M-Duo. The primary analysis was performed on the overlapping set of SNPs.
Stage 1
After QC, the UK replication totalled 1,105 cases and 6,819 controls. The controls were from the PoBI cohort (2,578)24 and 58C (4,241) samples that were not genotyped in the discovery phase. The case and 58C control samples were genotyped on the Illumina Immunochip and the PoBI samples were genotyped on the Illumina custom Human 1.2M-Duo array. The Immunochip is a custom-designed chip containing 196,524 SNPs in total, of which ~2,400 were selected on the basis of our discovery GWAS study.
Stage 2
The Dutch replication cohort consisted of 473 cases and 1,780 controls. These samples were all genotyped on the Illumina Immunochip but in two separate locations; the case samples were genotyped at WTSI and the control samples were genotyped as described in a previous report43.
See Supplementary Note for information on DNA sample preparation.
Circumferential and Maximal Extent Phenotypes
Length of the Barrett’s segment was available for a subset of discovery and replication phase samples. Where baseline measurements were not available, the earliest measurement taken after baseline was used. A small number of cases were excluded on the basis of reporting errors (if C >M or if either value exceeded 25cm). Of the discovery phase individuals after quality control, 1,744 had C measurements and 1,618 had M measurements, C mean=4.05 (range 0-22); M mean=4.60 (range 1-24). M measurements were available for 1,015 of the Stage 1 replication (M mean=4.66 (range 1-23)) and for 240 of the Stage 2 replication (M mean=4.44 (range 1-15)). Both C and M phenotypes were square-root transformed prior to analysis, to improve the fit of the linear regression model.
Stage 3
Irish Replication
The Irish replication cohort consisted of 245 cases and 473 controls. Cases were provided by St James’s Hospital and Mater Misericordiae University Hospital Dublin as part of EAGLE. Controls were provided by Trinity Biobank. 168 cases were genotyped on the Illumina Immunochip at WTSI. rs9257809 and rs9936833 were genotyped in 77 cases and all controls using competitive allele-specific PCR KASPar chemistry (KBiosciences Ltd, Hertfordshire, UK). Primers, probes and conditions used are available on request. Genotyping quality control was tested using duplicate DNA samples within studies and SNP assays, together with direct sequencing of subsets of samples to confirm genotyping accuracy. For all SNPs, >99% concordant results were obtained.
UK Replication 2
1,765 cases were ascertained using the diagnostic criteria and sampling from ASPECT as described above for discovery. 1586 controls were collected as part of the Colorectal Tumour Gene Identification (CoRGI) consortium44. Controls were spouses or partners unaffected by cancer and without a personal family history (to 2nd degree relative level) of colorectal neoplasia. All were of white UK ethnic origin, 45% male; mean age 45.1 years, SD±15.9. All samples were genotyped using KASPar competitive allele-specific PCR as described above.
BEACON Replication
2,398 cases and 2,167 controls were analyzed. Samples were collected as part of a GWAS study (BEAGESS) undertaken by the BEACON collaboration. Samples were collected from sites in Australia (cases n=325, controls n=561), Europe (England, Ireland, Sweden; cases n=363, controls n=333), and North America (Canada, United States, cases n=1,710, controls n=1,273). Samples were genotyped at the Fred Hutchinson Cancer Research Center (FHCRC) on the Illumina Omni1M Quad.
Quality control
Samples
As previously described21,45, we identified and removed samples whose genome-wide patterns of diversity differed from those of the collection at large, interpreting them as likely to be due to biases or artefacts. See Supplementary Note for further details. Following sample quality control our final discovery dataset consisted of 1,852 cases and 5,172 controls (Supplementary Table 3).
SNPs
For all arrays, normalised probe intensities were exported using the BeadStudio program and genotypes were called at the WTSI using Illuminus46. SNPs were excluded from analysis if in any of the data sets (58C, UKBS or cases) they had: a very low minor allele frequency (defined as <0.01%); extreme departures from Hardy-Weinberg equilibrium (P<10−20); showed a strong plate effect (P<10−6). SNPs were also excluded if the observed statistical (Fisher) information about the allele frequency was less than 98% of the information contained in a hypothetical sample of the same size and expected MAF but with no missing data. 45 SNPs were removed following visual inspection of cluster plots. In total 521,744 autosomal SNPs were available for analysis after quality control.
