Abstract
Background
Tumor necrosis factor (TNF) and TNF receptor superfamily (TNFR)-mediated immune response play an essential role in the pathogenesis of severe sepsis. Studies examining associations of TNF and lymphotoxin-α (LTA) single nucleotide polymorphisms (SNPs) with severe sepsis have produced conflicting results. The objective of this study was to investigate whether genetic variation in TNF, LTA, TNFRSF1A and TNFRSF1B was associated with susceptibility to or death from severe sepsis in Chinese Han population.
Methodology/Principal Findings
Ten SNPs in TNF, LTA, TNFRSF1A and TNFRSF1B were genotyped in samples of patients with severe sepsis (n = 432), sepsis (n = 384) and healthy controls (n = 624). Our results showed that rs1800629, a SNP in the promoter region of TNF, was significantly associated with risk for severe sepsis. The minor allele frequency of rs1800629 was significantly higher in severe sepsis patients than that in both healthy controls (Padj = 0.00046, odds ratio (OR)adj = 1.92) and sepsis patients (Padj = 0.002, ORadj = 1.56). Further, we investigated the correlation between rs1800629 genotypes and TNF-α concentrations in peripheral blood mononuclear cells (PBMCs) of healthy volunteers exposed to lipopolysaccharides (LPS) ex vivo, and the association between rs1800629 and TNF-α serum levels in severe sepsis patients. After exposure to LPS, the TNF-α concentration in culture supernatants of PBMCs was significantly higher in the subjects with AA+AG genotypes than that with GG genotype (P = 0.007). Moreover, in patients with severe sepsis, individuals with AA+AG genotypes had significantly higher TNF-α serum concentrations than those with GG genotype (Padj = 0.02). However, there were no significant associations between SNPs in the four candidate genes and 30 day mortality for patients with severe sepsis.
Conclusions/Significance
Our findings suggested that the functional TNF gene SNP rs1800629 was strongly associated with susceptibility to severe sepsis, but not with lethality in Chinese Han population.
Introduction
Sepsis is an infection-initiated and inflammation-induced syndrome. Despite progress in the development of antibiotics and other supportive care therapies, severe sepsis remains an unconquered challenge for the clinicians with an unacceptable high mortality rate of 30%–50% [1]. The response to infection is diverse among different individuals. Given the same therapies, most sepsis patients will recover and do well, while a small, but significant portion, will develop severe sepsis and multiple organ system failure, refractory hypotension and death [2], [3]. Currently, more and more evidence showed that genetic factors played an important role in the development and severity of sepsis [4], [5], [6], [7], [8], [9], [10], [11]. Common sequence variants within genes involved in pro-inflammatory response have received particular attention [12], [13].
Although the pathogenesis of sepsis remains incompletely understood, an excessive pro-inflammatory response has been established as a fundamental component of severe sepsis [14]. The proinflammatory cytokine TNF-α is an essential component in the host immune response to infection and has been widely reported to be an important mediator in severe sepsis and septic shock. High circulating levels of TNF-α were correlated with poor outcomes in sepsis patients [15]. TNF-α and lymphotoxin-α (LT-α) share the same receptors as well as many biological activities, and they are central mediators of immune responses [16]. TNF-α and LT-α are encoded by adjacent gene loci in the central or class III region of the human major histocompatibility complex (MHC), between the HLA class I and II genes on the short arm of chromosome 6 [17]. Several SNPs within the promoter region of TNF (−238, −308, −857, −863, −1031) and the first intron of LTA (+252) were thought to influence TNF-α and LT-α production, and have therefore been identified as candidate variants that might influence susceptibility to and/or outcomes from severe sepsis and infectious diseases [18], [19], [20], [21], [22], [23], [24], [25]. In particular, rs1800629 (TNF −308) and rs909253 (LTA +252) have been the focus of many investigations on sepsis. Although several studies have identified associations for rs1800629 and rs909253 with sepsis risk or outcomes [18], [19], [20], [23], [24], other studies have not replicated the associations [21], [26], [27], [28]. This inconsistency may be due to small samples size studied and ethnic differences [29].
TNF-α and LT-α exert their pleiotropic functions by activating intracellular signaling cascades via binding to two types of receptors, TNFR-1 (encoded by the TNFRSF1A gene) and TNFR-2 (encoded by the TNFRSF1B gene) [30]. TNFR1-deficient mice are resistant to endotoxic shock and have prolonged survival with less hypothermia [31]. TNFR2 influences the biological activity of TNF-α and LT-α both in a membrane-bound and a soluble form. Membrane-bound TNFR2 facilitates activation of nuclear factor (NF)-κB and mitogen-activated protein kinase signaling cascades upon binding with TNF-α and LT-α, whereas soluble TNFR2 is capable of binding and inactivating circulating TNF-α and LT-α [16], [30]. Moreover, animal studies showed that TNFR2 mediated protective effects in the development of severe sepsis [31]. Recent studies proposed that genetic variation in TNFRSF1A and TNFRSF1B was associated with susceptibility to inflammatory and autoimmune diseases, such as tuberculosis, systemic lupus erythematosus, rheumatoid arthritis and Crohn's disease [32], [33], [34], [35]. However, to date, only one study investigated the role of TNFRSF1A and TNFRSF1B polymorphisms in sepsis susceptibility and mortality [36].
Considering the important role of TNF-α, LT-α, TNFR1 and TNFR2 in the pathogenesis of severe sepsis, we hypothesized that genetic variation in TNF, LTA, TNFRSF1A and TNFRSF1B might be associated with susceptibility to and outcomes from severe sepsis in Chinese Han population. To test this hypothesis, we conducted a relatively large-scale case-control study enrolling 432 severe sepsis patients, 384 sepsis patients and 624 healthy individuals to investigate the association of genetic variants in TNF, LTA, TNFRSF1A and TNFRSF1B with severe sepsis susceptibility and prognosis in Chinese Han population. Furthermore, we investigated the association between the genotypes of the TNF gene SNP rs1800629 and TNF-α concentration in culture supernatants of LPS simulated PBMCs obtained from healthy donors and in serum from severe sepsis patients.
Results
Characteristics of Study Subjects
A total of 432 severe sepsis patients, 384 sepsis patients and 624 health volunteers were enrolled in this case-control study. According to the mortality within 30 days, severe sepsis patients were divided into survivor and non-survivor groups. The baseline characteristics and clinical data of all subjects are shown in Table 1. The average age and proportion of male among the severe sepsis, sepsis and healthy control groups did not show significant difference. The primary source of infection in severe sepsis patients was the lungs (69.9%), followed by abdomen (21.8%), blood stream (3.5%), urinary tract (2.5%) and others (2.3%). The overall 30-day mortality rate of severe sepsis patients was 36.1%. The mean APACHE II and SOFA score in non-survivor group was higher than that in survivor group (P<0.05).
