Abstract
16S rRNA gene (rrs) is considered of low taxonomic interest in the genus Aeromonas. Here, 195 Aeromonas strains belonging to populations structured by multilocus phylogeny were studied using an original approach that considered Ribosomal Multi-Operon Diversity. This approach associated pulsed-field gel electrophoresis (PFGE) to assess rrn operon number and distribution across the chromosome and PCR-temporal temperature gel electrophoresis (TTGE) to assess rrs V3 region heterogeneity. Aeromonads harbored 8 to 11 rrn operons, 10 operons being observed in more than 92% of the strains. Intraspecific variability was low or nul except for A. salmonicida and A. aquariorum suggesting that large chromosomic rearrangements might occur in these two species while being extremely rarely encountered in the evolution of other taxa. rrn operon number at 8 as well as PFGE patterns were shown valuable for taxonomic purpose allowing resolution of species complexes. PCR-TTGE revealed a high rate of strains (41.5%) displaying intragenomic rrs heterogeneity. Strains isolated from human samples more frequently displayed intragenomic heterogeneity than strains recovered from non-human and environmental specimens. Intraspecific variability ranged from 0 to 76.5% of the strains. The observation of species-specific TTGE bands, the recovery of identical V3 regions in different species and the variability of intragenomic heterogeneity (1–13 divergent nucleotides) supported the occurrence of mutations and horizontal transfer in aeromonad rrs evolution. Altogether, the presence of a high number of rrn operon, the high proportion of strains harboring divergent rrs V3 region and the previously demonstrated high level of genetic diversity argued in favor of highly adaptative capabilities of aeromonads. Outstanding features observed for A. caviae supported the ongoing process of adaptation to a specialized niche represented by the gut, previously hypothesized. 16S rRNA gene is an informative marker in the genus Aeromonas for both evolutionary and polyphasic taxonomic studies provided that multi-operon fingerprinting approaches are used.
Introduction
The genus Aeromonas groups waterborne gram-negative bacilli considered as opportunistic pathogens for a wide range of animals including humans. Currently, 25 species are recognized in the genus. 16S rRNA (rrs) gene is considered as a non-informative marker in the genus because most species shared more than 99% of their nucleotide positions [1]–[4]. Considering this low polymorphism, several attempts have been made to identify species by 16S rRNA gene-restriction fragment length polymorphism (RFLP) [5], [6] but atypical patterns [7], [8] that were later explained by sequence heterogeneities among 16S rRNA gene copies in a same genome [9], [10] impaired this approach. Due to these limits, 16S rRNA gene sequence was subsequently scarcely studied in the genus Aeromonas and multilocus sequence analysis is now the reference approach to structure populations and describe new species in the genus [2]–[4], [11]–[13].
Beside the sequence polymorphism, ribosomal operons display diversity in terms of copy number and copy heterogeneity in bacterial genomes [14], [15]. The genus Aeromonas is a valuable example that illustrates such diversity. Eight to ten copies of the 16S rRNA genes have been reported in totally sequenced Aeromonas genomes [16]–[19] and significant intragenomic microheterogeneities have been suggested using different approaches [9], [10], [20].
Here we evaluated the “Ribosomal Multi-Operon Diversity” (RiMOD), i.e., the diversity of the chromosomal rrn copy organisation and heterogenity, towards taxonomy and evolution of aeromonads. For this purpose, we used pulsed-field gel electrophoresis to determine rrn operon number and distribution across the chromosome and PCR-temporal temperature gel electrophoresis (TTGE) to assess rrs heterogeneity in a population of Aeromonas field isolates and reference strains from diverse origins previously structured by multilocus phylogenetic analysis (MLPA) [13]. We reported the features of rrs heterogeneity at the intrapopulation, intraspecific and intragenomic levels, and the variability of rrn number and chromosomal distribution at both inter- and intra-specific levels. Our approach showed that ribosomal diversity when studied at the multi-operon family level is an informative marker regarding evolution and taxonomy in the genus Aeromonas. In addition, we showed that RiMOD study results supported the structure in species complexes previously suggested for the genus Aeromonas on the basis of other genetic and genomic traits [13] and more generally related to “generalist” bacterial pathogens with sympatric lifestyles [21].
Materials and Methods
Bacterial Strains
Strain collection was as described in Roger et al. [13]. Briefly, a total of 195 strains of Aeromonas spp., including 62 type and reference strains were analyzed. Origin distribution of these strains reflected the different bacterial lifestyles in the genus with 115, 39 and 41 isolates recovered from human clinical samples, non-human animal samples and environmental samples, respectively. The strain collection included type strains of 25 species and a representative strain of hybridization group (HG) 11. Using MLPA, the population was distributed in eleven species corresponding to Aeromonas allosaccharophila (n = 3), Aeromonas aquariorum (n = 8), Aeromonas caviae (n = 34), Aeromonas hydrophila (n = 35), Aeromonas jandaei (n = 2), Aeromonas media (n = 6), Aeromonas piscicola (n = 3), Aeromonas salmonicida (n = 8), Aeromonas sobria (n = 5), Aeromonas tecta (n = 3), Aeromonas veronii (n = 71). Fourteen species were represented by their type strain only: Aeromonas bestiarum, Aeromonas bivalvium, Aeromonas diversa, Aeromonas encheleia, Aeromonas enteropelogenes/trota, Aeromonas eucrenophila, Aeromonas fluvialis, Aeromonas molluscorum, Aeromonas popoffii, Aeromonas sanarellii, Aeromonas simiae, Aeromonas schubertii, Aeromonas taiwanensis and Aeromonas rivuli. Finally, the strain CCM 1271 was representative of an unknown taxon (Table S1).
In silico Analysis
Complete genome sequences of strains A. hydrophila subsp. hydrophila ATCC 7966T (GenBank accession number NC_008570), A. salmonicida subsp. salmonicida A449 (NC_009348), and A. veronii B565 (NC_015424) were analyzed in silico for rrs copy number, rrs position determination and rrs sequence heterogeneity using Biological sequence aligment editor (BioEdit) programme version 7.0.9 [22] (http://www.mbio.ncsu.edu/bioedit/bioedit.html).
PFGE
Genomic DNA was prepared in agarose plugs as previously described by Miranda et al. [23] and adapted by Roger et al. [13]. DNA was digested at 37°C with 1 U of the intronic endonuclease I-CeuI (New England BioLabs, Hertfordshire, United Kingdom) according to the manufacturer’s recommendations. I-CeuI fragments were resolved using a CHEF-DRII apparatus (Bio-Rad Laboratories, Hercules, CA) in a 0.8% agarose gel in 0.5X Tris-Borate-EDTA (TBE) buffer added with 50 µM of thiourea at 4.5 V/cm and at 10°C. Pulse ramps were 60 to 10 s for 35 h. The gel was stained with ethidium bromide and photographed under UV light. Number of rrn copies was deduced from the number of I-CeuI-generated fragments. Sizes determined in this study were measured by comparison with lambda concatemer (New England BioLabs) used as size standard.
PCR-TTGE
Amplification by PCR of a 199 bp-fragment overlapping the 16S rDNA variable region V3 and TTGE were performed as described previously [24]. Bands were cut out of the gel, DNA was amplified using HDA1 and HDA2 primers as previously described and PCR products were sequenced on an Applied Biosystems automatic sequencer (Cogenics) by using the forward primer HDA1 [24]. Bands for which no interpretable sequences were obtained were subjected to a second run of PCR-TTGE followed by a re-sequencing as described above. TTGE bands were visually detected and analyzed by migration distance measuring after standardization. Each band with distinct distance of migration was designated by a number and TTGE profiles were indicated by a combination of band numbers separated by a + sign. A specifically designed ladder corresponding to a mix of V3 region amplification products from different strains showing PCR-TTGE pattern 1+4+15+30 was loaded on all the TTGE gels. The presence of more than one TTGE band, i.e., 16S rRNA gene V3 region copies with distinct sequences, in an isolate will be referred to hereafter as harboring intragenomic heterogeneity, while the presence of different TTGE bands in different isolates of a species will be referred to as intraspecific heterogeneity.
Statistical Analyses
Qualitative variables were compared with the Chi-square test with the Bonferroni’s correction where required; a P value ≤0.05 was considered to reflect significance. All computations were done with R project software (http://www.r-project.org).