To confirm genotyping accuracy of the different platforms used in the study, 5% of the UK, Irish and Dutch samples typed on each platform were re-genotyped at rs9257809 and rs9936833 using competitive allele-specific PCR KASPar. Concordance was >99% (Supplementary Table 11) suggesting genotyping robustness across platforms.
HLA Imputation
Classical HLA alleles were imputed using HLA*IMP 28. Further details of this can be found in Supplementary Note.
Statistical analysis
Genome-wide case-control analysis was performed using frequentist tests, under a missing data logistic regression model, as implemented in SNPTEST. Unless otherwise stated, we assumed a multiplicative model for allelic risk by encoding the genotypes at each SNP as a discrete explanatory variable with an indicator of case status as the binary response and the first principal component as a covariate (see Supplementary Note). Quantitative C and M measurements were analysed using frequentist tests under a missing data linear regression model, as implemented in SNPTEST. To combine the evidence of association across the discovery and replication datasets we conducted an inverse-variance weighted fixed effect meta-analysis in the statistical package R (see Supplementary Note). To test for interactions (see Supplementary Note), between SNPs, or between a SNP and sex, and to compare models which include additional SNPs or classical HLA alleles as predictors, we used logistic regression models implemented in R. These analyses used thresholded (posterior probability > 0.9) genotype calls.
SNP Imputation was performed using IMPUTE247, which adopts a two-stage approach using both a haploid reference panel and a diploid reference panel.
BEACON data was analysed under an additive logistic regression model including the first four principal components as covariates (see Supplementary Note). Genomic inflation λ was 1.037.
En Masse analysis was carried out on the discovery and Stage 1 data. In order to reduce possible population structure (such analyses are sensitive to this), we restricted the Stage 1 control set to the 58C individuals. SNPs with MAF > 0.01 which were genotyped in both the discovery (Illumina 670K and Illumina custom Human 1.2M-Duo) and the replication (Illumina Immunochip) were pruned to remove strong linkage disequilibrium. This was done by ranking the SNPs by Bayes factor calculated under an additive model in SNPTEST, and successively selecting SNPs from the top so that they were at least 0.125cM plus 25kb away from any SNPs that had already been selected. We obtained 7,673 SNPs from a total of 28,972 (after quality control) that were typed in discovery and UK Immunochip data. For the K SNPs showing the strongest signal of association, the sign test compares the direction of effect of each SNP in the discovery and replication samples. Using a likelihood-ratio test we compared the null model where the probability of the same direction of effect is assumed to be a half, to a model where the probability is not a half (two sided).
The disease-score test aims to measure indirectly the collective effect of many weakly associated alleles. We determined the risk allele and odds ratio for each pruned SNP from the discovery data as described above. Then, we used the top K SNPs to calculate the “score” for each individual in the replication data as the number of risk alleles carried by each individual weighted by the log of the odds ratio estimated from the discovery data. Under the null hypothesis, the risk alleles and odds ratios in the discovery and replication samples are independent. We tested a logistic regression model of disease status on the score in the replication data, conditioning on the first principal component, to control for population structure, and the number of missing genotypes (called with maximum probability < 0.9), to control for potential differences in genotyping rate, as covariates.
Supplementary Material
Acknowledgments
We acknowledge the patients who participated in the study and the physicians who helped in recruitment. We thank the many research nurses who also helped recruit including Sandra Prew (Sandwell General Hospital, UK). Funding for this study was provided by the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z) and the Wellcome Trust (072894/Z/03/Z, 090532/Z/09/Z and 075491/Z/04/B). P. Donnelly was supported in part by a Wolfson-Royal Society Merit Award. D, Whiteman is supported by a Future Fellowship from the Australian Research Council. S. Macgregor is supported by an Australian NHMRC Career Development Award. C. Spencer was supported by a Wellcome Trust Fellowship [097364/Z/11/Z]. We thank S. Bertrand, J. Bryant, S.L. Clark, J.S. Conquer, T. Dibling, J.C. Eldred, S. Gamble, C. Hind, A. Wilk, C.R. Stribling and S. Taylor of the Wellcome Trust Sanger Institute’s Sample and Genotyping Facilities for technical assistance. We also thank Jeff Barrett for design of the Immunochip array. We acknowledge use of the British 1958 Birth Cohort DNA collection funded by the Medical Research Council (grant G0000934) and the Wellcome Trust (grant 068545/Z/02), the UK National Blood Service controls funded by the Wellcome Trust and the People of the British Isles collection, funded by the Wellcome Trust. This work was also supported by the Esophageal Adenocarcinoma GenE consortia incorporating the ChOPIN project (grant C548/A5675) and IPOD project (grant MGAG1G7R), Cancer Research UK; (ASPECT grant - C548/A4584 and D9612L00090, HANDEL grant C548/A9085), Astrazeneca UK educational grant; University Hospitals of Leicester R and D grant; AspECT - T91 5211 University of Oxford grant- HDRMJQ0. The Barrett’s and Esophageal Adenocarcinoma Genetic Susceptibility Study (BEAGESS) within the BEACON consortium was supported by grant R01CA13672501.
