Neurospora and higher eukaryotes share a common mechanism for the signal transduction of environmental stimuli. A scenario is given in which the Neurospora WC-1 photoreceptor represents a function orthologous to that of vertebrate nuclear receptors, acting through the association with the HAT NGF-1 via a vertebrate-like LXXLL motif.
Abstract
In Neurospora crassa and other filamentous fungi, light-dependent–specific phenomena are regulated by transcription factors WC-1 and WC-2. In addition to its transcriptional activity, WC-1 is able to directly sense light stimuli through a LOV sensor domain. Its location in the nucleus and heterodimerization with WC-2, together with the presence of a zinc-finger DNA-binding domain and an environmental sensor domain, all resemble the functional evolutionary architecture adopted by vertebrate nuclear receptors (NRs). Here we describe a scenario in which WC-1 represents a functional orthologue of NRs and acts through association with the chromatin-modifying coactivator NGF-1, which encodes a homologue of the yeast Gcn5p acetyltransferase. To support this view, we show a direct association between WC-1 and NGF-1 that depends on a WC-1 region containing a conserved functional LXXLL motif, a signature previously described as being an exclusive feature of NR/coactivator interaction. Our data suggest that a WC-1/NGF-1 complex is preassembled in the dark on light-inducible promoters and that, after exposure to light stimulation, NGF-1–associated HAT activity leads to histone H3 acetylation and transcriptional activation. Finally, we provide evidence for a NGF-1–independent acetylated form of WC-1. Overall our data indicate that Neurospora and higher eukaryotes share a common mechanism for the signal transduction of environmental stimuli.
INTRODUCTION
All organisms need to respond to rapid changes in their environment. During evolution various mechanisms have been selected to enable an organism to rapidly sense and respond to environmental light cues.
In Neurospora, one of the most important extracellular cues is light, and a proper response to this stimulus is crucial for its survival. Light-signal transduction in Neurospora is mediated by the White Collar complex (WCC), a heterodimer of WC-1 (Ballario et al., 1996, 1998) and WC-2 (Linden and Macino, 1997), proteins that form a photoresponsive transcription factor complex (Crosthwaite et al., 1997; Talora et al., 1999; Cheng et al., 2002). WC-1 is both a photoreceptor and a zinc-finger DNA-binding protein, whereas its partner, WC-2, contains transcriptional activation domains essential for the WCC to function properly (He et al., 2002; Cheng et al., 2003). When WCC is activated upon light stimulus, a transient expression of light-responsive target genes ensues (Baima et al., 1991; Ruger-Herreros et al., 2011).
A whole-genome microarray of Neurospora revealed that ∼5.6% of transcript amounts rapidly increased after stimulation with a light pulse (Chen et al., 2009). Evidence suggests that such light-mediated transcription depends on White Collar proteins binding to responsive elements within the promoters of target genes (Carattoli et al., 1994; Froehlich et al., 2002; Liu et al., 2003).
Neurospora light signaling also involves chromatin modifications. Histone H3-K14 at light-inducible promoters (i.e., albino-3 and vivid) transiently increases its acetylation levels after photoinduction (Grimaldi et al., 2006). This histone modification has been shown to be under the control of the Neurospora histone acetyltransferase (HAT) Neurospora Gcn Five-1 (NGF-1), homologous to Saccharomyces cerevisiae HAT Gcn5p. Several of these features described for Neurospora light signaling resemble the evolutionary architecture adopted by vertebrate nuclear receptors (NRs). In higher eukaryotes, for example, the signal transduction of several stimuli is orchestrated by the NR superfamily of transcription factors (Gronemeyer et al., 2004). Canonical NRs seem to have appeared with the metazoans (Thornton et al., 2003).
NRs change their transcriptional activity upon the binding of small molecules such as hormones (Bain et al., 2007). Generally, NRs heterodimerize in the nucleus, and their association with coactivators or corepressors is essential for their functioning. NR and accessory proteins with chromatin-modifier activity are recruited to the promoter region of target genes (McDonnell et al., 1991; Wiench et al., 2011).
Among the coactivators, Gcn5-related N-acetyltransferase (GNAT) proteins (PCAF and GCN5) contribute to transcriptional activation in several physiological processes through their HAT enzymatic activity (Aoyagi and Archer, 2008).
A special signature represented by a consensus box (LXXLL motif, also called NR box, where L is a leucine and X is any amino acid) has been associated with coactivator–NR binding. The NR box can be present in single or multiple copies, and it can be found in the NR or its coregulator or both proteins (Savkur and Burris, 2004). Furthermore, in some cases the GNAT proteins also acetylate NRs, contributing to their response to signaling molecules (Wang et al., 2011).
Here we investigate the possibility that WC-1 exemplifies a functional orthologue of vertebrate NRs.
RESULTS
WC-1 binds to NGF-1 in dark and light conditions
Our previous data support a model in which NGF1 could be part of the WCC. We therefore wanted to see whether NGF-1 and WC-1 proteins would be able to interact in vivo and whether this interaction could be modulated by light.
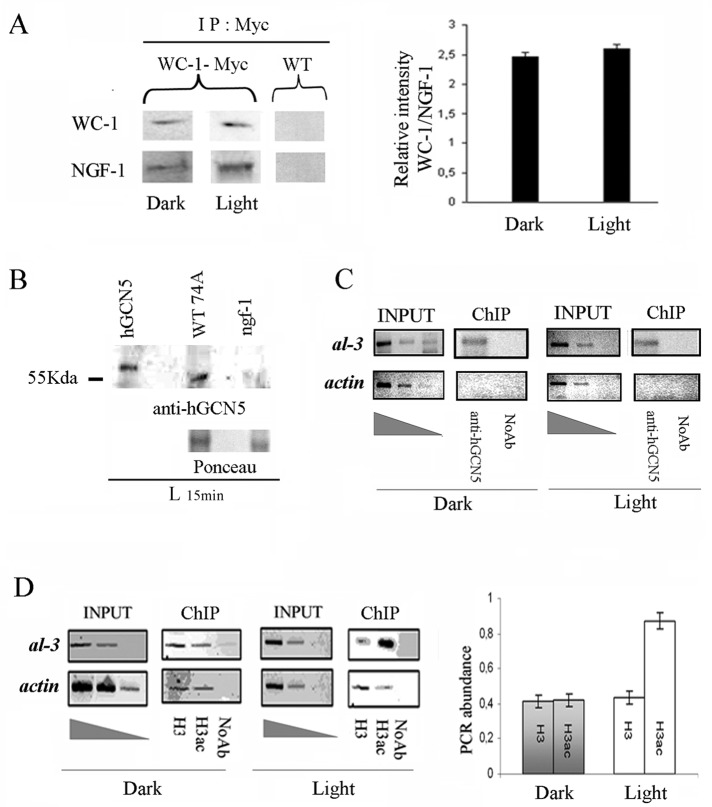
Protein extracts were prepared from a Neurospora strain expressing a Myc-tag version of WC-1 grown for 3 d in the dark, exposed to saturating white light for 5 min, and then returned to the dark for 25 min before harvesting (LP 30 min). WC-1 was then immunoprecipitated (IP) with an anti-Myc antibody, and the presence of NGF-1 in the IP samples was analyzed by Western blot with a commercial human GCN5 antibody. This analysis confirmed the in vivo association of NGF-1 with WC-1; of note, the two proteins coimmunoprecipitated in the dark (D) and after exposure to a light pulse (LP; Figure 1A, left). We also immunoprecipitated a wild-type (WT) strain (74A) but observed no signal with either the Myc or hGCN5 antibodies, confirming the specificity of the immunoprecipitation. Because we used a heterologous GCN5 (human) antibody, we tested this against a total ngf-1 extract to confirm lack of cross-hybridization. As expected, the hGCN5 revealed a signal only in the WT strain or hGCN5 purified protein and not in the ngf-1 mutant strain, confirming the specificity of the data (Figure 1B).