Results
rrs Sequence Diversity and Heterogeneity
The 195 strains of the collection were studied by 16S rRNA gene PCR-TTGE. Representative patterns are shown in Figure S1. Fifty-seven PCR-TTGE patterns associating bands with 34 different migration distances were observed in the population (Table 1, Table S1). Multiband TTGE patterns, revealing rrs heterogeneity, were more frequently observed (n = 45, 78.9%) than patterns composed by a unique band (n = 12, 21.1%). Among the 195 strains, 81 (41.5%) showed heterogeneity between their rrs copies. Up to four bands were combined within a pattern; however, the 2-band patterns were the most frequently observed (n = 27, 60% of the 45 heterogeneous patterns). Patterns with 3 or 4 different bands (13 and 5 patterns, respectively) were less frequently observed. Heterogeneity frequency differed significantly between the groups of isolates recovered from human, animal and environmental samples (P value = 0.01), V3 rrs heterogeneity being observed in 51.3% of isolates from human origin (59 of the 115 strains) compared to 28.2% of non-human animal isolates (11 of the 39 strains) and 26.8% of environmental isolates (11 of the 41 strains). Difference in heterogeneity frequency between strains purchased from collections and field isolates was not significant (P value = 0.078).
Table 1. Occurrence and distribution of the 34 PCR-TTGE bands observed among the Aeromonas population.
| PCR-TTGE banda | Occurrence in the population/number of taxa with band occurence (n/n) | Number of patterns with bandb | Occurence of band according to the taxon (n)c | |||||||||||||||||||||||||||
| A. veronii (n = 71) | A. hydrophila (n = 35) | A. caviae (n = 34) | A. aquariorum (n = 8) | A. salmonicida (n = 8) | A. media (n = 6) | A. sobria (n = 5) | A. allosaccharophila (n = 3) | A. piscicola (n = 3) | A. tecta (n = 3) | A. jandaei n = (2) | A. bestarium | A. bivalvium | A. diversa | A. encheleia | A. enteropelogenes | A. eucrenophila | A. fluvialis | A. molluscorum | A. popoffii | A. sanarellii | A. rivuli | A. simiae | A. schubertii | A. taiwanensis | A. trota | HG11 | Strain CCM 1271 | |||
| 1 | 42/4 | 15 | – | 35 | 5 | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | 1 | – | – | – | – | – | – | – | – | – |
| 2 | 6/4 | 2 | – | 3 | – | 1 | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – |
| 3 | 32/6 | 14 | 2 | – | 19 | 8 | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | 1 | – | – | – | 1 | – | – | – |
| 4 | 47/7 | 18 | 7 | 1 | 34 | 1 | – | – | – | – | – | – | 2 | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – | 1 | – | – |
| 5 | 3/1 | 1 | – | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 6 | 1/1 | 1 | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 8 | 2/2 | 2 | – | 1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 9 | 1/1 | 1 | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 10 | 2/1 | 2 | – | 2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 11 | 78/5 | 11 | 70 | – | 5 | – | – | 1 | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – |
| 12 | 2/1 | 2 | – | – | 2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 13 | 4/2 | 1 | 1 | – | – | – | – | – | – | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 14 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – | |
| 15 | 21/8 | 10 | 8 | – | 3 | – | – | 3 | – | – | – | 3 | – | – | – | – | – | – | 1 | – | – | 1 | – | – | – | – | – | 1 | 1 | – |
| 16 | 1/1 | 1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 17 | 3/1 | 1 | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 18 | 10/3 | 7 | – | 1 | 8 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – |
| 19 | 1/1 | 1 | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 20 | 3/1 | 3 | – | – | – | – | – | 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 21 | 4/1 | 4 | – | – | – | – | – | 4 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 24 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – |
| 25 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – |
| 27 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 29 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – |
| 30 | 20/8 | 4 | – | 4 | – | – | 8 | – | – | – | 3 | – | – | 1 | – | – | 1 | – | – | 1 | – | – | – | – | – | – | – | – | 1 | 1 |
| 31 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – |
| 32 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – |
| 33 | 1/1 | 1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 35 | 2/2 | 2 | – | 1 | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 36 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – |
| 37 | 1/1 | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 1 | – | – | – | – |
| 38 | 5/1 | 1 | – | – | – | – | – | – | 5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 39 | 1/1 | 1 | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 40 | 1/1 | 1 | – | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Number of pattern | 8 | 11 | 17 | 4 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| Number of heterogeneous pattern | 6 | 10 | 16 | 3 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | ||
| Number of strains with rrs V3 region heterogeneity | 22 | 17 | 26 | 3 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | ||
Bold type indicates bands which were successfully sequenced (GenBank accession numbers JX014439-JX014453 and JX453432-JX453445).
Bold type indicates that all patterns with the corresponding band showed heterogeneity.
Bold and italics, bands only found in one taxon; bold and underlined, bands found in all representatives of a taxon for taxa including more than one strain (bold, italics and underlined combined for band 38 in A. sobria).
Among the 34 TTGE bands with visually distinct migration distances observed for the overall population, 29 were confirmed as distinct by sequencing. For 15 of these bands, sequences were obtained after the first run of PCR-TTGE (GenBank accession numbers JX014439-JX014453) while for the 14 other bands a second run of PCR-TTGE was required (Genbank accession number JX453432-JX453445). Sequences could not be obtained for the 5 remaining bands because sequencings were either unrealizable due to very faint bands or resulted in uninterpretable bulk sequences despite the second run of PCR-TTGE. These 5 bands were observed in 8 strains belonging to 4 taxa (Table 1). The 49 patterns with full band sequencing displayed heterogeneity between V3 rrs copies ranging from 1 to 13 nucleotides (151 nt-alignment) (Table S1). The diversity of bands and profiles surpassed the number of taxa identified by MLPA in the collection, revealing variation of V3 rrs content in a same species.
When compared with multilocus phylotaxonomy, 23 of the 34 PCR-TTGE bands were specifically associated with all or some strains of a species while absent from all other taxa, indicating that some 16S rRNA gene copies can discriminate among Aeromonas species (Table 1 and Figure 1). The remaining 11 bands were shared by patterns of strains belonging to up to 8 taxa (e.g., bands 15 and 30) showing that strains from different species may harbor identical V3 region rrs copies. Diversity of patterns observed between strains belonging to a given species was mostly related to intragenomic V3 rrs copy heterogeneity. Among species represented by more than one strain, A. caviae and A. media showed the highest proportion of strains with heterogeneous TTGE patterns (76.5% and 66.7%, respectively), followed by A. hydrophila, A. aquariorum and A. veronii (48.6%, 37.5% and 29.6%, respectively) while none of the A. salmonicida, A. sobria, A. allosaccharophila, A. piscicola, A. tecta, and A. jandaei displayed rrs sequence heterogeneity (Table 1). The type strains of 7 other species displayed rrs V3 region sequence heterogeneity: A. encheleia, A. enteropelogenes/trota, A. fluvialis, A. molluscorum, A. popoffii, A. sanarellii and A. taiwanensis while no heterogeneity was found for the 7 remaining species A. bestiarum, A. bivalvium, A. diversa, A. eucrenophila, A. rivuli, A. simiae, and A. schubertii, as well as for the strains HG11 CECT 4253 and CCM 1271 (Table 1).
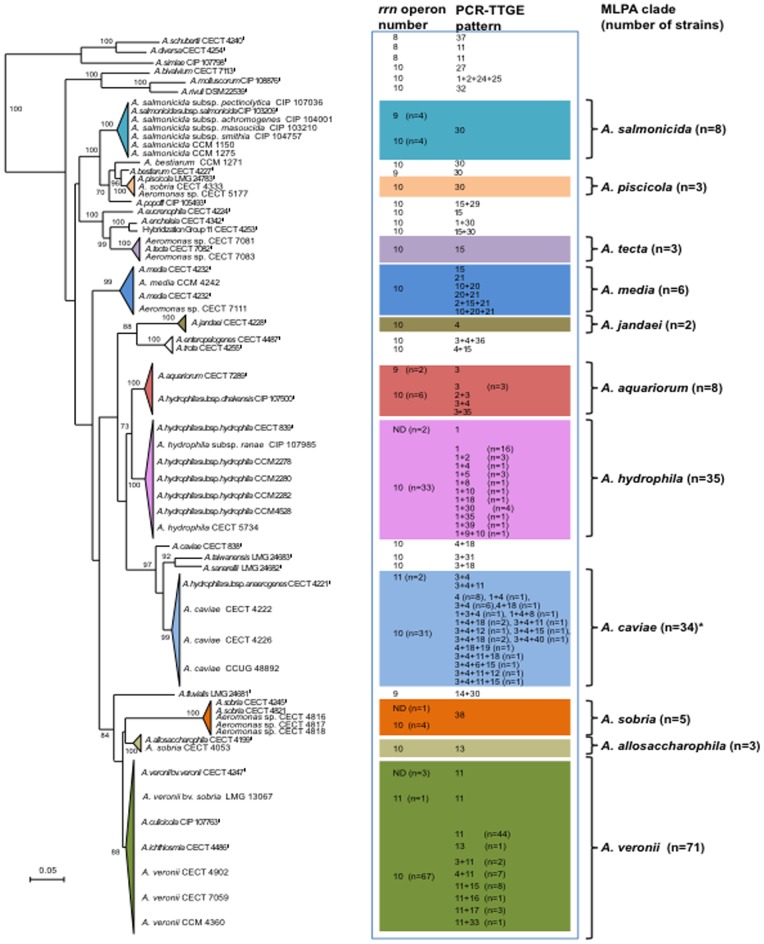
Figure 1. Number of rrn operons and PCR-TTGE patterns for the 195 Aeromonas strains of the study.