The authors of this paper are:
Zhan Su3,*, Laura J Gay4,*, Amy Strange3, Claire Palles3,5, Gavin Band3, David C Whiteman6, Francesco Lescai7, Cordelia Langford8, Manoj Nanji4, Sarah Edkins8, Anouk van der Winkel9, David Levine10, Peter Sasieni11, Céline Bellenguez3, Kimberley Howarth3, Colin Freeman3, Nigel Trudgill12, Art T Tucker13, Matti Pirinen3, Maikel P Peppelenbosch9, Luc JW van der Laan14, Ernst J Kuipers9, Joost PH Drenth15, Wilbert H Peters15, John V Reynolds16, Daniel A Kelleher17, Ross McManus17, Heike Grabsch18, Hans Prenen19, Raf Bisschops19, Kausila Krishnadath20, Peter D Siersema21, Jantine WPM van Baal21, Mark Middleton22, Russell Petty23, Richard Gillies22, Nicola Burch24, Pradeep Bhandari25, Stuart Paterson26, Cathryn Edwards27, Ian Penman28,29, Kishor Vaidya30, Yeng Ang31, Iain Murray32, Praful Patel33, Weimin Ye34, Paul Mullins35, Anna H Wu36, Nigel C Bird37, Helen Dallal38, Nicholas J Shaheen39, Liam J Murray40, Konrad Koss41, Leslie Bernstein42, Yvonne Romero43, Laura J Hardie44, Rui Zhang10, Helen Winter45, Douglas A Corley46, Simon Panter47, Harvey A Risch48, Brian J Reid49, Ian Sargeant50, Marilie D Gammon51, Howard Smart52, Anjan Dhar53, Hugh McMurtry54, Ali Haythem55, Geoffrey Liu56, Alan G Casson57, Wong-Ho Chow58, Matt Rutter59, Ashref Tawil60, Danielle Morris61, Chuka Nwokolo62, Peter Isaacs63, Colin Rodgers64, Krish Ragunath65, Chris MacDonald66, Chris Haigh67, David Monk68, Gareth Davies69, Saj Wajed70, David Johnston71, Michael Gibbons72, Sue Cullen73, Nicholas Church74, Ruth Langley75, Michael Griffin76, Derek Alderson77, Panos Deloukas8, Sarah E Hunt8, Emma Gray8, Serge Dronov8, Simon C Potter8, Avazeh Tashakkori-Ghanbaria8, Mark Anderson78, Claire Brooks79, Jenefer M Blackwell80,81, Elvira Bramon82,83, Matthew A Brown84, Juan P Casas85,86, Aiden Corvin87, Audrey Duncanson88, Hugh S Markus89, Christopher G Mathew90, Colin NA Palmer91, Robert Plomin92, Anna Rautanen3, Stephen J Sawcer93, Richard C Trembath90, Ananth C Viswanathan94, Nicholas Wood95, Gosia Trynka96, Cisca Wijmenga96, Jean-Baptiste Cazier3, Paul Atherfold97,98, Anna M Nicholson4, Nichola L Gellatly4, Deborah Glancy24, Sheldon C Cooper12, David Cunningham99, Tore Lind100, Julie Hapeshi101, David Ferry102, Barrie Rathbone24, Julia Brown103, Sharon Love104, Stephen Attwood105, Stuart MacGregor6, Peter Watson106, Scott Sanders107, Weronica Ek6, Rebecca F Harrison108, Paul Moayyedi109, John deCaestecker110, Hugh Barr111, Elia Stupka4,7,112, Thomas L Vaughan113, Leena Peltonen L8,†, Chris CA Spencer3, Ian Tomlinson3,5,^, Peter Donnelly3,114,+,^, Janusz AZ Jankowski4,22,24,115,+,^
Author Affiliations
Wellcome Trust Centre for Human Genetics Oxford UK.