Table 1. Demographic and clinical characteristics of the study subjects.
| Healthy controls | Sepsis patients | Severe sepsis patients | P1 value | Survivor | Nonsurvivor | P2 value | |
| Number | 624 | 384 | 432 | N.A | 276 | 156 | N.A |
| Age | 68.5±9.3 | 62.1±10.8 | 65.1±11.8 | 0.12 | 63.4±10.8 | 68.1±14.2 | 0.09 |
| Sex(Male/Female) | 363/261 | 220/164 | 256/176 | 0.57 | 163/113 | 93/63 | 0.91 |
| APACHE II score | N.A | 10.2±3.2 | 18.6±4.9 | 0.008 | 14.8±2.9 | 25.3±7.2 | 0.013 |
| Length of ICU stay (d) | N.A | 8.6±2.3 | 15.4±8.6 | 0.006 | 14.6±7.2 | 16.8±8.9 | 0.02 |
| Diabetes | N.A | 35 (9.1%) | 46 (10.6%) | 0.47 | 30 (10.8%) | 16 (10.2%) | 0.84 |
| Chronic liver disease | N.A | 9 (2.3%) | 15 (3.5%) | 0.34 | 8 (2.9%) | 7 (4.5%) | 0.39 |
| Chronic renal failure | N.A | 14 (3.6%) | 18 (4.2%) | 0.70 | 12 (4.3%) | 6 (3.8%) | 0.80 |
| Congestive heart failure | N.A | 22 (5.7%) | 28 (6.5%) | 0.66 | 18 (6.5%) | 10 (6.4%) | 0.96 |
| Chronic pulmonary disease | N.A | 28 (7.3%) | 37 (8.6%) | 0.50 | 22 (8.0%) | 15 (9.6%) | 0.56 |
| SOFA score | N.A | 1.4±0.3 | 8.2±1.6 | <0.001 | 6.9±1.8 | 10.5±1.9 | 0.011 |
| Failing organs (score >2 in SOFA scale) | |||||||
| Respiratory | N.A | N.A | 293 (67.8%) | N.A | 156 (56.5%) | 137 (87.8%) | <0.001 |
| Cardiovascular | N.A | N.A | 252 (58.3%) | N.A | 141 (51.1%) | 111 (71.2%) | <0.001 |
| Kidney | N.A | N.A | 112 (25.9%) | N.A | 46 (16.7%) | 66 (42.3%) | <0.001 |
| Neurologic | N.A | N.A | 59 (13.7%) | N.A | 29 (10.5%) | 30 (19.2%) | 0.01 |
| Liver | N.A | N.A | 42 (9.7%) | N.A | 18 (6.5%) | 24 (15.4%) | 0.003 |
| Hematologic | N.A | N.A | 35 (8.1%) | N.A | 14 (5.1%) | 21 (13.5%) | 0.002 |
| Infection Insult | |||||||
| Lung | N.A | 261 (68.0%) | 302 (69.9%) | 0.55 | 198 (71.7%) | 104 (66.7%) | 0.27 |
| Abdomen | N.A | 92 (24.0%) | 94 (21.8%) | 0.46 | 57 (20.7%) | 37 (23.7%) | 0.46 |
| Bloodstream | N.A | 11 (2.9%) | 15 (3.5%) | 0.62 | 7 (2.5%) | 8 (5.1%) | 0.16 |
| UTI | N.A | 9 (2.3%) | 11 (2.5%) | 0.85 | 8 (2.9%) | 3 (1.9%) | 0.52 |
| Others | N.A | 11 (2.9%) | 10 (2.3%) | 0.62 | 6 (2.2%) | 4 (2.6%) | 0.80 |
| Microbiology positive | N.A | 153 (39.8%) | 174 (40.3%) | 0.90 | 101 (36.6%) | 73 (46.8%) | 0.04 |
| Gram positive | N.A | 63 (41.2%) | 66 (37.9%) | 0.55 | 41 (40.6%) | 25 (34.2%) | 0.39 |
| Gram negative | N.A | 73 (47.7%) | 72 (41.4%) | 0.25 | 43 (42.6%) | 29 (39.7%) | 0.71 |
| Fungi | N.A | 7 (1.8%) | 17 (9.8%) | 0.07 | 9 (8.9%) | 8 (11.0%) | 0.65 |
| Mixed | N.A | 10 (6.5%) | 19 (10.9%) | 0.16 | 8 (7.9%) | 11 (15.1%) | 0.14 |
| Microbiology unknown | N.A | 231 (60.2%) | 258 (59.7%) | 0.90 | 175 (63.4%) | 83 (53.2%) | 0.04 |
N.A, not applicable; APACHE, acute physiology and chronic health evaluation; SOFA, sequential organ failure assessment. P1, sepsis group vs severe sepsis group. P2, survivor group vs non-survivor group.
Association Analyses of TNF, LTA, TNFRSF1A and TNFRSF1B SNPs with Susceptibility to Severe Sepsis
The genotyping success rates of all tested SNPs ranged from 95% to 99% and none of the ten SNPs diverged significantly from Hardy-Weinberg equilibrium (P>0.05) (Table 2). The allele and genotype distributions of all tested SNPs in severe sepsis patients, sepsis patients and healthy controls are listed in Table 3. Our results showed that rs1800629 (−308G/A), located in the promoter region of TNF, was associated with significantly increased risk for severe sepsis. The frequency of rs1800629A in severe sepsis patients was significantly higher than that in both the healthy control subjects (P = 0.00028, OR = 2.08) and the sepsis patients (P = 0.00035, OR = 2.39), and the difference remained significant after Bonferroni correction. Moreover, in multivariate analyses after adjustment for covariates, rs1800629A was still significantly associated with the development of severe sepsis when compared with healthy control group (Padj = 0.00046, ORadj = 1.92) and sepsis group (Padj = 0.002, ORadj = 1.56). The genotype distribution of rs1800629 in the severe sepsis group was also significantly different from that in the healthy control group (Padj = 0.003) and the sepsis group (Padj = 0.004), and the significance remained after Bonferroni correction. However, the difference of the allele and genotype frequencies of rs1800629 between subjects with sepsis and healthy controls were not statistically significant (P>0.05). When we analyzed the allele and genotype distributions of the other nine SNPs (rs361525, rs1799724, rs1799964, rs767455, rs4149570, rs1061622, rs3397, rs1800630 and rs909253), no significant difference was found between the severe sepsis, sepsis and healthy control groups (Table 3).
Table 2. Characteristics of all tested SNPs in TNF, LTA, TNFRSF1A and TNFRSF1B.
| Gene | SNP | Location | Major/minor allele | HWE P value |
| TNF | rs361525 (−238) | promoter | C/T | 0.46 |
| rs1800629 (−308) | promoter | G/A | 0.18 | |
| rs1799724 (−857) | promoter | G/A | 0.67 | |
| rs1800630 (−863) | promoter | C/A | 0.39 | |
| rs1799964 (−1031) | promoter | T/C | 0.49 | |
| LTA (TNF-β) | rs909253 (+252) | intron | A/G | 0.58 |
| TNFRSF1A | rs767455 (A36G) | exon | A/G | 0.22 |
| rs4149570 (G609T) | 5′UTR | G/T | 0.72 | |
| TNFRSF1B | rs1061622 (Met196Arg) | exon | T/G | 0.31 |
| rs3397 | 3′UTR | G/A | 0.44 |
SNP, single nucleotide polymorphism; HWE, Hardy-Weinberg equilibrium; UTR, untranslated region.
