Abstract
We identified nine FLOWERING LOCUS C homologues (BnFLC) in Brassica napus and found that the coding sequences of all BnFLCs were relatively conserved but the intronic and promoter regions were more divergent. The BnFLC homologues were mapped to six of 19 chromosomes. All of the BnFLC homologues were located in the collinear region of FLC in the Arabidopsis genome except BnFLC.A3b and BnFLC.C3b, which were mapped to noncollinear regions of chromosome A3 and C3, respectively. Four of the homologues were associated significantly with quantitative trait loci for flowering time in two mapping populations. The BnFLC homologues showed distinct expression patterns in vegetative and reproductive organs, and at different developmental stages. BnFLC.A3b was differentially expressed between the winter-type and semi-winter-type cultivars. Microsynteny analysis indicated that BnFLC.A3b might have been translocated to the present segment in a cluster with other flowering-time regulators, such as a homologue of FRIGIDA in Arabidopsis. This cluster of flowering-time genes might have conferred a selective advantage to Brassica species in terms of increased adaptability to diverse environments during their evolution and domestication process.
Introduction
FLOWERING LOCUS C (FLC) is a key regulator of flowering time in Arabidopsis thaliana. FLC and FRIGIDA (FRI) are important determinants of variation in the requirement for vernalization (i.e., the acceleration of flowering time in response to an extended period of low temperature) that is observed among natural ecotypes of Arabidopsis [1]–[3]. FLC encodes a MADS-domain transcriptional regulator that delays flowering by repressing the expression of floral integrators such as FT, SOC1, and FD in Arabidopsis [4]–[6]. FRI upregulates FLC expression through at least two distinct pathways, which involve increased methylation of H3K4 and a co-transcriptional process to regulate efficient splicing of the FLC locus [7]–[10]. FLC is negatively regulated by autonomous pathway regulators, which are thought to act through chromatin remodeling or RNA processing, and is also repressed by vernalization pathway regulators during and after cold exposure [11]. Recent studies have revealed that a number of chromatin modifiers including long noncoding RNAs and polycomb components are involved in vernalization-mediated epigenetic silencing of FLC [12]–[17]. A 272-bp region in the promoter of AtFLC (FLC in Arabidopsis thaliana) is required for the cold-induced repression of the gene, and a 75-bp segment in this region is essential for expression of AtFLC in the absence of vernalization [18].
Cultivated Brassica species comprise many important vegetable, oil, food, and feed crops, which have evolved as a result of single or multiple polyploidization events after divergence from Arabidopsis lineages. Monogenomic diploid Brassica species, such as B. rapa (AA), B. nigra (BB), and B. oleracea (CC), underwent whole-genome triplication and 24 conserved collinear blocks (from A to X) of the ancestral karyotype (n = 8) have been identified in Arabidopsis and Brassica lineages [19]. Allotetraploid rapeseed (Brassica napus L., 2n = 4x = 38, AACC), a major oilseed crop that originated from natural hybridization between B. rapa and B. oleracea, shows a high degree of collinearity to its diploid progenitors [20]–[22]. However, as a result of polyploidization, a number of genes in B. napus have undergone gene duplication and subsequent nonfunctionalization or functional divergence, such as neofunctionalization or alternative gene expression [23], [24].
The A. thaliana genome contains one FLC gene. However, the diploid members of Brassica contain multiple copies of FLC and several such homologues have been shown to be associated with flowering time variation, such as BrFLC.A10 (BrFLC1) and BrFLC.A2 (BrFLC2) in B. rapa [25], [26] and BoFLC.C2 (BoFLC2) in B. oleracea [27]. In B. napus, cDNA sequences of five FLC homologues (BnFLC1 to BnFLC5) have been isolated and their ectopic expression delays flowering in A. thaliana [28]. However, it is unknown whether additional FLC homologues are present in the B. napus genome. Molecular mapping studies have revealed a number of quantitative trait loci (QTL) that control flowering time, and might involve FLC homologues, in different biparental populations of B. napus [29], [30]. Recently, allelic variation of BnFRI.A3 (FRIGIDA, designated BnaA.FRI.a) was shown to be associated with flowering-time variation in a worldwide collection of B. napus accessions [31]. These findings suggest that the homologues of FLC and FRI play key roles in the regulation of flowering time in B. napus.
Various computational methods have been used to investigate potential gene regulatory elements from diverse organisms [32]. The comparison of promoter and intragenic regions of BoFLC genes in B. oleracea with AtFLC revealed extensive differences in structure and organization, but showed high levels of conservation within those segments essential for regulation of FLC expression [33]. This suggests that similar regulatory cis-acting elements were maintained during evolution even in duplicated genes that might have undergone functional divergence. In the present study, we characterized nine members of the BnFLC gene family and investigated their roles in the regulation of flowering time in B. napus via gene expression analysis. We also found that the BnFLC.A3b gene is tightly linked to a cold-responsive gene BnCBF.A3, and loosely linked to another flowering repressor gene, BnFRI.A3. Linkage of functionally related genes might have conferred a selective advantage during the evolution and domestication of Brassica species.
Results
Conservation and Divergence of FLC Homologues in Different Brassica Genomes
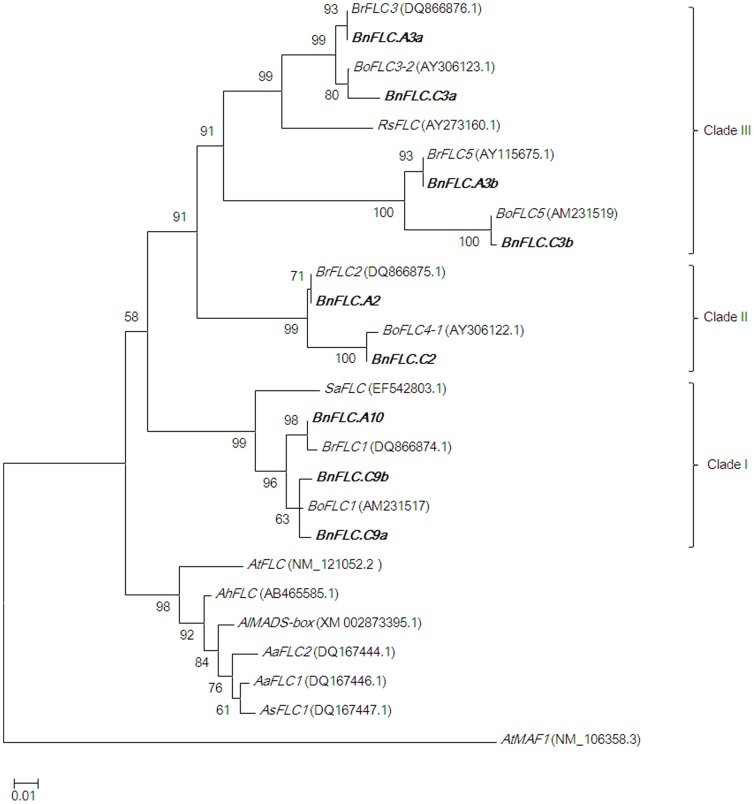
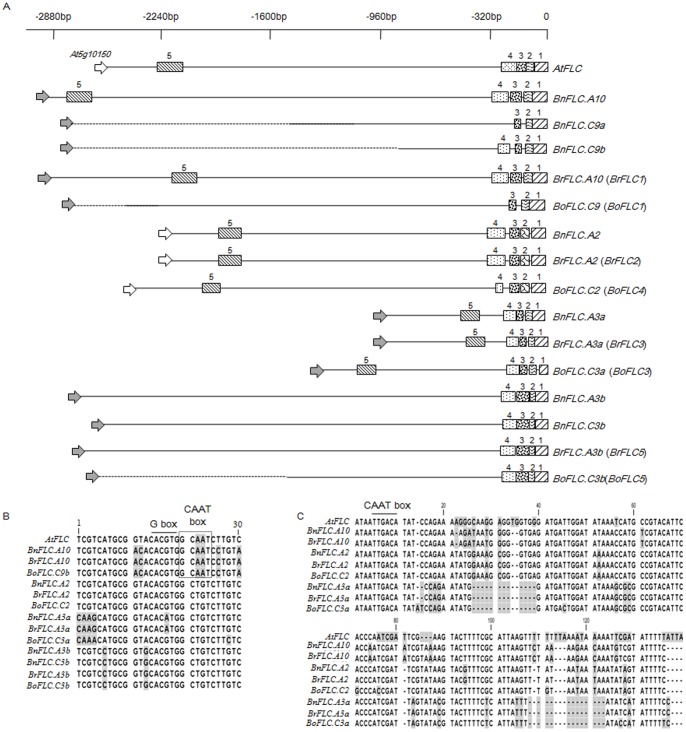
We cloned nine FLC homologues, four from the A genome and five from the C genome of B. napus, either from bacterial artificial chromosomes (BACs) or from PCR amplicons generated from genomic DNA (gDNA). We then compared their sequences to the homologues in their diploid progenitors B. rapa and B. oleracea (Table 1). Neighbour-joining cluster analysis using coding sequence data for FLC homologues among 10 species of Brassicaceae revealed that the BnFLC homologues were grouped into three distinct clades, which reflects the whole-genome triplication events that occurred during the evolution of the Brassica genome (Figure 1). Seven of the BnFLC homologues were located in the collinear ‘R’ block region (in which FLC is located in Arabidopsis) and included a tandem repeat on chromosome C9, whereas two homologues (BnFLC.A3b and BnFLC.C3b) were mapped to the noncollinear ‘J’ block (Table 1; Figure 2). Consistent with their homologues in other Brassicaceae, all BnFLC genes consisted of seven exons, which were interrupted by six introns and showed extremely high levels of similarity (95–100%) to the homologous sequences in the diploid progenitors B. rapa and B. oleracea. However, the promoter sequences of the BnFLC homologues were more divergent among each other and from AtFLC (Figure 3A). The 272-bp cis-regulatory region of the AtFLC promoter, which was reported to be conserved in B. oleracea [33], could be dissected further into four small blocks (referred to as cis-blocks) by sequence comparison in the present study. The first three cis-blocks were the most conserved among all Brassica FLC promoters, and the fourth cis-block, which corresponded to the crucial 75-bp segment for expression of AtFLC in nonvernalized plants [18], was absent from BnFLC.C9a and BoFLC.C9. A 30-bp segment that contained a G-box and a CAAT-box in cis-block 4 was highly conserved in some of the BnFLC homologues, as well as AtFLC (Figure 3B).