Centre for Digestive Diseases Blizard Institute Queen Mary University of London UK.
Oxford National Institute of Health Research Comprehensive Biomedical Research Centre UK.
Queensland Institute of Medical Research Brisbane Australia.
UCL Cancer Institute University College London UK.
Wellcome Trust Sanger Institute Cambridge UK.
Department of Gastroenterology and Hepatology Erasmus MC University Medical Centre Rotterdam The Netherlands.
Department of Biostatistics University of Washington Seattle USA.
Centre for Cancer Prevention Wolfson Institute of Preventive Medicine Barts and The London UK.
Department of Gastroenterology Sandwell General Hospital Lyndon West Bromwich UK.
Centre for Clinical Pharmacology William Harvey Research Institute Queen Mary University of London UK.
Department of Surgery Erasmus MC University Medical Centre Rotterdam Rotterdam The Netherlands.
Department of Gastroenterology and Hepatology Radboud University Nijmegen Medical Centre Nijmegen The Netherlands.
Department of Surgery Trinity Centre for Health Sciences Trinity College Dublin St James’ Hospital Ireland.
Department of Clinical Medicine Trinity Centre for Health Sciences Trinity College Dublin St James’s Hospital Ireland.
Pathology and Tumour Biology Leeds Institute of Molecular Medicine University of Leeds St James’s University Hospital UK.
Department of Digestive Oncology University Hospital Gasthuisberg Leuven Belgium.
Department of Gastroenterology and Hepatology Academic Medical Center Amsterdam The Netherlands.
Department of Gastroenterology and Hepatology University Medical Center Utrecht The Netherlands.
Department of Medical Oncology Churchill Hospital University of Oxford UK.
Institute of Medical Sciences School of Medicine and Dentistry University of Aberdeen UK.
Digestive Disease Academic Centre Leicester Royal Infirmary Leicester UK.
Gastroenterology Queen Alexandra Hospital Portsmouth UK.
Forth Valley Royal Hospital Larbert UK.
Department of Gastroenterology Torbay Hospital Torquay UK.
GI Unit Western General Hospital Edinburgh UK.
GI Unit Royal Infirmary of Edinburgh UK.
Victoria Hospital Kirkcaldy Fife UK.
Gastroenterology Royal Albert Edward Infirmary NHS Trust Wigan UK.
Department of Gastroenterology Royal Cornwall Hospital Truro Cornwall UK.
Southampton University Hospitals NHS Trust UK.
Department of Medical Epidemiology and Biostatistics Karolinska Institutet Stockholm Sweden.
Department of Gastroenterology East Lancashire Hospitals NHS Trust Royal Blackburn Hospital Lancashire UK.
Department of Preventive Medicine University of Southern California/Norris Comprehensive Cancer Center Los Angeles USA.
Department of Oncology The Medical School University of Sheffield UK.
South Tees NHS Foundation Trust UK.
Division of Gastroenterology and Hepatology UNC School of Medicine University of North Carolina Chapel Hill NC USA.
Centre for Public Health Queen’s University Belfast UK.
Macclesfield General Hospital UK.
Department of Population Sciences Beckman Research Institute and City of Hope Comprehensive Cancer Center Duarte USA.
Division of Gastroenterology and Hepatology Mayo Clinic Rochester MN USA.
Division of Epidemiology University of Leeds UK.
Great Western Hospital Swindon UK.
Division of Research and Oakland Medical Center Kaiser Permanente Oakland CA USA.
South Tyneside District Hospital South Shields UK.
Yale University School of Medicine Department of Epidemiology and Public Health New Haven CT USA.
Division of Human Biology Fred Hutchinson Cancer Research Center Seattle WA USA.