Table 3. Association analysis of SNPs in TNF, LTA, TNFRSF1A and TNFRSF1B between case and control groups.
| Gene | Healthy | Sepsis | Severe sepsis | Allelic Comparison | Genotypic Comparison | ||||
| SNP | controls | patients | patients | P1 adj | OR1 adj 95% CI | P2 adj | OR2 adj 95% CI | P1 adj | P2 adj |
| TNF | |||||||||
| rs1800629 | 0.00046 | 1.92 (1.26–2.92) | 0.002 | 1.56 (1.23–2.76) | 0.003 | 0.004 | |||
| GG | 560 (93.3%) | 352 (93.9%) | 369 (86.4%) | ||||||
| GA | 38 (6.3%) | 23 (6.1%) | 56 (13.1%) | ||||||
| AA | 2 (0.3%) | 0 (0%) | 2 (0.47%) | ||||||
| G | 1158 (96.5%) | 727 (96.9%) | 794 (93%) | ||||||
| A | 42 (3.5%) | 23 (3.1%) | 60 (7%) | ||||||
| rs361525 | 0.22 | 1.09 (0.98–1.87) | 0.88 | 1.02 (0.84–1.42) | 0.21 | 0.89 | |||
| CC | 550 (92%) | 333 (88.8%) | 382 (89.3%) | ||||||
| CT | 48 (8%) | 42 (11.2%) | 46 (10.7%) | ||||||
| C | 1148 (96%) | 708 (94.4%) | 810 (94.6%) | ||||||
| T | 48 (4%) | 42 (5.6%) | 46 (5.4%) | ||||||
| rs1799724 | 0.21 | 1.18 (0.98–2.01) | 0.52 | 1.06 (0.86–1.46) | 0.12 | 0.85 | |||
| GG | 444 (73.6%) | 263 (70.3%) | 285 (67.9%) | ||||||
| GA | 149 (24.7%) | 107 (28.6%) | 130 (31%) | ||||||
| AA | 10 (1.7%) | 4 (1.1%) | 5 (1.2%) | ||||||
| G | 1037 (86%) | 633 (84.6%) | 700 (83.3%) | ||||||
| A | 169 (14%) | 115 (15.4%) | 140 (16.7%) | ||||||
| rs1799964 | 0.62 | 1.02 (0.92–1.42) | 0.56 | 1.12 (0.84–1.46) | 0.87 | 0.52 | |||
| TT | 384 (64.4%) | 234 (62.2%) | 265 (62.6%) | ||||||
| TC | 188 (31.5%) | 130 (34.6%) | 140 (33.1%) | ||||||
| CC | 24 (4%) | 12 (3.2%) | 18 (4.3%) | ||||||
| T | 956 (80.2%) | 598 (79.5%) | 670 (79.2%) | ||||||
| C | 236 (19.8%) | 154 (20.5%) | 176 (20.8%) | ||||||
| rs1800630 | 0.32 | 1.09 (0.81–1.40) | 0.38 | 1.18 (0.72–1.43) | 0.12 | 0.42 | |||
| CC | 412 (69%) | 262 (70.6%) | 275 (66.3%) | ||||||
| AC | 179 (30%) | 102 (27.5%) | 128 (30.8%) | ||||||
| AA | 6 (1%) | 7 (1.9%) | 12 (2.9%) | ||||||
| C | 1003 (84%) | 626 (84.4%) | 678 (81.7%) | ||||||
| A | 191 (16%) | 116 (15.6%) | 152 (18.3%) | ||||||
| LTA | |||||||||
| rs909253 | 0.18 | 0.89 (0.79–1.21) | 0.36 | 0.82 (0.58–1.26) | 0.29 | 0.14 | |||
| AA | 178 (29.7%) | 103 (27.8%) | 140 (33.6%) | ||||||
| AG | 266 (44.4%) | 181 (48.9%) | 181 (43.5%) | ||||||
| GG | 155 (25.9%) | 86 (23.2%) | 95 (22.8%) | ||||||
| A | 622 (51.9%) | 387 (52.3%) | 461 (55.4%) | ||||||
| G | 576 (48.1%) | 353 (47.7%) | 371 (44.6%) | ||||||
| TNFRSF1A | |||||||||
| rs767455 | 0.16 | 1.20 (0.82–1.68) | 0.72 | 1.04 (0.76–1.23) | 0.27 | 0.76 | |||
| AA | 462 (76.4%) | 276 (73.4%) | 302 (71.4%) | ||||||
| GA | 131 (21.7%) | 90 (23.9%) | 112 (26.5%) | ||||||
| GG | 12 (2.0%) | 10 (2.7%) | 9 (2.1%) | ||||||
| A | 1055 (87.2%) | 642 (85.4%) | 716 (84.6%) | ||||||
| G | 155 (12.8%) | 110 (14.6%) | 130 (15.4%) | ||||||
| rs4149570 | 0.78 | 0.96 (0.67–1.09) | 0.41 | 1.09 (0.82–1.34) | 0.86 | 0.62 | |||
| GG | 178 (29.3%) | 118 (31.6%) | 123 (29.2%) | ||||||
| GT | 303 (49.9%) | 192 (51.3%) | 212 (50.4%) | ||||||
| TT | 126 (20.8%) | 64 (17.1%) | 86 (20.4%) | ||||||
| G | 659 (54.3%) | 428 (57.2%) | 458 (54.4%) | ||||||
| T | 555 (45.7%) | 320 (42.8%) | 384 (45.6%) | ||||||
| TNFRSF1B | |||||||||
| rs1061622 | 0.32 | 0.92 (0.87–1.28) | 0.32 | 1.02 (0.91–1.46) | 0.35 | 0.38 | |||
| TT | 398 (65.9%) | 262 (69.7%) | 297 (70.5%) | ||||||
| TG | 190 (31.5%) | 102 (27.1%) | 116 (27.6%) | ||||||
| GG | 16 (2.6%) | 12 (3.2%) | 8 (1.9%) | ||||||
| T | 986 (81.6%) | 626 (83.2%) | 710 (84.3%) | ||||||
| G | 222 (18.4%) | 126 (16.8%) | 132 (15.7%) | ||||||
| rs3397 | 0.48 | 1.04 (0.92–1.41) | 0.17 | 1.28 (0.94–1.68) | 0.30 | 0.32 | |||
| GG | 255 (41.9%) | 156 (41.8%) | 158 (37.6%) | ||||||
| AG | 304 (49.9%) | 194 (52%) | 232 (55.2%) | ||||||
| AA | 50 (8.2%) | 23 (6.2%) | 30 (7.2%) | ||||||
| G | 814 (66.8%) | 506 (67.8%) | 548 (65.2%) | ||||||
| A | 404 (33.2%) | 240 (32.2%) | 292 (34.8%) | ||||||
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval. Padj and ORadj came from multivariate logistic regression. P1 adj and OR1 adj, healthy control group vs severe sepsis group. P2 adj and OR2 adj, sepsis group vs severe sepsis group. A P-value of <0.005 (0.05/10) was considered statistically significant after Bonferroni correction.