FIGURE 1:
NGF-1 physically interacts with WC-1 in dark and light conditions. (A) Myc WC-1 was immunoprecipitated (IP) using a commercial anti-Myc antibody from 400 μg of total protein extracts from Neurospora mycelia grown for 3 d in the dark before harvesting (Dark) or exposed to saturating white light for 5 min and then returned to the dark for 20 min before harvesting (Light 25 min). Precipitated WC-1 and coprecipitated NGF-1 were detected with anti-Myc and anti-hGCN5, respectively. As control of the specificity of the signal obtained, the same experiment was performed on the WT strain without the Myc epitope (left). Quantification of the ratio between the relative optical density of WC-1 and NFG-1 bands from three independent experiments under Dark and Light conditions (right). The data are the mean ± SEM from three independent experiments. (B) Western blot with anti-hGCN5 against hGCN5 purified protein, Neurospora WT, and ngf-1 strains confirms the absence of cross-hybridization of the heterologous antibody. Ponceau staining was used to check the presence of similar protein amounts on the membrane. (C) ChIP assay on Neurospora mycelia grown for 3 d in the dark before harvesting (Dark) or exposed to saturating white light for 5 min and then returned to the dark for 20 min before harvesting (Light 25 min). DNA immunoprecipitated with anti-hGCN5 antibody was amplified for the al-3 light-responsive promoter region. A sequence for actin was used as negative control. INPUT, PCR on chromatin samples before immunoprecipitation. ChIP, PCR from anti-hGCN5 IP samples. Noab, PCR from samples without antibody. (D) ChIP analysis as described in C was performed with anti–histone H3 (H3) and anti–acetylated histone H3 (H3ac; left). Quantification of the PCR abundance for the al-3 promoter was calculated as described in Materials and Methods (right). The data are the mean ± SEM from three independent experiments.
In addition, the level of WC-1/NGF-1 binding was apparently similar in dark and light-pulse conditions (Figure 1A, right). This suggests the existence in the dark of a preassembled WC-1/NGF-1 complex on the promoters of the WCC target genes. To explore this possibility, we evaluated the recruiting of NGF-1 to the light-responsive region (LRR) of the WC-1 target gene albino-3 (al-3) in both D and LP conditions (Grimaldi et al., 2006). As revealed by chromatin immunoprecipitation (ChIP) with anti-hGCN5 antibody and subsequent PCR amplification of the al-3 LRR, a similar profile in the D and LP conditions was obtained (Figure 1C).
Remarkably, chromatin immunoprecipitation with anti-H3 and anti-AcH3 antibodies performed in the same samples showed a twofold increase in the acetylation of histone H3 at al-3 LRR in the LP condition (Figure 1D). From this we inferred that it is not the WC-1/NGF-1 interaction per se present in D and L conditions that induces histone H3 acetylation, according to our previous report (Grimaldi et al., 2006).
Overall these results support a scenario in which WCC-mediated gene activation is achieved by light-dependent modification of HAT activity of the NGF-1/WC-1 complex previously assembled in the dark on the target promoters.
This view is consistent with fast transcriptional response to light irradiation (He and Liu, 2005).
The C-terminal region of WC-1 dictates direct interaction with histone acetyltransferase
The in vivo interaction between WC-1 and NGF-1 (Figure 1A) indicates the presence in WC-1 of an as-yet-uncharacterized functional domain involved in NGF-1 binding. To identify this domain, we tested the ability of different recombinant WC-1 versions fused with glutathione S-transferase (GST) to interact with an S35-radiolabeled yeast Gcn5p in pull-down assays. GNATs are highly conserved in evolution; Gcn5p shares strong homology with NGF-1, especially in the catalytic HAT domain (residues 107–219; 78% identity) and the bromodomain (residues 316–401; 63% identity; Grimaldi et al., 2006). We used the yeast Gcn5p because the full-length NGF-1 product is not well characterized, thus raising the possibility of missing some regions important for its function.
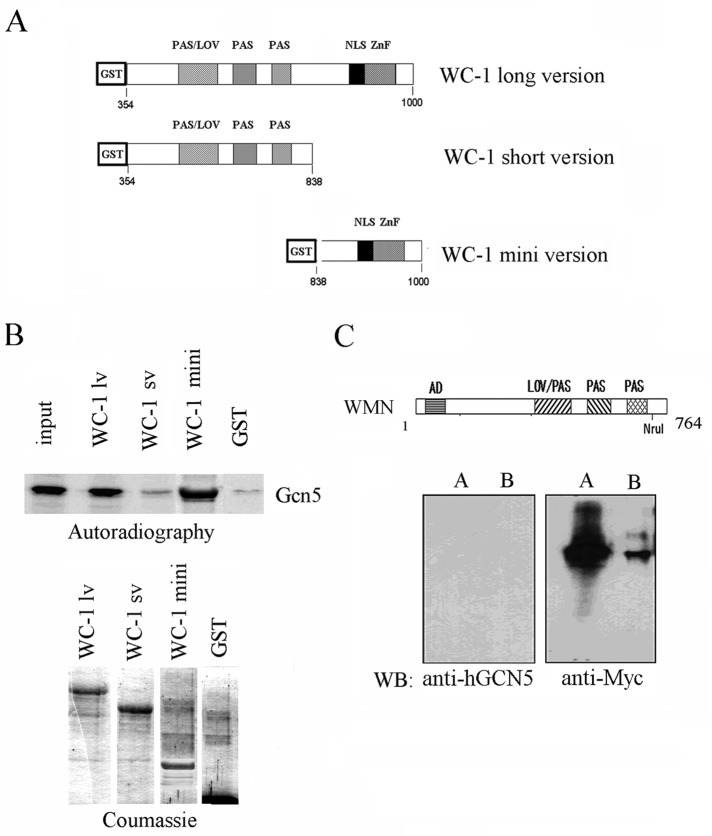
One of the recombinant WC-1 products is a long version of WC-1 (WC-1 lv, amino acids [aa] 354–1000) that includes the LOV domain, the two PAS domains, and the zinc-finger (ZnF) domain (Figure 2A). The other two WC-1 versions lack the C-terminal region containing the ZnF domain (WC-1 sv, aa 354–838) and the N−terminal region containing the LOV and the PAS domains (WC-1 mini, aa 838–1000), respectively (Figure 2A).
FIGURE 2:
WC-1 needs the C-terminal region between 838 and 1000 aa to bind Gcn5p in vitro and in vivo. (A) Schematic representation of the functional domains of the recombinant proteins used in the pull-down experiments: GST, glutathione S-transferase; LOV, light sensor domain; NLS, nuclear localization signal; PAS, protein–protein interaction domain; ZnF, zinc-finger DNA-binding domain. The numbers refer to the amino acid residues of WC-1 in relation to the initial methionine in the full-length protein. (B) Pull-down experiments using the same recombinant proteins as shown in A and [35S]Gcn5-radiolabeled protein. A representative autoradiography of the [35S]GCN5 protein retained by the GST fused proteins is shown (top). Input corresponds to 30% of total incorporation of [35S]methionine in the GCN5. Coomassie staining of the SDS–PAGE illustrates the amount of the recombinant WC-1 proteins used for the pull-down experiment shown in the top (bottom). (C) Scheme showing the NruI deletion in the Myc-tagged WC-1 protein (WMN; Cheng et al, 2003). Deleted WC-1 (aa 1–764) was immunoprecipitated with an anti-Myc antibody from 800 μg (lane A) and 400 μg (lane B) of total protein extract (bottom). IP samples were analyzed by Western blot with the indicated antibodies.