Lister Hospital Hertfordshire UK.
Department of Epidemiology University of North Carolina School of Public Health USA.
Royal Liverpool University Hospital UK.
County and Durham and Darlington NHS Foundation Trust UK.
Lancashire Teaching Hospitals NHS Foundation Trust UK.
Maidstone Hospital UK.
Department of Medicine Medical Biophysics and Epidemiology University of Toronto Canada.
Department of Surgery University of Saskatchewan Saskatoon SK Canada.
Division of Cancer Epidemiology and Genetics National Cancer Institute Bethesda MD USA.
University Hospital of North Tees UK.
Gastroenetrology North Devon Dsitrict Hospital Barnstaple North Devon UK.
Department of Gastroenterology QEII East & North Herts NHS Trust Welwyn Garden City Queen Elizabeth Hospital Welwyn Garden City UK.
Department of Gastroenterology University Hospital of Coventry UK.
Gastroenterology Blackpool Victoria Hospital Blackpool UK.
Gastroenterology Antrim and Whiteabbey United Hospitals Antrim UK.
Wolfson Digestive Diseases Centre Queens Medical Centre Nottingham UK.
Gastrolenterology Cumberland Infirmary Carlisle Cumbria UK.
Department of Gastroenterology Wansbeck General Hospital Ashington Northumberland UK.
General Surgery Countess of Chester Hospital Chester UK.
Gatroenterology Harrogate District Hospital Harrogate UK.
Department of Thoracic and Upper Gastrointestinal Surgery Royal Devon and Exeter NHS Foundation Trust Exeter UK.
Gastroenterology Ninewells Hospital Dundee UK.
Gastroenterology Craigavon Area Hospital Craigavon Northern Ireland.
Wycombe Hospital High Wycombe UK.
Edinburgh Royal Infirmary UK.
Medical Research Council Clinical Trials Unit London UK.
Northern Osophago Gastirc Unit Royal Victoria Infirmary Queen Victoria Road Newcastle upon Tyne UK.
University of Birmingham College of Medical and Dental Sciences School of Cancer Sciences Academic Department of Surgery Old Queen Elizabeth Hospital Birmingham UK.
GI Unit City Hospital Birmingham UK.
Oncology Clinical Trials Office Department of Oncology University of Oxford UK.
Telethon Institute for Child Health Research Centre for Child Health Research University of Western Australia.
Cambridge Institute for Medical Research University of Cambridge UK.
Department of Psychoisis Studies National Institute of Health Research Biomedical Research Centre for Mental Health at the Institute of Psychiatry King’s College London UK.
The South London and Maudsley NHS Foundation Trust London UK.
University of Queensland Diamantia Institute Princess Alexandra Hospital University of Queensland Brisbane Australia.
Department of Epidemiology and Public Health University College London UK.
Department of Epidemiology and Population Health London School of Hygiene and Tropical Medicine UK.
Neuropsychiatric Genetics Research Group Institute of Molecular Medicine Trinity College Dublin Ireland.
Molecular and Physiological Sciences The Wellcome Trust London UK.
Clinical Neurosciences St George’s University of London UK.
King’s College London Dept Medical and Molecular Genetics King’s Health Partners Guy’s Hospital London UK.
Biomedical Research Centre Ninewells Hospital and Medical School Dundee UK.
King’s College London Social Genetic and Developmental Psychiatry Centre Institute of Psychiatry London UK.
University of Cambridge Dept Clinical Neurosciences Addenbrooke’s Hospital Cambridge UK.
National Institute for Health Research Biomedical Research Centre for Ophthalmology Moorfields Eye Hospital NHS Foundation Trust and University College London Institute of Ophthalmology London UK.
Department of Molecular Neuroscience Institute of Neurology Queen Square London UK.
Department of Genetics University Medical Center Groningen and University of Groningen The Netherlands.
UCBPharma Ltd Slough UK.
Department of Clinical Pharmacology University of Oxford UK.
Medical Oncology Royal Marsden Hospital London UK.
Reasearch and Development AstraZeneca Lund Sweden.
Gloucestershire Research & Development Support Unit Gloucestershire Royal Hospital UK.
Department of Oncology New Cross Hosptial Royal Wolverhampton Hospitals NHS Trust Wolverhampton UK.
