Regulation of the formin Bni1p by Cdc42p in yeast does not require direct interaction between Bni1p and Cdc42p. The Cdc42p effector Gic2p can bind both Bni1p and GTP-Cdc42p, providing a novel regulatory input.
Abstract
Actin filaments are dynamically reorganized to accommodate ever-changing cellular needs for intracellular transport, morphogenesis, and migration. Formins, a major family of actin nucleators, are believed to function as direct effectors of Rho GTPases, such as the polarity regulator Cdc42p. However, the presence of extensive redundancy has made it difficult to assess the in vivo significance of the low-affinity Rho GTPase–formin interaction and specifically whether Cdc42p polarizes the actin cytoskeleton via direct formin binding. Here we exploit a synthetically rewired budding yeast strain to eliminate the redundancy, making regulation of the formin Bni1p by Cdc42p essential for viability. Surprisingly, we find that direct Cdc42p–Bni1p interaction is dispensable for Bni1p regulation. Alternative paths linking Cdc42p and Bni1p via “polarisome” components Spa2p and Bud6p are also collectively dispensable. We identify a novel regulatory input to Bni1p acting through the Cdc42p effector, Gic2p. This pathway is sufficient to localize Bni1p to the sites of Cdc42p action and promotes a polarized actin organization in both rewired and wild-type contexts. We suggest that an indirect mechanism linking Rho GTPases and formins via Rho effectors may provide finer spatiotemporal control for the formin-nucleated actin cytoskeleton.
INTRODUCTION
Actin filaments function as tracks, scaffolds, and force-generating devices for numerous cellular processes, including intracellular transport, morphogenesis, and cell motility (Campellone and Welch, 2010). Dynamic regulation of actin filament assembly and disassembly is critical to ensure that proper cellular functions occur at the right time and place. Such regulation is believed to rely to a large degree on nucleators of actin polymerization, including formins.
Formins are a conserved family of proteins that nucleate actin polymerization and facilitate barbed-end elongation (Chesarone et al., 2010). These catalytic activities are conducted by the formin homology 2 (FH2) domain, which dimerizes to form a donut-shaped catalytic core (Xu et al., 2004; Otomo et al., 2005b). The adjacent proline-rich formin homology 1 (FH1) domain recruits profilin-bound actin monomers, speeding barbed-end elongation (Sagot et al., 2002b; Romero et al., 2004; Kovar et al., 2006). Catalytic activity of many formin proteins can be suppressed by autoinhibitory binding of an N-terminal Diaphanous inhibitory domain (DID) to a C-terminal Diaphanous autoregulatory domain (DAD; Alberts, 2001; Li and Higgs, 2003; Wang et al., 2009). Autoinhibition can be disrupted by binding of Rho GTPases to the GTPase-binding domain (GBD), which partially overlaps with the DID (Lammers et al., 2005; Otomo et al., 2005a; Rose et al., 2005; Nezami et al., 2006; Martin et al., 2007). However, releasing the antoinhibitory effect of DID–DAD interactions in vitro requires very high concentrations of Rho proteins, raising the possibility that other mechanisms may help to activate formins in vivo (Li and Higgs, 2003; Seth et al., 2006).
Studies in the yeast Saccharomyces cerevisiae showed that formins were specifically required to nucleate and organize particular actin structures (Evangelista et al., 2002; Pruyne et al., 2002; Sagot et al., 2002a, b; Kovar and Pollard, 2004). Budding yeast have two formins, Bni1p and Bnr1p, which share a conserved modular domain organization with the 15 mammalian formins (Chesarone et al., 2010). Bni1p and Bnr1p nucleate and elongate filaments that are organized into two actin structures in yeast: actin cables that are polarized along the mother–bud axis and act as tracks for myosin-mediated delivery of vesicles and organelles to the bud, and the actomyosin ring at the mother–bud neck that contributes to cytokinesis. Bni1p and Bnr1p are each sufficient for cell survival, whereas deleting both formins leads to lethality (Vallen et al., 2000; Ozaki-Kuroda et al., 2001).
The polarized organization of actin cables in yeast relies on the conserved Rho-family GTPase Cdc42p (Adams et al., 1990; Chen et al., 1997; Howell and Lew, 2012). Cdc42p can bind to Bni1p and colocalizes with Bni1p at the bud tip, suggesting that Bni1p is a direct effector of Cdc42p (Evangelista et al., 1997, 2002; Sagot et al., 2002a). In addition, Bni1p interacts with the “polarisome” components, Spa2p and Bud6p (Evangelista et al., 1997; Fujiwara et al., 1998; Sheu et al., 1998). Spa2p is believed to contribute to Bni1p localization at the bud tip (Fujiwara et al., 1998). Bud6p binds actin monomers (Amberg et al., 1997) and promotes Bni1p-mediated actin filament nucleation in vitro (Graziano et al., 2011). Remarkably, however, it appears that a short catalytic FH1–FH2 domain construct from either Bni1p or Bnr1p suffices to provide essential formin functions in yeast (Gao and Bretscher, 2009). The catalytic fragment is diffusely localized throughout the cell and lacks the domains required for autoinhibition and for interaction with Rho GTPases and polarisome components. Although an excess of actin cables appears to be randomly oriented in these cells, some degree of cable orientation can occur via compensatory mechanisms, such as actin-capturing myosin II at the bud neck (Gao and Bretscher, 2009). The yeast cell's ability to cope with deregulated formins has hindered efforts to apply the power of yeast genetics to test the proposed role of the Cdc42p–Bni1p connection.
Polarity establishment in wild-type budding yeast involves a positive feedback loop that concentrates active Cdc42p at the incipient bud site (Johnson et al., 2011). The feedback mechanism involves recruitment of a rapidly diffusing cytoplasmic polarity complex by cortical GTP-Cdc42p. The complex contains a p21-activated kinase, the scaffold protein Bem1p, and the Cdc42p-directed guanine nucleotide exchange factor Cdc24p, which promotes GTP loading of Cdc42p (Kozubowski et al., 2008). Recruitment of the complex by GTP–Cdc42p therefore causes neighboring Cdc42p to become GTP bound, leading to recruitment of more complex and the formation of a polarity cluster. Cdc42p polarization by this mechanism is independent of F-actin (Ayscough et al., 1997; Irazoqui et al., 2003). A polarity cluster containing Cdc42p, Bem1p, and effectors is believed to form before actin polarization (Howell et al., 2009), allowing it to subsequently nucleate (via formins) or capture (via poorly characterized mechanisms) actin cables.
We recently generated a “rewired” strain, in which the positive-feedback mechanism is synthetically rewired to involve actin cables (Howell et al., 2009). In the rewired cell, the sole copy of Bem1p is fused to the vesicle soluble N-ethylmaleimide–sensitive factor attachment protein receptor (v-SNARE) Snc2p, which is concentrated on secretory and endocytic vesicles. The idea behind this manipulation is that it simultaneously blocks the normal positive-feedback mechanism (because the Bem1p complex is no longer able to diffuse rapidly in the cytoplasm to be captured by cortical GTP–Cdc42p) and enables a different type of positive feedback (because the vesicle-tethered Bem1p–Snc2p complex can now be delivered to cortical GTP–Cdc42p by actin cables). As predicted by this hypothesis, F-actin is required for polarity establishment in rewired cells (Howell et al., 2009).
We now show that in rewired cells, Bni1p activity and its regulation are essential for viability, allowing us to dissect the functional relevance of individual Bni1p regulatory inputs. Surprisingly, neither direct regulation by Rho proteins nor interactions with the polarisome components were necessary for Bni1p regulation in this context. Instead, other N-terminal regions were sufficient to connect formin activity to Cdc42p. We identified a novel interaction of a region overlapping the DID with the Cdc42p effector Gic2p, and fusion of the formin FH1-FH2 domain to the N-terminus of Gic2p was sufficient to provide regulated formin function in both rewired and wild-type cells. Our data suggest that rather than serving as a direct Cdc42p effector, Bni1p (and possibly other formins) can be regulated indirectly via other Cdc42p effectors.
RESULTS
Dynamics of polarity establishment in wild-type and rewired cells
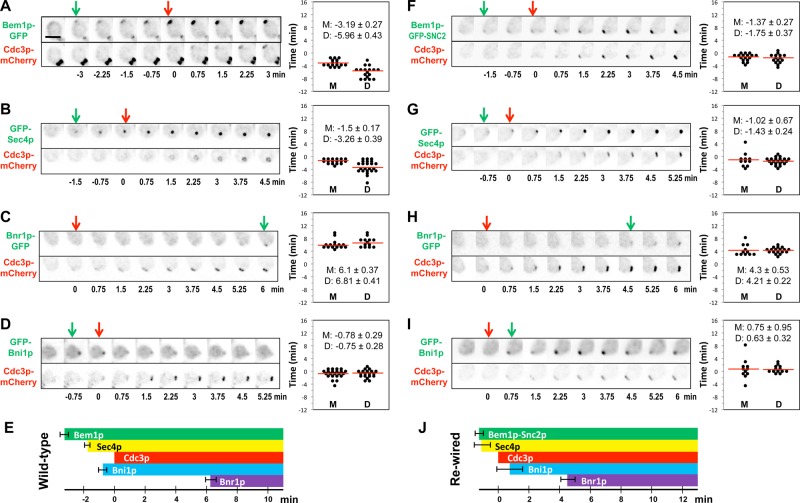
In wild-type cells, polarity proteins, including Bem1p and Cdc42p, are believed to become polarized before actin cables. In contrast, in rewired cells actin cables and the factors enabling their nucleation/capture by GTP–Cdc42p should all become concentrated at the polarization site with similar kinetics to the Bem1p-Snc2p protein. To investigate the timing of polarity-establishment events, we tagged various proteins with green fluorescent protein (GFP) at the endogenous locus in a strain that also expressed a red fluorescent septin marker, Cdc3p-mCherry. We quantified the intervals between green and red fluorescent markers’ first appearance at the incipient bud site. We focused on the polarization timing in mother cells because daughters (although displaying the same ordered sequence of events) exhibited greater variability in timing (Chen et al., 2011; Figure 1, right).
