Abstract
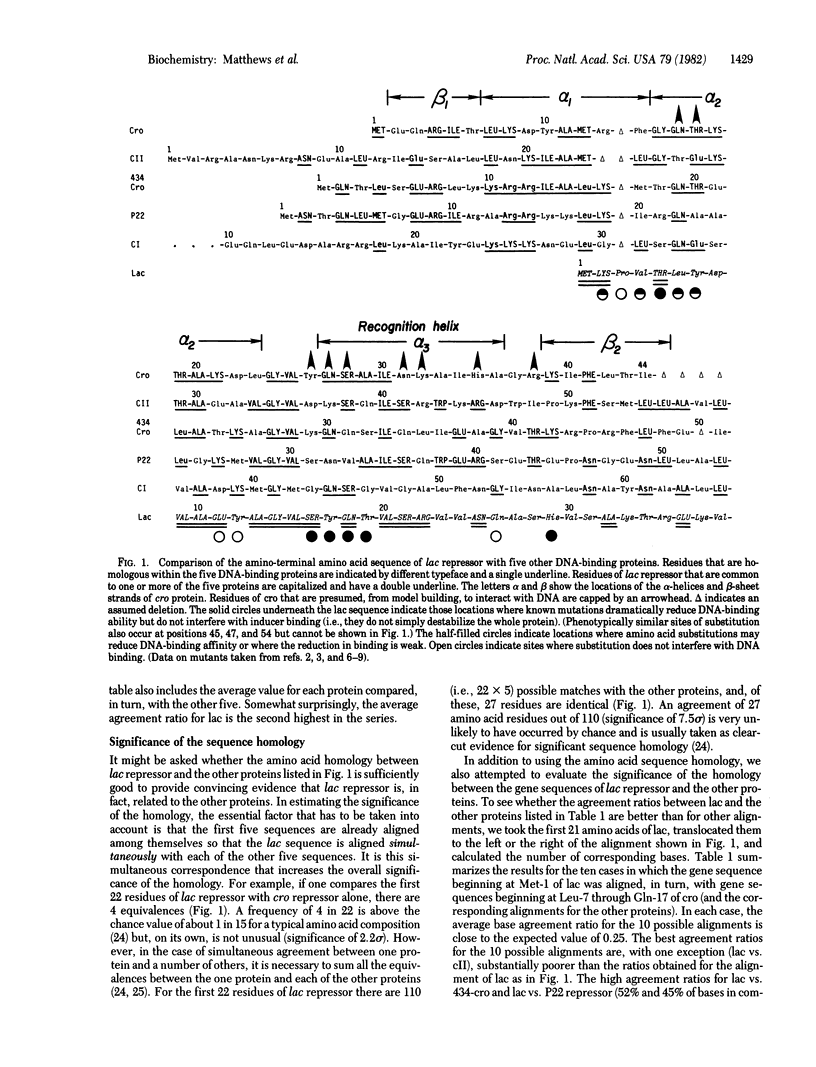
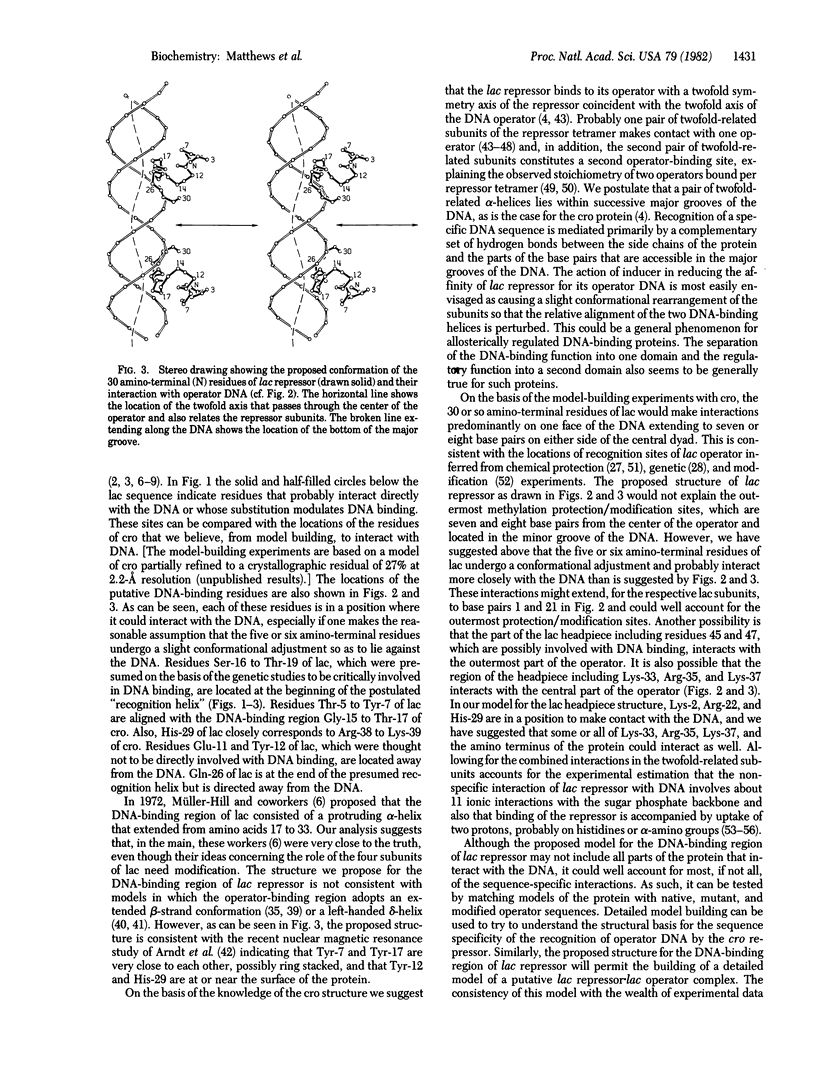
It is shown that the amino acid sequence and the DNA gene sequence of the 25 amino-terminal residues of the lac repressor protein of Escherichia coli are homologous with the sequences of five DNA-binding proteins: the cro repressor proteins from phage lambda and phage 434, the cI and cII proteins from phage lambda, and the repressor protein from Salmonella phage P22. The region of homology between lac repressor and the other proteins coincides with the principal DNA-binding region of cro repressor. In particular, residues Tyr-17 through Gln-26 of lac repressor correspond to the alpha-helix Gln-27 through Ala-36 of cro repressor, which we have postulated to bind within the major groove of the DNA and to be primarily responsible for the recognition of the DNA operator region by the protein [Anderson, W. F., Ohlendorf, D. H., Takeda, Y. & Matthews, B. W. (1981) Nature (London) 290, 754--758]. By analogy with cro repressor, we propose that residues 17--26 of lac repressor are alpha-helical and that this helix and a twofold-related alpha-helix in an adjacent subunit bind within successive major grooves of the lac operator, which is in a right-handed Watson--Crick B-DNA conformation. Also, by analogy with cro repressor, we suggest that residues Thr-5 through Ala-13 of lac repressor form a second alpha-helix and contribute, in part, to DNA binding. The proposed structure for the DNA-binding region of lac repressor is consistent with chemical protection data and with genetic experiments identifying the probable locations of a number of the residues of the repressor protein that either do or do not participate in DNA binding.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler K., Beyreuther K., Fanning E., Geisler N., Gronenborn B., Klemm A., Müller-Hill B., Pfahl M., Schmitz A. How lac repressor binds to DNA. Nature. 1972 Jun 9;237(5354):322–327. doi: 10.1038/237322a0. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Arndt K. T., Boschelli F., Lu P., Miller J. H. lac Repressor: a proton magnetic resonance look at the deoxyribonucleic acid binding fragment. Biochemistry. 1981 Oct 13;20(21):6109–6118. doi: 10.1021/bi00524a030. [DOI] [PubMed] [Google Scholar]