Clinical Trial Research Unit Leeds Institute of Molecular Medicine Leeds UK.
Centre for Statistics in Medicine and Oxford Clinical Trials Research Unit Oxford UK.
General Surgery North Tyneside General Hospital North Shields Tyne and Wear UK.
Department of Medicine Institute of Clinical Science Royal Vicotoria Hospital Belfast Northern Ireland.
Department of Cellular Pathology Warwick Hospital Warwick UK.
Department of Pathology Leicester Royal Infirmary Leicester UK.
Division of Gastroenterology Department of Medicine McMaster University Medical Centre Hamilton Canada.
Department of Gastroenterology Leicester General Hospital Leicester UK.
Department of Upper GI Surgery Gloucestershire Royal Hospital Gloucester UK.
Center for Translational Genomics and Bioinformatics San Raffaele Scientific Institute Milan Italy.
Division of Public Health Sciences Fred Hutchinson Cancer Research Center Seattle WA USA.
Department of Statistics University of Oxford UK.
Gastrointestinal Cancer Group University of Oxford UK.
Footnotes
Contributed equally to this manuscript
These authors jointly supervised this work
Deceased
AUTHOR CONTRIBUTIONS J.A.Z.J., I.T., L.J.G. and M.N. oversaw cohort collection for the discovery and replication datasets. N.T., N. Burch, P.B., S. Paterson, C.E., I.P., K.V., Y.A., I.M., P.P., P.Mullins, H.D., K. Koss, D.C., M. Griffin, D.A., H.W., S. Panter, I.S., H.S., A. Dhar, H. McMurtry, A.H., M.R., A. Tawil, D. Morris, C.N., R.L., P.I., C.R., K.R., C. MacDonald, C.H., D. Monk, G.D., S.W., D.J., M. Gibbons, S. Cullen, N.C., D.G., S.A., P.W., J. deCaestecker, H.B. and JAZJ recruited ≥50 cases to the AspECT and/or ChOPIN studies. L.J.G, M.N., K.H., P.A., A.M.N., N.L.G. processed AspECT/ChOPIN samples. The AspECT and ChOPIN management groups (P. Sasieni, A.T. Tucker, P.B., D.J., M.A., C.B., J.H., D.F., B. Rathbone, J.Brown, S.L., S.A., P.W., S.Sanders, R.F.H., P.Moayyedi, J. deCaestecker, H.B., J.A.Z.J.) monitored the appropriate use of samples and data from these studies. A. van der Winkel, N.T., M.P.Peppelenbosch, L.J.W.L., E.J.P, J.P.H.D, W.H.P, J.V.R., D.A.K., R.M., H.G., H.P., R.B., K. Krishnadath, P.D. Siersema, J.W.P.M.B., M.M., R. Petty, R.G. and S.C. Cooper provided samples as part of the EAGLE consortium. The BEACON consortium (D.C.W, D.L., W.Y., A.H. Wu, N.C. Bird, N.J.S, L.J.M, L.B., Y.M., L.J.H., R.Z., D.A.C., H.A.R., B.J. Reid, M.D. Gammon, G.L., A.G. Casson, W.H.C., S.M., W.E. and T.L.V.) provided data on the two lead SNPs for the second replication phase. G.T. and C.W. provided Dutch control samples for the first replication phase. The WTCCC2 DNA, genotyping, data quality control and informatics group (S.D., S.E.H., S.E., E.G., C.L., S.C.P., A.T-G. and L.P.) executed GWAS sample handling, genotyping and quality control. The WTCCC2 data and analysis group (Z.S., A.S., C.C.A.S., G.B., C.B., C.F., M.P. and P. Donnelly) led the statistical analyses. C.P., E.S., F.L., P. Sasieni and J.B.C. also undertook statistical analyses. A.S., C.P., I.T., J.A.Z.J, C.C.A.S. and P. Donnelly contributed to writing the manuscript. The WTCCC2 management committee (J.M.B., E.B., M.A.B., J.P.C., A.C., P. Deloukas, P. Donnelly (chairperson), A. Duncanson, J.A.Z.J., H.S.M., C.G.M., C.N.A.P., L.P., R.P., A.R., S.J.S., R.C.T., A.C.V. and N.W.) monitored the execution of the GWAS. All authors reviewed the final manuscript.