Table 1. Brassica napus FLC homologues, their map positions, and sequence identities compared with their orthologues in B. rapa or B. oleracea.
| BnFLC homologue | Sourcea | Isolated regionb | Orthologue in B. rapaor B. oleracea | Identity (%)c | ||
| Promoter region (approximately −1.5 kb) | Coding region | Intronic region | ||||
| BnFLC.A2 | JBnB035G21* | Promoter-3′ UTR | BrFLC2 (A2, R)d | 99 | 99 (0)e | 99 |
| BnFLC.A3a | JBnB50A15* | Promoter-3′ UTRf | BrFLC3 (A3, R) | 98 | 100 (0) | 99 |
| BnFLC.A3b | JBnB142O14* | Promoter-3′ UTR | BrFLC5 (A3, J) | 99 | 100 (0) | 99 |
| BnFLC.A10 | JBnB75D10* | Promoter-3′ UTR | BrFLC1 (A10, R) | 100 | 99 (0) | 99 |
| BnFLC.C2 | gDNA (T) | Exon 2-intron 6 | BoFLC4 (C2, R) | – | 100 (0) | 97 |
| BnFLC.C3a | gDNA (N) | Exon 1-intron 1 | BoFLC3 (C3, R) | – | – | 98 |
| BnFLC.C3b | JBnB090B17* | Promoter-3′ UTR | BoFLC5 (C3, J) | 100 | 99 (0) | 99 |
| BnFLC.C9a | JBnB003G07* | Promoter-intron 6 | BoFLC1 (C9, R) | 95 | 99 (0) | 98 |
| BnFLC.C9b | JBnB003G07* | Exon 1-intron 6 | BoFLC1 (C9, R) | – | 99 (2) | 98 |
The sequences isolated from BACs are indicated by an asterisk, and those isolated from PCR-amplified genomic DNA are designated ‘gDNA’ with the parent indicated in parentheses (T, B. napus cv. Tapidor; N, B. napus cv. Ningyou7).
The obtained sequences were compared with that of AtFLC to define the regions of each BnFLC gene that were isolated.
To calculate sequence identities, indels (insertions or deletions) were excluded and for partial sequences of BnFLC.C2 and BnFLC.C3a, the available region was used.
The letter and numeral in parentheses represent the linkage group followed by the ancestral block in which the FLC homologues are located.
The number of nonsynonymous nucleotide substitutions is shown in parentheses.
There was a fragment with higher-order structure in intron 1 of BnFLC.A3a for which we failed to obtain the sequence.
Figure 1. Phylogenetic tree of FLC homologues from Brassica, Arabidopsis, Raphanus, and Sinapis species.
BnFLC homologues are highlighted in bold, and AtMAF1 (MADS AFFECTING FLOWERING 1 of A. thaliana, an AtFLC-like gene) was used as the outgroup. Br, Brassica rapa; Bo, B. oleracea; Rs, Raphanus sativus (radish); Sa, Sinapis alba (white mustard); At, Arabidopsis thaliana; Al, A. lyrata; Ah, A. halleri; Aa, A. arenosa; As, A. suecica. GenBank accession numbers are given in parentheses. Bootstrap support values are shown beside the branches.
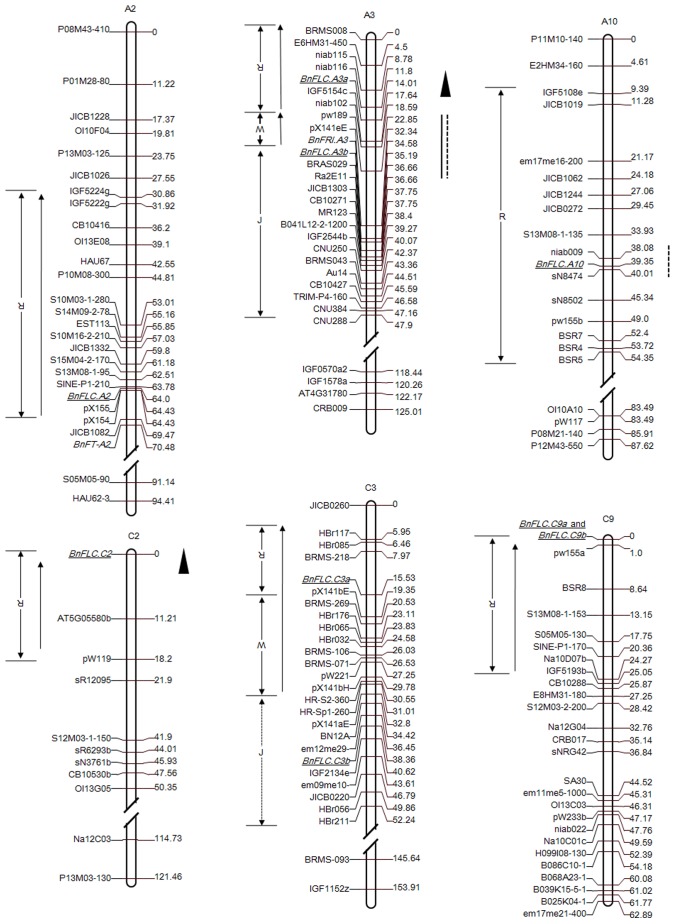
Figure 2. Location of BnFLC homologues and flowering-time QTL on the partial linkage map for TN-DH population.
Genome blocks related to BnFLC homologues (underlined) were defined by comparative mapping of B. napus and A. thaliana and using the locus boundaries of the blocks reported by Schranz et al. [19]. Arrows at left indicate an opposite orientation of the blocks, and a dashed line represents undefined boundaries. Solid and dashed vertical bars on the right represent the position of flowering-time QTL colocalized with BnFLC homologues under winter- and spring-cropping environments, respectively. Triangles indicate the approximate position of flowering-time QTL associated with BnFLC genes, which were detected from the Skipton/Ag-Spectrum population.
Figure 3. Analysis of the upstream regions of Brassica FLC homologues and AtFLC.
(A) Comparison of the potential conserved cis-blocks upstream of AtFLC and Brassica FLC (0 to −2880 bp) homologues. Different conserved segments (more than 75% sequence identity, and referred to as cis-blocks in the text) are shown as boxes with different shading and are numbered for comparison. Arrows at the 5′ end indicate the approximate position of the neighbour gene (At5g10150 and its homologues; unfilled) or the gene fragment (grey) upstream of FLC. Dashed lines represent upstream sequences that were not determined. (B) Alignment of 30 bp sequences that contain putative cis-regulatory elements (G-box and CAAT-box) in cis-block 4 among FLC homologues. (C) Alignment for the sequences in cis-block 5 among FLC homologues. The position of the putative CAAT-box (in the minus strand) is shown.