FIGURE 1:
Timing of protein polarization in wild-type and rewired cells. Wild-type (A–D) (DLY11909, 13890, 13344, 14014) and rewired (F–I) (DLY13072, 13884, 13945, 14026) cells were imaged and scored for the first appearance (arrows) of the indicated proteins at the incipient bud site. Inverted images are shown. Time intervals between polarization of GFP-tagged proteins and the septin Cdc3p-mCherry were quantified in mother and daughter cells (right; D, daughters; M, mothers). (E, J) Summary of the mean ± SEM timing in mother cells. Scale bar, 5 μm.
In wild-type cells, Bem1p-GFP recruitment preceded that of the septin marker Cdc3p-mCherry by ∼3 min (Chen et al., 2011; Figure 1A). GFP-Sec4p, a vesicle marker, was recruited 1.5 min before Cdc3p-mCherry (Figure 1B), indicating that actin cables polarize and deliver vesicles before septin recruitment. GFP-Bni1p was detected 0.8 min before Cdc3p-mCherry (Figure 1C), and Bnr1p-GFP was detected 6.1 min after Cdc3p-mCherry (Figure 1D). The fact that we detected Sec4p/vesicle accumulation before Bni1p accumulation was surprising, as we expected that Bni1p would be responsible for nucleating the actin cables that deliver the secretory vesicles. It could be that such cables are initially nucleated elsewhere and captured by polarity factors and that Bni1p recruitment does not occur until slightly later. However, the short lag between Sec4p and Bni1p recruitment to the polarization site may simply be due to the detection sensitivity for these probes, as the intensity of GFP-Bni1p signal is significantly weaker than that of GFP-Sec4p. Thus, during normal polarity establishment Bem1p accumulation is followed ∼1.5 min later by recruitment of actin cables and secretory vesicles, which is followed ∼1.5 min after that by recruitment of septins (Figure 1E). Recruitment of the formin Bnr1p occurred significantly later, once a septin ring was well established, suggesting that Bnr1p recruitment does not normally contribute to initial actin polarization.
In the rewired cells, Bem1p–GFP-Snc2p was detected at the incipient bud site only 0.9 min before Cdc3p-mCherry (Figure 1F), and GFP-Sec4p polarized 1.0 min before Cdc3p-mCherry (Figure 1G). We infer that Bem1p–Snc2p is recruited to the incipient bud site together with actin cables, as expected. GFP-Bni1p was detected 0.75 min after Cdc3p-mCherry (Figure 1H), whereas Bnr1p only arrived 4.3 min after Cdc3p-mCherry (Figure 1I). As in wild-type cells, the apparent lag between vesicle markers and Bni1p recruitment may stem from technical detection issues, or it may indicate that initial cables are nucleated elsewhere and then “captured” at the polarity site rather than locally nucleated at that site.
The timing data are summarized in Figure 1J. A striking feature is that Bni1p was recruited several minutes before Bnr1p. Thus, if local actin filament nucleation is important for rewired cell polarity establishment, then Bni1p would be predicted to play a critical role for polarization and survival of the rewired cells.
BNI1 is necessary for polarization in rewired cells
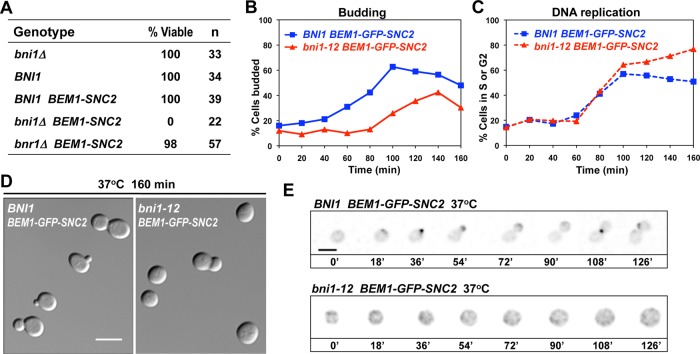
To test whether individual formins are essential in the rewired cells, we deleted one copy of BNI1 or BNR1 in a diploid strain and examined the viability of haploid segregants following sporulation and tetrad dissection. We found that bni1 mutants were synthetically lethal with BEM1-SNC2, indicating that BNI1 is essential in the rewired cells (Figure 2A). Deletion of BNR1, in contrast, was well tolerated by the rewired cells (Figure 2A), indicating that Bnr1p is neither necessary nor sufficient for formin functions in the rewired context. Based on the synthetic lethality and polarization timing, it seemed probable that BNI1 was the only formin present at the initial polarization site and needed for polarity establishment in the rewired cells; however, BNI1 has also been implicated in other cellular processes, such as cytokinesis and the proper organization of septin rings (Kohno et al., 1996; Kadota et al., 2004; Gladfelter et al., 2005). To investigate the essential role of Bni1p in rewired cells, we introduced the temperature-sensitive (ts) bni1-12 allele into the BEM1-GFP-SNC2 background and performed time-course experiments. G1-phase cells from permissive-temperature cultures were enriched by centrifugal elutriation and then shifted to restrictive temperature to inactivate bni1-12. Most bni1-12 BEM1-GFP-SNC2 cells failed to bud, and the remainder produced “fat” buds with wide necks (Figure 2B). The unbudded cells grew big and round (Figure 2D) and stayed unpolarized (Figure 2E), whereas the control BNI1 BEM1-GFP-SNC2 cells polarized and completed the budding cycle. Rewired BNI1 and bni1-12 cells replicated DNA with similar timing (Figure 2C), indicating that the G1–S transition did not require Bni1p. There was an accumulation of G2/M cells in the rewired bni1-12 strain at later times, consistent with the expectation that unbudded cells could not undergo cytokinesis. These results indicate that BNI1 is needed for efficient polarization in the rewired cells.
FIGURE 2:
BNI1 is essential in rewired cells. (A) Synthetic lethality of bni1 BEM1-SNC2. The spore viability of cells with the indicated genotypes was deduced from tetrad dissection of heterozygous diploids (DLY12648 and 12778). (B–E) Rewired cells carrying a temperature-sensitive Bni1p (DLY12226, compared with the control DLY8601) progress through the cell cycle but are defective in polarization and budding. G1-phase cells were enriched by centrifugal elutriation at 4°C and released into 37°C media to resume growth. Samples were fixed and scored for percentage budded (n > 100) (B) and percentage cells with S or G2 DNA content (n = 10,000) (C). (D) DIC images of cells released into 37°C media for 160 min. Most cells did not bud, and those that did formed very wide necks and did not complete cytokinesis. (E) Inverted time-lapse images of G1-phase cells, which were released and filmed at 37°C. The control BEM1-GFP-SNC2 cell (top) started to polarize and bud, whereas a similar-sized BEM1-GFP-SNC2 bni1-12 cell (bottom) grew big and round without establishing polarity. Scale bar, 5 μm.
Regulation of Bni1p is essential in rewired cells
Yeast cells lacking both BNI1 and BNR1 are able to survive if they express a formin catalytic fragment (FH1–FH2) believed to promote unregulated, delocalized actin nucleation (Gao and Bretscher, 2009). However, we found that the same Bni1p catalytic fragment (FH1–FH2) was not able to complement the requirement for BNI1 in rewired cells (Figure 3, A, B, and D). Thus, regulation of Bni1p is dispensable in normal cells but critical in rewired cells, making this genetic background ideal for dissecting the physiological significance of Bni1p regulatory inputs. Because the rewiring scheme does not involve changing the mechanism of actin regulation by Cdc42p, we expect that formin regulation in the rewired strain will be similar to that in the wild-type context.