- Beyreuther K., Adler K., Geisler N., Klemm A. The amino-acid sequence of lac repressor. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3576–3580. doi: 10.1073/pnas.70.12.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois S., Jernigan R. L., Szu S. C., Kabat E. A., Wu T. T. Composite predictions of secondary structures of lac repressor. Biopolymers. 1979 Oct;18(10):2625–2643. doi: 10.1002/bip.1979.360181017. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran R., Jardetzky T. S., Jardetzky O. The delta helix--a possible left-handed stable polypeptide structure in the N-terminal segment of the lac repressor. FEBS Lett. 1979 May 1;101(1):11–14. [PubMed] [Google Scholar]
- Charlier M., Maurizot J. C., Zaccai G. Neutron scattering studies of lac repressor. Nature. 1980 Jul 24;286(5771):423–425. doi: 10.1038/286423a0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Adler A. J., Fasman G. D. Conformational prediction and circular dichroism studies on the lac repressor. J Mol Biol. 1975 Jul 25;96(1):29–45. doi: 10.1016/0022-2836(75)90180-1. [DOI] [PubMed] [Google Scholar]
- Culard F., Maurizot J. C. Lac repressor - lac operator interaction. Circular dichroism study. Nucleic Acids Res. 1981 Oct 10;9(19):5175–5184. doi: 10.1093/nar/9.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F. Similar amino acid sequences: chance or common ancestry? Science. 1981 Oct 9;214(4517):149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- Dunaway M., Matthews K. S. Hybrid tetramers of native and core lactose repressor protein. Assessment of operator and nonspecific DNA binding parameters and their relationship. J Biol Chem. 1980 Nov 10;255(21):10120–10127. [PubMed] [Google Scholar]
- Farabaugh P. J. Sequence of the lacI gene. Nature. 1978 Aug 24;274(5673):765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- Files J. G., Weber K. Limited proteolytic digestion of lac repressor by trypsin. Chemical nature of the resulting trypsin-resistant core. J Biol Chem. 1976 Jun 10;251(11):3386–3391. [PubMed] [Google Scholar]
- Fitch W. M. An improved method of testing for evolutionary homology. J Mol Biol. 1966 Mar;16(1):9–16. doi: 10.1016/s0022-2836(66)80258-9. [DOI] [PubMed] [Google Scholar]
- Folkmanis A., Takeda Y., Simuth J., Gussin G., Echols H. Purification and properties of a DNA-binding protein with characteristics expected for the Cro protein of bacteriophage lambda, a repressor essential for lytic growth. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2249–2253. doi: 10.1073/pnas.73.7.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N., Weber K. Isolation of a set of hybrid lac repressors made in vitro between normal lac repressor and its homogeneous tryptic core. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3103–3106. doi: 10.1073/pnas.73.9.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N., Weber K. Isolation of amino-terminal fragment of lactose repressor necessary for DNA binding. Biochemistry. 1977 Mar 8;16(5):938–943. doi: 10.1021/bi00624a020. [DOI] [PubMed] [Google Scholar]