Interestingly, another region (cis-block 5) was identified in the promoter of nine of the 16 Brassica FLC homologues. The region was located 2.3 kb upstream from the transcription start site of AtFLC and was well conserved between most Brassica FLC genes and AtFLC. Its size ranged from 115 to 142 bp (Figure 3C). A CAAT-box was found within this region, which is an indication of transcription factor binding activity.
Transcriptional Divergence of BnFLC Homologues
BnFLC cDNA was amplified with gene-specific primers (Table S1) and subsequently sequenced. In total, eight different sequences were identified by multiple sequence comparison. Six of the sequences were isolated from both Tapidor and Ningyou7. The transcripts of BnFLC.C2 and BnFLC.C3a were only isolated from a spring-type cultivar, Westar (Table S2). No BnFLC.C3b transcript was identified in Tapidor, Ningyou7 or Westar. BnFLC.C3b might be a pseudogene because it contains stop codons created by inserted nucleotides in exon 2 and exon 7 (Figure S1A).
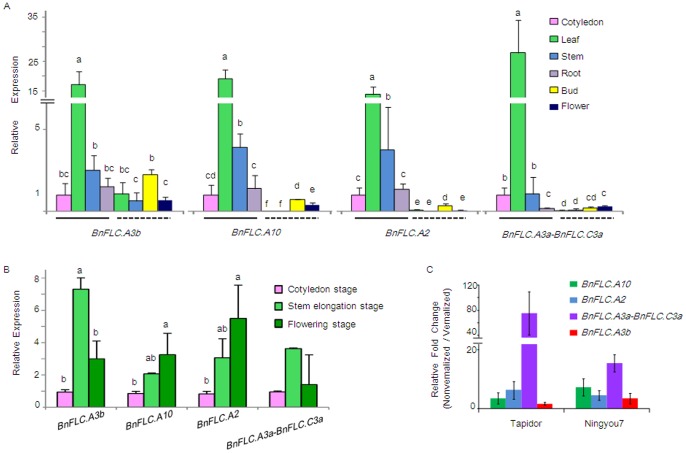
We selected four BnFLC homologues, BnFLC.A2, BnFLC.A3b, BnFLC.A10, and BnFLC.A3a-BnFLC.C3a for further analyzes. We would not design gene-specific primers for the other copies which were suitable for quantitative real-time PCR (qRT-PCR) due to the high sequence similarity among multiple copies. First, the relative expression levels of these BnFLC homologues were analyzed in different organs of the winter cultivar Tapidor at three developmental stages. Distinct expression patterns were distinguished. Without vernalization, the BnFLC homologues were highly transcribed in leaves, moderately transcribed in stems, and weakly transcribed in cotyledons and roots (Figure 4A). Different copies showed distinct relative expression levels in different organs. The expression of BnFLC.A3a-BnFLC.C3a was extremely high in leaves, and very low in roots, whereas BnFLC.A10 and BnFLC.A2 were highly expressed in leaves and stems. After vernalization, BnFLC transcriptional activity dropped sharply to very low levels in vegetative organs but was still detectable in reproductive organs (flower buds and fully-opened flowers).
Figure 4. Quantitative real-time PCR analysis of expression patterns for four BnFLC homologues.
Different letters above a bar indicate a significant difference (P<0.05). Expression levels at the cotyledon stage were considered to be the control. Relative expression values of each BnFLC homologue were normalized with the reference gene β-Actin. (A) Comparison of the relative expression levels of BnFLC homologues in different tissues of the winter cultivar Tapidor. Samples underlined with a solid or dashed line were collected from nonvernalized and vernalized plants, respectively. (B) Relative expression levels of BnFLCs in leaves and cotyledons at different developmental stages in the semi-winter cultivar Ningyou7. No cold treatment was applied throughout all of the developmental stages. (C) Vernalization responsiveness of BnFLC homologues in Tapidor and Ningyou7. The relative fold change between four-week-old leaves (without vernalization) and seven-week-old leaves (four-week-old plants followed by three weeks of cold treatment) was measured.
Next, we analyzed the temporal expression profile of BnFLC homologues in cotyledons and leaves at different developmental stages [34] in the cultivar Ningyou7 (a semi-winter type), because it flowers without vernalization under long-day conditions. Compared with the cotyledon stage, BnFLC transcript abundance increased markedly at the stem elongation stage (Figure 4B). It was notable that, at the flowering stage, the BnFLC homologues were still highly expressed in nonvernalized Ningyou7 plants, which was in contrast to their repression in leaves at the flowering stage of vernalized Tapidor plants. The transcriptional activities of BnFLC.A2 and BnFLC.A10 were increased slightly at the flowering stage, whereas BnFLC.A3b was downregulated significantly compared with the stem elongation stage. Moreover, we found differences in the transcriptional activities of the BnFLC homologues after cold treatment for three weeks; all BnFLC genes were transcriptionally downregulated. However, BnFLC.A3b expression decreased at the slowest rate under cold treatment for 3 weeks in both Tapidor and Ningyou7, which suggested that BnFLC.A3b responds differently to cold than the other BnFLC homologues (Figure 4C).
Sequence Differences between BnFLC.A3b Alleles and Differential Splicing
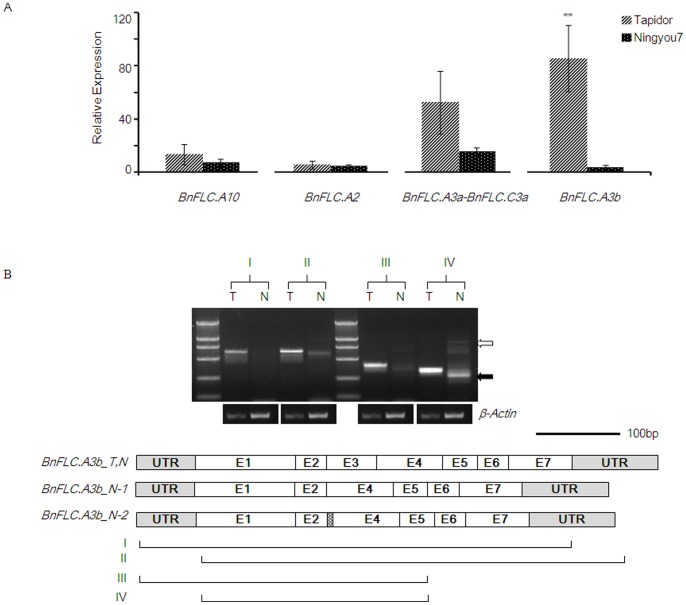
Four QTL that control flowering time have been identified in the vicinity of BnFLC.A3b and BnFLC.A10 (TN-DH population), and of BnFLC.A3a and BnFLC.C2 (Skipton/Ag-Spectrum DH population), respectively (Figure 2). In the present study, we compared the expression levels of BnFLC alleles in four-week-old leaves from Tapidor and Ningyou7 plants to test whether the alleles were expressed differentially between the two parents of the TN-DH population (Figure 5A). However, only BnFLC.A3b allele showed significant expression differences. Twenty-five-fold higher expression of BnFLC.A3b was observed in the winter-type rapeseed parent compared with that in Ningyou7. It was notable that both BnFLC.A3b and BnFRI.A3 were located within the confidence intervals of flowering-time QTL under both conditions, namely with and without cold treatment. To investigate the basis of the differential expression of the BnFLC.A3b alleles, we compared genomic BnFLC.A3b sequences from Tapidor and Ningyou7. Although a few indels and nonsynonymous single nucleotide polymorphisms were detected in intronic and regulatory regions (Figure S1B), these variations did not seem to contribute to the differences in expression of the two BnFLC.A3b alleles.
Figure 5. Differential expression of BnFLCs in four-week-old nonvernalized leaves from B. napus cultivars Tapidor and Ningyou7.