When analyzed in the pull-down assays, the WC-1 lv was able to retain 30% of the input S35-radiolabeled Gcn5p (Figure 2B). In marked contrast, the WC-1 version lacking the C-terminal region (WC-1 sv) retained only an amount similar to that obtained with the GST protein used as a negative control (Figure 2B). Of note, the WC-1 mini product expressing only the C-terminal domain preserved its ability to retain the labeled protein (Figure 2B).
These results identify the WC-1 portion between 838 and 1000 aa as the region directly involved in the association of WC-1 with HATs.
To further confirm that the C-terminal region of WC-1 dictates its interaction with Gcn5p also in vivo, we used a mutant of the Neurospora deletion mutant collection provided by Y. Liu (Cheng et al., 2003). This collection contains a mutant strain that express a C-terminal–deleted, Myc-tagged WC-1 (WC-1 Myc NruI [WMN]) corresponding to the WC-1 short-version recombinant protein, which was unable to bind Gcn5p in our pull-down assays (Figure 2C, top). Coimmunoprecipitation assays on protein samples from the WMN strain clearly showed that the WC-1 C-terminal region was required for association with NGF-1 in vivo (Figure 2C, bottom). In fact, the mutated WC-1 protein was unable to coimmunoprecipitate NGF-1 starting from two different input concentrations (800 ng; Figure 2C, lane A; and 400 ng; Figure 2C, lane B), although the amount of the WC-1 protein was high, as revealed by the anti-Myc antibody.
WC-1 C-terminal region is required for light-induced H3 acetylation
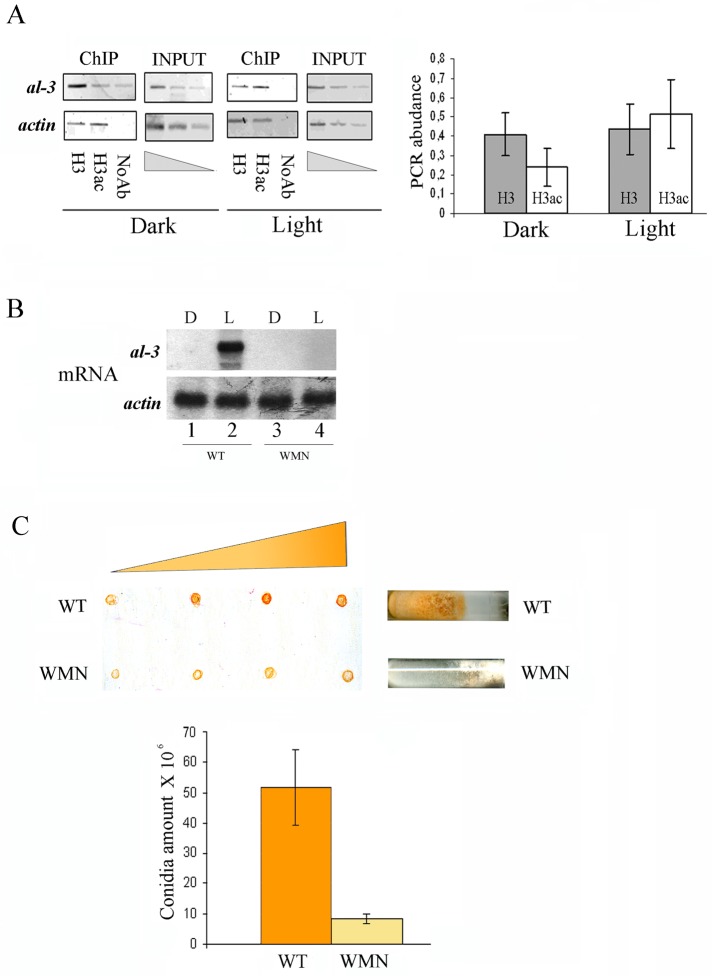
The C-terminal–deleted WC-1 protein failed to interact with NGF-1; accordingly, this HAT should not be recruited on light-induced promoters in the WMN mutant strain, and a light pulse should not increase H3 acetylation levels. To test this possibility, we performed ChIP assays with anti-H3 and anti-acH3 on the WMN strain kept in the dark or after exposure to a pulse of 5 min of light followed by 25 min of dark. The negligible differences in the H3 acetylation levels on the al-3 LRR observed between dark and light-pulse conditions confirmed that the WC-1/NGF-1 association is required for light-induced HAT activity (Figure 3A).
FIGURE 3:
Impairment of light-mediated H3 acetylation in the WMN strain. (A) ChIP assay on Neurospora WMN mycelia grown for 3 d in the dark before harvesting (Dark) or exposed to saturating white light for 5 min and then returned to the dark for 20 min before harvesting (Light 25 min). DNA immunoprecipitated with the anti–histone H3 (H3) and anti–acetyl histone H3 (H3ac) antibodies was amplified for the al-3 light-responsive promoter region. A sequence for actin was used as negative control. INPUT, PCR on chromatin samples before immunoprecipitation. ChIP, PCR from anti–hGCN5 IP samples. Noab, PCR from samples without antibody. Quantification of the PCR abundance for the al-3 promoter was calculated as described in Materials and Methods (right). The data are the mean ± SEM from three independent experiments. (B) Northern blot analysis of al−3mRNA in Neurospora WT and WMN strains kept in the dark (Dark) or after photoinduction (Light 25 min). The membrane was rehybridized using a probe against the actin mRNA as loading control (bottom). (C) WT and WMN strains were grown in slant for 4 d in the dark, illuminated for 5 h, and then kept in the dark for 8 h at 4°C to accumulate carotenoids. Increasing amounts of the conidia resuspended in water (5, 10, 20, and 25 μl) were spotted on a nitrocellulose filter to observe the increment of the red color corresponding to carotenoid content (top). Conidia count in WT and WMN was measured using a Burker cell counter (bottom). The data are the mean ± SEM from three independent experiments.
To further assess whether the lack of WC-1/NGF-1 interaction and light-induced H3 acetylation impairs the activation of carotenoid biosynthesis and conidia formation, we evaluated the al-3 transcription levels and the carotenoid production in the WMN and the isogenic wild-type strains.
Northern blot analysis of the two strains with a specific probe against al-3 mRNA showed that light-induced activation of the al-3 transcript is completely abolished in the WMN strain (Figure 3B). In addition, progressive dilutions of conidia from the WT and WMN strains spotted on a nitrocellulose filter showed a dramatic inhibition of conidiation in the strain harboring the WC-1–deleted mutant (Figure 3C, top). Cell count by hemocytometer revealed a 10-fold reduction of conidia in the WMN strain (Figure 3C, bottom). Overall these results indicate that the proper formation of a WC-1/NGF-1 complex on light-induced promoters is required for transcriptional responsiveness to light.
WC-1 is a fungal nuclear receptor–like transcription factor with a functional vertebrate NR box
Key to adaptation and survival has been the evolution of mechanisms that enable organisms to sense and rapidly respond to sudden environmental changes. In metazoans, the nuclear receptor superfamily of transcription factors comprises direct one-step signaling sensors that typically bind and respond to small molecules to regulate gene expression programs governing numerous cellular processes (for review see van de Wijngaart et al., 2011).
Although fungi and plants do not exhibit nuclear receptor orthologues, bioinformatics sequence analysis has shown that the transcription activity of members of the fungal zinc-finger transcription factor family is also modulated by small molecules, potentially representing environmental cues (for review see Näär and Thakur, 2009).
Although a photon cannot be considered a physical compound like nutrients or cellular metabolites, light constitutes an important environmental stimulus that can modulate the activity of the zinc-finger transcription factor WC-1 through a specific domain.