FIGURE 3:
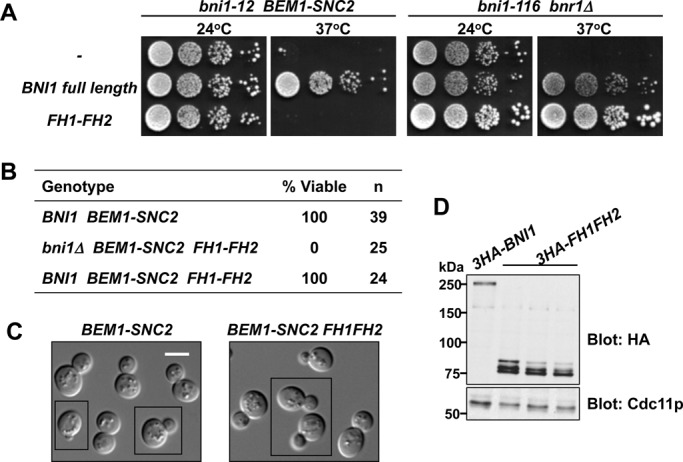
Bni1p regulation is essential in rewired cells. (A) A catalytic fragment of Bni1p (FH1–FH2) can provide formin function in wild-type but not rewired cells. Full-length Bni1p or the FH1–FH2 fragment was tested for the ability to rescue temperature-sensitive formin function in rewired (bni1-12 BEM1-SNC2; DLY12226, 14024, 12620) and otherwise wild-type (bni1-116 bnr1; DLY7924, 14025, 12627) strains at 37°C. Serial dilutions of cell cultures (containing ∼104, 103, 102, and 101 cells) were spotted on plates with rich media and incubated at the indicated temperature for 2.5 d. (B) FH1–FH2 does not rescue the synthetic lethality of bni1 BEM1-SNC2. Spore viability of cells with the indicated genotypes was deduced from tetrad dissection of heterozygous diploids (DLY12848, 14272). (C) FH1–FH2 is not dominantly deleterious in rewired cells. DIC images of exponentially growing rewired cells expressing (right; DLY14811) or not expressing (left; DLY13095) FH1–FH2 in addition to full-length Bni1p. Images were assembled from multiple fields (denoted with black boxes). Scale bar, 5 μm. (D) Immunoblotting shows expression of the full-length Bni1p (DLY14024) and the FH1-FH2 fragment (DLY12620, 12848, 14272) in the strains used in A and B. Cdc11p (septin) serves as loading control.
In principle, regulation of Bni1p might be needed for localizing the nucleation activity to the forming polarization site and/or for tuning (e.g., by autoinhibition) the catalytic activity to avoid producing an excessive number of actin filaments. In the latter scenario, unregulated nucleation by the FH1–FH2 fragment would be toxic even in the presence of wild-type Bni1p. We tested this hypothesis but found that rewired cells containing the FH1–FH2 fragment in addition to the endogenous Bni1p were viable and showed normal morphology (Figure 3, B–D). These results suggest that in rewired cells, unregulated formin activity is not catastrophic; rather, regulation is needed to target formin activity to the growing cluster of Cdc42p during polarity establishment.
The GTPase-binding domain is not necessary for Bni1p regulation
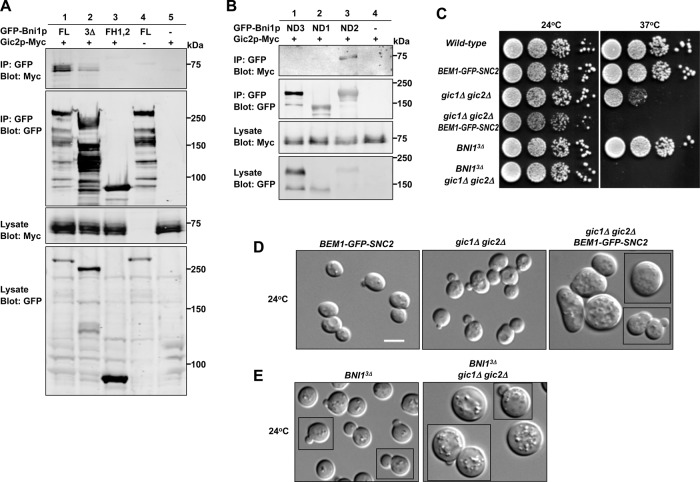
The prevailing view of formin regulation by Rho-family GTPases, based on biochemical dissection of diaphanous-family formins, is that direct interaction between the GTPase (in this case Cdc42p) and the formin mediates both targeting and activation through relief of autoinhibition (Alberts, 2001; Li and Higgs, 2003; Lammers et al., 2005; Otomo et al., 2005a; Rose et al., 2005; Nezami et al., 2006; Seth et al., 2006; Martin et al., 2007). However, we found that a construct lacking the Bni1p GTPase-binding domain (GBD) remained fully competent to rescue the formin requirement in rewired cells and yielded cells with normal morphology (Figure 4, A and B). This finding suggests that direct Cdc42p–Bni1p interaction is dispensable for formin regulation.
FIGURE 4:
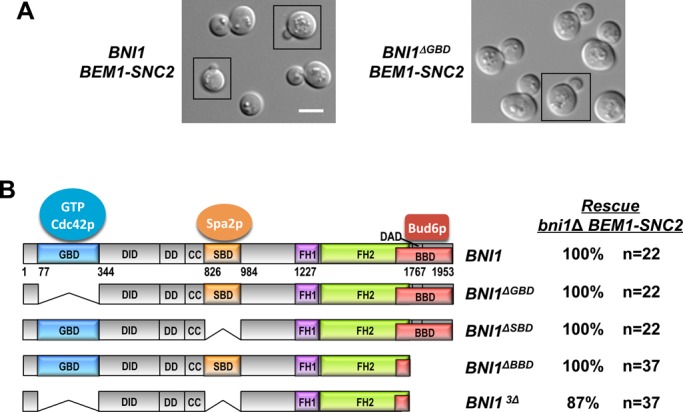
The GTPase-binding domain is not necessary for Bni1p regulation. (A) The GBD is not required for Bni1p regulation in rewired cells. DIC images of rewired cells expressing full-length (left; DLY13095) Bni1p or a version lacking the GBD (right; DLY14223). Scale bar, 5 μm. (B) Left, schematic of Bni1p indicating GBD, SBD, and BBD domains that interact with GTP-Cdc42p, Spa2p, and Bud6p, respectively. CC, coiled coil; DAD, Diaphanous autoregulatory domain; DD, dimerization domain; DID, Diaphanous inhibitory domain; FH1, formin homology 1; FH2, formin homology 2. Right, spore viability of cells with the indicated genotypes was deduced from tetrad dissection of heterozygous diploids (DLY14194, 14195, 12959, 12960).
In addition to the GBD, Bni1p harbors binding domains that interact with Spa2p (Spa2p-binding domain [SBD]) and Bud6p (Bud6p-binding domain [BBD]), polarisome components that can also bind to each other (Evangelista et al., 1997; Fujiwara et al., 1998; Sheu et al., 1998). Both Spa2p and Bud6p are localized to polarity sites (Snyder, 1989; Amberg et al., 1997; Arkowitz and Lowe, 1997), raising the possibility that they might mediate regulatory inputs from Cdc42p to Bni1p. Deleting either the SBD or the BBD did not compromise viability of the rewired cell (Figure 4B). Remarkably, even simultaneously deleting the GBD, SBD, and BBD did not prevent Bni1p3Δ from rescuing the bni1Δ BEM1-SNC2 synthetic lethality (Figure 4B). These results demonstrate that the direct interactions of Bni1p with Cdc42p, Spa2p, and Bud6p are all dispensable for Bni1p regulation and suggest that the critical regulation of Bni1p required in rewired cells can be mediated by N-terminal regions that were not known to receive regulatory inputs.
N-terminal regions can mediate Bni1p localization
To investigate the contributions of Bni1p N-terminal regions to Bni1p localization at the polarization site, we constructed wild-type (i.e., not rewired) strains in which the only copy of Bni1p was tagged with GFP. Bni1p lacking the BBD and Bni1p3Δ were able to localize both to the bud tip in small- and medium-budded cells and to the bud neck in large-budded cells (Figure 5A). To test whether the localization of these Bni1p constructs requires actin cables, we treated cells with latrunculin A to depolymerize actin. The constructs were still able to polarize, and colocalized with the polarity marker Bem1p-tdTomato (Figure 5B). In contrast, the catalytic fragment (FH1–FH2) was diffusely localized in the cytoplasm and nucleus in the presence or absence of F-actin. Thus, N-terminal regions of Bni1p outside the known GBD and SBD can target the formin to polarity sites independent of F-actin.
FIGURE 5:
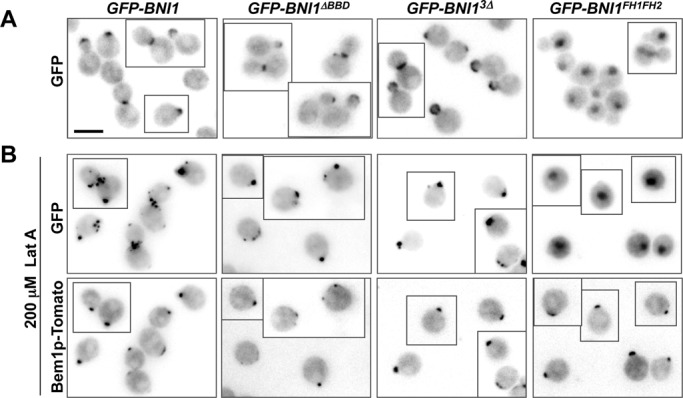
Localization of Bni1p to polarity sites does not require known regulatory domains. (A) The catalytic fragment (FH1–FH2; DLY13426) localizes diffusely in the nucleus and cytoplasm, but addition of N-terminal Bni1p sequences suffices to target the fragment to the bud tip and mother–bud neck, whether or not the GBD and SBD regions are included (DLY13916, 13433, 13431). Representative images of the indicated GFP-tagged constructs are shown. (B) To assess actin-independent localization, cells were treated with 200 μM Lat A at 24°C for 2.5 h (DLY13916, 13433, 13431, 13426). Bem1p-Tomato marks the polarization sites, and the indicated Bni1p constructs (except for the catalytic fragment alone) colocalize with Bem1p. Additional spots of GFP-Bni1p staining remain at old mother–bud necks, presumably reflecting problems with cytokinesis in the absence of F-actin. Scale bar, 5 μm.