(A) Relative expression levels for four BnFLC homologues. β-Actin was used as a reference gene. Error bars represent standard errors among the three biological replicates; **represents significantly different (p<0.05). (B) PCR products of BnFLC.A3b amplified from total RNA of Tapidor (T) and Ningyou7 (N). The major band amplified from Tapidor comprises correctly spliced BnFLC.A3b transcripts, whereas incompletely (unfilled arrow) and incorrectly spliced BnFLC.A3b transcripts (usually lack exon 3; filled arrow) were amplified in Ningyou7. The schematic diagram of a BnFLC.A3b transcript shows exons (E1 to E7) and untranslated regions (UTR). BnFLC.A3b_T, N represents correctly spliced transcript in Tapidor and Ningyou7, BnFLC.A3b_N-1 and BnFLC.A3b_N-2 represent the two kinds of alternative spliced transcripts in Ningyou7 which lacked partial or complete exon 3, respectively. Shadow box in BnFLC.A3b_N-1 indicates the last four nucleotides of exon 3 which were retained. I, II, III and IV, which represent four amplifications from different transcriptional regions of correctly spliced BnFLC.A3b_T, N transcript, are shown below the schematic diagram.
Next, we analyzed the DNA methylation status of BnFLC.A3b from −393 bp, in the promoter region, to +202 bp, at the 5′ end of the first intron, which covers the two CG islands of BnFLC.A3b. We used leaves from nonvernalized plants of Tapidor and Ningyou7. No obvious differences in DNA methylation were observed between the BnFLC.A3b alleles, and only a few cytosine sites at the 5′ end of the analyzed sequence were methylated (Table S3).
Finally, the BnFLC.A3b transcripts were analyzed with regard to splice site variations. Differential splicing was observed in nonvernalized leaves between Tapidor and Ningyou7 at the seedling stage. The transcripts from Tapidor were usually spliced correctly, but a greater number of incomplete or incorrectly spliced transcripts were identified in Ningyou7 than in Tapidor by semi-quantitative RT-PCR (Figure 5B) and sequencing of the PCR products. Further sequence analysis revealed that the mature BnFLC.A3b transcripts in Ningyou7 often lacked the complete or partial exon 3 (just retained the last four nucleotides). The latter transcript contained a premature stop codon, which suggests that the polypeptide might be non-functional (Figure S2). In conclusion, the amount of functional transcripts differed between both rapeseed types because of inefficient pre-mRNA processing in the semi-winter rapeseed.
BnFLC.A3b Resides within a Cluster of Cold Responsive Genes
We analyzed the sequences that bordered the BnFLC.A3b and BnFRI.A3 genes with an aim to investigate the genomic structure between these two flowering-time genes. In contrast to their close linkage in B. napus, these genes are unlinked in A. thaliana and A. lyrata (Figure S3). BnFLC.A3b and BnFRI.A3 were located in close proximity to each other on chromosome A3 (Figure 2). This genomic region is collinear to neither the ‘R’ block, which contains AtFLC (At5_3.5 Mb), nor the ‘O’ block, which contains AtFRI (At4_0.2 Mb).
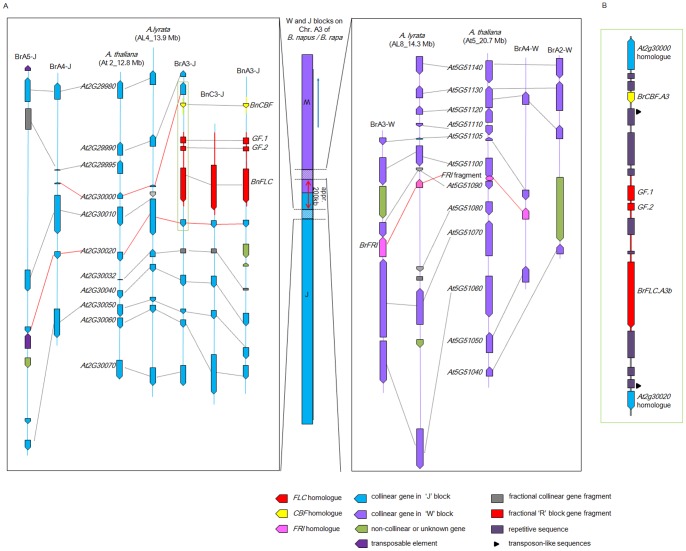
First, we compared the BnFLC.A3b and BnFLC.C3b loci with the corresponding triplicated segments from the ‘J’ block in B. rapa, and with the orthologous genomic regions in A. thaliana and A. lyrata (Figure 6A). Significant collinearity at the sequence level was observed, except for a region between At2G30000 and At2G30020. The homologue of At2G30010 was lost from the A3/C3 homoeologous region of B. napus and B. rapa, and a part of the ‘R’ block that contained the FLC homologue and a small part of the ‘U’ block that contained a homologue of CBF1 (C-REPEAT/DRE BINDING FACTOR 1, a cold-response gene) were inserted into this region. BLASTN searches against coding sequences of A. thaliana revealed no further evidence for homologous fragments of FLC and CBF1 in those regions (Table S4). This interruption of microsynteny indicates that a multiple rearrangement occurred after the whole-genome triplication of the Brassica genomes. We further analyzed a B. rapa BAC (KBrH038M21) that contained BrFLC.A3 from 24.3–24.4 Mb of chromosome A3 [35]. This BAC contained many repetitive sequences, including a few transposon-like sequences (Figure 6B), which suggests that the complex structural rearrangement involved transposon activation.
Figure 6. Microsynteny analysis of homologous regions related to BnFLC.A3b, BnFRI.A3, and BnCBF.A3.
Collinear gene models are connected by dotted lines. (A) The colored bar in the center represents the portion of chromosome 3 of B. napus that contains the three genes. The blue arrow indicates the opposite orientation of the ‘W’ block. The left-hand box represents a region of the ‘J’ block (BnA3-J) in which BnFLC.A3b and BnCBF.A3 are located, and alignment of conserved gene models of BnC3-J of B. napus, the triplicated regions (BrA3-J, BrA4-J and BrA5-J) of B. rapa, and the homologous regions in A. thaliana and A. lyrata. Red lines connect the collinear gene models in the ‘J’ block next to FLC and CBF homologues. The right-hand box represents a region of the ‘W’ block in which BnFRI.A3 is located, and alignment of conserved gene models in the triplicated regions (BrA2-W, BrA3-W and BrA4-W) of B. rapa, and the homologous regions in A. thaliana and A. lyrata. Red lines connect the FRI homologues and the FRI homologous fragments. (B) Detailed structure of a region in BrA3-J (indicated by a green box in the left-hand box of (a) from the BAC clone KBrH038M21; [35]). Repeat sequences were identified between the homologues of At2g30000 and At2g30020. GF-1 and GF-2 represent two gene fragments homologous to the ‘R’ block genes, At5G10200 and At5G10150, respectively.
The genomic sequences of B. rapa, A. thaliana, and A. lyrata were used to examine the microsynteny structure around the FRI locus. Surprisingly, in contrast to FRI which is located within the ‘O’ block of A. thaliana, all the other FRI homologues were found in the ‘W’ blocks either on chromosome 8 of A. lyrata (AL8_14.3 Mb), which is thought to be more similar to the ancestral karyotype of Brassicaceae (n = 8), or chromosomes A3 and A4 of B. rapa (BrA3_W and BrA4_W), respectively (Figure 6A). BrFRI.A3 was localized approximately 200 kb away from BrFLC.A3b according to the genomic sequence (version 1.1) of B. rapa database [36]. However, a fragment between the gene models of At5G51090 and At5G51000 showed high similarity (approximately 85–98%) to AtFRI but lacked exon 1 was found in the‘W’block of A. thaliana (At5_20.7 Mb). Consistently, a previous study on comparative mapping of FRI loci between A. lyrata and A. thaliana showed that AlFRI was unexpectedly mapped to A. lyrata chromosome AL8 rather than AL6, which contained the homologous ‘O’ block [37]. Moreover, a recent research on comparative analysis of FRI among B. oleracea (BoFRI) and the Arabidopsis lineage also showed that the two BoFRI homologues are located within the regions collinear to the ‘W’ blocks [38]. The results of previous and present studies suggested that the FRI homologues located in the ‘W’ block is the same as the gene inherited from the common ancestor of Brassicaceae, and that the FRI copy on the ‘W’ block had probably been translocated to the ‘O’ block during the evolution of A. thaliana.