- Goeddel D. V., Yansura D. G., Caruthers M. H. How lac repressor recognizes lac operator. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3578–3582. doi: 10.1073/pnas.75.8.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Schwarz E. Nucleotide sequence of the cro-cII-oop region of bacteriophage 434 DNA. Nucleic Acids Res. 1979 Mar;6(3):867–881. doi: 10.1093/nar/6.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I., Hagen D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- Hsiang M. W., Cole R. D., Takeda Y., Echols H. Amino acid sequence of Cro regulatory protein of bacteriophage lambda. Nature. 1977 Nov 17;270(5634):275–277. doi: 10.1038/270275a0. [DOI] [PubMed] [Google Scholar]
- JACOB F., MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961 Jun;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Kania J., Brown D. T. The functional repressor parts of a tetrameric lac repressor-beta-galactosidase chimaera are organized as dimers. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3529–3533. doi: 10.1073/pnas.73.10.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim V. I. Algorithms for prediction of alpha-helical and beta-structural regions in globular proteins. J Mol Biol. 1974 Oct 5;88(4):873–894. doi: 10.1016/0022-2836(74)90405-7. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Hofer M., Schmeissner U., Sommer H., Schmitz A., Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979 Jun 25;131(2):191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B. Lac repressor and lac operator. Prog Biophys Mol Biol. 1975;30(2-3):227–252. doi: 10.1016/0079-6107(76)90011-0. [DOI] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. DNA-binding site of lac repressor probed by dimethylsulfate methylation of lac operator. J Mol Biol. 1979 Aug 25;132(4):709–728. doi: 10.1016/0022-2836(79)90384-x. [DOI] [PubMed] [Google Scholar]
- Platt T., Files J. G., Weber K. Lac repressor. Specific proteolytic destruction of the NH 2 -terminal region and loss of the deoxyribonucleic acid-binding activity. J Biol Chem. 1973 Jan 10;248(1):110–121. [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, deHaseth P. L., Lohman T. M. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977 Nov 1;16(22):4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- Revzin A., von Hippel P. H. Direct measurement of association constants for the binding of Escherichia coli lac repressor to non-operator DNA. Biochemistry. 1977 Nov 1;16(22):4769–4776. doi: 10.1021/bi00641a002. [DOI] [PubMed] [Google Scholar]