Results are presented according to the structure of the population observed in multilocus phylogenetic analysis (MLPA). Unrooted maximum-likelihood tree was based on concatenated sequences of five housekeeping genes (gltA, gyrB, rpoB, tsf and zipA genes), as described in Roger et al. [13]. Only type and references strains are indicated within clades on the tree. The horizontal lines show genetic distance, the scale bar indicating the number of substitutions per nucleotide position. The numbers at the nodes are support values estimated with 100 bootstrap replicates. Only bootstrap values >70 are indicated on the tree. When all members of a clade shared identical features, the common rrn operon number and/or PCR-TTGE pattern was indicated. * The A. caviae clade included the type strain A. caviae CECT 838T displaying an external position to other members of the A. caviae clade in the tree.
Regarding the 3 most represented species in our population, all A. hydrophila isolates harbored the band 1 that was associated or not with 10 different other bands (Table 1). Similarly, all A. caviae isolates shared the band 4 associated or not with 10 other bands. For A. veronii, band 11 was present in all the strains but one (strain AK241), associated or not with 6 other bands. A. caviae displayed the highest level of profile diversity, 17 patterns being observed among the 34 strains (0.5 pattern per strain) compared to A. hydrophila (11 patterns, 0.31 pattern per strain) and A. veronii (8 patterns, 0.11 pattern per strain) (Table 1). Heterogeneity frequency differed significantly between the three main species (P value = 0.00008), particularly when A. caviae was compared to A. veronii (P value = 0.000078) while difference was slightly significant between A. hydrophila and A. veronii (P value = 0.046). In spite of the low number of strains studied, it is noteworthy that none of the 6 isolates belonging to A. media shared an identical pattern (Table 1, Table S1) and that the 8 isolates of A. aquariorum showed a diversity of TTGE patterns similar to that of A. caviae (0.5 pattern per strain).
In addition to the high rate of A. caviae isolates with heterogeneous rrs copies, we observed among strains displaying V3 region rrs heterogeneity that TTGE profiles formed of 3 or 4 bands were significantly more frequent in A. caviae (16 out of 21 strains) than in other taxa (P value = 0.00048). These patterns were particularly found in A. caviae isolated from human clinical samples (12 of the 16 strains), mainly from gut-related specimens (10 out of 12 strains). However, strains recovered from other origins were underrepresented among the A. caviae isolates of our collection (10 strains from non-human and environmental samples, and 6 strains from human specimens not related to the gut) (Table S1). Identical complex PCR-TTGE patterns, e.g., 1+4+18 for A. caviae strains BVH63 and BVH84 and 3+4+11 for A. caviae strains BVH19 and BVH4 were observed in strains isolated from geographically distant French regions. Another outstanding example is represented by the pattern 3+4+18 observed for the two environmental A. caviae strains CCUG 48892 isolated from water in Sweden in 2004 and AK234 recovered from wastewater treatment lagoon in France in 2006. Beside A. caviae members, the strains displaying 3- or 4-band TTGE patterns were A. molluscorum and A. enteropelogenes type strains, one A. hydrophila strain from human wound and two A. media strains from snail and oyster (Table S1).
High rrn Copy Number with Low Structural Variability in Aeromonads
PFGE patterns were obtained for 189 strains giving for the first time a whole perspective of rrn copy number and distribution across the chromosome for about 20 not yet investigated Aeromonas species, as well as intrapecific variability data. PFGE patterns obtained for all the type and reference strains of this study are shown in Figure S2. The population displayed 8 to 11 I-CeuI-restricted DNA fragments corresponding to 8 to 11 rrn operons. However, the variability in the whole genus was low, more than 92% of the strains displaying 10 rrn operons (n = 175). Species and/or strains harboring 8 (n = 3), 9 (n = 8) or 11 (n = 3) rrn operons were rare and no particular feature could be identified for these bacteria being either recovered from human (n = 5), non-human (n = 6) or environmental (n = 3) samples (Table S1). Intraspecific variability in rrn operon number was observed for A. salmonicida, A. aquariorum, A. caviae and A. veronii strains (Figure 1, Table S1) but the most notable variability was observed for A. salmonicida and A. aquariorum with 50% and 25% of the strains harboring 9 rrn operons, respectively. Finally, no correlation could be drawn here between rrn operon number and rrs V3 region heterogeneity due to the low number of strains harboring 8, 9 or 11 operons.
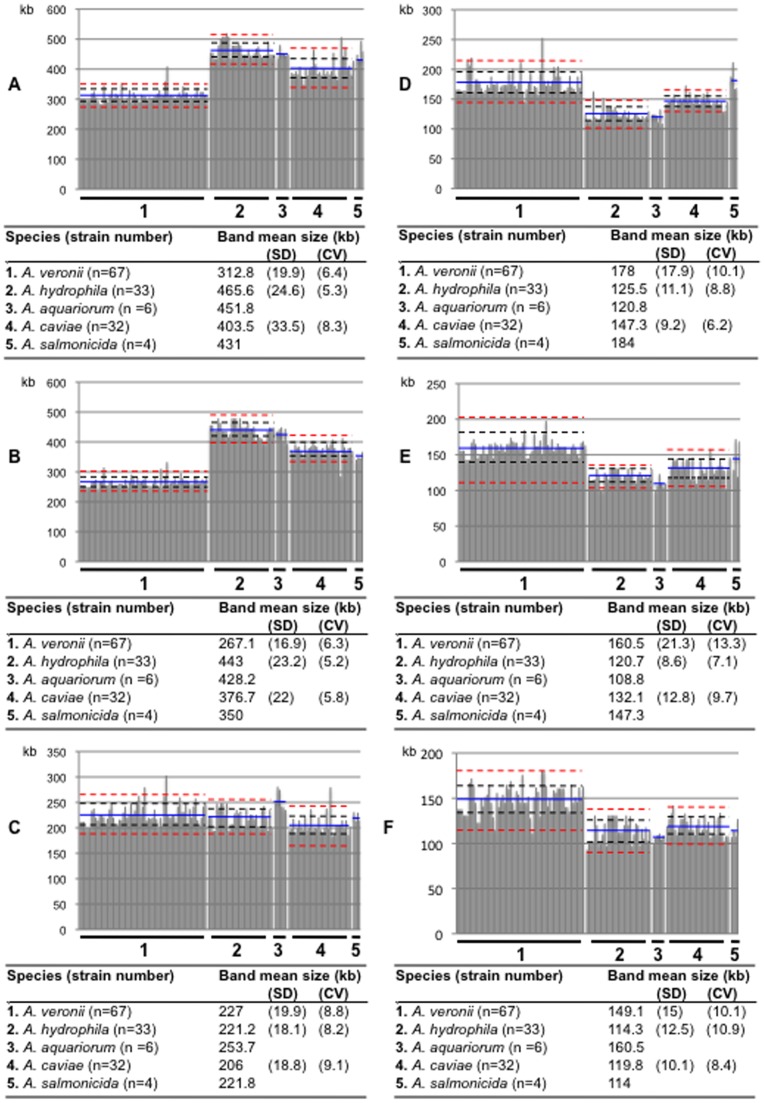
Beside variability in copy number, variability in size of I-CeuI fragments, i.e., variability in location of rrn on the chromosome, was also observed in the population. Variability in the size of the largest I-CeuI fragment could not be precisely measured using the migration conditions applied herein. Considering the 3 complete genome sequences, the largest fragment size variation was 2950±70 kb (2.4%) (3019 kb for the A. hydrophila, 2885 kb for the A. salmonicida and 3001 kb for the A. veronii strains), a value in agreement with sizes previously evaluated to 2900 kb for A. hydrophila strain JMP636 [25] using PFGE-based experiment. Based on this low variability in size, we assumed that excluding this fragment from pattern comparison would not greatly affect both intra- and interspecific diversity analysis. Despite this limitation, we showed that most Aeromonas species had a quite specific I-CeuI fingerprint with exception of A. aquariorum, which displayed a roughly similar pattern to the one observed for A. hydrophila (Figure 2, Figure S2, Table S1). Another notable exception was A. media that displayed highly diverse rrn chromosomal repartition (Figure S2, Table S1). We also evaluated the variability of rrn chromosomal distribution at the species level for the main species represented by a large number of strains, A. hydrophila, A. caviae and A. veronii (Figure 2). For the 6 genomic fragments analyzed, the coefficient of variation of the mean size varied from 5.2% to 13.3%. The fragment with the most variable size varied according to the species. A. veronii was the most variable species for the size of fragments B, D and E, A. caviae for fragments A and C, and A. hydrophila for fragment F. Regardless of the species, A and B fragments appeared the most stable in size. Despite this intraspecific variability, the size of most genomic fragments enabled to distinguish a species from others (Figure 2).