To obtain a deeper insight into the coevolution of FLC and FRI homologues in B. napus, we calculated the correlation between the expression of different pairs of FLC and FRI homologues during different developmental stages. Since we could only design suitable qRT-PCR primers for BnFRI.A3 and BnFRI.X (termed as BnaA.FRI.a and BnX.FRI.c by Wang et al. [31]), the two BnFRI homologues along with the four BnFLC homologues were examined for their expression levels by qRT-PCR. A significant correlation was observed within different BnFLC and BnFRI gene pairs (BnFRI.A3 and BnFLC.A3b, BnFRI.X and BnFLC.A2, and BnFRI.X and BnFLC.A10) in leaves of nonvernalized Ningyou7 plants (Table 2). Among these tested gene pairs, the closely linked genes BnFRI.A3 and BnFLC.A3b showed a highly significant correlation (Pearson’s correlation coefficient, r = 0.965, P<0.001) in relative expression levels from the cotyledon stage to the flowering stage in nonvernalized Ningyou7. We investigated whether the correlation of expression was significant in the winter-type cultivar, Tapidor, in the absence of vernalization. Given that Tapidor has an obligate requirement for cold to enter the reproductive phase, we analyzed the correlation of expression during leaf development (the cotyledon to four-leaf stages). Although all BnFRI and BnFLC pairs showed positive correlation of expression in Tapidor, none of them had significant correlation (p<0.05).
Table 2. Correlation coefficients between BnFLC and BnFRI loci in the Tapidor and Ningyou7 during different developmental stages.
| BnFRI.A3 | BnFRI.X | BnFRI.A3 | BnFRI.X | |
| Ningyou7a | Ningyou7 | Tapidor | Tapidor | |
| BnFLC.A2 | 0.599 (0.1249)b | 0.945 (0.0004) | 0.518 (0.1025) | 0.313 (0.3492) |
| BnFLC.A3b | 0.965 (0.0001) | 0.658 (0.0760) | 0.370 (0.2634) | 0.453 (0.1613) |
| BnFLC.A10 | 0.544 (0.1702) | 0.883 (0.0036) | 0.347 (0.2961) | 0.149 (0.6624) |
Pearson’s correlation coefficients were calculated by relative expression values within different BnFLC and BnFRI gene pairs in cotyledons or leaves which harvested from the cotyledon, the stem elongation and the flowering stages of nonvernalized Ningyou7, or from the cotyledon, the first-, two-, and four-leaf stages of nonvernalized Tapidor plants.
The value in parentheses represents the P-value.
Discussion
We cloned nine FLC homologues from the B. napus genome and identified their map positions. We analyzed the promoter sequences of these genes in comparison to their homologues from the diploid progenitors B. rapa and B. oleracea, and in comparison to the A. thaliana gene FLC. The BnFLC homologues were expressed differentially. One FLC homologue (BnFLC.A3b) was of particular interest because it colocalized with a major QTL for flowering time. Moreover, it was expressed differentially between the parents of our mapping population and was located in the vicinity of the FRI homologue BnFRI.A3. The progenitor of BnFLC.A3b was duplicated ectopically into the ‘J’ block on chromosome A3 and was part of a gene cluster with two functionally related genes, BnFRI.A3 and BnCBF.A3. This gene cluster is present specifically in the Brassica lineages.
FLC Homologues in B. napus Contribute Mainly to Variation in Flowering Time
We identified nine FLC homologues in B. napus. In addition to the non-functional BnFLC.C3b, six cDNA sequences were found in the winter cultivar Tapidor and in the semi-winter cultivar Ningyou7, and the remaining two (BnFLC.C2 and BnFLC.C3a) were only isolated from the spring cultivar Westar (Table S2). This suggests that most of the homologues were transcriptionally active in different cultivars. Interestingly, flowering-time QTL have been found to coincide with the map positions of FLC homologues in different mapping populations, e.g. BnFLC.A3b and BnFLC.A10 (TN-DH population) and BnFLC.A3a and BnFLC.C2 (Skipton/Ag-Spectrum DH population), which clearly suggests a role of these homologues in controlling flowering time in B. napus. However, direct links between the expression of FLC homologues and each of the flowering-time QTL need to be investigated further.
Four BnFLC homologues tested in the present study were expressed ubiquitously before vernalization and were repressed significantly after vernalization, which suggested that they act in a similar manner to the FLC gene in Arabidopsis [4], [39], [40]. However, the expression patterns of different BnFLC copies were quite different. BnFLC transcripts were detected in buds and flowers of vernalized Tapidor plants, especially for BnFLC.A3b (Figure 4); similar observations were reported for transgenic Arabidopsis plants that harboured the pBoFLC4-1::BoFLC4-1::GUS construct [41]. These results indicated that reactivation of Brassica FLC genes during gametogenesis was similar to that of FLC in Arabidopsis. In Arabidopsis, FLC activity is reset in the male reproductive tissue and repressed in maturing pollen in vernalized flower buds [39], although the precise biological roles of FLC in these tissues remain unknown. In addition, the expression level of BnFLC homologues in leaves of nonvernalized Ningyou7 plants at the stage of inflorescence emergence was as high as during the seedling stage, which suggested that in the semi-winter cultivar Ningyou7, other genes might overcome the repression by BnFLC to activate the downstream floral integrators. The high level of expression of BnFLC genes even after flowering indicates that Brassica FLC homologues might have multiple functions in the regulation of plant development, as reported in Arabidopsis [42]. Additionally, BnFLC.A10 showed more decrease of fold change in Ningyou7 as compared to Tapidor at the early stage of cold-treatment, which is consistent with functional differences of BnFLC.A10 alleles between Tapidor and Ningyou7, which are located in the major flowering-time QTL (TN-DH population). Their expression decreased rapidly in Ningyou7 leaves during vernalization (Hou et al., unpublished data), suggesting that some BnFLC homologues respond in a more sensitive way to vernalization in the early flowering cultivar (Ningyou7).
Although FLC homologues were highly conserved among B. napus and its diploid progenitors B. rapa and B. oleracea, the promoter sequences among Brassica FLC genes that belonged to different clades were quite variable and also diverged substantially from that of AtFLC. Variations in cis-regulatory sequences that affect their function might contribute to phenotypic diversity among different species [43]. In the present study, only five cis-blocks were identified in the promoter of the BnFLC homologues and AtFLC, which suggested that these segments might require for common regulation of FLC homologues. Furthermore, we identified a more highly conserved 30-bp region within cis-block 4, which together with the presence of the newly discovered cis-block 5 in most FLC homologues indicated that important cis-regulatory element(s) might be present in these regions. The BnFLC sequences were highly conserved among the cultivars Columbus [28], Tapidor, and Ningyou7 at the coding sequence level; thus, the divergent promoter sequences might contribute to the differential regulation of Brassica FLC homologues and explain their functional divergence. Consequently, a more sophisticated network of interactions among FLC homologues and their targets might have evolved in B. napus as compared with Arabidopsis.
Functionally Related Flowering-time Genes are Tightly Linked in Brassica Species
Gene homologues usually maintain collinearity among closely related plant species, for example the extensive sequence conservation identified within the Brassicaceae [19], [20]. In the present study, BnFLC.A3b/BnFLC.C3b was located at one end of the ‘J’ block on chromosome 3, in contrast to AtFLC and other BnFLCs, which were located within the ‘R’ block. A microsynteny analysis of FLC-related segments in Brassica and Arabidopsis lineages suggested that the ancestral homologue of BnFLC.A3b in Brassica was inserted into the ‘J’ block on chromosome 3. In addition, a neighbour-joining analysis revealed that BnFLC.A3b/BnFLC.C3b and BnFLC.A3a/BnFLC.C3a belong to the same clade (Figure 1). These results suggest that BnFLC.A3b/BnFLC.C3b might be derived from a common ancestor of BnFLC.A3a/BnFLC.C3a, and was duplicated ectopically at the present loci during the period between the genome triplication of Brassica and the divergence of B. rapa and B. oleracea. Similarly, a homologue of CBF1 was also translocated from the ‘U’ block; CBF1 is a cold- and dehydration-responsive gene [44], is regulated negatively by SOC1, and probably interacts with FLC [42], [45]. The present study and recent comparative analysis of FRI homologous regions between B. oleracea and the Arabidopsis lineage [38] revealed that the ancestral FRI was located originally in the ‘W’ block. Chromosomal rearrangements during the evolution of Brassica genome, led to separated blocks from different chromosomes of the ancestral karyotype which are recombined among each other and which are not shared with A. thaliana genome [46], e.g. the blocks ‘W’ and ‘J’ on the homologous chromosome A3/C3 from the Brassica genome. Thus, the three unlinked functionally related homologues of FLC, FRI, and CBF1 were typically clustered within a short distance in the Brassica A genome, and the clustering event involved at least two translocations and a reorganization of the ‘J’ and ‘W’ blocks on Brassica chromosome A3 (Figure 6).