Bni1p interacts with the Cdc42p effector Gic2p
If none of the known formin interactions is required for the localization and regulation of Bni1p, then how is Bni1p activity tied to Cdc42p in rewired cells? We took a candidate approach to ask whether known Cdc42p effectors might interact with Bni1p. The related Cdc42/Rac interactive binding (CRIB)–domain effectors Gic1p and Gic2p have been implicated in both actin and septin polarization (Brown et al., 1997; Chen et al., 1997; Bi et al., 2000; Jaquenoud and Peter, 2000; Iwase et al., 2006). We found that myc-tagged Gic2p could be coimmunoprecipitated with both full-length Bni1p and Bni1p3Δ (Figure 6A). Binding to full-length Bni1p was stronger than binding to Bni1p3Δ, consistent with previous reports that Gic2p can bind to Bud6p and possibly Spa2p (Jaquenoud and Peter, 2000). Gic2p-myc did not bind to the FH1–FH2 fragment (Figure 6A), suggesting that the interaction requires N-terminal regions of Bni1p. We narrowed down the Gic2p-binding region of Bni1p to the portion between the GBD and SBD (ND2, residues 343–825; Figure 6B).
FIGURE 6:
The Cdc42p effector Gic2p may regulate Bni1p. (A, B) Gic2p binds to a region of Bni1p distinct from the GBD, SBD, or BBD. (A) Gic2p-Myc was coimmunoprecipitated with overexpressed GFP-Bni1p full-length (FL; DLY14515) and Bni1p3Δ (DLY14516) but not with FH1–FH2 (DLY14517). The stronger binding to full-length Bni1p is consistent with previous findings that Gic2p can also bind to Bud6p/Spa2p. Negative controls: DLY 14513, 13857. (B) The uncharacterized N-terminal Bni1p regions upstream of the GBD (ND1), between the GBD and SBD (ND2), and between the SBD and FH1 (ND3) were fused to the catalytic fragment and expressed as GFP-tagged proteins from the GAL1 promoter (DLY14885, 14886, 14887). The ND2 construct (but not the others) coimmunoprecipitated Gic2p-Myc. (C) Gic1p and Gic2p become critical in rewired cells and when the Bni1p GBD, SBD, and BBD are deleted. The gic1Δ gic2Δ growth defect (DLY14169) at 37°C is greatly exacerbated when combined with BEM1-GFP-SNC2 (DLY14170) or with BNI13Δ (DLY14601). Serial dilutions of the indicated strains were spotted on yeast extract/peptone/dextrose media and grown for 2.5 d at the indicated temperature. (D, E) DIC images of exponentially growing cells of indicated genotypes at 24°C, showing strong synthetic polarity defects even at permissive temperature. Scale bar, 5 μm.
Deletion of GIC1 and GIC2 is tolerated at 24°C but causes severe growth defects at 37°C (Figure 6, C and D; Brown et al., 1997; Chen et al., 1997). We found that in the rewired cell context, deletion of GIC1 and GIC2 was lethal at 37°C (Figure 6C) and caused dramatic morphological defects at 24°C (Figure 6D). Many of the cells were big and round, suggestive of a polarization defect (Figure 6D) and consistent with the possibility that Gic2p mediates critical Bni1p regulation in rewired cells. Moreover, although otherwise wild-type cells expressing Bni1p3Δ as the only copy of Bni1p grow well even at 37°C, deleting GIC1 and GIC2 in BNI13Δ cells was lethal at 37°C (Figure 6C). Even at 24°C, many of these cells were unbudded, big, and round (Figure 6E), suggestive of compromised polarization. These findings suggest that Gic1p and Gic2p act in parallel with other regulators of Bni1p function in wild-type as well as in rewired cells.
Regulation of Bni1p using the Gic2p GTP-Cdc42p–binding domain
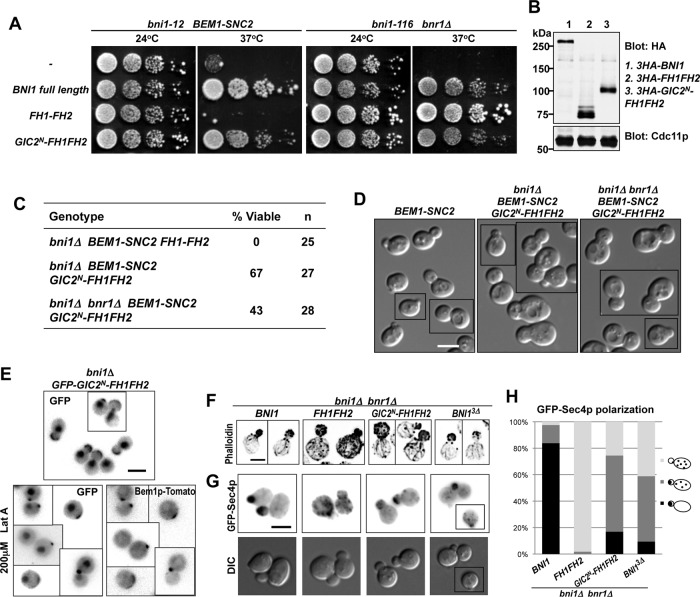
If Bni1p regulation occurs predominantly via a GTP-Cdc42p-Gic2p-Bni1p linkage, it might be possible to bypass the need for Bni1p N-terminal regulatory domains by directly fusing a Cdc42p-binding domain to the Bni1p catalytic domain. Indeed, a construct in which the N-terminus of Gic2p (containing the GTP-Cdc42p–binding domain) was fused to the Bni1p FH1–FH2 was able to function in rewired cells (Figure 7, A–D). This fusion could support proliferation and proper bud growth in rewired cells even when it was the sole formin (Figure 7, C and D), although these cells had wide necks suggestive of a septin defect (Figure 7D). The Gic2p N-terminus was sufficient to concentrate the Bni1p catalytic domain at the polarization site of wild-type cells in the presence or absence of actin filaments (Figure 7E). These results suggest that localizing the formin catalytic activity to active Cdc42p can bypass or mimic the normal Bni1p regulation essential for rewired cells.
FIGURE 7:
The Gic2p N-terminus can replace the Bni1p regulatory regions. (A) The Gic2p N-terminal Cdc42p-binding domain is sufficient for Bni1p regulation in rewired cells (DLY12621, 12626). Function of the indicated constructs was tested as in Figure 3A. (B) 3HA-tagged Bni1p and the GIC2N-FH1-FH2 construct are expressed at comparable levels. Blot probed with anti-HA and anti-Cdc11p (loading control). (C) GIC2N-FH1FH2 can function as the sole formin in rewired cells. Spore viability of cells with the indicated genotypes was deduced from tetrad dissection of heterozygous diploids (DLY 12848, 12594, 12744). (D) DIC images of rewired cells expressing full-length Bni1p (left; DLY12888) or Gic2N-FH1-FH2 (middle, right; DLY12892, 12887) in place of endogenous Bni1p (left, middle) or both Bni1p and Bnr1p (right). (E) GFP-Gic2N-FH1-FH2 (DLY13430) localizes to the bud tip (top) and to the polarization site (marked with Bem1p-tdTomato) in the absence of F-actin (bottom). (F) F-actin distribution in cells containing the indicated construct as the sole formin (DLY13703, 13446, 13444, 13452). (G) GFP-Sec4p (vesicle marker) localization in the same strains as in F. (H) GFP-Sec4p localization categories (cartoon) were scored in small- and medium-budded cells of the strains in F and G. Scale bar, 5 μm.