Figure 2. Schematic representation of I-CeuI-restricted DNA fragment sizes for Aeromonas spp. strains with 10 rrn operons.
Data were given for the 142 A. veronii, A. hydrophila, A. aquariorum, A. caviae and A. salmonicida strains with 10 rrn operons with the aim to illustrate interspecific variability between the 5 taxa and the intraspecific variability for the 3 main species. I-CeuI-restricted DNA fragments were numbered in size descending order; the size of the largest fragment 1 was not measured (as discussed in the text). A to F, size distribution for fragments 2 to 7, respectively. Fragment mean sizes (in kilobases) were indicated in the corresponding tables according to the species and by a blue line in the corresponding scheme A to F. Standard deviation values (SD) and coefficient of variation (CV) were indicated in kb and %, respectively, in the corresponding tables according to the species except for species with less than 10 representatives; +/−1 SD were represented by black dotted lines and +/−2 SD by red dotted lines in the corresponding scheme A to F. Schematic representations for the 3 smallest fragments with mean size lower than 97 kb (mean size and SD of 93.7±23 kb, 66.7±9.1 kb and 40.5±7.5 kb, respectively) were not presented here because they were not informative.
RiMOD Fits the Taxonomy of Aeromonads
RiMOD has been confronted to the current taxonomy of the genus Aeromonas including recent taxonomic changes. Firstly, several recently characterized species such as A. taiwanensis, A. sanarellii and A. fluvialis [2], [3] displayed clearly distinct features in both PFGE and TTGE-based analyses (Table S1, Figure 1, Figure S2). The proposal of A. diversa including Aeromonas sp. HG13, so-called Aeromonas group 501, as a distinct species from A. schubertii [26] was supported in PCR-TTGE by different rrs V3 regions. These 2 species displayed 8 rrn operons, as observed for the MLPA-related species A. simiae (Figure 1), but their PFGE fingerprints were clearly individualized (Table S1, Figure S2). Moreover, the suggestion that A. hydrophila subsp. anaerogenes and A. caviae are conspecific [27] was supported by the RiMOD results, A. hydrophila subsp. anaerogenes strain CECT 4221 displaying a rrn operon PFGE fingerprint related to those observed in members of the A. caviae clade and the 3+4 PCR-TTGE pattern never observed in the A. hydrophila clade members (Figure 1, Figure S2).
Several taxonomic reappraisals were supported by one method of the RiMOD approach. In particular, we distinguished A. hydrophila from A. aquariorum, a species proposed in 2008 on the basis of gyrB and rpoD gene phylogeny to accomodate members of the subspecies A. hydrophila subsp. dhakensis [28]. Although PFGE fingerprinting did not distinguish A. aquariorum from A. hydrophila, PCR-TTGE clearly showed that rrs V3 region was discriminative between the 2 species, band 3 being found in all A. aquariorum isolates while absent from all the A. hydrophila strains (Table 1, Figures 1 and 2, Table S1, Figure S2).
Additional arguments were also given here on controversial issues recently reviewed by Janda & Abbott [29]. A. ichthiosmia and A. culicicola considered as later synonyms of A. veronii [30] shared similar PFGE patterns and displayed PCR-TTGE patterns mainly (pattern 11) or exclusively (pattern 4+11) observed in the A. veronii clade. (Table S1, Figure S2). Previous suggestions to unify the two pairs of taxa A. enteropelogenes and A. trota on one hand, and A. encheleia and HG11 on the other hand could hardly be addressed and discussed here because one strain only for each taxon was included in the study. We noted that A. enteropelogenes and A. trota displayed closely related rrn operon PFGE fingerprints and specific TTGE patterns, i.e., 3+4+36 and 4+15, respectively, not shared by any other strain of the collection despite all bands forming these patterns with the exception of the band 36 were present in other species. Similarly, A. encheleia and strain representative of HG11 respectively displayed TTGE patterns 1+30 and 15+30 formed by bands found in other taxa but showed distinct rrn operon PFGE patterns (Table 1, Figure S2). For A. allosaccharophila, our results supported the unification of the species with A. veronii as suggested by DNA-DNA hybridization experiment and contrarily to MLPA results because strains shared similar rrn operon PFGE patterns and harbored main TTGE bands differing by a unique variable position (band 13 in the 3 A. allosaccharophila strains and band 11 found in 70 out of the 71 A. veronii isolates of the study) (Table 1, Figure 1). Of note, the only A. veronii isolate lacking band 11 had a PCR-TTGE pattern 13. Although the number of A. allosaccharophila strains studied in this work was too low to be conclusive, these observations together with previous results leading to controversy on the taxonomic status of this species [29] may suggest that A. allosaccharophila is at the starting time of speciation.
In addition, previous MLPA results suggested that the species A. media could be polyphyletic containing yet-undescribed taxa [13] and that strain CCM 1271 probably represent an unknown Aeromonas taxon [11], [13]. Here, we confirmed the heterogeneity of genetic and genomic features of the 6 studied A. media strains (Figure 1, Table S1, Figure S2) and showed that strain CCM 1271, referenced in the Czech Collection of Microorganisms as A. bestiarum, shared the TTGE pattern 30 with A. bestiarum, A. salmonicida and A. piscicola (Table 1) but presented a PFGE fingerprint distinct from those observed for these 3 species by the number and/or the chromosomal repartition of rrn operons (data not shown).
Results obtained for A. sharmana, a species further shown not to belong to the genus Aeromonas [31], [32] confirmed its distant relationship to members of the studied population by showing a specific and unique PCR-TTGE band (data not shown) and a rrn skeleton not clearly resolved in the conditions appropriate to the study of Aeromonas spp. but confidently formed by 6 or 7 operons (Figure S2).
Discussion
Striking Diversity in Ribosomal Genes of Aeromonads
We described an outstanding diversity in ribosomal genes in a representative population of aeromonads. This result is paradoxal considering that the majority of species in the genus shared more than 99% of their nucleotide positions when 16S rRNA gene is directly sequenced from whole genomic DNA. The diversity described herein concerns both the number and the polymorphism of rrs copies and is therefore underestimated when the whole 16S rRNA gene sequence that reflects the mix of sequences from a multigenic family is studied. Indeed, we confirmed on a large panel of strains the high rrn copy number in Aeromonas spp. previously published, i.e., 9 operons in most A. salmonicida [16], [18] and 10 operons in A. hydrophila subsp. hydrophila strain ATCC 7966T [17], A. culicicola strain MTCC 3249T [33] and A. veronii strain B565 [19].
The rrn operons in aeromonads are to be considered as a multi-operon family given the number of operon copies and their diversity. As shown in a previous study, we confirmed that PCR-TTGE is a powerful approach to study the rate of bacterial isolates with rrs heterogeneity in large natural populations [24]. In this previous study conducted on Veillonella spp. that displayed 4 rrn copies, all the bands observed in TTGE were successfully sequenced after a single run of PCR-TTGE [24]. For aeromonads, interpretable sequences were obtained for 44% of the bands only after a single run of PCR-TTGE and 15% of the bands remained unsequenced after a double run. We assumed that this was related to the high number of rrn operons in aeromonads leading to band cross-contamination during the electrophoresis step and thereby constituted a limitation of the method when applied to bacteria harboring a high number of rrn operons. Sequences obtained for 29 TTGE bands showed up to 13 variable positions between rrs V3 region copies in a genome, a number exceeding the 1 to 10 microheterogeneities (0.06 to 0.66% of divergence) observed through chromatograms of the nearly complete 16S rRNA gene sequences by Alperi et al. [10] and the number of divergent nucleotides (2 to 5) observed between rrs copies for the 3 strains with complete genome sequences.