Increasing evidence demonstrates that functionally related genes tend to cluster together in several eukaryotes and this clustering might be maintained by selection pressure [47], [48]. Recently, several gene clusters involved in secondary metabolic pathways that presumably confer a selective advantage in nature have been discovered in plants [49]–[51]. The clustering of FLC–FRI–CBF1 in Brassica might be subjected to selection to adjust the flowering time in biennial plants. Although we identified a major flowering-time QTL adjacent to the FLC–FRI–CBF1 region (in the TN-DH population) and detected higher expression of the BnFLC.A3b allele from the later-flowering parent (Tapidor), neither sequence variation in the regulatory regions, nor different DNA methylation level from the core promoter to the 5′ end of intron 1 were observed between the BnFLC.A3b alleles of Tapidor and Ningyou7. In Arabidopsis, FRI upregulates expression and promotes efficient splicing of FLC via a cotranscriptional mechanism [10]. Although the expression levels of each BnFRI homologue were similar between Tapidor and Ningyou7, nonsynonymous nucleotide substitutions, which are located within the N-terminal putative coiled-coil domain of the BnFRI.A3 allele in Ningyou7, probably contribute to functional differences between Tapidor and Ningyou7 [31]. Therefore, the splicing variants of BnFLC.A3b probably arise because of the presence of the weaker functional allele of BnFRI.A3 in Ningyou7, which is less capable of upregulating FLC and promoting accurate splicing. Although the precise role of BnFRI.A3 in altering the expression of BnFLC.A3b remains unknown, adjacent genes that are localized within the same chromatin domain can be regulated coordinately [52]. A tendency for coexpression of BnFRI.A3 and BnFLC.A3b at different developmental stages in nonvernalized Ningyou7 plants further suggested that the adjacent BnFRI.A3 and BnFLC.A3b genes probably interact, although it was not obvious in Tapidor. This result probably reflected the fact that the samples of Tapidor used for analysis could only be harvested at stages of leaf development, but alternatively the winter-type cultivar might harbour more strongly functional genes that affect the expression of BnFRI and BnFLC homologues. Thus, regardless of whether the FLC–FRI–CBF1 cluster in the Brassica lineage was assembled randomly or driven by evolution, this cold-responsive gene cluster in B. napus might be maintained by selection pressure and thus enable adaptation to the extremely diverse environments under which Brassica crops are cultivated worldwide.
Materials and Methods
Plant Materials
The B. napus winter-type cultivar Tapidor which has an obligate vernalization requirement, semi-winter-type cultivar Ningyou7 which has a weak vernalization requirement, and spring-type cultivar Westar were used for cloning and/or expression analysis of BnFLC genes. The TN-DH population, which comprised 202 doubled-haploid lines, was derived from the F1 cross between Tapidor and Ningyou7 [53]. The Skipton/Ag-Spectrum DH population which comprised 186 doubled-haploid lines was developed from the cross between spring-type Australian cultivars, Skipton and Ag-Spectrum at the Wagga Wagga Agricultural Institute, Australia [54]. Both DH populations were used for genetic mapping and detection of QTL for flowering time. Plants were grown in the field that are either belong to Huazhong Agricultural University or to NSW Department of Primary Industries and only used for phenotyping or DNA/RNA extraction.
Cloning and Sequence Analysis of BnFLC Homologues
To isolate genomic sequences of the BnFLC homologues, a BAC library (JBnB) constructed from the gDNA of Tapidor [20] was used. A fragment of BnFLC.A3a (exon 2 to exon 6) was used as a probe to screen the JBnB BAC library by Southern blot hybridization. Different BnFLC homologues were identified by sequencing the PCR products that were amplified from the BAC clones. The PCR primers used are listed in Table S1.
To isolate the cDNA sequences of the BnFLC homologues, 2 µg of total RNA that had been extracted from four-week-old leaves of Tapidor, Ningyou7, or Westar was used to synthesize first-strand cDNA with the First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie, MD, USA). Gene-specific primer pairs were designed on the basis of an alignment of the Brassica FLC cDNA sequences deposited in GenBank (AY036888–AY036892, DQ866874–DQ866876, AY115678, AM231517, AM231519, AY306122, and AY306123). The existing nomenclature for Brassica FLC genes is unsuitable to distinguish different FLC homologues from the two subgenomes (AA and CC); therefore, we used a new nomenclature to describe the Brassica FLC genes on the basis of their map position on different linkage groups. For example, BnFLC.A3a denotes the FLC gene of Brassica napus (Bn) that maps to linkage group (chromosome) 3 of the A genome; an additional suffix (a or b) is used if the same linkage group contains more than one FLC homologue.
The alternatively spliced transcript of BnFLC.C3a was isolated from leaves of B. napus cv. Westar with GeneRacer kit (Invitrogen Corporation, Carlsbad, CA, USA) using degenerate 3′ RACE primers (Table S1). KOD-Plus DNA polymerase (Toyobo Co., Osaka, Japan) was used for PCR amplification, and additional deoxyadenosine nucleotides were introduced at the 3′ end of amplicons using Taq DNA polymerase (Fermentas). Amplicons were ligated to the pGEM-T vector (Promega Corporation, Madison, WI, USA) and transformed into Escherichia coli strain DH5α. Positive clones were selected and sequenced by the Nanjing GenScript Co., Jiangsu, China. Sequence data from this article have been deposited in the GenBank database with the following accession numbers: JQ255381 to JQ255389 for genomic sequences of BnFLC homologues, and JQ255390 to JQ255397 for the corresponding mRNA sequences.
Potential conserved cis-blocks and cis-elements of FLC homologues were identified by Dna Block Aligner (http://www.ebi.ac.uk/Tools/Wise2/dbaform.html, validated on 30th July, 2012), after removal of the simple repetitive sequences by Repeatmasker (http://www.repeatmasker.org), and potential cis-elements were predicted by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, validated on 30th July, 2012). The GenBank accession numbers of the BrFLC and BoFLC genes that were used for analysis are: BrFLC.A10 (BrFLC1), AC155344; BrFLC.A2 (BrFLC2), AC155341; BrFLC.A3a (BrFLC3), AC155342; BrFLC.A3b (BrFLC5), AC232559; BoFLC.C9 (BoFLC1), AM231517; BoFLC.C2 (BoFLC4), AY306124; BoFLC.C3a (BoFLC3), AM231518; BoFLC.C3b (BoFLC5), AM231519.
Genetic Mapping of BnFLC Homologues and QTL Detection
On the basis of allelic polymorphisms between Tapidor and Ningyou7, gene-specific PCR primers for BnFLC.A3a and BnFLC.A3b were designed, and a single-strand conformation polymorphism marker for BnFLC.C2 was developed on the basis of the sequence of the BrFLC2-containing BAC (KbrH004D11, Table S1). The genotypic data for BnFLC.A3a, BnFLC.A3b, and BnFLC.C2, and those reported previously for BnFLC.A10, BnFLC.C3a, and BnFLC.C3b [29], were incorporated into the map data for the TN-DH population [55] using JoinMap 3.0 software (http://www.kyazma.nl/index.php/mc.JoinMap). Common markers between a consensus genetic linkage map [56] and the TN-DH genetic map were used as anchors to integrate the RFLP markers of FLC2aH (BnFLC.A2) and FLC1aH (BnFLC.C9) into the TN-DH genetic map and for alignment with the physical position of FLC in Arabidopsis. The integrated map, based upon 786 molecular markers, was used to locate QTL for flowering time by the composite interval method with WinQTL Cartographer 2.5 software [57], [58] using flowering-time data collected previously from 12 winter- (with cold treatment) and two spring-crop (without cold treatment) environments [29], [59].
For the Skipton/Ag-Spectrum DH population, the published molecular linkage map comprising 674 markers was utilized to detect flowering time QTL associated with BnFLC homologues. Flowering time was scored under glasshouse and field conditions as described previously [60], [61].