Association Analyses of TNF, LTA, TNFRSF1A and TNFRSF1B SNPs with Severe Sepsis Outcomes
We next investigated the association between all tested SNPs and 30-day mortality. The overall 30-day mortality rate among severe sepsis patients was 36.1%. We compared the allele and genotype distributions of all tested SNPs between survivors and non survivors of severe sepsis patients. No association was observed between TNF, LTA, TNFRSF1A and TNFRSF1B variants and 30-day mortality in the severe sepsis cohort in either the unadjusted or adjusted models (Table 4).
Table 4. Association analysis of SNPs in TNF, LTA, TNFRSF1A and TNFRSF1B between survivors and non-survivors of severe sepsis patients.
| Gene | Allelic Comparison | Genotypic Comparison | |||
| SNP | Nonsurvior | Survior | Padj | ORadj 95% CI | Padj |
| TNF | |||||
| rs1800629 | 0.43 | 0.72 (0.58–1.32) | 0.56 | ||
| GG | 137 (89%) | 232 (85%) | |||
| GA | 17 (11%) | 39 (14.3%) | |||
| AA | 0 (0%) | 2 (0.7%) | |||
| G | 291 (94.5%) | 503 (92.1%) | |||
| A | 17 (5.5%) | 43 (7.9%) | |||
| rs361525 | 0.68 | 1.15 (0.59–1.98) | 0.62 | ||
| CC | 135 (87.7%) | 247 (90.1%) | |||
| CT | 19 (12.3%) | 27 (9.9%) | |||
| C | 289 (93.8%) | 521 (95.1%) | |||
| T | 19 (6.2%) | 27 (4.9%) | |||
| rs1799724 | 0.21 | 0.76 (0.49–1.21) | 0.12 | ||
| GG | 109 (71.7%) | 176 (65.7%) | |||
| GA | 40 (26.3%) | 90 (33.6%) | |||
| AA | 3 (2%) | 2 (0.7%) | |||
| G | 258 (84.9%) | 442 (82.5%) | |||
| A | 46 (15.1%) | 94 (17.5%) | |||
| rs1799964 | 0.68 | 1.04 (0.72–1.53) | 0.78 | ||
| TT | 93 (60.8%) | 172 (63.7%) | |||
| TC | 52 (34%) | 88 (32.6%) | |||
| CC | 8 (5.2%) | 10 (3.7%) | |||
| T | 238 (77.8%) | 432 (80%) | |||
| C | 68 (22.2%) | 108 (20%) | |||
| rs1800630 | 0.42 | 1.18 (0.81–1.67) | 0.76 | ||
| CC | 97 (63%) | 178 (68.2%) | |||
| AC | 52 (33.7%) | 76 (29.1%) | |||
| AA | 5 (3.3%) | 7 (2.7%) | |||
| C | 246 (80%) | 432 (82.8%) | |||
| A | 62 (20%) | 90 (17.2%) | |||
| LTA | |||||
| rs909253 | 0.26 | 0.82 (0.54–1.12) | 0.38 | ||
| AA | 58 (37.4%) | 82 (31.4%) | |||
| AG | 62 (40%) | 119 (45.6%) | |||
| GG | 35 (22.6%) | 60 (23%) | |||
| A | 178 (57.4%) | 283 (54.2%) | |||
| G | 132 (42.6%) | 239 (45.8%) | |||
| TNFRSF1A | |||||
| rs767455 | 0.16 | 1.26 (0.87–1.86) | 0.55 | ||
| AA | 106 (69.7%) | 196 (72.3%) | |||
| GA | 41(27%) | 71 (26.2%) | |||
| GG | 5 (3.3%) | 4 (1.5%) | |||
| A | 253 (83.2%) | 463 (85.4%) | |||
| G | 51 (16.8%) | 79 (14.6%) | |||
| rs4149570 | 0.65 | 1.15 (0.86–1.56) | 0.26 | ||
| GG | 48 (31.4%) | 75 (28%) | |||
| GT | 70 (45.6%) | 142 (53%) | |||
| TT | 35 (23%) | 51 (19%) | |||
| G | 166 (54.2%) | 292 (54.5%) | |||
| T | 140 (45.8%) | 244 (45.5%) | |||
| TNFRSF1B | |||||
| rs1061622 | 0.64 | 1.18 (0.78–1.82) | 0.66 | ||
| TT | 101 (67.3%) | 196 (72.3%) | |||
| TG | 46 (30.7%) | 70 (25.8%) | |||
| GG | 3 (2%) | 5 (1.9%) | |||
| T | 248 (82.7%) | 462 (85.2%) | |||
| G | 52 (17.3%) | 80 (14.8%) | |||
| rs3397 | 0.46 | 1.12 (0.82–1.61) | 0.21 | ||
| GG | 52 (34.4%) | 106 (39.4%) | |||
| AG | 90 (59.6%) | 142 (52.8%) | |||
| AA | 9 (6%) | 21 (7.8%) | |||
| G | 194 (64.2%) | 354 (65.8%) | |||
| A | 108 (35.8%) | 184 (34.2%) | |||
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval. Padj and ORadj came from multivariate logistic regression. A P-value of <0.005 (0.05/10) was considered statistically significant after Bonferroni correction. The comparator group was survivors.
Rs1800629 Genotypes were Associated with Elevated TNF-α Concentrations
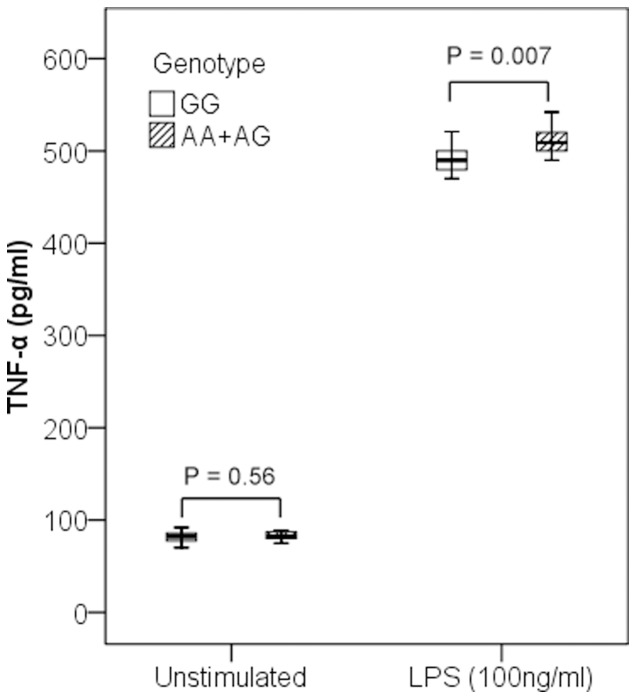
To determine whether rs1800629 genotypes influenced TNF-α production, we investigated TNF-α levels in culture supernatants of PBMCs obtained from 24 healthy volunteers. We observed a significant association between TNF-α levels and rs1800629 genotypes under the LPS-stimulated condition. AA+AG genotypes were associated with higher levels of TNF-α compared with GG genotype after LPS stimulation (P = 0.007) (Figure 1). However, no significant association was observed under the unstimulated condition.