V3 rrs region analysis revealed an unexpectedly high rate of Aeromonas strains displaying rrs intragenomic heterogeneity compared with previously published studies based on other methodological approaches [9], [10]. Indeed, we showed that 41.5% of the strains included in our study displayed rrs V3 region heterogeneity while microheterogeneities were suspected from atypical RFLP patterns in only 8.1% of the 999 Aeromonas strains examined by Alperi et al. [10], and in 21% of the 82 strains studied by Morandi et al. [9]. We suspected this rate to be even higher because the PCR-TTGE approach is limited by the size of the amplified fragment that can migrate in TTGE, i.e., approximately 200 bp. Although the V3 region is one of the most variable regions among the 16S rRNA gene [34], intragenomic heterogeneity may still be underestimated when the V3 region alone is studied as shown by the in silico analysis of the three totally sequenced genomes. Indeed, rrs microheterogeneity was observed in the 3 strains, A. hydrophila subsp. hydrophila strain ATCC7966T harbored 3 different rrs copies among 10 resulting from up to 2 divergent nucleotides; A. veronii strain B565 harbored 5 different rrs copies among 10 (3 divergent nucleotides), and A. salmonicida subsp. salmonicida strain A449 showed the highest level of intragenomic heterogeneity with 5 distinct rrs copies among 9 differing by up to 5 nucleotides. Using V3 region rrs PCR-TTGE, intragenomic heterogeneity would have been observed in the A. salmonicida strain only, one of the 5 polymorphic nucleotides being located in the V3 region while overlooked in the other two strains because none of the divergent nucleotides were located in the targeted variable region. This was confirmed here by the one band PCR-TTGE pattern observed for the type strain of A. hydrophila subsp. hydrophila purchased from another strain collection. Of note, A. salmonicida complete genome sequence showed the existence of rrs heterogeneity within the V3 region for strain A449 not detected in strains included in our study.
Diverse RFLP patterns have been observed for A. veronii, A. media and A. encheleia in a study investigating 62 reference strains belonging to 13 species while other species included in the study were thought to harbor conserved 16S rRNA genes [7]. Further studies suspected or demonstrated that 16S rRNA gene intragenomic heterogeneity is encountered in a wider range of species within the genus (for a review, see [10]). We demonstrated that intragenomic rrs heterogeneities occurred in half of the taxa included in our study and firstly described the occurrence of such microheterogeneities in the recently described taxa A. fluvialis, A. sanarelli, and A. taiwanensis. We also confirmed microheterogeneities previously suspected in some taxa such as A. trota [10], [35]. The rate of isolates displaying diversity of the rrs V3 region copies varied according to the taxon considered. Among the 3 main clades, A. caviae displayed the highest number of isolates with V3 rrs heterogeneity in agreement with a previous RFLP-based study [10]. However, we found that A. media was the species displaying the highest intraspecific rrs V3 region diversity, each of the 6 strains displaying its own pattern, before A. aquariorum, A. caviae, A. hydrophila and then A. veronii, thereby differing from the distribution observed by Alperi et al. after RFLP patterns analysis [10].
RiMOD and Aeromonad Evolution
rrn distribution across the chromosome, mainly stable within and different between species, reflected evolutionary relationships between aeromonads. Among the 3 main clades, patterns observed for A. hydrophila and A. caviae were more similar while those observed for A. veronii were more distinct and this reflected the relationships observed in the MLPA-based tree. In contrast, closely related species in the MLPA-based tree that may represent more recently emerging species, such as A. allosaccharophila compared to A. veronii and A. aquariorum compared to A. hydrophila, were indistinguishable using this approach. This suggested that comparison of I-CeuI fingerprints may reflect phylogenetic relationships in the genus Aeromonas and allow recognition and/or delineation of species complexes, from which novel lineages or novel species may emerge.
It is generally admitted that intragenomic 16S rRNA gene heterogeneity is due to mutations after gene duplication or to horizontal gene transfer (HGT) [36], [37], followed by a lack of intragenomic concerted evolution [38]. In the genus Streptomyces, it has been suggested that copies differing by one or two nucleotides originate from mutations while copies differing by more than 5 nucleotide positions and found in another species more likely originate from HGT [39]. Besides, the occurrence of hybrid events in 16S rRNA gene sequences was previously demonstrated in the form of gene crossovers [9], [40].
All together, the observation of species-specific bands, the recovery of identical V3 region in different species and intragenomic heterogeneity of rrs V3 copies differing by one to 13 nucleotides argue for the existence of the two modes of rrs evolution in the genus Aeromonas. As an example, bands 3 and 18 found in some A. caviae strains, each differing by one nucleotide from the band 4 found in all A. caviae strains, were probably generated by mutation while band 1 differing by 12 nucleotides from band 4 was probably transferred to A. caviae by HGT. We showed that rrs V3 region HGT defined by more than 5 divergent nucleotidic positions, was detected in 23 PCR-TTGE patterns observed in 7 species (Table S1). Our results also supported a non-random occurrence of some particular heterogeneous patterns because unrelated isolates shared identical patterns made of a combination of bands generated by mutations or by mutations and HGT, e.g., patterns 3+4+18 and 1+4+18 in A. caviae.
It is noteworthy that the concerted evolution as a mode of homogenization of copies in multigenic families is not efficient for most strains in aeromonads. Homogenization of copies is a spontaneous mechanism that occurs by homologous recombination among closely related sequences. The rate of isolates harboring divergent rrs copies in aeromonads suggested a selective pressure that maintains differences in their sequences. This is also the case for the maintenance of a high number of rrn operons in spite of the associated metabolic cost [41].
RiMOD and Lifestyles in Aeromonads
Harboring about 10 rrn operons is highly atypical compared with bacteria in general and particularly with other well-known waterborne human opportunistic pathogens such as Pseudomonas aeruginosa for which genome sequencing revealed 4 rrn operons only. According to the ribosomal RNA operon copy number database (rrndb, consulted on February the 16th, 2012) [42], 8 to 11 operons have been reported in only 46 out of 782 species (6%). Focusing on bacteria harboring 10 rrn operons other than Aeromonas revealed 13 species distributed among 7 genera and clustering in few phylogenetic lineages, i.e., 6 families (Clostridiaceae, Peptococcaceae, Bacillaceae, Aeromonadaceae, Psychromonadaceae and Shewanellaceae) belonging to 2 phyla (Firmicutes and Proteobacteria). Within the class Gammaproteobacteria, Aeromonas spp. shared common characteristics with members of the genus Shewanella (8 to 11 rrn operons recorded among the 16 entries available in the rrndb for the genus), a genus found in highly diverse aquatic environments and considered as a human emerging opportunistic pathogen [43]–[45]. It has been proposed that sequence modifications among 16S rRNA gene copies provide a fine-tuning of the ribosome function to optimize bacterial niche fitness [46]. Similarly, it could be hypothesized that a high number of 16S rRNA gene copies and a high rate of isolates with intragenomic heterogeneity could be features involved in maintaining functional diversity of Aeromonas spp. in environment, animal and human microbiotae.
Here, A. caviae displayed outstanding characteristics, i.e., the highest number of isolates with V3 rrs heterogeneity and the highest diversity in the heterogeneity types. These features significantly different from those observed in other species may be related to a particular lifestyle and an ongoing process of adaptation of this species to a particular niche that could be represented, as previously hypothesized, by the gut [13]. It was also noteworthy that among the 71 A. veronii studied strains, the 22 A. veronii isolates displaying rrs heterogeneity were all recovered from human samples except for two strains recovered from animal samples while all strains isolated from environmental specimens (n = 20) had identical rrs V3 region copies. More generally, we showed that isolates recovered from human samples more frequently displayed V3 rrs heterogeneity than isolates recovered from samples from non-human origin according to previous results of Alperi et al. [10].
RiMOD data confirmed and completed the vision of aeromonads as a bacterial population structured in species complexes, an organization associated with bacteria displaying sympatric lifestyles, i.e., a high level of horizontal gene transfers, a large pan-genome, a large genome, and a significant number of ribosomal operons, and a behavior of generalist opportunistic pathogen [21].
Taxonomic Considerations
Taxonomy of the genus Aeromonas has greatly evolved over the past decades following advances in methods applied to infer relationships between members of the genus. The bulk 16S rDNA sequencing was shown to be insufficiently discriminatory between tightly related species [1]–[4]. The use of 16S rRNA gene PCR-RFLP has then been proposed for species identification [5], [6]. However, considering the endonucleases used in 16S rRNA gene PCR-RFLP, the intragenomic divergent positions identified in this study within the rrs V3 region modified the number of AluI restriction sites in 16% of the strains (32 strains belonging to 7 taxa and displaying 15 different TTGE patterns), as well as the number of NarI restriction sites in the type strain of A. encheleia (data not shown), thereby leading to atypical restriction patterns and possible misidentification of the strains as also previously underlined [7]–[10]. Thus, the high intragenomic heterogeneity of the different rrs sequences and the intraspecific rrs variability, sometimes exceeding the interspecific variability, blurred the 16S rRNA-based studies that were progressively replaced or implemented by single housekeeping gene analysis and further by multilocus sequence analysis [2]–[4], [11]–[13]. However, some conflicting results may arise from the use of different approaches. For example, A. allosaccharophila existence is still controversial despite DNA-DNA hybridization values to A. veronii strain CECT 4247T ( = ATCC 35624T) have been reevaluated to 78–82% by Huys et al. [30]. Based on single housekeeping gene (gyrB, cpn60, tsf or zipA)-based studies, the taxon occupied a closely related but robust external position to the A. veronii clade supporting the separation of the two taxa [13], [47], [48]. Similar results have been observed with MLPA studies [11], [13]. Recently, a hypothesis that could support at least in part some controversial taxonomic data was proposed based on the evolution mode of Aeromonas spp. Some taxa subjected to controversy may correspond to lineages emerging from species complexes and for which signs of starting speciation are detected or not according to the characteristic investigated and to the method used [13], [21], [49]. RiMOD study supported this hypothesis for A. allosaccharophila.