Gene Expression Analysis and Quantitative Real-time PCR (qPCR)
To analyze the spatial expression patterns in response to vernalization of different BnFLC homologues, Tapidor plants were grown in a greenhouse at approximately 25°C under long-day conditions (14 h light/10 h dark). For vernalization treatment, four-week-old plants were incubated for three weeks in a climate chamber maintained at approximately 5°C under the same long-day conditions. Cotyledons were harvested 5 days after sowing; leaves, stems, and roots from nonvernalized Tapidor plants were harvested at the four-leaf stage; leaves, stems, floral buds, and flowers from vernalized Tapidor plants were harvested at the flowering stage under the field conditions. To analyze changes in the expression of BnFLC homologues among different developmental stages and the correlation in expression between BnFLC and BnFRI genes, we used the same set of cDNA samples from leaves of Ningyou7 plants that was used by Wang et al. [31]. For the expression correlation analysis in Tapidor, cotyledons and leaves at the first-, two-, and four-leaf stages from nonvernalized Tapidor plants were used.
Gene-specific primers were designed on the basis of the cDNA sequences of the BnFLC homologues with Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA; Table S1), and tested by melting curve analysis and sequencing. Total RNA was extracted from different samples with the Quick Extract™ RNA isolation kit (BioTeke Corporation, China). qRT-PCR was performed on a CFX96 Real-Time System (Bio-Rad Laboratories, Hercules, CA, USA). Reactions were performed in a final volume of 20 µL with SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen Corporation, Carlsbad, CA, USA) as described previously [31]. Three to four technical replications and three biological replicates were performed for each sample. For data analysis, the relative standard curve method was used [62]. A standard curve was prepared on each PCR plate for each primer pair using five 10-fold serial dilutions of plasmid DNA to determine the amplification efficiency. Quantification cycle (Cq) values were calculated and the relative expression values of target genes were normalized with the reference gene β-Actin [31] using the CFX Manager 1.5 software (Bio-Rad). Subsequently, the data was exported to SAS 8.0 (SAS Institute, 1999) for statistical analysis.
Detection of DNA Methylation
Genomic DNA was extracted from leaves of nonvernalized plants. Approximately 1 µg of DNA was treated with the EpiTect Bisulfite Kit (Qiagen, Germany) in accordance with the manufacturer’s instructions. Gene-specific primers (Table S1) were designed with MethPrimer [63]. Stable unmethylated endogenous gene controls [64] were used to estimate whether the bisulfite treatment was completed. At least 10 clones were sequenced for each PCR product.
Comparative Genome Analysis
Two BACs (JBnB142O14 and JBnB090B17), which contained BnFLC.A3b and BnFLC.C3b, respectively, were sequenced in different pools that contained several B. napus and B. oleracea BACs. Each pool was sequenced with a Genome Sequencer FLX system (Roche, Germany). The obtained raw sequence data were assembled using the GS De Novo Assembler (Newbler) 2.5 software (-v pBACsacB1 -s ecoli). Assembled contigs were submitted to a BLAST search against KBrH038M21 (which contains BrFLC5) and highly homologous contigs were selected for further analysis in this context.
The genomic sequences of A. thaliana, A. lyrata, and B. rapa were downloaded from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/), the Department of Energy Joint Genome Institute (JGI; A. lyrata version 1.0 http://genome.jgi-psf.org/Araly1/Araly1.home.html), and the Brassica Database (BRAD; http://brassicadb.org/brad/index.php). The sequences of the four BACs that contained BrFLC (KBrH080A08, KBrH004D11, KBrH117M18, and KBrH052O08) were used. The sequences of the analyzed segments were reannotated in detail using Genscan (http://genes.mit.edu/GENSCAN.html) and submitted to a BLAST search against TAIR to identify homologous genes in Arabidopsis. Predicted proteins that were less than 100 amino acids in length and had no hits in the target database were excluded from the final annotation. Homologous fragments that were similar to coding sequences in Arabidopsis but were not incorporated into gene models were identified using the BLASTN tool [65]. Repetitive sequences were identified by lower-case letters and were submitted to a search against the Plant Repeat Databases (http://plantrepeats.plantbiology.msu.edu/index.html) and Repeatmasker (http://www.repeatmasker.org) for comparison with known repeat sequences in plants.
Phylogenetic Analysis
A multiple alignment of relative sequences was generated with CLUSTALW [66]. Ambiguously aligned regions were modified manually to minimize the number of inserted gaps. Phylogenetic trees were constructed with MEGA 5.0 software [67] using the neighbour-joining method with the ‘partial deletion’ option for analysis of gaps. A bootstrap analysis with 2000 replicates was used to evaluate the robustness of the tree topology.
Supporting Information
(A) Alignment of cDNA sequences of BnFLC homologues and their relative orthologues in B. rapa and B. oleracea. The accession numbers of the B. rapa and B. oleracea sequences are shown in parentheses. Additional nucleotides in exon 2 and exon 7 of BnFLC.C3b and BoFLC.C3b (BoFLC5; GenBank accession No. AM231519) are highlighted with arrows. (B) Gene structure of BnFLC.A3b. The positions of exons and untranslated regions (UTRs) are represented by boxes.
(TIF)
(A) cDNA and (B) amino acid sequence alignment of alternatively spliced variants and their predicted polypeptides of BnFLC.A3b in Tapidor and Ningyou7. BnFLC.A3b_T, N represents a correctly spliced transcript in Tapidor and Ningyou7, BnFLC.A3b_N-1 and BnFLC.A3b_N-2 represent the two kinds of alternatively spliced transcripts in Ningyou7 which are missing exon 3 partly or completely.
(TIF)
Genome blocks and the location of FRI , FLC , and CBF homologues in Arabidopsis thaliana (At) and Arabidopsis lyrata (Al). Arrows indicate the opposite orientation of the blocks.
(TIF)
Primers used for cloning, mapping, expression, and DNA methylation analysis.
(XLS)
Information for isolated cDNA sequences of BnFLC homologues.
(XLS)
Comparison of methylated sites in the predicted CG islands of BnFLC.A3b between the Tapidor and Ningyou7 alleles.
(XLS)
Summary of BLASTN analysis to detect similar coding sequences in the Arabidopsis genome.
(XLS)
Acknowledgments
We thank Jinhua Sun (Chinese Academy of Tropical Agricultural Science, Haikou, China) for RNA extraction and phylogenetic analysis, and Ruiyuan Li and Xiaoxue Jiang (Huazhong Agricultural University, Wuhan, China) for assembly of the BAC sequences.
Funding Statement
This work was supported by the National Basic Research and Development Programme (No. 2006CB101600) and National Natural Science Foundation of China (No. 30900788), Project Based Personnel Exchange Program with China Scholarship Council and German Academic Exchange Service (No. 2008–3070), and New South Wales Agricultural Genomics Centre funded under the BioFirst Initiative of New South Wales Government, Australia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Michaels SD, He Y, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis . Proc Natl Acad Sci USA 100: 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, et al. (2005) Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192. [DOI] [PubMed] [Google Scholar]
- 6. Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, et al. (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michaels SD, Bezerra IC, Amasino RM (2004) FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis . Proc Natl Acad Sci USA 101: 3281–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi K, Kim J, Hwang H-J, Kim S, Park C, et al. (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bezerra IC, Michaels SD, Schomburg FM, Amasino RM (2004) Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis . Plant J 40: 112–119. [DOI] [PubMed] [Google Scholar]
- 10. Geraldo N, Bäurle I, Kidou S-i, Hu X, Dean C (2009) FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol 150: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Angel A, Song J, Dean C, Howard M (2011) A polycomb-based switch underlying quantitative epigenetic memory. Nature 476: 105–108. [DOI] [PubMed] [Google Scholar]
- 13. De Lucia F, Crevillen P, Jones AME, Greb T, Dean C (2008) A PHD-Polycomb Repressive Complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802. [DOI] [PubMed] [Google Scholar]
- 15. Turck F, Roudier F, Farrona S, Martin-Magniette M-L, Guillaume E, et al. (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, et al. (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heo JB, Sung S (2011) Vernalization-mediated eigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- 18. Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11: 535–542. [DOI] [PubMed] [Google Scholar]
- 20. Rana D, van den Boogaart T, O’Neill CM, Hynes L, Bent E, et al. (2004) Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J 40: 725–733. [DOI] [PubMed] [Google Scholar]
- 21. Suwabe K, Morgan C, Bancroft I (2008) Integration of Brassica A genome genetic linkage map between Brassica napus and B. rapa . Genome 51: 169–176. [DOI] [PubMed] [Google Scholar]
- 22. Cho K, O’Neill CM, Kwon S-J, Yang T-J, Smooker AM, et al. (2010) Sequence-level comparative analysis of the Brassica napus genome around two stearoyl-ACP desaturase loci. Plant J 61: 591–599. [DOI] [PubMed] [Google Scholar]
- 23. Osborn TC, Chris Pires J, Birchler JA, Auger DL, Jeffery Chen Z, et al. (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet 19: 141–147. [DOI] [PubMed] [Google Scholar]
- 24. Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- 25. Yuan Y-X, Wu J, Sun R-F, Zhang X-W, Xu D-H, et al. (2009) A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J Exp Bot 60: 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao J, Kulkarni V, Liu N, Pino Del Carpio D, Bucher J, et al. (2010) BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa . J Exp Bot 61: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okazaki K, Sakamoto K, Kikuchi R, Saito A, Togashi E, et al. (2007) Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea . Theor Appl Genet 114: 595–608. [DOI] [PubMed] [Google Scholar]
- 28. Tadege M, Sheldon CC, Helliwell CA, Stoutjesdijk P, Dennis ES, et al. (2001) Control of flowering time by FLC orthologues in Brassica napus . Plant J 28: 545–553. [DOI] [PubMed] [Google Scholar]
- 29. Long Y, Shi J, Qiu D, Li R, Zhang C, et al. (2007) Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis . Genetics 177: 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osborn TC, Kole C, Parkin I, Sharpe AG, Kuiper M, et al. (1997) Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thaliana . Genetics 146: 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang N, Qian W, Suppanz I, Wei L, Mao B, et al. (2011) Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA.FRI.a . J Exp Bot 62: 5641–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oleksyk TK, Smith MW, O’Brien SJ (2010) Genome-wide scans for footprints of natural selection. Phil Trans R Soc B January 365: 185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Razi H, Howell E, Newbury H, Kearsey M (2008) Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor Appl Genet 116: 179–192. [DOI] [PubMed] [Google Scholar]
- 34.Meier U (2001) Growth stages of mono-and dicotyledonous plants. BBCH Monograph Federal Biological Research Centre for Agriculture and Forestry, Germany.