- Ribeiro A. A., Wemmer D., Bray R. P., Wade-Jardetzky N. G., Jardetzky O. High-resolution nuclear magnetic resonance studies of the Lac repressor. 1. Assignments of tyrosine resonances in the N-terminal headpiece. Biochemistry. 1981 Feb 17;20(4):818–823. doi: 10.1021/bi00507a025. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Anderegg R. Primary structure of the lambda repressor. Biochemistry. 1978 Mar 21;17(6):1092–1100. doi: 10.1021/bi00599a024. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Pan J., Hopper P., Hehir K., Brown J., Poteete A. R. Primary structure of the phage P22 repressor and its gene c2. Biochemistry. 1981 Jun 9;20(12):3591–3598. doi: 10.1021/bi00515a044. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Scherer G., Hobom G., Kössel H. Nucleotide sequence of cro, cII and part of the O gene in phage lambda DNA. Nature. 1978 Mar 30;272(5652):410–414. doi: 10.1038/272410a0. [DOI] [PubMed] [Google Scholar]
- Simatake H., Rosenberg M. Purified lambda regulatory protein cII positively activates promoters for lysogenic development. Nature. 1981 Jul 9;292(5819):128–132. doi: 10.1038/292128a0. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Richmond T. J., Wise D., Engelman D. The lac repressor protein: molecular shape, subunit structure, and proposed model for operator interaction based on structural studies of microcrystals. Proc Natl Acad Sci U S A. 1974 Mar;71(3):593–597. doi: 10.1073/pnas.71.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Folkmanis A., Echols H. Cro regulatory protein specified by bacteriophage lambda. Structure, DNA-binding, and repression of RNA synthesis. J Biol Chem. 1977 Sep 10;252(17):6177–6183. [PubMed] [Google Scholar]
- Weber K., Platt T., Ganem D., Miller J. H. Altered sequences changing the operator-binding properties of the Lac repressor: colinearity of the repressor protein with the i-gene map. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3624–3628. doi: 10.1073/pnas.69.12.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHaseth P. L., Gross C. A., Burgess R. R., Record M. T., Jr Measurement of binding constants for protein-DNA interactions by DNA-cellulose chromatography. Biochemistry. 1977 Nov 1;16(22):4777–4783. doi: 10.1021/bi00641a003. [DOI] [PubMed] [Google Scholar]
- deHaseth P. L., Lohman T. M., Record M. T., Jr Nonspecific interaction of lac repressor with DNA: an association reaction driven by counterion release. Biochemistry. 1977 Nov 1;16(22):4783–4790. doi: 10.1021/bi00641a004. [DOI] [PubMed] [Google Scholar]