Here, we showed that rrs heterogeneity patterns might inform on taxonomic relationships between species because some PCR-TTGE bands were specific of some species despite HGT. On the other hand, large-scale chromosome structure has previously been described as a sensitive indicator of phylogenetic relationships between bacteria [50]–[53] but it received little attention in the polyphasic strategy used for taxonomic studies in the genus Aeromonas. In this study, rrn operon number at 8 as well as rrn operon PFGE fingerprints were valuable for taxonomic purpose, allowing taxonomic resolution of species complexes. To our knowledge, only the study by Umelo & Trust on A. salmonicida previously showed that I-CeuI digestion fingerprints could help identify related strains and thus help to better classify and subdivide the atypical strains [16]. Combining both approaches for studying RiMOD supported current taxonomy including recent taxonomic changes in the genus Aeromonas and was especially useful here to identify heterogeneity within MLPA clades such as A. salmonicida and A. media clades. The latter species warrant further investigations to search whether they are robust groups including atypical strains or groups submitted to a tempo of evolution associated to a high rate of speciation or a strong selective pressure generating diversity within the group, or polyphyletic taxa. We therefore assumed that RiMOD approach proposed herein could be useful to complement the polyphasic strategy used for isolate characterization within the genus Aeromonas.
The 16S rRNA gene is often criticized because this marker provides a limited point of view on the bacterial evolution. However, 16S rRNA-based hierarchical system is considered to be the backbone of prokaryote taxonomy that was globally confirmed by phylogenomics. The main pitfalls of 16S rRNA-based phylogeny is the low amount of genetic information explored regarding genome size and the multigenic organisation that lead to blurred information when bulk DNA is sequenced. However, multigene family is an exception in bacterial genome and therefore the maintaining of several heterogeneous 16S rRNA loci as observed in aeromonads is probably not trivial. In this study, we showed that 16S rRNA multigenic organisation should no longer be considered as a pitfall that confuse relationships among aeromonads but rather be exploited in all the dimensions of its diversity, which probably reflects major mechanisms involved in the generalist pathogen lifestyle [21], as previously observed for other Aeromonas genetic and genomic traits [54].
Supporting Information
TTGE profiles of amplified 16S rRNA gene V3 region for Aeromonas spp. strains. A) Lanes 1–12, A. veronii isolates ADV103, BVH61, ADV109, ADV119, ADV125, ADV127, ADV129, ADV130, ADV131, ADV133, ADV135, and ADV137b; lane 13, A. bivalvium CECT 7113T. Profiles are indicated at the bottom of the figure. Arrows indicated the position of the four TTGE bands constituting the A. molluscorum type strain pattern. B) 13 Aeromonas spp. type and reference strains. Names of strains are indicated at the top of the figure. Each TTGE band number is noted on the band. L, ladder with 1+4+15+30 profile.
(TIF)
PFGE migration of genomic DNA digested by I- Ceu I for Aeromonas spp. strains. A) 37 type and reference strains of Aeromonas spp. and A. sharmana DSM 17445T. Names of strains and names of main clades are indicated at the top and bottom of the figure, respectively. Lambda ladder was used as molecular size marker (lanes M). Sizes are indicated in kilobases (kb). B) Lane 1, A. hydrophila subsp. dhakensis CIP 107500 (now A. aquariorum); lane 2, A. hydrophila subsp. hydrophila CECT 839T; lane 3, A. hydrophila subsp. ranae CIP 107985; lane 4, A. aquariorum CECT 7289T; lane 5, A. caviae strain ADV118; lane 6, A. caviae strain ADV121; lane 7, A. caviae CECT 838T; lane 8, A. media CECT 4232T; lane 9, A. allosaccharophila CECT 4199T; lane 10, A. culicicola CIP 107763T (now A. veronii); lane 11, A. ichtiosma CECT 4486T (now A. veronii); lane 12, A. veronii biovar veronii CECT 4247T; lane 13, A. veronii biovar sobria LMG 13067; lane 14, A. media strain BVH40; lane 15, A. media strain AK202; lane 16, A. media strain AK211. Lambda ladder was used as molecular size marker (lanes M). Sizes are indicated in kilobases (kb).
(TIF)
PCR-TTGE pattern, rrs V3 region heterogeneity and rrn operon number for the 195 Aeromonas strains of the study according to multilocus phylogenetic clade determined in Roger et al . [13] .
(DOCX)
Acknowledgments
We are particularly indebted to the microbiology laboratory team of the Montpellier academic hospital for providing some clinical isolates and to Fabien Aujoulat for technical assistance.
Contributors
Members of the colBVH (college de Bactériologie, de Virologie et d’Hygiène des hôpitaux) study group who participated in this study: F. Carmagnol (Centre Hospitalier de Cannes), E. Chachaty (Institut Gustave Roussy), C. Alba-Sauviat (Centre Hospitalier de Chaumont), C. Auvray (Centre Hospitalier de Charleville-Mézières), D. Barraud (Centre Hospitalier de Gonesse), Z. Benseddik (Centre Hospitalier de Chartres), A. Bertrou (Centre Hospitalier de Carcassone), F. Bessis (Centre Hospitalier de Cherbourg), H. Biessy (Centre Hospitalier de La Rochelle), V. Blanc (Centre Hospitalier d’Antibes-Juan-les-pins), Y. Boucaud-Maitre (Centre Hospitalier Saint Joseph-Saint Luc, Lyon), P. Brunet & A. Michel (Hôpital Saint Joseph, Marseille), B. Cancet (Centre Hospitalier de Villeneuve/Lot), J. Carrere (Centre Hospitalier de Hyères), A. Cecille (Centre Hospitalier de Digne-les-bains), G. Chambreuil (Centre Hospitalier de La Roche/Yon), P. Chantelat (Centre Hospitalier de Vesoul), H. Chardon (Centre Hospitalier d’Aix-en-Provence), C. Charrel (Centre Hospitalier de Salon de Provence), H. De Montclos (Centre Hospitalier de Bourg-en-Bresse), J.W. Decousser (Hôpital Antoine Beclère, Paris), J. M. Delarbre/A. Gravet (Centre Hospitalier de Mulhouse), D. Deligne (Centre Hospitalier de Remiremont), C. Denoix (Centre Hospitalier Universitaire de La Réunion), J. Deregnaucourt (Hôpital Léopold Bellan, Paris), F. Desroys du Roure (Centre Hospitalier de Chatellerault), S. Dubourdieu (Centre Hospitalier de Gisors), Z. El Harrif (Centre Hospitalier de Libourne), C. Eloy (Centre Hospitalier de Troyes), A. Evers (Centre Hospitalier de Annonay), C. Febvre (Centre Hospitalier de Montbéliard), D. Fevre (Centre Hospitalier de Vienne), S. Gabriel (Centre Hospitalier Princesse Grace, Monaco), M. J. Galanti (Centre Hospitalier de Coulommiers), E. Garnotel (Hôpital d’Instruction des Armées Lavean à Marseille), M. Gavignet (Centre Hospitalier de Lavaur), F. Geffroy (Centre Hospitalier de Quimper), G. Grise (Centre Hospitalier de Elbeuf-Louviers), I. Gros (Centre Hospitalier de St Denis), I. Hermes (Centre Hospitalier de St-Malo), J. Heurte (Centre Hospitalier de Beauvais), E. Heusse (Centre Hospitalier de Bayeux), D. Jan (Centre Hospitalier de Laval), E. Jaouen (Centre Hospitalier de Sablé/Sarthe), S. Laluque (Centre Hospitalier de Montluçon), R. Lamarca (Centre Hospitalier de Narbonne), E. Laurens (Centre Hospitalier de Belfort), A. Le Coustumier (Centre Hospitalier de Cahors), E. Lecaillon (Centre Hospitalier de Perpignan), C. Lemble (Centre Hospitalier de Selestat), M. Leneveu (Centre Hospitalier Intercommunal de Poissy Saint-Germain-en-Laye), S. Leotard (Centre Hospitalier de Grasse), M. N. Letouzey (Centre Hospitalier de Villefranche/saone), C. Malbrunot (Centre Hospitalier de Corbeil-Essonnes), O. Menouni (Centre Hospitalier de Montceau-les-Mines), M. Morel (Centre Hospitalier de Le Havre), C. Olive (Centre Hospitalier Universitaire de Fort de France), B. Pangon (Centre Hospitalier de Versailles), J. G. Paul (Centre Hospitalier de Boulogne/mer), J. M. Perez (Centre Hospitalier Universitaire de Pointe-à-Pitre), P. Pouedras (Centre Hospitalier de Vannes), D. Pressac (Centre Hospitalier de Tulle), R. Sanchez (Centre Hospitalier de Périgueux), Y. Scat (Centre Hospitalier de Pontivy), A. Secher (Centre Hospitalier de Dreux), J. Semon (Centre Hospitalier de Chalon-sur-Saone), D. Simeon (Centre Hospitalier de Langres), C. Simonin (Centre Hospitalier de Macon), J. P. Thellier (Centre Hospitalier de Château-Thierry), B. Tourand (Centre Hospitalier d’Alès), A. Vachée (Centre Hospitalier de Roubaix), C. Varache (Centre Hospitalier du Mans), J. Vaucel (Centre Hospitalier de Saint-Brieux), A. C. Vautrin (Centre Hospitalier Universitaire de Saint-Etienne), A. Verhaeghe (Centre Hospitalier de Dunkerke), M. Villemain (Centre Hospitalier d’Aurillac) and L. Villeneuve (Centre Hospitalier d’Aubagne).