- 35. Mun J-H, Kwon S-J, Seol Y-J, Kim J, Jin M, et al. (2010) Sequence and structure of Brassica rapa chromosome A3. Genome Biol 11: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X, Wang H, Wang J, Sun R, Wu J, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa . Nat Genet 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- 37. Kawabe A, Hansson B, Forrest A, Hagenblad J, Charlesworth D (2006) Comparative gene mapping in Arabidopsis lyrata chromosomes 6 and 7 and A. thaliana chromosome IV: evolutionary history, rearrangements and local recombination rates. Genet Res 88: 45–56. [DOI] [PubMed] [Google Scholar]
- 38. Irwin J, Lister C, Soumpourou E, Zhang Y, Howell E, et al. (2012) Functional alleles of the flowering time regulator FRIGIDA in the Brassica oleracea genome. BMC Plant Biol 12: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sheldon CC, Hills MJ, Lister C, Dean C, Dennis ES, et al. (2008) Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc Natl Acad Sci USA 105: 2214–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi J, Hyun Y, Kang M-J, In Yun H, Yun J-Y, et al. (2009) Resetting and regulation of FLOWERING LOCUS C expression during Arabidopsis reproductive development. Plant J 57: 918–931. [DOI] [PubMed] [Google Scholar]
- 41. Lin S-I, Wang J-G, Poon S-Y, Su C-l, Wang S-S, et al. (2005) Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis . Plant Physiol 137: 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, et al. (2011) FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis . Proc Natl Acad Sci USA 108: 6680–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wittkopp PJ, Kalay G (2012) Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13: 59–69. [DOI] [PubMed] [Google Scholar]
- 44. Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106. [DOI] [PubMed] [Google Scholar]
- 45. Seo E, Lee H, Jeon J, Park H, Kim J, et al. (2009) Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC . Plant Cell 21: 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trick M, Kwon S-J, Choi S, Fraser F, Soumpourou E, et al. (2009) Complexity of genome evolution by segmental rearrangement in Brassica rapa revealed by sequence-level analysis. BMC Genomics 10: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Al-Shahrour F, Minguez P, Marqués-Bonet T, Gazave E, Navarro A, et al. (2010) Selection upon genome architecture: conservation of functional neighborhoods with changing genes. PLoS Comput Biol 6: e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee JM, Sonnhammer ELL (2003) Genomic gene clustering analysis of pathways in eukaryotes. Genome Res 13: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Field B, Fiston-Lavier A-S, Kemen A, Geisler K, Quesneville H, et al. (2011) Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc Natl Acad Sci USA 108: 16116–16121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70: 1645–1651. [DOI] [PubMed] [Google Scholar]
- 51. Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ (2009) CYP76M7 Is an ent-Cassadiene C11α-Hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hurst LD, Pal C, Lercher MJ (2004) The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310. [DOI] [PubMed] [Google Scholar]
- 53. Qiu D, Morgan C, Shi J, Long Y, Liu J, et al. (2006) A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114: 67–80. [DOI] [PubMed] [Google Scholar]
- 54. Raman H, Raman R, Nelson MN, Aslam MN, Rajasekaran R, et al. (2012) Diversity array technology markers: genetic diversity analyses and linkage map construction in rapeseed (Brassica napus L.). DNA Res 19: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jiang C, Ramchiary N, Ma Y, Jin M, Feng J, et al. (2011) Structural and functional comparative mapping between the Brassica A genomes in allotetraploid Brassica napus and diploid Brassica rapa . Theor Appl Genet 123: 927–941. [DOI] [PubMed] [Google Scholar]
- 56. Udall JA, Quijada PA, Osborn TC (2005) Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics. 169: 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang S, C. J Basten, Zeng Z-B (2006) Windows QTL cartographer 2.5. Bioimformatics Research Center website of Department of Statistics, North Carolina State University, Raleigh, NC. Available: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Accessed 2012 Aug 28.
- 59. Shi J, Li R, Qiu D, Jiang C, Long Y, et al. (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus . Genetics 182: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Raman R, Taylor B, Marcroft S, Stiller J, Eckermann P, et al. (2012) Molecular mapping of qualitative and quantitative loci for resistance to Leptosphaeria maculans causing blackleg disease in canola (Brassica napus L.). Theor Appl Genet 125: 405–418. [DOI] [PubMed] [Google Scholar]
- 61.Raman H, Raman R, Eckermann P, Coombes N, Manoli S, et al. (2012) Genetic and physical mapping of flowering time loci in oilseed rape (Brassica napus L.). Theor Appl Genet doi:10.1007/s00122-012-1966-8. [DOI] [PubMed]
- 62. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li L-C, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427–1431. [DOI] [PubMed] [Google Scholar]
- 64. Wang J, Wang C, Long Y, Hopkins C, Kurup S, et al. (2011) Universal endogenous gene controls for bisulphite conversion in analysis of plant DNA methylation. Plant Methods 7: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Town C, Cheung F, Maiti R, Crabtree J, Haas B, et al. (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Alignment of cDNA sequences of BnFLC homologues and their relative orthologues in B. rapa and B. oleracea. The accession numbers of the B. rapa and B. oleracea sequences are shown in parentheses. Additional nucleotides in exon 2 and exon 7 of BnFLC.C3b and BoFLC.C3b (BoFLC5; GenBank accession No. AM231519) are highlighted with arrows. (B) Gene structure of BnFLC.A3b. The positions of exons and untranslated regions (UTRs) are represented by boxes.
(TIF)
(A) cDNA and (B) amino acid sequence alignment of alternatively spliced variants and their predicted polypeptides of BnFLC.A3b in Tapidor and Ningyou7. BnFLC.A3b_T, N represents a correctly spliced transcript in Tapidor and Ningyou7, BnFLC.A3b_N-1 and BnFLC.A3b_N-2 represent the two kinds of alternatively spliced transcripts in Ningyou7 which are missing exon 3 partly or completely.
(TIF)
Genome blocks and the location of FRI , FLC , and CBF homologues in Arabidopsis thaliana (At) and Arabidopsis lyrata (Al). Arrows indicate the opposite orientation of the blocks.
(TIF)
Primers used for cloning, mapping, expression, and DNA methylation analysis.
(XLS)
Information for isolated cDNA sequences of BnFLC homologues.
(XLS)
Comparison of methylated sites in the predicted CG islands of BnFLC.A3b between the Tapidor and Ningyou7 alleles.
(XLS)
Summary of BLASTN analysis to detect similar coding sequences in the Arabidopsis genome.
(XLS)