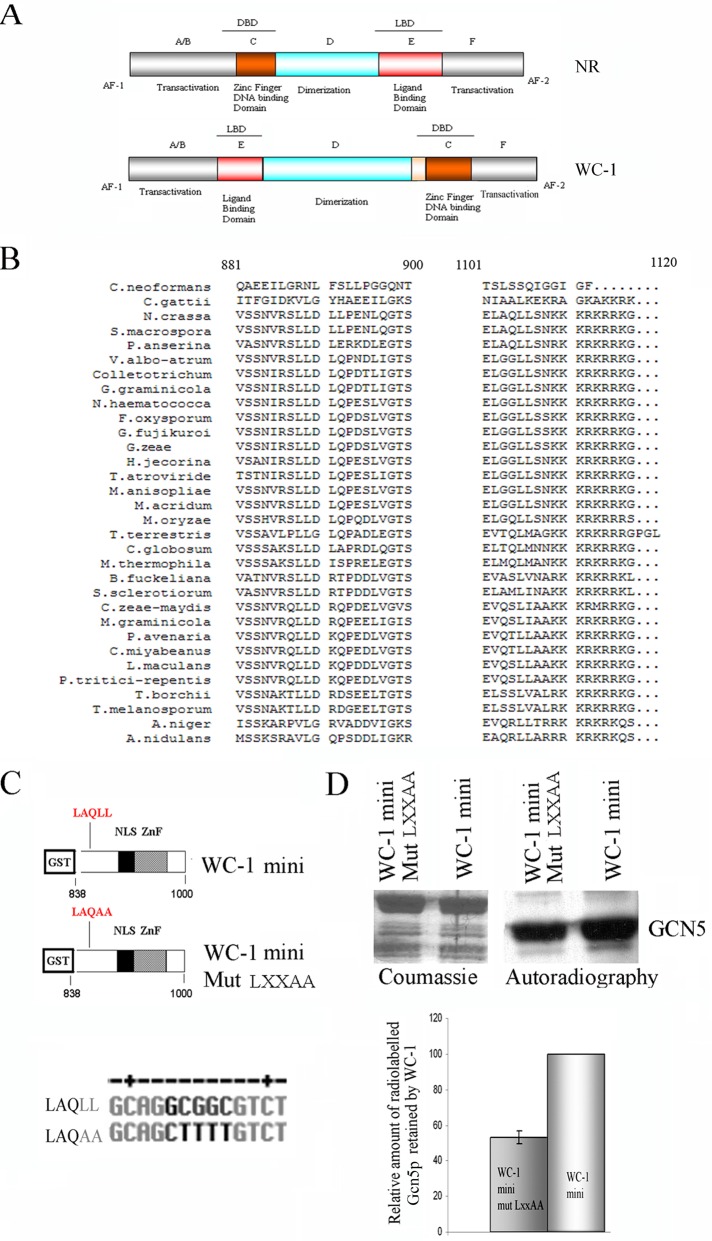
When the LOV domain is viewed as a ligand-binding domain (LBD) for light, extraordinary similarities emerge between the structural organization of metazoan nuclear receptors and Neurospora WC-1 (Figure 4A; Schaufele et al., 2005). Indeed, both transcriptional factors present N- and C-terminal domains involved in promoting gene activation, a zinc-finger DNA-binding domain and an LBD domain accountable for response to stimuli (Figure 4A).
FIGURE 4:
Identification of a WC-1 LXXLL motif involved in the association with HAT. (A) Schematic representation of a generic nuclear receptor (NR) and WC-1 structural organization, evidencing architectural and functional similarities. (B) Multialignment of the regions containing the LXXLL motifs within Neurospora WC-1 and several WC-1–like proteins present in the National Center for Biotechnology Information databank (LLDLL and LAQLL, indicated by a dashed line). Consensus analysis reveals conservation only for the LXXLL motif present at position 1008–1012 in Neurospora WC-1. Organisms and protein accession numbers: Cryptococcus neoformans, XP_567995.1; Cryptococcus gattii, XP_003193431.1; Neurospora crassa, CAA63964; Sordaria macrospora, XP_003351224.1; Podospora anserina, CAD60767; Verticillium albo-atrum, XP_003001986.1; Colletotrichum higginsianum, CCF31796.1; Glomerella graminicola, EFQ30482.1; Nectria haematococca, XP_003040715.1; Fusarium oxysporum, ABY47609; Gibberella fujikuroi, CAO85915; Gibberella zae, XP_388117; Hypocrea jecorina, AAV80185; Trichoderma atroviride, AAU14171.1; Metarhizium anisopliae, EFY99524.1; Metarhizium acridum, EFY92414; Magnaporthe oryzae, XP_360995.2; Thielavia terrestris, XP_003654033; Chaetomium globosum XP_001219613; Myceliophthora thermophila, XP_003665493.1; Botryotinia fuckeliana, XP_001547999.1; Sclerotinia sclerotiorum, XP_001586924.1; Cercospora zeae-maydis, AEH41590.1; Mycosphaerella graminicola, EGP83257.1; Phaeosphaeria avenaria, ACS74812.1; Cochliobolus miyabeanus, BAF35570.1; Leptosphaeria maculans, CBX98033.1; Pyrenophora tritici-repentis, XP_001933567.1; Tuber borchii, CAE01390.1; M., Tuber melanosporum, CAZ83744.1; Aspergillus niger, XP_001395039.1; Aspergillus nidulans, XP_661040.1. (C) Schematic representation of the domains in the two WC-1 mini versions and WT and point mutation mutant (Mut LXXAA) used in the pull-down experiments. The LAQLL mutagenized version is changed in LAQAA. (D) Recombinant proteins (C) were used in pull-down assays to evaluate their interaction with a radiolabeled Sc-Gcn5p. A representative Coomassie-stained gel and its autoradiography are shown (top). Quantification of the relative amount of 35S-ScGcn5p retained by the two recombinant products. The signal corresponding to WC-1 mini/ScGcn5p interaction was set to 100% (bottom). The data are the mean ± SEM from three independent experiments.
Of note, the molecular mechanism of action we describe here for WC-1 is also used by several NR members. It involves the direct interaction with chromatin-remodeling coregulators to execute their transcriptional activity (Green and Han, 2011). In vertebrates, several NRs, as well as several coactivators, harbor a leucine-X-X-leucine-leucine (LXXLL, where X is any amino acid) motif, also known as an NR box, which regulates the association of the transcription factors with the coregulators (Savkur and Burris, 2004; Jiang et al., 2010).
Although NR-like analogues in nonvertebrate organisms have been shown to lack the LXXLL motif, our attention was attracted by the presence of two LXXLL sequences (LLDLL, aa 727–732, and LAQLL, aa 911–916) within the primary sequence of WC-1. Figure 4B shows the alignment of most WC-1– or WC-1–like identified proteins; ∼50% of WC proteins examined contained the canonical LXXLL sequence and were members of Sordariomycetes, the largest class of Ascomycetes. Another 30% of WC-1 proteins were characterized by conservative substitution of polar neutral amino acids (leucine substituted by alanine or valine). The conserved LAQLL sequence is located in the C-terminal region of the WC-1 identified here to dictate its interaction with coregulator NGF-1. In contrast with the second conserved LXXLL, the first putative motif (LLDLL), located in the WC-1 truncated version lacking the NGF-1–interacting region (Figure 2A), appears not to be conserved (Figure 4B) and was not further investigated.
To assess whether the WC-1 LAQLL conserved region could work as an NR box mediating the association of WC-1 with its coactivator, we constructed a mutated version of the WC-1 mini protein used in the in vitro pull-down assay shown in Figure 2, where the two leucines were substituted by two alanines (Figure 4C, bottom). GST pull-down assays demonstrated that mutation of the LXXLL motif resulted in a 45% decrease in the ability of WC-1 mini protein to retain labeled Gcn5 protein (Figure 4D), clearly indicating that the integrity of the LXXLL motif identified in Neurospora WC-1 is required for maximal association with the coactivator NGF-1 and confirming previous observations (Fan et al., 2004).