Funding Statement
This work was supported by the Association des Biologistes de l’Ouest and by ADEREMPHA (Association pour la Recherche et le Développement en Microbiologique & Pharmacie). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
the colBVH study group:
Carmagnol F., Chachaty E., Alba-Sauviat C., Auvray C., Barraud D., Benseddik Z., Bertrou A., Bessis F., Biessy H., Blanc V., Boucaud-Maitre Y., Brunet P., Michel A., Cancet B., Carrere J., Cecille A., Chambreuil G., Chantelat P., Chardon H., Charrel C., De Montclos H., Decousser J.W., Delarbre J. M., Gravet A., Deligne D., Denoix C., Deregnaucourt J., Desroys du Roure F., Dubourdieu S., El Harrif Z., Eloy C., Evers A., Febvre C., Fevre D., Gabriel S., Galanti M. J., Garnotel E., Gavignet M., Geffroy F., Grise G., Gros I., Hermes I., Heurte J., Heusse E., Jan D., Jaouen E., Laluque S., Lamarca R., E. Laurens, Le Coustumier A., Lecaillon E., Lemble C., Leneveu M., Leotard S., Letouzey M. N., Malbrunot C., Menouni O., Morel M., Olive C., Pangon B., Paul J. G., Perez J. M., Pouedras P., Pressac D., Sanchez R., Scat Y., Secher A., Semon J., Simeon D., Simonin C., Thellier J. P., Tourand B., Vachée A., Varache C., Vaucel J., Vautrin A. C., Verhaeghe A., Villemain M., and Villeneuve L.
References
- 1. Martínez-Murcia AJ, Benlloch S, Collins MD (1992) Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int J Syst Bacteriol 42: 412–421. [DOI] [PubMed] [Google Scholar]
- 2. Alperi A, Martínez-Murcia AJ, Ko WC, Monera A, Saavedra MJ, et al. (2010) Aeromonas taiwanensis sp. nov. and Aeromonas sanarellii sp. nov., clinical species from Taiwan. Int J Syst Evol Microbiol 60: 2048–55. [DOI] [PubMed] [Google Scholar]
- 3. Alperi A, Martínez-Murcia AJ, Monera A, Saavedra MJ, Figueras MJ (2010) Aeromonas fluvialis sp. nov., isolated from a Spanish river. Int J Syst Evol Microbiol 60: 72–77. [DOI] [PubMed] [Google Scholar]
- 4. Figueras MJ, Alperi A, Beaz-Hidalgo R, Stackebrandt E, Brambilla E, et al. (2011) Aeromonas rivuli sp. nov., isolated from the upstream region of a karst water rivulet. Int J Syst Evol Microbiol 61: 242–248. [DOI] [PubMed] [Google Scholar]
- 5. Borrell N, Acinas SG, Figueras MJ, Martínez-Murcia AJ (1997) Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S rRNA genes. J Clin Microbiol 35: 1671–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Figueras MJ, Guarro J, Martínez-Murcia A (2000) Use of restriction fragment length polymorphism of the PCR-amplified 16S rRNA gene for the identification of Aeromonas spp. J Clin Microbiol 38: 2023–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graf J (1999) Diverse restriction fragment length polymorphism patterns of the PCR-amplified 16S rRNA genes in Aeromonas veronii strains and possible misidentication of Aeromonas species. J Clin Microbiol 37: 3194–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinetti Lucchini G, Altwegg M (1992) rRNA gene restriction patterns as taxonomic tools for the genus Aeromonas . Int J Syst Bacteriol 42: 384–399. [DOI] [PubMed] [Google Scholar]
- 9. Morandi A, Zhaxybayeva O, Gogarten JP, Graf J (2005) Evolutionary and diagnostic implications of intragenomic heterogeneity in the 16S rRNA gene in Aeromonas strains. J Bacteriol 187: 6561–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alperi A, Figueras MJ, Inza I, Martínez-Murcia AJ (2008) Analysis of 16S rRNA gene mutations in a subset of Aeromonas strains and their impact in species delineation. Int Microbiol 11: 185–194. [PubMed] [Google Scholar]
- 11. Martinez-Murcia AJ, Monera A, Saavedra MJ, Oncina R, Lopez-Alvarez M, et al. (2011) Multilocus phylogenetic analysis of the genus Aeromonas . Syst Appl Microbiol 34: 189–199. [DOI] [PubMed] [Google Scholar]
- 12. Martino ME, Fasolato L, Montemurro F, Rosteghin M, Manfrin A, et al. (2011) Determination of microbial diversity of Aeromonas strains on the basis of multilocus sequence typing, phenotype, and presence of putative virulence genes. Appl Environ Microbiol 77: 4986–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roger F, Marchandin H, Jumas-Bilak E, Kodjo A (2012) the colBVH study group, et al (2012) Multilocus genetics to reconstruct aeromonad evolution. BMC Microbiol 12: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coenye T, Vandamme P (2003) Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol Lett 228: 45–49. [DOI] [PubMed] [Google Scholar]
- 15. Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF (2004) Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 186: 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Umelo E, Trust TJ (1998) Physical map of the chromosome of Aeromonas salmonicida and genomic comparisons between Aeromonas strains. Microbiol 144: 2141–2149. [DOI] [PubMed] [Google Scholar]
- 17. Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, et al. (2006) Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol 188: 8272–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, et al. (2008) The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Liu Y, Zhou Z, Huang H, Ren Y, et al. (2011) Complete genome sequence of Aeromonas veronii strain B565. J Bacteriol 193: 3389–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tacão M, Moura A, Alves A, Henriques I, Saavedra MJ, et al. (2005) Evaluation of 16S rDNA- and gyrB-DGGE for typing members of the genus Aeromonas . FEMS Microbiol Lett 246: 11–18. [DOI] [PubMed] [Google Scholar]
- 21. Georgiades K, Raoult D (2010) Defining pathogenic bacterial species in the genomic era. Front Microbiol 1: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 23. Miranda G, Kelly C, Solorzano F, Leanos B, Coria R, et al. (1996) Use of pulsed-field gel electrophoresis typing to study an outbreak of infection due to Serratia marcescens in a neonatal intensive care unit. J Clin Microbiol 34: 3138–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Michon AL, Aujoulat F, Roudière L, Soulier O, Zorgniotti I, et al. (2010) Intragenomic and intraspecific heterogeneity in rrs may surpass interspecific variability in a natural population of Veillonella . Microbiol 156: 2080–2091. [DOI] [PubMed] [Google Scholar]
- 25. Dodd HN, Pemberton JM (1998) Construction of a physical and preliminary genetic map of Aeromonas hydrophila JMP636. Microbiology 144: 3087–3096. [DOI] [PubMed] [Google Scholar]
- 26. Miñana-Galbis D, Farfán M, Lorén GJ, Fusté MC (2010) Proposal to assign Aeromonas diversa sp. nov. as a novel species designation for Aeromonas group 501. Syst Appl Microbiol 33: 15–19. [DOI] [PubMed] [Google Scholar]
- 27. Lamy B, Laurent F, Kodjo A (2010) Validation of a partial rpoB gene sequence as a tool for phylogenetic identification of aeromonads isolated from environmental sources. Can J Microbiol 56: 217–228. [DOI] [PubMed] [Google Scholar]
- 28. Martínez-Murcia AJ, Saavedra MJ, Mota VR, Maier T, Stackebrandt E, et al. (2008) Aeromonas aquariorum sp. nov., isolated from aquaria of ornamental fish. Int J Syst Evol Microbiol 58: 1169–1175. [DOI] [PubMed] [Google Scholar]
- 29. Janda JM, Abbott SL (2010) The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23: 35–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huys G, Kämpfer P, Swings J (2001) New DNA-DNA hybridization and phenotypic data on the species Aeromonas ichthiosmia and Aeromonas allosaccharophila: A. ichthiosmia Schubert, et al. 1990 is a later synonym of A. veronii Hickman-Brenner, et al. 1987. Syst Appl Microbiol 24: 177–182. [DOI] [PubMed] [Google Scholar]