Figure 1. Association of TNF-α levels and rs1800629 genotypes in healthy volunteers.
Concentrations of TNF-α in culture supernatants of PBMCs were expressed as the median, interquartile range and extremes. The TNF-α levels were significantly different between individuals with AA+GA and GG genotypes under the LPS-stimulated condition (P = 0.007). However, no significant difference was observed under the unstimulated condition (P = 0.56).
Furthermore, we measured TNF-α serum concentrations in 120 severe sepsis patients, including 104 patients with rs1800629GG genotype, 14 patients with GA genotype and 2 patients with AA genotype. Our results showed that rs1800629A allele was associated with higher TNF-α serum concentrations on the first day of severe sepsis. As shown in Figure 2, the serum concentration of TNF-α in severe sepsis patients with AA+AG genotypes was significantly higher than that of patients with GG genotype (550.4±73.6 pg/mL vs. 488.0±68.5 pg/mL, P = 0.001). To control confounding variables, we used the possible confounding factors (age, gender and APACHE II scores) as covariates in a linear regression model and found that the rs1800629 genotypes remained associated with TNF serum concentration (Padj = 0.02).
Figure 2. Association of TNF-α levels and rs1800629 genotypes in severe sepsis patients.
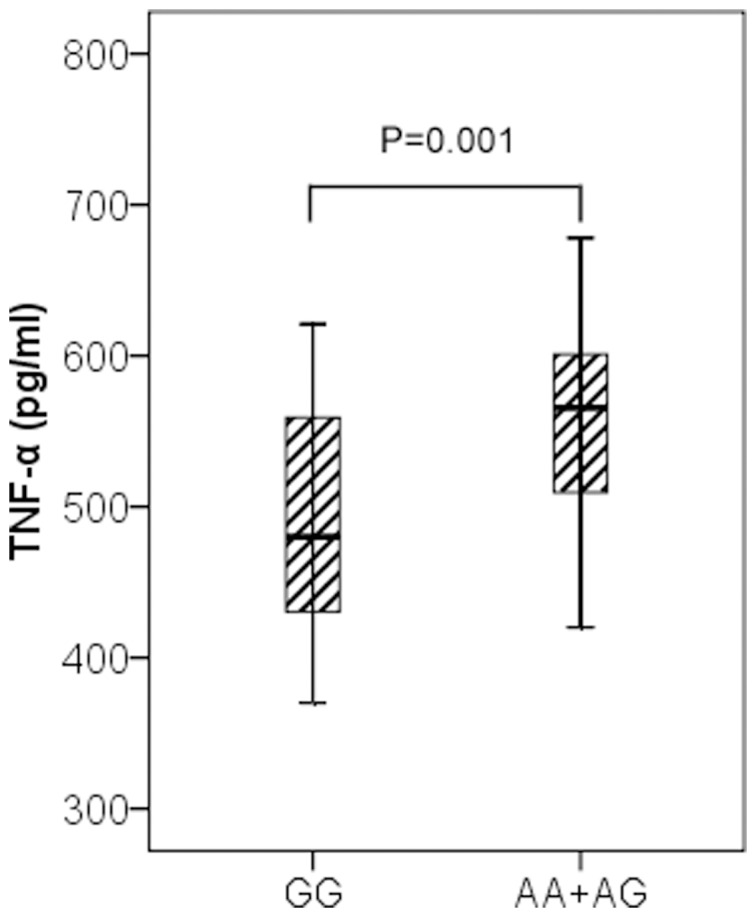
TNF-α serum levels in severe sepsis patients were expressed as the median, interquartile range and extremes. The TNF-α levels were significantly different between individuals with AA+GA and GG genotypes (P = 0.001). The difference remained significant (Padj = 0.02) after adjustment for age, gender and APACHE II scores in a linear regression model.
Discussion
Several genetic variants within genes involved in pro-inflammatory response have been associated with morbidity and mortality in patients with severe sepsis or septic shock, which is a complex and multifactorial syndrome [2]. TNF-α is an important pro-inflammatory cytokine involved in sepsis; several functional SNPs in TNF and LTA have been extensively studied in sepsis [29], [37]. However, results from previous studies were inconsistent. Discrepancies among previous studies may have resulted from differences in the populations studied, sepsis phenotype or imprecise definition of phenotype and limited sample size [38]. Considering these factors that might affect the results, we designed the present study with large samples size to achieve greater statistical power. Furthermore, all the samples were recruited from central Chinese Han population, thus the ethnic heterogeneity could be eliminated.
To our knowledge, this was the first relatively large-scale study investigating associations of genetic variants within TNF, LTA, TNFRSF1A and TNFRSF1B with severe sepsis in Chinese Han population. Our results provided evidence that rs1800629, a functional SNP in the promoter region of TNF, was significantly associated with susceptibility to severe sepsis in Chinese Han population. The association between rs1800629 and severe sepsis risk may be explained by its influence on the expression of TNF-α. In this study, we found that the risk allele (rs1800629A) is associated with increased TNF-α production in PBMCs from healthy subjects after stimulation with LPS. Moreover, we found that TNF-α serum levels in severe sepsis patients with AA+AG genotypes for rs1800629 were significantly higher than those in the individuals with GG genotype. Previous studies also showed that rs1800629A allele was associated with a six fold higher expression of both basal and induced TNF mRNA [39], [40]. Menges et al. found that the plasma TNF-α concentrations in patients with sepsis secondary to severe traumatic injury were significantly elevated in rs1800629A carriers on the first day after admission and for the following 14 days [41]. As TNF-α plays a pivotal role in the pathogenesis of severe sepsis in response to infection, it is reasonable to assume that patients with rs1800629A allele might produce a higher amount of TNF-α, and therefore become more susceptible to severe sepsis. In contrast to the TNF −308G/A polymorphism, the LTA +252A/G was not associated with the development of severe sepsis in our study. Our data showed that −308G/A of TNF and +252A/G of LTA were in weak linkage disequilibrium (LD) (D′ = 0.118) in Chinese Han population. The LD pattern is quite dissimilar to Caucasian population, which might result from the racial difference [24].
Rs1800629 was not associated with mortality among subjects with severe sepsis in our study. This was consistent with the study by Stuber et al., which demonstrated that the rs1800629 genotypes were not associated with poorer prognosis in severe sepsis. However, they did not find an association between rs1800629 genotypes and plasma TNF-α levels [26]. Recently, Teuffel et al. conducted a systematic review and meta-analysis, which also concluded that rs1800629 (TNF −308 AA/AG, TNF2) was associated with susceptibility to sepsis, but not with sepsis mortality [29].