URLs SNPTEST. http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html
IMPUTE2. mathgen.stats.ox.ac.uk/impute/impute_v2.html
1000 Genomes. www.1000genomes.org/
ENCODE. http://genome.ucsc.edu/encode/
Conflicts of interest: JAZJ consultant to Astrazeneca and Chief Investigator of AspECT and ChOPIN trial
References
- 1.Ronkainen J, et al. Prevalence of Barrett’s esophagus in the general population: an endoscopic study. Gastroenterology. 2005;129:1825–31. doi: 10.1053/j.gastro.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 2.Jankowski JA, Harrison RF, Perry I, Balkwill F, Tselepis C. Barrett’s metaplasia. Lancet. 2000;356:2079–85. doi: 10.1016/S0140-6736(00)03411-5. [DOI] [PubMed] [Google Scholar]
- 3.Kulig M, et al. Risk factors of gastroesophageal reflux disease: methodology and first epidemiological results of the ProGERD study. J Clin Epidemiol. 2004;57:580–9. doi: 10.1016/j.jclinepi.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Koek GH, Sifrim D, Lerut T, Janssens J, Tack J. Multivariate analysis of the association of acid and duodeno-gastro-oesophageal reflux exposure with the presence of oesophagitis, the severity of oesophagitis and Barrett’s oesophagus. Gut. 2008;57:1056–64. doi: 10.1136/gut.2006.119206. [DOI] [PubMed] [Google Scholar]
- 5.Sikkema M, de Jonge PJ, Steyerberg EW, Kuipers EJ. Risk of esophageal adenocarcinoma and mortality in patients with Barrett’s esophagus: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2010;8:235–44. doi: 10.1016/j.cgh.2009.10.010. quiz e32. [DOI] [PubMed] [Google Scholar]
- 6.Jankowski JA, Provenzale D, Moayyedi P. Esophageal adenocarcinoma arising from Barrett’s metaplasia has regional variations in the west. Gastroenterology. 2002;122:588–90. doi: 10.1053/gast.2002.31599. [DOI] [PubMed] [Google Scholar]
- 7.Thomas T, Abrams KR, De Caestecker JS, Robinson RJ. Meta analysis: Cancer risk in Barrett’s oesophagus. Aliment. Pharmacol. Therapy. 2007;26:1465–1477. doi: 10.1111/j.1365-2036.2007.03528.x. [DOI] [PubMed] [Google Scholar]
- 8.Corley DA, Buffler PA. Oesophageal and gastric cardia adenocarcinomas: analysis of regional variation using the Cancer Incidence in Five Continents database. International Journal of Epidemiology. 2001;30:1415–25. doi: 10.1093/ije/30.6.1415. [DOI] [PubMed] [Google Scholar]
- 9.Jankowski J, Barr H, Wang K, Delaney B. Diagnosis and management of Barrett’s oesophagus. BMJ. 2010;341:c4551. doi: 10.1136/bmj.c4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allum WH, Stenning SP, Bancewicz J, Clark PI, Langley RE. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol. 2009;27:5062–7. doi: 10.1200/JCO.2009.22.2083. [DOI] [PubMed] [Google Scholar]
- 11.Winberg H, Lindblad M, Lagergren J, Dahlstrand H. Risk factors and chemoprevention in Barrett’s esophagus - an update. Scand J Gastroenterology. 2012;47:397–406. doi: 10.3109/00365521.2012.667145. [DOI] [PubMed] [Google Scholar]
- 12.Chak A, et al. Familial aggregation of Barrett’s oesophagus, oesophageal adenocarcinoma, and oesophagogastric junctional adenocarcinoma in Caucasian adults. Gut. 2002;51:323–8. doi: 10.1136/gut.51.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drovdlic CM, et al. Demographic and phenotypic features of 70 families segregating Barrett’s oesophagus and oesophageal adenocarcinoma. J Med Genet. 2003;40:651–6. doi: 10.1136/jmg.40.9.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chak A, et al. Familiality in Barrett’s esophagus, adenocarcinoma of the esophagus, and adenocarcinoma of the gastroesophageal junction. Cancer Epidemiol Biomarkers Prev. 2006;15:1668–73. doi: 10.1158/1055-9965.EPI-06-0293. [DOI] [PubMed] [Google Scholar]
- 15.Trudgill NJ, Kapur KC, Riley SA. Familial clustering of reflux symptoms. Am J Gastroenterol. 1999;94:1172–8. doi: 10.1111/j.1572-0241.1999.01060.x. [DOI] [PubMed] [Google Scholar]