To our knowledge, the box LAQLL is the first example of a nonvertebrate LXXLL motif mediating coactivator/NR-like interaction.
WC-1 is an acetylated protein
Because HATs acetylate many transcription factors and regulators, including NRs (Sterner and Berger, 2000; Roth et al., 2001; Wang et al., 2011), we decided to investigate whether WC-1 could be acetylated by the interacting acetyltransferase NGF-1.
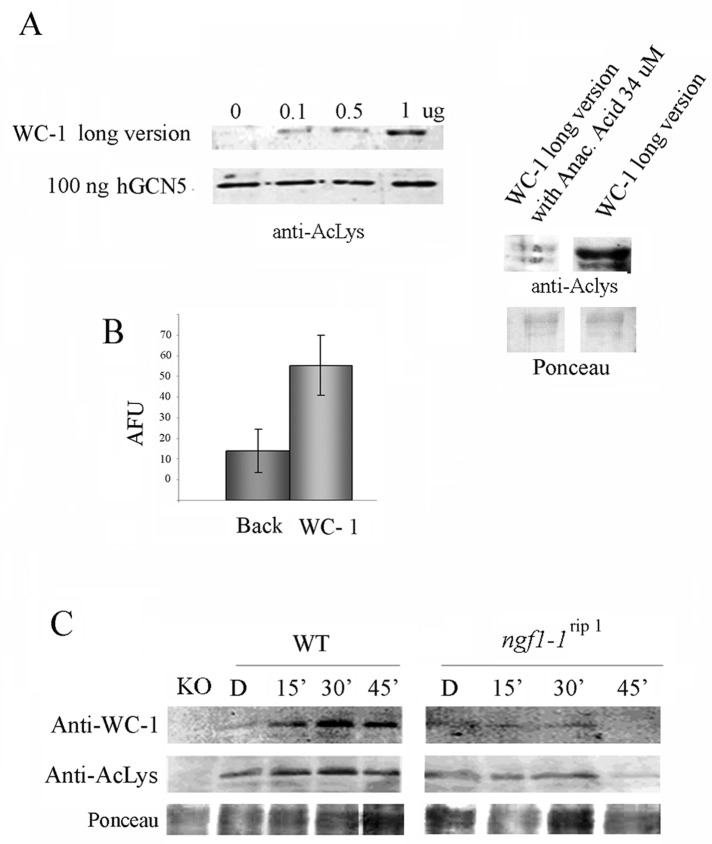
We first evaluated the ability of a recombinant WC-1 protein to be a substrate for an in vitro acetylation assay. Different amounts of the recombinant long-version GST–WC-1 were tested as a substrate for acetylation in the presence of human GCN5 acetyltransferase. As expected (see technical data sheet hGCN5 n.cat. 31204; www.activemotif.com/catalog/details/31204/gen5-active), hGCN5 was autoacetylated in each sample (Figure 5A, left). In addition, recombinant WC-1 was acetylated in this assay (Figure 5A). Addition of the HAT inhibitor anacardic acid (34 μM) decreased the WC-1 acetylated band, confirming that the acetylated signal corresponding to WC-1 was produced in the in vitro reaction (Figure 5A, right).
FIGURE 5:
WC-1 is an acetylated protein. (A) Progressive amounts of GST–WC-1 long version (Figure 2A) were tested in an in vitro acetylation assay in the presence of human GCN5, as described in Materials and Methods, using an anti–Pan acetylated-lysine antibody. Anacardic acid, a common HAT inhibitor, was used to prove the dependence of the acetylated signal on the acetyltransferase reaction. Ponceau staining was used to check the presence of similar WC-1 amounts on the membrane (right). (B) WC-1 acetylation was confirmed by a fluorescence in vitro acetylation assay. The data are the mean ± SEM from three independent experiments. AFU, arbitrary fluorescence units. (C) Western blot analysis with anti–WC-1 and anti–Pan acetylated- lysine antibodies of WT and ngf-1 RIP strains grown for 3 d in the dark before harvesting (Dark) or exposed to saturating white light for 5 min and then returned to the dark for 10, 25, and 55 min before harvesting (Light 15, 30, 60 min). A wc-1–null strain (KO) was used as a control of the specificity of the anti–WC-1 antibody. Ponceau staining was used to check the presence of similar WC-1 amounts on the membrane.
To rule out the possibility that the observed WC-1 acetylation was an artifact of our in vitro acetylation assay conditions, we further used a different assay based on fluorescence emission (Trievel et al., 2000), which, in the presence of recombinant WC-1 as substrate, showed a significantly higher degree of acetylation with respect to background (Figure 5B). Overall these results indicate that WC-1 can be a substrate for HAT enzymatic activity.
To further evaluate whether WC-1 can be acetylated in vivo by NGF-1, we performed Western blot analysis on WT and ngf-1RIP mutant strains exposed to different time intervals of light stimulus (from 0 to 60 min; Figure 5C). As reported previously (Lee et al., 2000), the profile of WC-1 expression showed a transient kinetic activation in the WT strain. No signal was detected in the protein extract from a wc-1 null strain (wc-1KO), confirming the specificity of the WC-1 antibody (Figure 5C). When the same filter was tested with an antibody against anti–Pan acetylated- lysine, a band corresponding to WC-1 was detected for all time intervals in the WT but not in the wc-1KO sample (Figure 5C).
In the ngf-1RIP mutant strain, the WC-1 protein signal was lower than that of the WT strain (Figure 5C), further confirming the role of NGF-1 in the activation of wc-1 expression (Grimaldi et al., 2006). Nevertheless, the band revealed by the anti–acetylated lysine antibody was also very faintly detected in the ngf-1RIP mutant strain (Figure 5C), indicating that NGF-1 could not be entirely responsible for WC-1 acetylation in vivo and other HATs might compensate for this function in the ngf-1RIP mutant background.
DISCUSSION
In the eukaryotic model organism Neurospora crassa, blue light signal transduction is mediated by a photoreceptor formed by the two zinc-finger transcription factors WC−1 and WC−2 (WCC). The heterodimeric WCC controls a light-dependent gene expression program by binding specific GATA sequences within LRRs of target promoters (Talora et al., 1999).
Chromatin modification and signal transduction have been extensively studied in several model organisms, including S. cerevisiae and filamentous fungi; however, the relevance of chromatin alterations in response to external stimuli was analyzed only relatively recently (Belden et al., 2007, 2011). In Neurospora the acetylation of histone H3-K14 at the light-inducible albino-3 promoter is transiently induced by light (Grimaldi et al., 2006). NGF-1, the Neurospora HAT homologue to S. cerevisiae Gcn5p, was shown to be responsible for this light-induced acetylation, and WC-1 was necessary for this process (Grimaldi et al., 2006).
In addition, we reported that WC-1 directly forms a complex with NGF-1, switching HAT activity on the target promoters (Figures 1 and 3). Remarkably, we observed that the association of WC-1 with NGF-1 does not depend on the light stimulus (Figure 1A) and that NGF-1 is present at the promoter of the light-inducible al-3 gene already in the dark (Figure 1C). Therefore the light stimulus seems to mediate a switching of HAT activity of the preassembled NGF-1 associated with the WCC, as indicated by the chromatin immunoprecipitation results under dark and light conditions (Figure 1D).
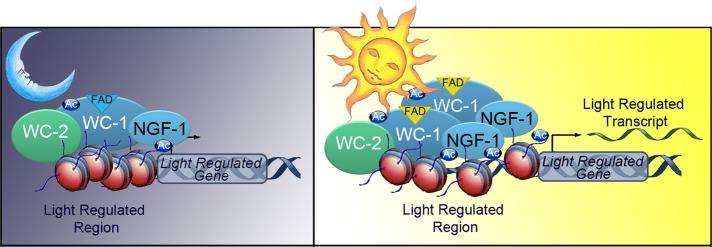
One intriguing possibility is that light determines a conformational change that exposes NGF-1 to the histone regions to be acetylated (Figure 6). This idea takes into account the proposed model in which a light-dependent conformational change of the LOV sensor domain can regulate the activity of LOV-containing proteins such as WC-1 (Crosson and Moffat, 2002).
