Abstract
Tumor microenvironment (TME) is important in tumor development and may be a target for anti-cancer therapy. The genesis of TME is a dynamic process that is regulated by intrinsic and extrinsic factors and coordinated by multiple genes, cells, and signal pathways. Cancer anaerobic metabolism and various oncogenes may stimulate the genesis of TME. Tumor cells and cancer stem cells actively participate in the genesis of the cancer stem cell niche and tumor neovascularization, important in the initiation of the TME. Various cancer-associated stromal cells, derived niche factors, and tumor-associated macrophages may function as promoters in the genesis of the TME. Dicer1 gene-deleted stromal cells can induce generation of cancer stem cells and initiate tumorigenesis, suggesting that stromal cells also may promote the genesis of the TME. Therefore, the key features of TME include niche-driving oncogenes, cancer anaerobic metabolism, niche-driving cancer stem cells, neovascularization, tumor-associated inflammatory cells, and cancer-associated stromal cells. These features are potential targets for normalization of the malignant TME and effective anti-cancer therapy.
Keywords: Stem cell, Niche, Oncogene, Metabolism, Angiogenesis, Stromal cells
Introduction
Cancers account for the death of more than 10 million people per year globally. However, cancer treatments frequently fail owing to limited understanding of the cancers, and new strategies for the study of malignant tumors are necessary. In the past 3 decades, extensive cancer genetic and epigenetic studies have accumulated much data about cancers. During the past decade, thousands of targets were selected for anti-tumor drug research, but only several new drugs have successfully passed clinical trials to treat malignant tumors. The development of cancer is not just the result of an abnormality of a single gene, signal pathway, or cell type, but usually, multiple genes, signal pathways, cells, and environmental cues are involved. Much cancer research has been devoted to genetic and epigenetic events in cancer cells to understand malignant transformation and develop anti-cancer drugs. Although progress has been achieved in anti-cancer therapy, the understanding of tumor initiation, progression, and metastasis is limited.
A malignant tumor microenvironment (TME) or niche is an important factor in tumor growth, progression, metastasis, and drug resistance [1–3]. The genesis of TME is a dynamic, complex process that involves various intrinsic and extrinsic factors [1, 2]. In the historical “seeds and soil” theory of tumorigenesis, tumor cells were regarded as “seeds” and stoma as “soil” [4], and a more-recent theory of TME has been developed [5]. However, the initiation and genesis of the TME are poorly understood, and the primary features of the TME have not been well defined. It is unknown how a tumor initiates its own microenvironment; which key genes and cells initiate the genesis of the TME; which key stromal cells and derived niche factors trigger the genesis of the TME; whether stromal cells may initiate the generation of the TME; how tumor cells together with stromal cells initiate and develop a malignant TME; and what the key features of a malignant TME are.
Abnormal cancer cell metabolism and a diversity of oncogenes may stimulate the genesis of TME [6–8]. In addition, aggressive tumor cells (such as cancer stem cells) are important in the initiation and development of a malignant TME by active participation in tumor neovascularization and generation of a cancer stem cell niche [9–11]. Although stromal cells are considered a promoter during the genesis of the TME [12], dysfunction of stromal cells may induce generation of cancer stem cells and malignant tumors [13]. Rapid progress in the study of the TME has changed the approach to cancer research. The purpose of this article is to decipher the microenvironmental dimension of the TME in cancers, to explore possible key factors in the genesis of the malignant TME, and to propose new strategies for targeting the TME to normalize the malignant microenvironment in tumors for effective anti-cancer therapy.
Niche-Driving Oncogenes and Tumor Microenvironment
Generation of the TME is driven by intrinsic and extrinsic factors including multiple genes, signal transduction pathways, and cells [1–3]. Niche-driving oncogenes may function as key initiators of development of the TME. Gene mutations and epigenetic alterations result in the formation of oncogenes that stimulate tumorigenesis [6–8]. Oncogenes may activate key intracellular signal transduction pathways and gene transcription to promote over-expression of multiple genes responsible for cell metabolism, proliferation, migration, and immunity, resulting in the initiation and development of the TME (Fig. 1). There are 181 oncogenes that have been identified and recorded in a genetic sequence database (GenBank®, National Institutes of Health, Bethesda, MD), including 94 well-characterized oncogenes associated with genetic mutations and others that have shown over-expression owing to abnormal epigenetic alterations. These oncogenes present in a variety of cells, including cancer stem cells (CSC), malignant tumor cells, some of embryonic stem cells, and induced pluripotent stem cells (iPSC), which have potential tumorigenecity as we addressed recently [14]. Oncogenes regulate cell plasma membrane receptors, nonreceptor protein kinases, intracellular signaling proteins, metabolic enzymes, and transcriptional factors [6–8]. There are 18 niche-driving oncogenes that contribute to the genesis of the TME and tumorigenesis (Table 1 and Fig. 1).