- 31. Saha P, Chakrabarti T (2006) Aeromonas sharmana sp. nov., isolated from a warm spring. Int J Syst Evol Microbiol 56: 1905–1909. [DOI] [PubMed] [Google Scholar]
- 32. Martínez-Murcia AJ, Figueras MJ, Saavedra MJ, Stackebrandt E (2007) The recently proposed species Aeromonas sharmana sp. nov., isolate GPTSA-6T, is not a member of the genus Aeromonas . Int Microbiol 10: 61–64. [PubMed] [Google Scholar]
- 33. Pidiyar VJ, Jangid K, Patole MS, Shouche YS (2003) Analysis of 16S–23S intergenic spacer regions and rrn operon copy number of Aeromonas culicicola MTCC 3249T . DNA Seq 14: 183–194. [DOI] [PubMed] [Google Scholar]
- 34. Chakravorty S, Helb D, Burday M, Connell N, Alland D (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69: 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soler L, Yanez MA, Chacon MR, Aguilera-Arreola MG, Catalan VM, et al. (2004) Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int J Syst Evol Microbiol 54: 1511–1519. [DOI] [PubMed] [Google Scholar]
- 36. Yap WH, Zhang Z, Wang Y (1999) Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J Bacteriol 181: 5201–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Berkum P, Terefework Z, Paulin L, Suomalainen S, Lindström K, et al. (2003) Discordant phylogenies within the rrn loci of Rhizobia . J Bacteriol 185: 2988–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Santoyo G, Romero D (2005) Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol Rev 29: 169–183. [DOI] [PubMed] [Google Scholar]
- 39. Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M (1999) Two distinct mechanisms cause heterogeneity of 16S rRNA. J Bacteriol 181: 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sneath PH (1993) Evidence from Aeromonas for genetic crossing-over in ribosomal sequences. Int J Syst Bacteriol 43: 626–629. [DOI] [PubMed] [Google Scholar]
- 41. Klappenbach JA, Dunbar JM, Schmidt TM (2000) rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol 66: 1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee ZMP, Bussema III C, Schmidt TM (2009) rrnDB: Documenting the number of rRNA and tRNA genes in Bacteria and Archaea. Nucleic Acids Research 37: D489–D493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Holt HM, Gahrn-Hansen B, Bruun B (2005) Shewanella algae and Shewanella putrefaciens: clinical and microbiological characteristics. Clin Microbiol Infect 11: 347–352. [DOI] [PubMed] [Google Scholar]
- 44. Dikow RB (2011) Genome-level homology and phylogeny of Shewanella (Gammaproteobacteria: lteromonadales: Shewanellaceae). BMC Genomics 12: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sharma KK, Kalawat U (2010) Emerging infections: Shewanella - a series of five cases. J Lab Physicians 2: 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jensen S, Frost P, Torsvik VL (2009) The nonrandom microheterogeneity of 16S rRNA genes in Vibrio splendidus may reflect adaptation to versatile lifestyles. FEMS Microbiol Lett 294: 207–215. [DOI] [PubMed] [Google Scholar]
- 47. Saavedra MJ, Perea V, Fontes MC, Martins C, Martínez-Murcia A (2007) Phylogenetic identification of Aeromonas strains isolated from carcasses of pig as new members of the species Aeromonas allosaccharophila . Antonie Van Leeuwenhoek 91: 159–167. [DOI] [PubMed] [Google Scholar]
- 48. Miñana-Galbis D, Urbizu-Serrano A, Farfán M, Fusté MC, Lorén JG (2009) Phylogenetic analysis and identification of Aeromonas species based on sequencing of the cpn60 universal target. Int J Syst Evol Microbiol 59: 1976–1983. [DOI] [PubMed] [Google Scholar]
- 49. Silver AC, Williams D, Faucher J, Horneman AJ, Gogarten JP, et al. (2011) Complex evolutionary history of the Aeromonas veronii group revealed by host interaction and DNA sequence data. PLoS One 6: e16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu SL, Schryvers AB, Sanderson KE, Johnston RN (1999) Bacterial phylogenetic clusters revealed by genome structure. J Bacteriol 181: 6747–6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marchandin H, Jumas-Bilak E, Gay B, Teyssier C, Jean-Pierre H, et al. (2003) Phylogenetic analysis of some Sporomusa sub-branch members isolated from human clinical specimens: description of Megasphaera micronuciformis sp. nov. Int J Syst Evol Microbiol 53: 547–553. [DOI] [PubMed] [Google Scholar]
- 52. Jumas-Bilak E, Jean-Pierre H, Carlier JP, Teyssier C, Bernard K, et al. (2005) Dialister micraerophilus sp. nov. and Dialister propionicifaciens sp. nov., isolated from human clinical samples. Int J Syst Evol Microbiol 55: 2471–2478. [DOI] [PubMed] [Google Scholar]
- 53. Zheng JF, Liu GR, Liu SL (2006) Phylogenetically clustering of rhizobia by genome structure: application to unclassified Rhizobium . J Environ Sci 18: 530–536. [PubMed] [Google Scholar]
- 54. Aujoulat F, Roger F, Bourdier A, Lotthé A, Lamy B, et al. (2012) From environment to man: genome evolution and adaptation of human opportunistic bacterial pathogens. Genes 3: 191–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lamy B, Kodjo A (2009) colBVH study group, Laurent F (2009) Prospective nationwide study of Aeromonas infections in France. J Clin Microbiol 47: 1234–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TTGE profiles of amplified 16S rRNA gene V3 region for Aeromonas spp. strains. A) Lanes 1–12, A. veronii isolates ADV103, BVH61, ADV109, ADV119, ADV125, ADV127, ADV129, ADV130, ADV131, ADV133, ADV135, and ADV137b; lane 13, A. bivalvium CECT 7113T. Profiles are indicated at the bottom of the figure. Arrows indicated the position of the four TTGE bands constituting the A. molluscorum type strain pattern. B) 13 Aeromonas spp. type and reference strains. Names of strains are indicated at the top of the figure. Each TTGE band number is noted on the band. L, ladder with 1+4+15+30 profile.
(TIF)
PFGE migration of genomic DNA digested by I- Ceu I for Aeromonas spp. strains. A) 37 type and reference strains of Aeromonas spp. and A. sharmana DSM 17445T. Names of strains and names of main clades are indicated at the top and bottom of the figure, respectively. Lambda ladder was used as molecular size marker (lanes M). Sizes are indicated in kilobases (kb). B) Lane 1, A. hydrophila subsp. dhakensis CIP 107500 (now A. aquariorum); lane 2, A. hydrophila subsp. hydrophila CECT 839T; lane 3, A. hydrophila subsp. ranae CIP 107985; lane 4, A. aquariorum CECT 7289T; lane 5, A. caviae strain ADV118; lane 6, A. caviae strain ADV121; lane 7, A. caviae CECT 838T; lane 8, A. media CECT 4232T; lane 9, A. allosaccharophila CECT 4199T; lane 10, A. culicicola CIP 107763T (now A. veronii); lane 11, A. ichtiosma CECT 4486T (now A. veronii); lane 12, A. veronii biovar veronii CECT 4247T; lane 13, A. veronii biovar sobria LMG 13067; lane 14, A. media strain BVH40; lane 15, A. media strain AK202; lane 16, A. media strain AK211. Lambda ladder was used as molecular size marker (lanes M). Sizes are indicated in kilobases (kb).
(TIF)
PCR-TTGE pattern, rrs V3 region heterogeneity and rrn operon number for the 195 Aeromonas strains of the study according to multilocus phylogenetic clade determined in Roger et al . [13] .
(DOCX)