To examine the organization of actin filaments nucleated by the Bni1p constructs, we used phalloidin to visualize F-actin in wild-type cells containing specific constructs as the only source of formin activity. Phalloidin labels both actin cables and cortical actin patches, which are nucleated by Arp2/3p and function in endocytosis (Winter et al., 1997). Full-length Bni1p nucleated actin cables in the bud, a few of which extended into the mother cell, whereas the unregulated Bni1p catalytic domain (FH1–FH2) generated a much larger number of actin cables, many of which appeared randomly oriented within the mother cell (Figure 7F). The Gic2N-FH1-FH2p fusion and Bni1p3Δ both generated a pattern of actin cables that more closely resembled that generated by full-length Bni1p (Figure 7F). However, misoriented cables were also detected in cells carrying these constructs. To obtain a more quantitative measure of cable orientation in the cells expressing the formin mutants, we used the vesicle marker GFP-Sec4p. As vesicles are delivered to the cable-barbed ends by myosin V, the vesicle distribution reflects the functional orientation of cables. GFP-Sec4p localized exclusively to the buds of small- and medium-budded cells expressing full-length Bni1p, whereas cells expressing the FH1–FH2 fragment showed a dispersed GFP-Sec4p distribution (Figure 7, G and H). In cells expressing Gic2N-FH1-FH2p or Bni1p3Δ as the sole formin, GFP-Sec4p was polarized in 75 and 59% of the cells, respectively (Figure 7, G and H). Thus, consistent with our findings in the rewired-cell context, the previously described regulatory inputs to Bni1p are largely dispensable for generating functional, oriented actin cables in wild-type cells, and fusing the Bni1p catalytic fragment to the Gic2p N-terminus bypasses normal Bni1p regulation and generates actin cables that enable polarized delivery of secretory vesicles.
DISCUSSION
The formin Bni1p plays a critical role in polarity establishment by rewired cells
Direct Cdc42p-Bni1p interaction has been proposed for more than a decade as a key step linking Cdc42p polarization to a polarized actin cytoskeleton (Evangelista et al., 1997). This hypothesis has been difficult to test in vivo because wild-type yeast contain redundant compensatory mechanisms to polarize actin cables that render Bni1p regulation dispensable. We addressed this question with a rewired yeast strain in which the redundancy is eliminated and BNI1 is essential for viability.
The rewiring scheme predicts that polarity establishment in rewired cells needs to coordinate actin-cable and secretory-vesicle polarization. Consistent with this notion, we found that Bni1p and the vesicle marker GFP-Sec4p polarized at approximately the same time as Bem1p–Snc2p (within ∼1 min) in rewired cells. In addition (and unexpectedly), the time interval between Bem1p–Snc2p and septin polarization in rewired cells was shorter than the interval between Bem1p and septin polarization in wild-type cells. It is conceivable that precocious actin polarization in rewired cells facilitates precocious septin recruitment. Although F-actin is not required for septin polarization (Ayscough et al., 1997), the absence of F-actin can lead to defective septin ring assembly (Kozubowski et al., 2005) and exacerbate septin defects in sensitized mutants (Kadota et al., 2004), suggesting that actin filaments may promote septin ring assembly (Park and Bi, 2007).
The two formins in yeast, Bni1p and Bnr1p, can each support bud formation in wild-type cells, but only Bni1p can do so in rewired cells. We detected Bni1p at the polarization site several minutes before Bnr1p, in both wild-type and rewired cells. Bnr1p, whose localization is septin dependent (Pruyne et al., 2004; Buttery et al., 2007; Gao et al., 2010), was not polarized until several minutes after septins were polarized, suggesting that some septin maturation process may be needed for Bnr1p recruitment. This delay may render Bnr1p inactive during initial polarization, explaining why it cannot sustain polarization in the rewired cells. We took advantage of the unique requirement for Bni1p in these cells to dissect the importance of the various regulatory domains in connecting Bni1p to polarity factors.
Bni1p regulation does not require the domains that mediate binding to Cdc42p, Spa2p, or Bud6p
The prevailing model of formin regulation is that binding of Rho GTPases (in this context, Cdc42p) to the GBD of the formin releases autoinhibition and targets the formin to the site of action (path 1 in Figure 8; Li and Higgs, 2003; Lammers et al., 2005; Otomo et al., 2005a; Rose et al., 2005; Nezami et al., 2006; Seth et al., 2006; Martin et al., 2007). However, we found that the GBD of Bni1p was dispensable for the critical Bni1p function in rewired cells, suggesting that additional factors connect Bni1p to the polarity site.
FIGURE 8:
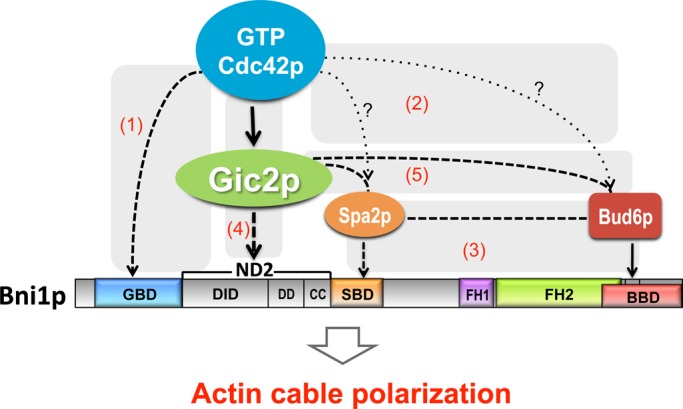
Schematic of the Bni1p regulatory network. Cdc42p interacts directly with Bni1p (1), but this interaction is not necessary for Bni1p regulation. Two other “polarisome” proteins (Spa2p and Bud6p) are localized to sites of Cdc42p action (2) and interact with Bni1p (3), but even in combination these pathways (1–3) are dispensable for Bni1p regulation. We identified a novel path (4) from Cdc42p to Bni1p via the Cdc42p effector Gic2p, which was previously found to interact with Spa2p and Bud6p (5). Genetic interactions suggest that paths 1, 2–3, and 4 act in parallel, and biochemical findings suggest that paths 4 and 5 operate in parallel. Solid lines indicate interactions shown by in vitro biochemical assays. Dashed lines indicate interactions identified with immunoprecipitation and/or cofractionation. Dotted lines with question marks indicate interactions suggested by colocalization.
Spa2p and Bud6p, two other Bni1p-binding proteins that localize to the polarization site (Evangelista et al., 1997; Fujiwara et al., 1998; Sheu et al., 1998), could potentially mediate the communication between Rho GTPases and Bni1p (paths 2 and 3 in Figure 8). However, neither the BBD nor the SBD was required for Bni1p function in rewired cells. Even when the binding domains for Rho GTPases, Spa2p, and Bud6p were simultaneously deleted, Bni1p was still able to connect actin nucleation to the growing polarity site and promote polarized growth of rewired cells. Thus, additional mechanisms must exist to regulate Bni1p.
Regulation of Bni1p by the Cdc42p effector Gic2p
A version of Bni1p lacking the three known regulatory interaction domains (GBD, BBD, and SBD) was still localized to polarity sites, even in the absence of F-actin, suggesting the presence of additional localization signals in the N-terminal domain. Consistent with this conclusion, a recent study identified a total of four localization signals in Bni1p (Liu et al., 2012). Three corresponded to the known GBD, SBD, and BBD regions, but the fourth lay in an uncharacterized domain between GBD and the SBD (residues 334–834, referred to as ND2 in Figure 6). Each of the four regions was sufficient to localize GFP to the bud cortex and bud neck. Our finding that Bni1p3Δ was able to localize properly and rescue the viability of rewired cells indicates that the novel localization determinant in ND2 is capable of conferring regulated formin functionality. We took the study of this new regulatory region one step further by showing that ND2 interacts with the Cdc42p effector Gic2p (path 4 in Figure 8; Brown et al., 1997; Chen et al., 1997).
Gic2p and its homologue, Gic1p, contain a CRIB domain (Burbelo et al., 1995) and a basic-rich domain that binds membranes (Takahashi and Pryciak, 2007); their closest mammalian homologues are the Binder of Rho GTPases (BORG) proteins (Joberty et al., 1999). Gic1p and Gic2p have been implicated in polarization of both the actin and septin cytoskeletons (Brown et al., 1997; Chen et al., 1997; Bi et al., 2000; Jaquenoud and Peter, 2000; Iwase et al., 2006). Gic2p was shown to interact with Bud6p and to cofractionate with both Bud6p and Spa2p (path 5 in Figure 8; Jaquenoud and Peter, 2000). We found that Gic2p could interact with Bni1p even if the Bud6p- and Spa2p-binding domains were deleted and that this interaction was mediated by the ND2 region of Bni1p (path 4 in Figure 8). ND2 (residues 334–834) overlaps most of the DID, which is believed to confer autoinhibition, raising the possibility that Gic2p binding could also lead to formin activation.
Genetic interactions support the conclusion that Gic1p and Gic2p are important for connecting Cdc42p to Bni1p. In rewired cells, deletion of GIC1 and GIC2 produced severe polarity defects even at permissive temperatures where their loss is well tolerated in wild-type cells. Moreover, deletion of GIC1 and GIC2 produced severe polarity defects when combined with removal of the Bni1p GBD, SBD, and BBD domains in wild-type context. These findings, combined with previous studies, suggest that a redundant web of indirect interactions connects Cdc42p to Bni1p (paths 1–5 in Figure 8).