Several studies have proposed that genetic variation in TNFRSF1A and TNFRSF1B was associated with susceptibility to inflammatory and autoimmune diseases, such as tuberculosis, systemic lupus erythematosus, rheumatoid arthritis and Crohn's disease [32], [33], [34], [35]. However, up to now, only one case control study investigated associations between TNFRSF1A and TNFRSF1B polymorphisms and sepsis susceptibility [36]. Four potentially functional SNPs in TNFRSF1A and TNFRSF1B were genotyped in our study. However, none showed association with susceptibility to or death from severe sepsis in Chinese Han population. Our findings were consistent with the results of Gordon et al. that five functional SNPs in TNFRSF1A and TNFRSF1B were not associated with susceptibility to or outcomes from sepsis in Caucasian population [36].
Potential limitations of this study should be addressed. First, although we knew that different pathogens had different impact on severity and outcomes of sepsis, we did not perform stratification analysis by different pathogens due to small number of cases with a definite microbiologic diagnosis. Second, we did not resequence these genes or select tag SNPs for genotyping. Instead, only ten potentially functional SNPs in TNF, LTA, TNFRSF1A and TNFRSF1B were included in our study, which was far from comprehensive. Indeed, these four genes are highly polymorphic. Therefore, it was possible that some important SNPs might be missed or the observed association might be due to other polymorphisms in LD with the studied ones. Additionally, assuming the prevalence of 0.01 for severe sepsis and using a significance level of 0.05, our study with 432 severe sepsis patients and 624 healthy controls had about 80% power to detect a 5% risk allele with an odds ratio of 1.63. Variant with an effect size smaller than this cannot convincingly be excluded based on these results. Therefore, our results cannot exclude variant associations with weaker effects between severe sepsis and the other three candidate genes (LTA, TNFRSF1A, TNFRSF1B). A more highly powered study involving thousands of subjects may yet exclude the role of these variants in severe sepsis susceptibility and outcomes.
In conclusion, our relatively large scale association study demonstrated that individuals with a functional variant in the promoter region of TNF may confer susceptibility to severe sepsis. However, common functional genetic variants in TNF, LTA, TNFRSF1A and TNFRSF1B were not associated with severe sepsis mortality in Chinese Han population.
Materials and Methods
Ethics Statement
This study was approved by the Ethics Study Board of Zhongshan Hospital, Fudan University, Shanghai, China (Record no: 2006-23). Written informed consent was obtained from patients or the next of kin, carers or guardians on the behalf of the participants before enrollment.
Study Design and Enrollment
From May 2005 to March 2011, a total of 432 severe sepsis patients, 384 sepsis patients and 624 ethnic-matched healthy controls were enrolled in this study (Table 1). The severe sepsis patients were those admitted to the Emergency, Surgical and Respiratory ICU at Zhongshan Hospital. The sepsis patients were those admitted to Zhongshan Hospital, but did not develop severe sepsis during hospital stay. The sepsis patients were considered as at risk controls for severe sepsis. Of 384 sepsis patients, 174 patients overlapped with that from our previous study [42]. Another 210 sepsis patients were collected between May 2008 and March 2011, and these patients were not included in our previous study. Sepsis patients recruited in the current study included multi-trauma subjects and patients with a history of chronic heart, renal, liver or pulmonary failure, thus they spent a long time (more than 8 days on average) on ICU (Table 1). Sex- and age-matched controls were selected from healthy blood donors. To reduce the potential confounding from ethnic backgrounds, only central Han Chinese individuals were recruited in this study.
The diagnosis of sepsis was based on the criteria presented at the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference in 1992 [43]. Severe sepsis was defined as sepsis in combination with infection-induced acute organ dysfunction in at least one organ. Acute organ dysfunction was defined as Sequential Organ Failure Assessment (SOFA) scores more than 2 for the organ in question. Baseline characteristics (age, gender and previous health status), as well as clinical data including Acute Physiology and Chronic Health Evaluation II (APACHE II) and SOFA scores, source of infection, microbiology and ICU mortality were obtained after the patient met sepsis criteria. The APACHE II and SOFA scores were calculated in the first 24 hours after the diagnosis of sepsis and severe sepsis. All patients included in the protocol were followed up for 30 days or hospital discharge. When cultures were absent or negative, the source of infection was determined by two senior physicians. Exclusion criteria included age below 18 years, pregnancy, severe chronic respiratory disease, severe chronic liver disease (defined as a Child–Pugh score of >10), malignancy, using of high-dose immunosuppressive therapy and AIDS diagnosis. Questionnaires were obtained from all control subjects to document smoking status, and history of chronic illness or severe sepsis. Healthy controls were defined as individuals without any recent acute illness, chronic illness or history of sepsis and severe sepsis.
SNPs Selection and Genotyping
Previous studies found that several functional SNPs in TNF, LTA, TNFRSF1A and TNFRSF1B were associated with inflammatory and autoimmune diseases. In our study, SNPs in TNF, LTA, and TNFRSF1A and TNFRSF1B were selected based on the following criteria: (1) location within the gene region (promoter, intron, exon, 3′UTR and 5′UTR); (2) association with inflammatory and autoimmune diseases such as sepsis, asthma, tuberculosis, systemic lupus erythematosus, rheumatoid arthritis and Crohn's disease in more than two studies. A total of ten SNPs were selected and genotyped in our study. Location and characterization of all selected SNPs were listed in Table 2.
Genomic DNA was extracted from whole blood with a FlexiGene DNA Kit (Qiagen, Hilden, Germany) in accordance with the protocol of the manufacturer. Six SNPs (rs1800629, rs1799724, rs361525, rs1800630, rs1799964 and rs909253) in TNF and LTA were selected and genotyped by direct sequencing. The sequencing reactions were performed using Applied Biosystems BigDye (version 3.1) chemistry (Applied Biosystem, Foster City, CA, USA), and the sequences were resolved using an ABI 3730 Genetic Analyzer. Analyses of the sequence traces were performed using the Staden package and were double scored by a second operator. The primers and PCR protocols used were shown in Table S1. Four SNPs in TNFRSF1A (rs767455, rs4149570) and TNFRSF1B (rs1061622, rs3397) were selected and genotyped on the GenomeLab SNPstream high-throughput 12-plex genotyping platform (Beckman Coulter, Fullerton, CA) following the manufacturer's instructions. The primers for PCR and single base extension were performed with Beckman Coulter Autoprimer software and shown in Table S2.
Isolation and Stimulation of Cells from Healthy Subjects
To determine the associations between rs1800629 genotypes and TNF-α levels in PBMCs, we investigated 15 subjects with rs1800629GG genotype, 8 subjects with GA genotype and 1 subject with AA genotype. PBMCs were derived by using Ficoll gradient density centrifugation method. Isolated PBMCs were plated at a density of 1×106 cells/ml in 24-well plates and cultured in RPMI 1640 medium with 10% FBS at 37 °C with 5% CO2. The cells were then incubated for 6 hours in presence or absence of 100 ng/ml Escherichia coli 0111:B4 LPS (Sigma, USA). After incubation, supernatants were harvested and stored at −80°C until use.