FIGURE 6:
Diagram showing a possible working model. WC-1 and NGF-1 are preassembled in the dark on the promoter of light-inducible genes, but the histone tails are not accessible for NGF-1–mediated acetylation. (B) On photoinduction, the White Collar complex undergoes a conformational change that allows NGF-1 to acetylate histone H3 and promote transcriptional activation.
To confirm the importance of WC-1/NGF-1 interaction in light signaling and to localize the region involved, we performed GST pull-down experiments and identified a carboxy-terminal region in WC-1 responsible for its association with NGF-1 in vitro (Figure 2, A and B). This region also dictates WC-1/NGF-1 association in vivo, as indicated by the failure of the mutant WMN protein, lacking the C-terminus, to coimmunoprecipitate NGF-1 (Figure 2C).
Removal of the NGF-1–interacting region of WC-1 in the WMN strain completely impairs light-induced histone acetylation (Figure 3A) and abolishes light-mediated gene transcription (Figure 3B). As a result, the WMN strain presents a strong reduction of carotenoid biosynthesis (Figure 3C).
The association of WC-1 with the HAT NGF-1 reveals a functional architecture of the Neurospora photoreceptor with extraordinary similarities to the members of the nuclear receptor superfamily, as highlighted in Figure 4A.
As previously pointed out by Näär and Takum (2009), bioinformatics-based evolutionary studies were ineffective in recognizing NRs in lower eukaryotes. However, the study of functional domains of fungal transcription factors activated by environmental cues revealed the existence of a structure very similar to that of NRs. This finding can be interpreted as the product of the independent coevolution of vertebrate NRs and fungal one-step signal-effector proteins, leading to the hypothesis that blue light behaves in WC-1–mediated photoreception signaling similarly to the small effector molecules in NR–mediated transduction.
Remarkably, WC-1 region responsible for NGF-1 association contains a conserved LXXLL motif (Figure 4B), also known as an NR box, reported to be involved in the interaction between NRs and coregulators (Savkur and Burris, 2004). Mutagenesis experiments confirmed the importance of WC-1 LXXLL-motif integrity in the association with the coactivator (in this case Gcn5p; Figure 4C). Our results show that upon the substitution of the terminal LL residues with the amino acid AA there is a 45% reduction of NGF-1/WC-1–binding activity, in accord with previous observations (Fan et al., 2004) and partially in accord with others (Jiang et al., 2010). VIVID is the other Neurospora protein containing a LOV domain (Heintzen et al., 2001). However, it does not contain structural features reminiscent of nuclear receptor and the NR box. This suggests that the NR box marks only a subset of proteins involved in signal transduction. Other fungal and metazoan nonvertebrate zinc-finger factors have been reported to display functional similarity with vertebrate NRs. The lack of NR boxes in these proteins suggests that the LXXLL motif appeared with the evolution of higher eukaryotes (Näär and Takur, 2009).
For the first time, our findings revealed the involvement of the LXXLL motif in the association of transcription factors and coregulators in nonvertebrates. Overall our data indicate a conserved evolutionary mechanism in Neurospora and higher eukaryotes for one-step signal transduction responding to environmental stimuli, which involves the direct association of sensor proteins and chromatin-modifier coregulators.
Finally, we provide evidence, using two different HAT assays (Figure 5, A and B), that WC-1 can be acetylated in vitro and that this modification is blocked by the HAT inhibitor anacardic acid (Figure 5A).
Supporting the in vivo acetylation of WC-1, Western blot analysis with anti–WC-1 and anti–Pan acetylated lysine antibodies of WT protein extract showed an overlapping signal (Figure 5C). Although we cannot rule out the possibility that the acetylated band corresponds to a protein with the same molecular weight as WC-1, its absence in the wc-1KO strain strongly suggests the existence in vivo of a WC-1–acetylated form. Of note, this acetylation does not change upon photoinduction and is unlikely to depend on Neurospora NGF-1 HAT, considering its presence in the ngf-1RIP strain (Figure 5). However, further evidence that the acetylated band corresponds to WC-1 is that its signal is reduced in the ngf-1 mutant background, where WC-1 protein levels decrease due to defective photoinduction in the absence of NGF-1 (Figure 5). Taken together, the data suggest that WC-1 may be acetylated in vivo. Further studies are needed to elucidate the role of this modification in light signaling.
Figure 6 illustrates our working model for light-dependent transcriptional activation in Neurospora. It is a simply basal model for genes dependent on light activation.
The peculiarity of our model is the role of the histone acetyltransferase NGF-1 and of chromatin modification in blue light signal transduction.
MATERIALS AND METHODS
Neurospora strains and growth conditions
Neurospora wild-type strain 74OR23-1A (FGSC 987) was obtained from the Fungal Genetic Stock Center (Kansas City, KS). The wc- 1KO mutant (matA; his-3; bd; wc-1null; FGSC 3081) was a gift from J. Dunlap (Dartmouth Medical School, Hanover, NH).
The WC-1 Myc strain was a wc-1KO transformed with pDE3dBH derivative (Cheng et al., 2001) containing the entire wc-1 Myc tagged under the qa2 promoter. The WMN strain expresses a WC-1 protein truncated at the carboxy-terminus (from aa 764 to the end of the protein; Cheng et al., 2003). The strain ngf-1RIP has been described previously (Grimaldi et al., 2006). For liquid growth, 1 × 106 conidia were inoculated in Vogel's minimal medium supplemented with 1.5% sucrose, and cultures were incubated for 48–72 h in the dark at 28°C. For the preparation of dark-grown samples, mycelia were collected by filtration under a red safety lamp and frozen in liquid nitrogen. For the photoinduction experiments, mycelia were induced for 5 min with saturating light (10 W/cm2) and further grown in the dark for different time intervals before collection under a red safety lamp. The time intervals indicated in the figures refer to the time elapsed after exposure to the light-pulse stimulus.
Conidia count
Neurospora strains were grown in 250-ml flasks containing 100 ml of solid Vogel's minimal medium supplemented with 1.5% sucrose and then recovered in 25 ml of water. A dilution of conidia was observed with Burker cell counter, and the count was performed according to the manufacturer's formula:
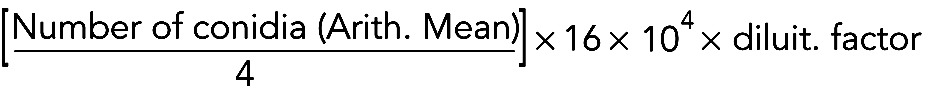 |
The chromatic effects (qualitative data) of the dilution were observed by spotting the conidia on a nitrocellulose filter.
Immunoprecipitation assay
Total protein samples from frozen mycelia were extracted with lysis buffer A (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 137 mM KCl, 10% glycerol containing 1 mM phenylmethylsulfonylfluoride [PMSF], 1 mM EDTA, and 1× protease inhibitor cocktail tablets [Roche, Indianapolis, IN]). For the immunoprecipitation assay, 5 μl of affinity-purified anti-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was previously incubated with 10 μl of protein A–Sepharose for 1 h at 4°C in 500 μl of lysis buffer A. After centrifugation, the antibody/protein A–Sepharose complex was washed twice with buffer A and then added to 400–600 of μg protein extract precleaned with protein A–Sepharose for 1 h at 4°C. After incubation for 2 h at 4°C, the beads were washed five times with 500 μl of lysis buffer containing 0.05% Triton X-100. The beads were then resuspended in Laemmli's sample buffer, boiled for 10 min at 95°C, and loaded on 10% SDS–PAGE gel.