Generality of formin regulation via Rho/Cdc42 effectors
Our findings suggest that regulation of formins by Rho-family GTPases may occur predominantly via Rho effectors like Gic2p. This idea is supported by recent findings that the Cdc42 effector Pob1 helps connect Cdc42 to For3 in fission yeast (Rincon et al., 2009), that IQGAP1 promotes Dia1 localization to phagocytic cups (Brandt et al., 2007), and that anillin promotes mDia2 localization to cleavage furrows (Watanabe et al., 2010). Although the Pob1-binding site of For3 has yet to be narrowed down, it was shown that both IQGAP and anillin bind to the armadillo repeat-dimerization domain region (analogous to ND2 in Bni1p) of Dia1 and mDia2, respectively (Brandt et al., 2007; Watanabe et al., 2010). Thus, partially overlapping effector-binding domains and the DID domain may represent a common theme in formin regulation. Of interest, in vitro actin assembly assays call for very high concentrations of Rho proteins to release the antoinhibitory effect of DID–DAD interactions (Li and Higgs, 2003; Seth et al., 2006), raising the possibility that Rho-effector binding may facilitate formin activation in vivo.
Complexity in formin regulation
Why would a web of indirect interactions be advantageous compared with a single direct interaction between the GTPase and the formin? Studies on Bni1p (Buttery et al., 2007) and on For3p in fission yeast (Martin et al., 2007) indicate that cable formation involves a repeated cycle of formin-mediated nucleation, elongation, and capping of individual actin filaments that are then stitched together side by side to form cables. Proper dynamic switching between nucleation, elongation, and capping may be important for generating a well-organized cable or for regulating the length, orientation, or lifetime of the cable. However, regulators that affect the speed and duration of elongation can in some cases bind directly to the FH1–FH2 catalytic core (Chesarone et al., 2009; Chesarone-Cataldo et al., 2011), and it is unclear whether and how the dynamics of the intricate formin cycle are affected by the various Bni1p-interacting proteins discussed here. Further work is required to elucidate the interplay between the proteins in the formin regulatory network.
MATERIALS AND METHODS
Yeast strains
Yeast strains used in this study are listed in Table 1. All strains are in the YEF473 background (his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52). Standard media and procedures were used for yeast genetic manipulations. To generate strains expressing Bem1p-GFP, a plasmid (pDLB2968) (Kozubowski et al., 2008) containing a C-terminal fragment of BEM1 fused to GFP was cut with PstI to target integration at BEM1. Strains expressing Bem1-GFP-Snc2p were generated by integrating a PstI-cut plasmid (pDLB2823) (Howell et al., 2009) containing a C-terminal fragment of BEM1 fused to GFP and SNC2 at BEM1. BEM1-SNC2 strains (lacking the GFP) were generated by integrating PstI-cut pDLB3267, which was made by deleting GFP from pDLB2823, at BEM1. Strains expressing Cdc3p-mCherry were generated by integrating BglII-cut pDLB3138 (YIp128-CDC3-mCherry, (Tong et al., 2007)) at CDC3. To express GFP-Sec4p, pDLB2776 (YIp211-GFP-SEC4) (a gift from E. Bi, University of Pennsylvania) was cut with StuI to target integration at URA3. BNR1-GFP and BEM1-tdTomato were generated by the PCR-based C-terminal tagging method (Longtine et al., 1998) using pFA6a-GFP-HIS3MX6 and pFA6a-tdTomato-HIS3MX6, respectively, as templates. GIC2-13myc was created by integrating a HindIII-cut plasmid pDLB2132 (pRS306-GIC2(C-term)-13myc) at GIC2.
TABLE 1:
Yeast strains.
| Strain (DLY) | Relevant genotype | Source |
|---|---|---|
| 7924 | a bni1-116 bnr1::HIS3 | E. Bi |
| 8155 | a | Howell et al. (2009) |
| 8601 | a BEM1-GFP-SNC2:LEU2 | Howell et al. (2009) |
| 11909 | a/α CDC3-mCherry:LEU2/CDC3-mCherry:LEU2 | Chen et al. (2011) |
| BEM1-GFP:LEU2/BEM1-GFP:LEU2 | ||
| 12226 | a bni1-12 BEM1-GFP-SNC2:TRP1 | This study |
| 12594 | a/α bni1::HIS3:3HA-GIC2N-FH1FH2:URA3/BNI1 | This study |
| BEM1-GFP-SNC2:LEU2/BEM1 | ||
| 12620 | a bni1-12:3HA-FH1FH2:URA3 BEM1-GFP-SNC2:TRP1 | This study |
| 12621 | a bni1-12:3HA-GIC2N-FH1FH2:URA3 BEM1-GFP-SNC2:TRP1 | This study |
| 12622 | a bni1-12:3HA-GBD-FH1FH2:URA3 BEM1-GFP-SNC2:TRP1 | This study |
| 12626 | a bni1-116:3HA-GIC2N-FH1FH2:URA3 bnr1::HIS3 | This study |
| 12627 | a bni1-116:3HA-FH1FH2:URA3 bnr1::HIS3 | This study |
| 12629 | a bni1-116:3HA-GBD-FH1FH2:URA3 bnr1::HIS3 | This study |
| 12648 | a/α BEM1-GFP-SNC2:LEU2/BEM1 bni1::TRP1/BNI1 | This study |
| 12744 | a/α bni1::HIS3:3HA-GIC2N-FH1FH2:URA3/BNI1 | This study |
| BEM1-GFP-SNC2:LEU2/BEM1 bnr1::TRP1/BNR1 | ||
| 12788 | a/α bnr1::URA3/BNR1 | This study |
| BEM1-GFP-SNC2:LEU2/BEM1-GFP-SNC2:LEU2 | ||
| 12848 | a/α BEM1-GFP-SNC2:LEU3/BEM1 | This study |
| bni1::TRP1:3HA-FH1FH2:URA3/BNI1 | ||
| 12887 | a bni1::HIS3:3HA-GIC2N-FH1FH2:URA3 | This study |
| BEM1-GFP-SNC2:LEU2 bnr1::TRP1 | ||
| 12888 | a BEM1-GFP-SNC2:LEU2 | This study |
| 12892 | a bni1::HIS3:3HA-GIC2N-FH1FH2:URA3 | This study |
| BEM1-GFP-SNC2:LEU2 | ||
| 12959 | a/α bni1::HIS3:3HA-BNI1ΔBBD:URA3/BNI1 | This study |
| BEM1-GFP-SNC2:LEU2/BEM1 | ||
| 12960 | a/α bni1::HIS3:3HA-BNI13Δ:URA3/BNI1 | This study |
| BEM1-GFP-SNC2:LEU2/BEM1 | ||
| 13072 | a/α CDC3-mCherry:LEU2/CDC3-mCherry:LEU2 | This study |
| BEM1-GFP-SNC2:LEU2/BEM1-GFP-SNC2:LEU2 | ||
| 13095 | α BEM1-SNC2:LEU2 | This study |
| 13140 | a bni1::HIS3:3HA-GFP-GBD-FH1FH2:URA3 | This study |
| 13344 | a/α CDC3-mCherry:LEU2/CDC3-mCherry:LEU2 | Chen et al. (2011) |
| BNR1-GFP:HIS3/BNR1-GFP:HIS3 | ||
| 13426 | a bni1::HIS3:3HA-GFP-FH1FH2:URA3 BEM1-tdTomato:HIS3 | This study |
| 13430 | α bni1::HIS3:3HA-GIC2N-FH1FH2:URA3 BEM1-tdTomato:HIS3 | This study |
| 13431 | a bni1::HIS3:3HA-GFP-BNI13Δ:URA3 BEM1-tdTomato:HIS3 | This study |
| 13433 | a bni1::HIS3:3HA-GFP-BNI1ΔBBD:URA3 BEM1-tdTomato:HIS3 | This study |
| 13444 | a bni1::HIS3:3HA-GIC2N-FH1FH2:URA3 bnr1::HIS3 | This study |
| GFP-SEC4:URA3 | ||
| 13446 | a bni1::HIS3:3HA-FH1FH2:URA3 bnr1::HIS3 GFP-SEC4:URA3 | This study |
| 13452 | α bni1::HIS3:3HA-BNI13Δ:URA3 bnr1::HIS3 GFP-SEC4:URA3 | This study |
| 13703 | a bni1::HIS3:3HA-BNI1:URA3 bnr1::HIS3 GFP-SEC4:URA3 | This study |
| 13884 | a/α GFP-SEC4:URA3/GFP-SEC4:URA3 | This study |
| BEM1-SNC2:LEU2/BEM1-SNC2:LEU2 | ||
| CDC3-mCherry:LEU2/CDC3-mCherry:LEU2 | ||
| 13890 | a/α CDC3-mCherry:LEU2/CDC3-mCherry:LEU2 | This study |
| GFP-SEC4:URA3/GFP-SEC4:URA3 | ||
| 13916 | a bni1::HIS3:3HA-GFP-BNI1:URA3 BEM1-tdTomato:HIS3 | This study |
| 13945 | a/α BEM1-SNC2:LEU2/BEM1-SNC2:LEU2 | This study |
| BNR1-GFP:HIS3/BNR1-GFP:HIS3 | ||
| CDC3-mCherry:LEU2/CDC3-mCherry:LEU2 | ||
| 14014 | a/α bni1::HIS3:3HA-GFP-BNI1:URA3/bni1::HIS3::3HA-GFP-BNI1::URA3 CDC3-mCherry:LEU2/CDC3-mCherry:LEU2 | This study |
| 14024 | a bni1-12:3HA-BNI1:URA3 BEM1-GFP-SNC2:TRP1 | This study |
| 14025 | a bni1-116:HIS3:3HA-BNI1:URA3 bnr1::HIS3 | This study |
| 14026 | a/α bni1::HIS3:3HA-GFP-BNI1:URA3/bni1::HIS3:3HA-GFP-BNI1:URA3 BEM1-SNC2:LEU2/BEM1-SNC2:LEU2 | This study |
| CDC3-mCherry:LEU2/CDC3-mCherry:LEU2 | ||
| 14169 | a gic1::TRP1 gic2::HIS3 | This study |
| 14170 | α gic1::TRP1 gic2::HIS3 BEM1-GFP-SNC2:LEU2 | This study |