Serum Collection and TNF-α Level Measurement
Blood samples (5 mL) were collected within 24 hours of meeting criteria for severe sepsis. Samples were centrifuged at 4°C for 10 min at 3200 rpm within 60 min after collection. Then the serum was stored at −80°C until use. TNF-α level was determined by human ELISA kit (R&D Systems, USA) according to the manufacturer's protocol.
Statistical Analysis
The genotype data of cases and controls was analyzed for deviations from Hardy-Weinberg equilibrium by the Haploview v4.1 software [44]. The differences in allele and genotype distributions between severe sepsis and control groups were compared using χ2-test or Fisher's exact test when appropriate. The test for association with genotypes used the global genotype test in the 3×2 contingency table. Allele frequencies of cases and controls were used to calculate the OR and the 95% CI. Multivariate logistic regression was used to adjust for potential confounding factors. When comparing severe sepsis group to sepsis group, we entered the genotypes or alleles in the multivariate models controlling for the confounding variables including age, gender, history of diseases, source of infection, APACHE II and SOFA scores. When comparing severe sepsis patients to healthy controls, age and gender were included in the multivariate models. The Bonferroni method was used to correct for multiple comparisons where applicable. The power analysis was performed using the Genetic Power Calculator web tool [45]. A two tailed P-value of <0.05 was considered statistically significant, whereas a value of corrected P<(0.05/number of tests) was considered significant after Bonferroni correction. Continuous variables were described as either a mean ± standard deviation, or as a median with interquartile range. TNF-α serum levels between individuals with different rs1800629 genotypes (AA+GA vs. GG) were compared by Student's t-test. To determine whether an association with rs1800629 genotypes might depend on other potential confounding factors for TNF-α serum levels, we investigated the association of rs1800629 genotypes by adding the polymorphisms to a linear regression model controlling for age, gender and APACHE II scores. The software used for statistical calculations was SPSS 15.0 (SPSS Inc., Chicago, IL, USA) unless specified.
Supporting Information
Primers and PCR protocols for six SNPs in TNF and LTA .
(DOC)
The primers of SNPs in TNFRSF1A and TNFRSF1B .
(DOC)
Acknowledgments
We gratefully thank Drs Jae Woo Lee, Keyong Li and Zhiqiang Dong for their helpful comments on this manuscript.
Funding Statement
This work was supported by grants from the Major Program of the National Natural Science Foundation of China (30930090), the National Natural Science Foundation of China (81000023 and 81171837), the Shanghai Committee of Science and Technology (09411960400) and the Shanghai Public Health Fund for Distinguished Young Scholars (08GWQ026). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Martin GS, Mannino DM, Eaton S, Moss M (2003) The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 348: 1546–1554. [DOI] [PubMed] [Google Scholar]
- 2. Dahmer MK, Randolph A, Vitali S, Quasney MW (2005) Genetic polymorphisms in sepsis. Pediatr Crit Care Med 6: S61–73. [DOI] [PubMed] [Google Scholar]
- 3. Holmes CL, Russell JA, Walley KR (2003) Genetic polymorphisms in sepsis and septic shock: role in prognosis and potential for therapy. Chest 124: 1103–1115. [DOI] [PubMed] [Google Scholar]
- 4. Song Z, Yin J, Yao C, Sun Z, Shao M, et al. Variants in the Toll-interacting protein gene are associated with susceptibility to sepsis in the Chinese Han population. Crit Care 15: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hamann L, Kumpf O, Schuring RP, Alpsoy E, Bedu-Addo G, et al. (2009) Low frequency of the TIRAP S180L polymorphism in Africa, and its potential role in malaria, sepsis, and leprosy. BMC Med Genet 10: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arcaroli J, Silva E, Maloney JP, He Q, Svetkauskaite D, et al. (2006) Variant IRAK-1 haplotype is associated with increased nuclear factor-kappaB activation and worse outcomes in sepsis. Am J Respir Crit Care Med 173: 1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Toubiana J, Courtine E, Pene F, Viallon V, Asfar P, et al. (2010) IRAK1 functional genetic variant affects severity of septic shock. Crit Care Med 38: 2287–2294. [DOI] [PubMed] [Google Scholar]
- 8. Ferwerda B, Alonso S, Banahan K, McCall MB, Giamarellos-Bourboulis EJ, et al. (2009) Functional and genetic evidence that the Mal/TIRAP allele variant 180L has been selected by providing protection against septic shock. Proc Natl Acad Sci U S A 106: 10272–10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pino-Yanes M, Corrales A, Casula M, Blanco J, Muriel A, et al. (2010) Common variants of TLR1 associate with organ dysfunction and sustained pro-inflammatory responses during sepsis. PLoS ONE 5: e13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barber RC, Aragaki CC, Rivera-Chavez FA, Purdue GF, Hunt JL, et al. (2004) TLR4 and TNF-alpha polymorphisms are associated with an increased risk for severe sepsis following burn injury. J Med Genet 41: 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng X, Zheng H, Lan R, Ye C, Wang Y, et al. (2011) Identification of genes and genomic islands correlated with high pathogenicity in Streptococcus suis using whole genome tiling microarrays. PLoS ONE 6: e17987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jessen KM, Lindboe SB, Petersen AL, Eugen-Olsen J, Benfield T (2007) Common TNF-alpha, IL-1 beta, PAI-1, uPA, CD14 and TLR4 polymorphisms are not associated with disease severity or outcome from Gram negative sepsis. BMC Infect Dis 7: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reid CL, Perrey C, Pravica V, Hutchinson IV, Campbell IT (2002) Genetic variation in proinflammatory and anti-inflammatory cytokine production in multiple organ dysfunction syndrome. Crit Care Med 30: 2216–2221. [DOI] [PubMed] [Google Scholar]
- 14. Hotchkiss RS, Karl IE (2003) The pathophysiology and treatment of sepsis. N Engl J Med 348: 138–150. [DOI] [PubMed] [Google Scholar]
- 15. Debets JM, Kampmeijer R, van der Linden MP, Buurman WA, van der Linden CJ (1989) Plasma tumor necrosis factor and mortality in critically ill septic patients. Crit Care Med 17: 489–494. [DOI] [PubMed] [Google Scholar]