Purification of GST–WC-1 proteins
The WC-1 long version, which is a PCR fragment corresponding to aa residues 354–100 of WC-1, was cloned in pGex 2T in a BamHI cloning site. The primers were Bat1 (5′-GGGAATTCGATGAAGGGGCGACGAGTG-3′) forward and Bat3 (5′-GGG AATTCGGACTTCTCGATG-3′) reverse, both with BamHI restriction enzyme box as tails. The WC-1 short version and mini version, described in Ballario et al. (1998; WC-1 short version, GST–WC-1 LOV+PAS; WC-1 mini version, GST–WC-1 Zn), were transformed in Escherichia coli BL21(DE3 pLys). Isopropyl β-d-1-thiogalactopyranoside was added for 3 h, and the cells were collected by centrifugation. The cell pellets were resuspended in a GST protein buffer containing an anti-protease cocktail inhibitor (Roche) and 0.5 mM EDTA and then sonicated. Insoluble material was removed by centrifugation. The GST-fusion proteins were purified from the soluble extracts in a glutathione–Sepharose 4B column (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's instructions.
LXXLL mutagenesis
The protocol for mutagenesis followed the indications given in the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). For mutagenesis of LAQLL in LAQAA, we performed PCR using as template WC-1 mini DNA and the following mutagenesis primers:
Primer 1: 5′-GAAGAGCTGGCGCAGGCGGCGTCTAACAAGAAAAAACGG-3′
Primer 2: 5′-CCGTTTTTTCTTGTTAGACGCCGCCTGCGCCAGCTCTTC-3′
In vitro pull-down assay
GST–WC-1 long, short, mini, and mutated LXXAA constructs were expressed in the BL21 E. coli strain and purified to homogeneity with glutathione–agarose beads (Amersham) as described previously (Ballario et al., 1996). Plasmid pGem 4-Z containing the full cDNA sequence of S. cerevisiae Gcn5 under the control of the T7 promoter was previously described (Ornaghi et al., 1999). In vitro–radiolabeled Gcn5p was synthesized using reticulocyte reaction (Promega, Madison, WI) and incubated for 2 h with a similar amount of recombinant GST–WC-1 proteins resuspended in extraction buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 1 mM dithiothreitol, 2 mM MgCl2, 0.5 mM EDTA, pH 7.5, 0.5% NP40, 1 mM PMSF, and 1× protease inhibitor). The glutathione–Sepharose beads were sedimented by centrifugation and washed five times with washing buffer (50 mM Tris-HCl, pH 8.0, 300 mM KCl, 1 mM EDTA, 1× protease inhibitor, 1 mM PMSF, 5% glycerol). After the final wash, the beads were resuspended in Laemmli's sample buffer, and the proteins were run on 10% SDS–PAGE electrophoresis gel. After staining with Coomassie blue reagent, the gel was dried and subjected to autoradiography.
Chromatin immunoprecipitation assays
The ChIP experiments were performed as described in Grimaldi et al. (2006). The antibodies for immunoprecipitation were anti H3 (06-755; Upstate Biotechnology, Charlottesville, VA), anti–acetyl H3 (06-599; Upstate Biotechnology), and anti-hGCN5 (Santa Cruz Biotechnology). The obtained template was subjected to PCR using the following primers: 30 (5′-AGA TAG ATC TCT TGG CCT TG-3′) and 31 (5′-CGA TTA TTG GAA ACC CGT CGG TA-3′) for the promoter region of al-3, and ACT1 (5′-CCT CTC TCA GCC AAA GCA TC-3′) and ACT2 (5′-GAA AGC TTA CCC CAT TGT CG-3′) for the promoter region of the actin gene used for normalization. PCR products were amplified for 25 cycles and resolved on 2% agarose gel. To ensure that the amplified PCR products were in the linear range, the PCR conditions were calibrated with different amounts of immunoprecipitated samples and input DNA (derived from cross-linked chromatin without immunoprecipitation). Band intensities were quantified by optical density analysis with OptiQuant Software (PerkinElmer Life and Analytical Sciences, Boston, MA). As negative controls, mock precipitations were performed in the absence of antibody. PCR products from these negative control samples were not detectable by ethidium bromide staining. The histograms in Figures 1C and 3A show the ratios between the values for the al-3 and actin PCR products immunoprecipitated, divided by the same ratio obtained by PCR with input DNA [(al-3/act)IP/(al-3/act)input]. Three ChIP replicates were performed on different preparations for each experiment.
Protein extraction and Western blot analysis of proteins
Western blot analysis was performed as previously described (Crosthwaite et al., 1997). Tissue was ground in liquid nitrogen with mortar and pestle and suspended in ice-cold lysis buffer A (50 mM HEPES, pH 7.4, 137 mM KCl, 10% glycerol containing 1 mM PMSF, 1 mM EDTA, 1× protease inhibitor) at a ratio of 0.5 ml of buffer to 0.1 g of tissue. Extracts were homogenized with three strokes of a Teflon/glass homogenizer, and cellular debris was removed by centrifugation at 10,000 relative centrifugal force. The protein concentration of the supernatant was determined with the Bio-Rad (Hercules, CA) reagent according to the manufacturer's instructions. Equal amounts of proteins (100 μg) for different extracts were denatured in Laemmli's sample buffer and separated by 8% SDS–PAGE for WC-1 analysis. After transfer to nitrocellulose (Amersham Pharmacia Biotech), the blots were incubated for 2 h at room temperature (RT) with anti–Myc affinity-purified immunoglobulin G (IgG; diluted 1:300 in phosphate-buffered saline [PBS] 1×, Tween 0.5%, and milk 5%), anti-Gcn5 (Santa Cruz Biotechnology) purified IgG (1:500 in PBS 1×, Tween 0.5%, and milk 5%), or anti–Pan acetylated-lysine (Santa Cruz Biotechnology) purified IgG (1:300 in PBS 1×, Tween 0.5%, and milk 5%). Horseradish peroxidase–conjugated antibodies (1:10,000) were used as secondary antibody (Bio-Rad), and membranes were developed using chemiluminescence reaction (ECL; Amersham Pharmacia Biotech).
Northern blot analysis
Total RNA from the WT and WMN strains was isolated according to a previously published method (Baima et al., 1991). For Northern analyses, a volume of 20 μg of total RNA was fractionated on 1.2% denaturing agarose gels in 5% formaldehyde-3-(N-morpholino) propanesulfonic acid buffer and transferred to positively charged nylon membranes (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Hybridization was carried out according to the manufacturer's instructions. The al-3 transcript was detected with a 32P–labeled PCR fragment amplified with primers AL3F (5′-GTCCCTCCAAGGACCTCTTC-3′) and AL3R (5′-CCAGGAGGCTGTGTGTAGCA-3′). For normalization, the membranes were stripped in 0.1% SDS at 95°C and hybridized with an actin-1, 32P-labeled probe obtained by PCR amplification with primers ACTF (5′-GCCTTCTACGTCTCCACCA-3′) and ACTR (5′-GTCGGAGAGACCAGGGTACA-3′).
In vitro protein acetylation assay
Different amounts of recombinant GST WC-1 were incubated at 30°C for 60 min in 30 μl with 6 μl of 5× HAT assay buffer (250 mM Tris, pH 8.0, 25% glycerol, 0.5 mM EDTA, 250 mM KCl, and 10 mM sodium butyrate), 0.5 mM acetyl-CoA, and 100 ng of human GCN5. After incubation, the reactions were stopped by adding 6 μl of 5× Laemmli's buffer, and the proteins were separated on SDS–PAGE gel. After transfer to a nitrocellulose membrane, the filter was probed with an anti–Pan acetylated-lysine (Santa Cruz Biotechnology) antibody.