| 14194 | a/α bni1::HIS3:3HA-BNI1ΔGBD:URA3/BNI1 | This study |
| BEM1-SNC2:LEU2/BEM1 | ||
| 14195 | a/α bni1::HIS3:3HA-BNI1ΔSBD:URA3/BNI1 | This study |
| BEM1-SNC2:LEU2/BEM1 | ||
| 14223 | a bni1::HIS3:3HA-BNI1ΔGBD:URA3 BEM1-SNC2:LEU2 | This study |
| 14272 | a/α BEM1-SNC2:LEU2/BEM1 rsr1::TRP1/RSR1 | This study |
| BNI1:3HA-FH1-FH2:URA3/BNI1 | ||
| 14461 | α GIC2-13myc:URA3 Gal4BD-hER-VP16::Trp1 | This study |
| 14513 | α Gal4BD-hER-VP16:URA3 bni1::KanR:PGal1:GFP-BNI1:URA3 | This study |
| 14515 | a Gal4BD-hER-VP16:URA3 bni1:KanR:PGal1:GFP-BNI1:URA3 GIC2-13myc:URA3 | This study |
| 14516 | α Gal4BD-hER-VP16:URA3 | This study |
| bni1:KanR:PGal1:GFP-BNI13Δ:URA3 GIC2-13myc:URA3 | ||
| 14517 | α Gal4BD-hER-VP16:URA3 | This study |
| bni1:KanR:PGal1:GFP-FH1FH2:URA3 GIC2-13myc:URA3 | ||
| 14601 | α bni1:HIS3:3HA-GFP-BNI3Δ:URA3 SPA2-mCherry:KanR | This study |
| gic1::TRP1 gic2::HIS3 | ||
| 14602 | a bni1:HIS3:3HA-GFP-BNI3Δ:URA3 SPA2-mCherry:KanR | This study |
| 14811 | α BEM1-SNC2:LEU2 BNI1:3HA-FH1-FH2:URA3 | This study |
| 14885 | a KanR:GALp-GFP-BNI1 ND1-FH1,2:URA | This study |
| GIC2-13myc:URA3 Gal4BD-hER-VP16:Trp1 | ||
| 14886 | a KanR:GALp-GFP-BNI1 ND2-FH1,2:URA | This study |
| GIC2-13myc:URA3 Gal4BD-hER-VP16:Trp1 | ||
| 14887 | a KanR:GALp-GFP-BNI1 ND3-FH1,2:URA | This study |
| GIC2-13myc:URA3 Gal4BD-hER-VP16:Trp1 |
All strains are in the YEF473 background (his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52).
The one-step PCR-based method (Baudin et al., 1993) was used to generate bni1::HIS3, bni1::TRP1, bnr1::TRP1, and gic2::HIS3, using pRS303, pRS304, pRS304, and pRS403, respectively, as templates. To generate bni1-12 in the YEF473 background, a PCR product covering the temperature-sensitive mutations (using template DLY8421, a gift from A. Bretscher, Cornell University) was cotransformed with pRS315 into a bni1::URA3 strain. Transformants were selected on media lacking leucine and screened for replacement of bni1::URA3 with bni1-12 using 5-fluoroorotic acid plates.
Various BNI1 constructs were generated by modifying a backbone plasmid pDLB3393, which contains the following elements inserted in the pRS306 polylinker between the ApaI and NotI sites: 1) the BNI1 5′-untranslated region (UTR (488 base pairs upstream of ATG); 2) an ATG followed by a triple-hemagglutinin (3HA) epitope tag; 3) a linker sequence (ACGCGTGGACCCGGG) containing MluI and XmaI recognition sites; 4) the FH1–FH2 fragment of BNI1 (3679–5298 base pairs) in the same reading frame; and 5) a STOP codon followed by a BamHI site and the BNI1 3′-UTR (500 base pairs downstream of the STOP). BNI1 coding sequence N-terminal to FH1–FH2 (1–3678 base pairs, pDLB3575), ND1 (4–228 base pairs, pDLB3545), ND2 (1030–2465 base pairs, pDLB3547), ND3 (2950–3678 base pairs, pDLB3507), and GIC2N (1–624 base pairs, pDLB3397) were inserted between the 3HA and FH1–FH2 using MluI and XmaI sites. BNI1 coding sequences N-terminal to FH1–FH2 but lacking the GBD (∆229–1029 base pairs) or SBD (∆2476–2949 base pairs) were amplified from appropriate templates (gifts from K. Kono and D. Pellman, Dana-Farber/Harvard Cancer Center) and inserted in the same way, yielding pDLB3633 (ΔGBD) and pDLB3634 (ΔSBD). A similar construct in which both the GBD and SBD were deleted was constructed by subcloning using the individual deletion constructs, yielding pDLB3476 (ΔGBD ΔSBD ΔBBD, or BNI13Δ). All of the plasmids discussed thus far lack the coding sequences C-terminal to FH1–FH2 (i.e., the BBD). To add back the BBD, a C-terminal fragment from BNI1 was cloned into pDLB3393 using ClaI (in the FH2) and SpeI (in the 3′-UTR) sites. To make fluorescent versions of the constructs, we inserted GFP coding sequences at the MluI site (between the 3HA and Bni1p). Digestion with SpeI (which cuts at a single site within the 3′-UTR) was used to target integration to the BNI1 3′-UTR next to a deletion or a temperature-sensitive allele of the endogenous BNI1.
To generate PGAL1:GFP-BNI1 strains for immunoprecipitation experiments, we amplified the GAL1 promoter and a kanR marker from pFA6a-kanMX6-PGAL1 (Longtine et al., 1998) and transformed them into yeast containing GFP-tagged Bni1p variants. This procedure removes the bni1 deletion, the BNI1 promoter, and the 3HA tag, so that the constructs begin with the GFP coding sequence. PGAL1-dependent expression of BNI1 constructs in response to β-estradiol was achieved using the synthetic transcription factor GAL4BD-hER-VP16 (pDLB3103; Takahashi and Pryciak, 2008), which was integrated at URA3.
Microscopy
Imaging was performed as previously described (Howell et al., 2009). Except for the phalloidin-stained cells, which were imaged with a Zeiss 780 confocal microscopy system consisting of an Axio Examiner (Carl Zeiss, Thornwood, NY) and a 63x/1.4 Plan Apochromat oil immersion objective, all the other images were acquired using an AxioObserver Z1 (Carl Zeiss) with a 100×/1.46 Plan Apochromat oil immersion objective and a QuantEM backthinned electron-multiplying (EM)-charge-coupled device camera (Photometrics, Ottobrunn, Germany). Exponentially growing cells were mounted on a slide with a slab of complete synthetic medium solidified with 2% agarose (Denville Scientific, Metuchen, NJ) and sealed with Vaseline (Unilever, London, United Kingdom). For time-lapse images, EM gain of the camera was set to 750, and cells were exposed to 2% excitation light for 250 ms for most GFP-tagged proteins, 500 ms for GFP-Bni1p and Bnr1-GFP, and 150 ms for all red fluorescent protein (RFP)-tagged proteins during each image acquisition. Time-lapse images were taken with 25–30 z-planes at 0.25-μm steps, deconvolved (see later discussion), and displayed as maximum projections. For Figure 1, cells were synchronized with 200 mM hydroxyurea (HU; Sigma-Aldrich, St. Louis, MO) in complete synthetic medium (MP Biomedicals, Irvine, CA) for 3 h at 30°C, washed, and released into fresh medium for 1 h and 5 min before filming at 30°C. Synchronization with HU shortens the filming time required to acquire enough polarization events for quantification and fortuitously increased cell tolerance to light exposure without altering polarization kinetics (Howell et al., 2012). For single-image acquisition, EM gain of the camera was set to 250, and cells were exposed to 100% excitation light for 250 ms for GFP and 150 ms for RFP. These images were taken with nine z-planes at 0.5-μm steps, deconvolved, and displayed as maximum projections for fluorescent proteins and the best-focused plane for differential interference contrast (DIC).
Image analysis
Time-lapse and confocal images were deconvolved with Huygens Essential software (Scientific Volume Imaging, Hilversum, Netherlands), using the classic maximum likelihood estimation and predicted point-spread function with a background value set constant across all images from the same time lapse, with a signal-to-noise ratio of 10 and a maximum of 40 iterations. The deconvolved images were compiled with MetaMorph (Molecular Devices, Sunnyvale, CA) and scored visually for the initial time points of GFP and RFP polarization at the incipient bud site. Images for presentation were processed using MetaMorph and ImageJ (National Institutes of Health, Bethesda, MD).