- 16. Douni E, Kollias G (1998) A critical role of the p75 tumor necrosis factor receptor (p75TNF-R) in organ inflammation independent of TNF, lymphotoxin alpha, or the p55TNF-R. J Exp Med 188: 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nedwin GE, Naylor SL, Sakaguchi AY, Smith D, Jarrett-Nedwin J, et al. (1985) Human lymphotoxin and tumor necrosis factor genes: structure, homology and chromosomal localization. Nucleic Acids Res 13: 6361–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mira JP, Cariou A, Grall F, Delclaux C, Losser MR, et al. (1999) Association of TNF2, a TNF-alpha promoter polymorphism, with septic shock susceptibility and mortality: a multicenter study. Jama 282: 561–568. [DOI] [PubMed] [Google Scholar]
- 19. Appoloni O, Dupont E, Vandercruys M, Andriens M, Duchateau J, et al. (2001) Association of tumor necrosis factor-2 allele with plasma tumor necrosis factor-alpha levels and mortality from septic shock. Am J Med 110: 486–488. [DOI] [PubMed] [Google Scholar]
- 20. O'Keefe GE, Hybki DL, Munford RS (2002) The G→A single nucleotide polymorphism at the −308 position in the tumor necrosis factor-alpha promoter increases the risk for severe sepsis after trauma. J Trauma 52: 817–825; discussion 825–816. [DOI] [PubMed] [Google Scholar]
- 21. Calvano JE, Um JY, Agnese DM, Hahm SJ, Kumar A, et al. (2003) Influence of the TNF-alpha and TNF-beta polymorphisms upon infectious risk and outcome in surgical intensive care patients. Surg Infect (Larchmt) 4: 163–169. [DOI] [PubMed] [Google Scholar]
- 22. Wang S, Wei M, Han Y, Zhang K, He L, et al. (2008) Roles of TNF-alpha gene polymorphisms in the occurrence and progress of SARS-Cov infection: a case-control study. BMC Infect Dis 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang GJ, Huang SL, Yien HW, Chen WS, Chi CW, et al. (2000) Tumor necrosis factor gene polymorphism and septic shock in surgical infection. Crit Care Med 28: 2733–2736. [DOI] [PubMed] [Google Scholar]
- 24. Duan ZX, Gu W, Zhang LY, Jiang DP, Zhou J, et al. (2011) Tumor necrosis factor alpha gene polymorphism is associated with the outcome of trauma patients in Chinese Han population. J Trauma 70: 954–958. [DOI] [PubMed] [Google Scholar]
- 25. Majetschak M, Flohe S, Obertacke U, Schroder J, Staubach K, et al. (1999) Relation of a TNF gene polymorphism to severe sepsis in trauma patients. Ann Surg 230: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stuber F, Udalova IA, Book M, Drutskaya LN, Kuprash DV, et al. (1995) −308 tumor necrosis factor (TNF) polymorphism is not associated with survival in severe sepsis and is unrelated to lipopolysaccharide inducibility of the human TNF promoter. J Inflamm 46: 42–50. [PubMed] [Google Scholar]
- 27. Garnacho-Montero J, Aldabo-Pallas T, Garnacho-Montero C, Cayuela A, Jimenez R, et al. (2006) Timing of adequate antibiotic therapy is a greater determinant of outcome than are TNF and IL-10 polymorphisms in patients with sepsis. Crit Care 10: R111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duan ZX, Gu W, Zhang LY, Jiang DP, Zhou J, et al. (2011) Tumor necrosis factor alpha gene polymorphism is associated with the outcome of trauma patients in Chinese Han population. J Trauma 70: 954–958. [DOI] [PubMed] [Google Scholar]
- 29. Teuffel O, Ethier MC, Beyene J, Sung L (2010) Association between tumor necrosis factor-alpha promoter −308 A/G polymorphism and susceptibility to sepsis and sepsis mortality: a systematic review and meta-analysis. Crit Care Med 38: 276–282. [DOI] [PubMed] [Google Scholar]
- 30. Secher T, Vasseur V, Poisson DM, Mitchell JA, Cunha FQ, et al. (2009) Crucial role of TNF receptors 1 and 2 in the control of polymicrobial sepsis. J Immunol 182: 7855–7864. [DOI] [PubMed] [Google Scholar]
- 31. Ebach DR, Riehl TE, Stenson WF (2005) Opposing effects of tumor necrosis factor receptor 1 and 2 in sepsis due to cecal ligation and puncture. Shock 23: 311–318. [DOI] [PubMed] [Google Scholar]
- 32. Moller M, Flachsbart F, Till A, Thye T, Horstmann RD, et al. (2010) A functional haplotype in the 3′untranslated region of TNFRSF1B is associated with tuberculosis in two African populations. Am J Respir Crit Care Med 181: 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horiuchi T, Washio M, Kiyohara C, Tsukamoto H, Tada Y, et al. (2009) Combination of TNF-RII, CYP1A1 and GSTM1 polymorphisms and the risk of Japanese SLE: findings from the KYSS study. Rheumatology (Oxford) 48: 1045–1049. [DOI] [PubMed] [Google Scholar]
- 34. Waschke KA, Villani AC, Vermeire S, Dufresne L, Chen TC, et al. (2005) Tumor necrosis factor receptor gene polymorphisms in Crohn's disease: association with clinical phenotypes. Am J Gastroenterol 100: 1126–1133. [DOI] [PubMed] [Google Scholar]
- 35. Constantin A, Dieude P, Lauwers-Cances V, Jamard B, Mazieres B, et al. (2004) Tumor necrosis factor receptor II gene polymorphism and severity of rheumatoid arthritis. Arthritis Rheum 50: 742–747. [DOI] [PubMed] [Google Scholar]
- 36. Gordon AC, Lagan AL, Aganna E, Cheung L, Peters CJ, et al. (2004) TNF and TNFR polymorphisms in severe sepsis and septic shock: a prospective multicentre study. Genes Immun 5: 631–640. [DOI] [PubMed] [Google Scholar]
- 37. Tiancha H, Huiqin W, Jiyong J, Jingfen J, Wei C (2011) Association between lymphotoxin-alpha intron +252 polymorphism and sepsis: a meta-analysis. Scand J Infect Dis 43: 436–447. [DOI] [PubMed] [Google Scholar]
- 38. Clark MF, Baudouin SV (2006) A systematic review of the quality of genetic association studies in human sepsis. Intensive Care Med 32: 1706–1712. [DOI] [PubMed] [Google Scholar]
- 39. Louis E, Franchimont D, Piron A, Gevaert Y, Schaaf-Lafontaine N, et al. (1998) Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin Exp Immunol 113: 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huizinga TW, Westendorp RG, Bollen EL, Keijsers V, Brinkman BM, et al. (1997) TNF-alpha promoter polymorphisms, production and susceptibility to multiple sclerosis in different groups of patients. J Neuroimmunol 72: 149–153. [DOI] [PubMed] [Google Scholar]
- 41. Menges T, Konig IR, Hossain H, Little S, Tchatalbachev S, et al. (2008) Sepsis syndrome and death in trauma patients are associated with variation in the gene encoding tumor necrosis factor. Crit Care Med 36: 1456–1462, e1451–1456. [DOI] [PubMed] [Google Scholar]
- 42. Song Z, Tong C, Sun Z, Yao C, Shao M, et al. (2009) Association study of TLR4 polymorphisms with severe community-acquired pneumonia susceptibility and outcome. Chinese Journal of Emergency Medicine 18: 956–959. [Google Scholar]
- 43. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med 20: 864–874. [PubMed] [Google Scholar]
- 44. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 45. Purcell S, Cherny SS, Sham PC (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers and PCR protocols for six SNPs in TNF and LTA .
(DOC)
The primers of SNPs in TNFRSF1A and TNFRSF1B .
(DOC)