Acetyltransferase fluorescence assay
The HAT assay kit is a fluorescence method for assaying samples for histone or non–histone acetyltransferase (HAT) activity or to screen HAT inhibitors (Trievel et al., 2000). A volume of 30 μl of reaction mix (1× buffer HAT, 0.5 mM acetyl-CoA, 125 ng of hGCN5) is needed per assay well. In brief, 10 μl (1 μg) of WC-1 long version is added to the assay well, without adding protein substrate to background wells so that autoacetylation activity of the acetyltransferase is included in the background measurement. The plate is covered and incubated for 30 min at room temperature. A 50-μl amount of stop solution is added per well, followed by addition of 1 μg of WC-1 to the background well. Then 100 μl of developing solution is added to each well, followed by incubation for 15 min in the dark at room temperature. Fluorescence is read with excitation at 360–390 nm and emission at 450–470 nm.
Sequence analysis of WC-1 putative NR boxes
The sequences of WC-1 proteins from the different species (Figure 5) were aligned with GeneBee Multialignment software (www.genebee.msu.su). The local supermotif containing the putative NR boxes is shown in Figure 5.
Acknowledgments
We thank Yi Liu for the generous gift of Neurospora mutant WC-1 Myc NruI. We thank Alberto Gualtieri and Raffaele Gerace for their assistance in setting up the acetylation assay and bioinformatic analysis, respectively. We also thank Francesca Spadaro for scientific collaboration. A.B. is a Teresa Ariaudo Fellowship scholar.
Abbreviations used:
- NGF-1
Neurospora GCN Five-1
- WCC
white collar complex
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-02-0142) on August 8, 2012.
*Present address: Drug Discovery and Development, Italian Institute of Technology, 16163 Genoa, Italy.
REFERENCES
- Aoyagi S, Archer TK. Dynamics of coactivator recruitment and chromatin modifications during nuclear receptor mediated transcription. Mol Cell Endocrinol. 2008;280:1–5. doi: 10.1016/j.mce.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S, Macino G, Morelli G. Photoregulation of the albino-3 gene in Neurospora crassa. J Photochem Photobiol. 1991;11:107–115. doi: 10.1016/1011-1344(91)80253-e. [DOI] [PubMed] [Google Scholar]
- Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annu Rev Physiol. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- Ballario P, Talora C, Galli D, Linden H, Macino G. Roles in dimerization and blue light photoresponse of the PAS and LOV domains of Neurospora crassa white collar proteins. Mol Microbiol. 1998;29:719–729. doi: 10.1046/j.1365-2958.1998.00955.x. [DOI] [PubMed] [Google Scholar]
- Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Lewis ZA, Selker EU, Loros JJ, Dunlap JC. CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet. 2011;7:e1002166. doi: 10.1371/journal.pgen.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Carattoli A, Cogoni C, Morelli G, Macino G. Molecular characterization of upstream regulatory sequences controlling the photoinduced expression of the albino-3 gene of Neurospora crassa. Mol Microbiol. 1994;13:787–795. doi: 10.1111/j.1365-2958.1994.tb00471.x. [DOI] [PubMed] [Google Scholar]
- Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;28:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Gardner KH, Liu Y. PAS domain–mediated WC-1/WC-2 interaction is essential for maintaining the steady state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol Cell Biol. 2002;22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Liu Y. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc Natl Acad Sci USA. 2001;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Wang L, He Q, Liu Y. WHITE COLLAR-1, a multifunctional Neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J Biol Chem. 2003;278:3801–3808. doi: 10.1074/jbc.M209592200. [DOI] [PubMed] [Google Scholar]
- Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, Jäeger S, Erdjument-Bromage H, Tempst P, Spiegelman BM. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- Green CD, Han JD. Epigenetic regulation by nuclear receptors. Epigenomics. 2011;3:59–72. doi: 10.2217/epi.10.75. [DOI] [PubMed] [Google Scholar]
- Grimaldi B, Coiro P, Filetici P, Bergem E, Dobosy JR, Freitag M, Selkerm EU, Ballario P. The Neurospora crassa White Collar-1 dependent blue light response requires acetylation of histone H3 lysine 14 by NGF-1. Mol Biol Cell. 2006;17:4576–4583. doi: 10.1091/mbc.E06-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson JA, Laudet V. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y. White collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- He Q, Liu Y. Degradation of the Neurospora circadian clock protein FREQUENCY through the ubiquitin-proteasome pathway. Biochem Soc Trans. 2005;33:953–956. doi: 10.1042/BST20050953. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- Jiang P, Hu Q, Ito M, Meyer S, Waltz S, Khan S, Roeder RG, Zhang X. Key roles for MED1 LxxLL motifs in pubertal mammary gland development and luminal-cell differentiation. Proc Natl Acad Sci USA. 2010;107:6765–6770. doi: 10.1073/pnas.1001814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Loros JJ, Dunlap JC. Interconnected feedback loops in the Neurospora circadian system. Science. 2000;289:107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]
- Linden H, Macino G. White collar 2, a partner in blue light signal transduction, controlling expression of light regulated genes in Neurospora crassa. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He Q, Cheng P. Photoreception in Neurospora: a tale of two White Collar proteins. Cell Mol Life Sci. 2003;60:2131–2138. doi: 10.1007/s00018-003-3109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Nawaz Z, O'Malley BW. In situ distinction between steroid receptor binding and transactivation at a target gene. Mol Cell Biol. 1991;11:4350–4355. doi: 10.1128/mcb.11.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär AM, Thakur JK. Nuclear receptor–like transcription factors in fungi. Genes Dev. 2009;23:419–432. doi: 10.1101/gad.1743009. [DOI] [PubMed] [Google Scholar]
- Ornaghi P, Ballario P, Lena AM, González A, Filetici P. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J Mol Bio. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Ruger-Herreros C, Rodríguez-Romero J, Fernández-Barranco R, Olmedo M, Fischer R, Corrochano LM, Canovas D. Regulation of conidiation by light in Aspergillus nidulans. Genetics. 2011;188:809–822. doi: 10.1534/genetics.111.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Burris TP. The coactivator LXXLL nuclear receptor recognition motif. J Pept Res. 2004;63:207–212. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- Schaufele F, Carbonell X, Guerbadot M, Borngraeber S, Chapman MS, Ma AA, Miner JN, Diamond MI. The structural basis of androgen receptor activation: intramolecular and intermolecular amino–carboxy interactions. Proc Natl Acad Sci USA. 2005;102:9802–9807. doi: 10.1073/pnas.0408819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talora C, Franchi L, Linden H, Ballario P, Macino G. Rule of a white collar-1–white collar-2 complex in blue-light signal transduction. EMBO J. 1999;18:4961–4968. doi: 10.1093/emboj/18.18.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- Trievel RC, Li FY, Marmorstein R. Application of a fluorescent histone acetyltransferase assay to probe the substrate specificity of the human p300/CBP–associated factor. Anal Biochem. 2000;287:319–328. doi: 10.1006/abio.2000.4855. [DOI] [PubMed] [Google Scholar]
- van de Wijngaart DJ, Dubbink HJ, van Royen ME, Trapman J, Jenster G. Androgen receptor coregulators: recruitment via the coactivator binding groove. Mol Cell Endocrinol. 2011;352:57–69. doi: 10.1016/j.mce.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Wang C, Lifeng T, Popov VM, Pestell RG. Acetylation and nuclear receptor action. Steroid Biochem Mol Biol. 2011;123:91–100. doi: 10.1016/j.jsbmb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiench M, Miranda TB, Hager GL. Control of nuclear receptor function by local chromatin structure. FEBS J. 2011;278:2211–2230. doi: 10.1111/j.1742-4658.2011.08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]