- 16.Mohammed I, Cherkas LF, Riley SA, Spector TD, Trudgill NJ. Genetic influences in gastro-oesophageal reflux disease: a twin study. Gut. 2003;52:1085–9. doi: 10.1136/gut.52.8.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson EV, Jankowski JA. Genetics of gastroesophageal cancer: paradigms, paradoxes, and prognostic utility. Am J Gastroenterol. 2008;103:443–9. doi: 10.1111/j.1572-0241.2007.01574.x. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, et al. A segregation analysis of Barrett’s esophagus and associated adenocarcinomas. Cancer Epidemiol Biomarkers Prev. 2010;19:666–74. doi: 10.1158/1055-9965.EPI-09-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett C, et al. Consensus statements for management of Barrett’s Dysplasia and early-stage esophageal adenocarcinoma, based on a Delphi process. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.04.032. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jankowski J, Barr H. Improving surveillance for Barrett’s oesophagus: AspECT and BOSS trials provide an evidence base. BMJ. 2006;332:1512. doi: 10.1136/bmj.332.7556.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 23.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winney B, et al. People of the British Isles: preliminary analysis of genotypes and surnames in a UK control population. European Journal of Human Genetics. 2012;20:203–210. doi: 10.1038/ejhg.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin V, Shaw-Smith C. Review of genetic factors in intestinal malrotation. Pediatr Surg Int. 2010;26:769–81. doi: 10.1007/s00383-010-2622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Z, Cardin N, WTCCC, Donnelly P, Marchini J. A Bayesian method for detecting and characterizing allelic heterogeneity and boosting signals in genome-wide association studies. Statistical Science. 2009;24:430–450. [Google Scholar]
- 27.Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP--an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–72. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shalauta MD, Saad R. Barrett’s esophagus. Am Fam Physician. 2004;69:2113–8. [PubMed] [Google Scholar]
- 30.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akiyama T, et al. Visceral obesity and the risk of Barrett’s esophagus. Digestion. 2011;83:142–5. doi: 10.1159/000321810. [DOI] [PubMed] [Google Scholar]
- 33.Lagergren J. Influence of obesity on the risk of esophageal disorders. Nat Rev Gastroenterol Hepatol. 2011;8:340–7. doi: 10.1038/nrgastro.2011.73. [DOI] [PubMed] [Google Scholar]
- 34.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindgren CM, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heid IM, et al. Meta-analysis of the INSIG2 association with obesity including 74,345 individuals: does heterogeneity of estimates relate to study design? PLoS Genet. 2009;5:e1000694. doi: 10.1371/journal.pgen.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heard-Costa NL, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moayyedi P, et al. Mortality rates in patients with Barrett’s oesophagus. Aliment Pharmacol Ther. 2008;27:316–20. doi: 10.1111/j.1365-2036.2007.03582.x. [DOI] [PubMed] [Google Scholar]
- 40.Sharma P, et al. The development and validation of an endoscopic grading system for Barrett’s esophagus: the Prague C & M criteria. Gastroenterology. 2006;131:1392–9. doi: 10.1053/j.gastro.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 41.Watson A, Heading RC, Shepherd NA. Guidelines for the diagnosis and management of Barrett’s columnar-lined oesophagus. A Report of the Working Party of the British Society of Gastroenterology. 2005 [Google Scholar]
- 42.Sampliner RE. Updated guidelines for the diagnosis, surveillance, and therapy of Barrett’s esophagus. Am J Gastroenterol. 2002;97:1888–95. doi: 10.1111/j.1572-0241.2002.05910.x. [DOI] [PubMed] [Google Scholar]
- 43.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houlston RS, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13 33. Nat Genet. 2010;42:973–7. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spencer CC, et al. Dissection of the genetics of Parkinson’s disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet. 2011;20:345–53. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teo YY, et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics. 2007;23:2741–6. doi: 10.1093/bioinformatics/btm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.