Latrunculin A treatment
Exponentially growing cells were treated with 200 μM latrunculin A (Invitrogen, Carlsbad, CA) in complete synthetic medium with 2% dextrose at 24°C for 2.5 h and then mounted onto agarose slabs containing 200 μM latrunculin A for live-cell imaging.
Phalloidin staining
Electron microscopy–grade formaldehyde (Polyscience, Warrington, PA) was added to exponentially growing cultures to a final concentration of 4% for 10 min. Cells were then spun down and fixed in 4% electron microscopy–grade formaldehyde in phosphate-buffered saline (PBS) for 1 h at room temperature. After two quick washes with PBS, cells were stained with a 1:10 dilution of 6.6 μM rhodamine phalloidin (Invitrogen) in the dark for 1 h, washed five times with PBS, and mounted for imaging.
Centrifugal elutriation and fluorescence-activated cell sorting analysis
Small G1 cells were isolated from exponentially growing cultures by centrifugal elutriation as preciously described (Lew and Reed, 1993). Cells were grown in yeast extract/peptone (YEP) with 2% sucrose and 0.1% dextrose overnight to (1–2) × 107 cells/ml before being elutriated and concentrated by centrifugation at 4°C and then resuspended to a density of 107 cells/ml in prewarmed media at 37°C. For live-cell imaging, cells were released into complete synthetic media for 30 min before mounted on a slab for filming. To distinguish the two strains (DLY8601, 12226) filmed simultaneously, DLY8601 was stained briefly with a red fluorescent cell wall dye Alexa Fluor 594–conjugated concanavalin A (Invitrogen). For the time-course experiment, cells were released in YEP medium with 2% dextrose and collected every 20 min for 160 min, fixed, and scored for budded ratio and DNA content. Fluorescence-activated cell sorting analysis was preformed as described previously (Haase and Reed, 2002). Cells were fixed overnight in 70% ethanol at 4°C, washed once with H2O, and then incubated in 2 mg/ml RNaseA (Sigma-Aldrich) in 50 mM Tris-HCl (pH 8.0) for 2 h. Cells were treated with 5 mg/ml pepsin (Sigma-Aldrich) in 0.45% HCl for 15 min. DNA was stained with Sytox Green (Invitrogen) in 50 mM Tris-HCl (pH 7.5). The DNA content of 10,000 cells was measured for each sample with a Becton Dickinson FACScan and analyzed with Cell Quest software (Becton Dickinson Biosciences, San Jose, CA).
Western blotting and immunoprecipitation
Yeast cell lysis, SDS–PAGE, and immunoblotting were performed as previously described (Keaton et al., 2008). Mouse anti-HA (Roche Applied Science, Penzberg, Germany) was used at a 1:1000 dilution. Mouse anti-Myc (Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:5000 dilution. Rabbit anti-Cdc11p (Santa Cruz Biotechnology) was used at 1:10,000 dilution. Rabbit anti-GFP (Invitrogen) was used at 1:5000 dilution. For immunoprecipitation, cells were treated with 10 nM β-estradiol (Sigma-Aldrich) for 3 h to induce GAL1 promoter–driven expression of GFP-Bni1p constructs with the synthetic transcription factor GAL4BD-hER-VP16. Cells were lysed by vortexing with acid-washed glass beads (Sigma-Aldrich) in lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.5% NP40, 5% glycerol, and 1:50 diluted protease inhibitors (P8215) (Sigma-Aldrich). Cell lysates were incubated with agarose beads conjugated with camelid anti-GFP (Chromotek-GFP-TRAP; Allele Biotechnology, San Diego, CA) for 3 h. The beads were then washed in lysis buffer and boiled with SDS sample buffer. Samples were analyzed by SDS–PAGE and Western blotting.
Acknowledgments
We thank Erfei Bi, Tony Bretscher, Bruce Goode, and David Pellman for generously sharing plasmid constructs and information before publication. Thanks go to Lew lab members for stimulating interactions. This work was supported by National Institutes of Health Grants GM53050 and GM62300 to D.J.L.
Abbreviations used:
- BBD
Bud6p-binding domain
- DAD
Diaphanous autoregulatory domain
- DID
Diaphanous inhibitory domain
- FH1
formin homology 1
- FH2
formin homology 2
- GBD
GTPase-binding domain
- SBD
Spa2p-binding domain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-05-0400) on August 23, 2012.
*Present address: Department of Biology, Stanford University, Stanford, CA 94305.
REFERENCES
- Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkowitz RA, Lowe N. A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J Cell Biol. 1997;138:17–36. doi: 10.1083/jcb.138.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Chiavetta JB, Chen H, Chen GC, Chan CS, Pringle JR. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol Biol Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Jaquenoud M, Gulli MP, Chant J, Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo PD, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- Buttery SM, Yoshida S, Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell. 2007;18:1826–1838. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GC, Kim YJ, Chan CS. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Howell AS, Robeson A, Lew DJ. Dynamics of septin ring and collar formation in Saccharomyces cerevisiae. Biol Chem. 2011;392:689–697. doi: 10.1515/BC.2011.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16:292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chesarone-Cataldo M, Guerin C, Yu JH, Wedlich-Soldner R, Blanchoin L, Goode BL. The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev Cell. 2011;21:217–230. doi: 10.1016/j.devcel.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Bretscher A. Polarized growth in budding yeast in the absence of a localized formin. Mol Biol Cell. 2009;20:2540–2548. doi: 10.1091/mbc.E09-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Liu W, Bretscher A. The yeast formin Bnr1p has two localization regions that show spatially and temporally distinct association with septin structures. Mol Biol Cell. 2010;21:1253–1262. doi: 10.1091/mbc.E09-10-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Kozubowski L, Zyla TR, Lew DJ. Interplay between septin organization, cell cycle and cell shape in yeast. J Cell Sci. 2005;118:1617–1628. doi: 10.1242/jcs.02286. [DOI] [PubMed] [Google Scholar]
- Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22:4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- Howell AS, Jin M, Wu CF, Zyla TR, Elston TC, Lew DJ. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 2012;149:322–333. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AS, Lew DJ. Morphogenesis and the cell cycle. Genetics. 2012;190:51–77. doi: 10.1534/genetics.111.128314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139:731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, Caruso C, Tong Z, Pringle JR, Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell. 2006;17:1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M, Peter M. Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p) Mol Cell Biol. 2000;20:6244–6258. doi: 10.1128/mcb.20.17.6244-6258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G, Perlungher RR, Macara IG. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol Cell Biol. 1999;19:6585–6597. doi: 10.1128/mcb.19.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Jin M, Lew DJ. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr Opin Genet Dev. 2011;21:740–746. doi: 10.1016/j.gde.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton MA, Szkotnicki L, Marquitz AR, Harrison J, Zyla TR, Lew DJ. Nucleocytoplasmic trafficking of G2/M regulators in yeast. Mol Biol Cell. 2008;19:4006–4018. doi: 10.1091/mbc.E08-03-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, et al. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Larson JR, Tatchell K. Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:3455–3466. doi: 10.1091/mbc.E04-09-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol. 2008;18:1719–1726. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M, Rose R, Scrima A, Wittinghofer A. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 2005;24:4176–4187. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Liu W, Santiago-Tirado FH, Bretscher A. Yeast formin Bni1p has multiple localization regions that function in polarized growth and spindle orientation. Mol Biol Cell. 2012;23:412–422. doi: 10.1091/mbc.E11-07-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Martin SG, Rincon SA, Basu R, Perez P, Chang F. Regulation of the formin for3p by cdc42p and bud6p. Mol Biol Cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami AG, Poy F, Eck MJ. Structure of the autoinhibitory switch in formin mDia1. Structure. 2006;14:257–263. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005a;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005b;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Ozaki-Kuroda K, Yamamoto Y, Nohara H, Kinoshita M, Fujiwara T, Irie K, Takai Y. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:827–839. doi: 10.1128/MCB.21.3.827-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Gao L, Bi E, Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol Biol Cell. 2004;15:4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon SA, Ye Y, Villar-Tajadura MA, Santos B, Martin SG, Perez P. Pob1 participates in the Cdc42 regulation of fission yeast actin cytoskeleton. Mol Biol Cell. 2009;20:4390–4399. doi: 10.1091/mbc.E09-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002a;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002b;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Seth A, Otomo C, Rosen MK. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol. 2006;174:701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Pryciak PM. Identification of novel membrane-binding domains in multiple yeast Cdc42 effectors. Mol Biol Cell. 2007;18:4945–4956. doi: 10.1091/mbc.E07-07-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr Biol. 2008;18:1184–1191. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J Cell Biol. 2007;179:1375–1384. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Neo SP, Cai M. Regulation of the yeast formin Bni1p by the actin-regulating kinase Prk1p. Traffic. 2009;10:528–535. doi: 10.1111/j.1600-0854.2009.00893.x. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, Arakawa T, Kinoshita M, Ishizaki T, Narumiya S. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell. 2010;21:3193–3204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Podtelejnikov AV, Mann M, Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]