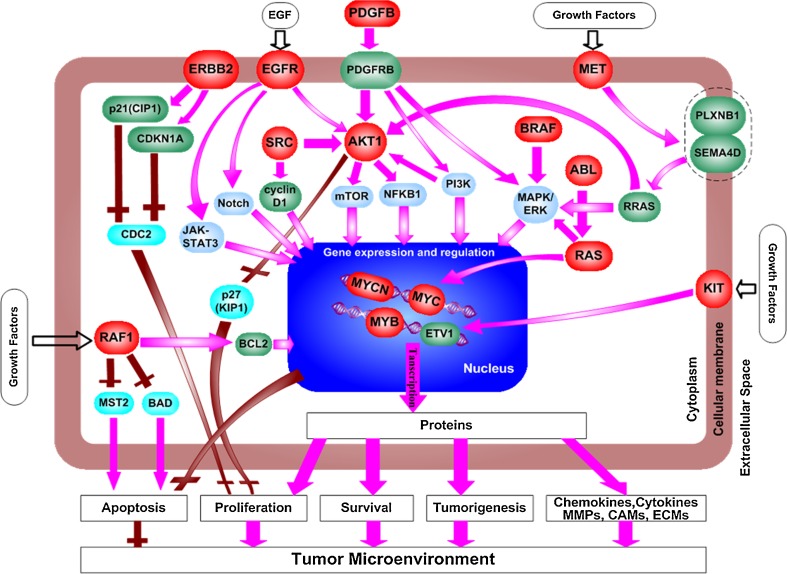
Fig. 1.
Activation of multiple signal transduction pathways by niche-driving oncogenes. Niche-driving oncogenes activate key signal transduction pathways at multiple levels, including the cell membrane, cytoplasm, and nucleus. The oncogenes activate transcriptome and secretome in cancer cells and initiate a tumor microenvironment (TME) with distinct features, including cancer aerobic metabolism, cancer stem cell niche, tumor neovascularization, tumor-associated macrophages, cancer-associated stromal cells, and derived niche factors. ( , niche-driving oncogenes;
, niche-driving oncogenes;  , normal signaling proteins;
, normal signaling proteins;  , key signal pathways;
, key signal pathways;  , normal proteins that regulate cell proliferation or apoptosis;
, normal proteins that regulate cell proliferation or apoptosis;  , promotion of signal transduction;
, promotion of signal transduction;  , inhibition of signal transduction; and
, inhibition of signal transduction; and  , mutated growth factors activating proto-oncogenes)
, mutated growth factors activating proto-oncogenes)
Table 1.
Tumor microenvironment stimulator genes
| Oncogene | Function |
|---|---|
| (1) Membrane receptors | |
| ERBB2 (amplification, small insertions, and Leu755Pro) | The mutation-activating ERBB2 involves in the activation of down-stream signaling pathways, such as MAPK and PI3K, and contributes to tumor growth by inhibiting the CDC2-mediated apoptosis. |
| KIT (Asp816Val, Lys550Ile) | Mutations in KIT result in constitutive activation of KIT that contributes to tumor development, including leukemia and gastrointestinal tumors. |
| MET (amplification and MET1149THR) | Mutations in MET gene lead to constitutive activation of the MET protein, which is important in tumorigenesis of papillary renal cell carcinoma and hepatocellular carcinoma. |
| ROS1/FIG fusion gene | The ROS1/FIG fusion protein has a constitutive tyrosine kinase activity which promotes oncogenesis in glioblastoma cell lines. |
| EGFRvI and EGFRvIII (large deletions) | EGFRvI and EGFRvIII have a constant EGFR tyrosine kinase activity that leads to carcinogenesis, such as in glioblastoma and lung cancer. |
| FGFR1 (Asn546Lys and Arg576Trp); Fusion genes of FGFR1 with ZNF198, BCR | The point mutations in FGFR1 lead to the constitutive kinase activity in glioblastomas. ZNF198/FGFR1, BCR/FGFR1, and FGFR1OP2/FGFR1 fusion genes contain the FGFR1 tyrosine-kinase domain and effect constitutive FGFR1 activation in both lymphoma and leukemia cells. |
| (2) Non-receptor protein kinases | |
| BCR/ABL and ABL/NUP214 fusion genes | The BCR/Abl and Abl/NUP214 fusion genes contain the Abl tyrosine kinase activity domain which leads to abnormal activation of down-stream signal cascades in leukemia. |
| AKT1 (Glu17Lys and amplification) | Mutation-activating AKT1 promotes tumorigenesis by activating down-stream signaling pathways, such as mTOR and NFκB, and inhibiting apoptosis, in breast cancer, colorectal cancer, and ovarian cancer. |
| BRAF (Val600Glu, Arg462Ile) | Mutations lead to elevation of the BRAF kinase activity (MAPK/ERK-dependent) in melanoma, colorectal cancer, and lung cancer. |
| FES (translocation) | The t(15;17)(q24;q21.1) translocation of FES was found in acute promyelocytic leukemia and may be related to neoplasia. |
| SRC (Gln531Ter) | A mutation in SRC elevates SRC activity in human colon cancers and functions in the progression of human colon cancer. |
| (3) Signal transducers | |
| RAF1/SRGAP3 fusion gene | RAF1/SRGAP3 fusion gene contains the entire RAF1 kinase domain and shows higher activity than wild-type RAF1 in astrocytoma. |
| PDGFB/COL1A1 fusion gene | The PDGFB/COL1A1 fusion gene induces tumor formation through the abnormal activation of the PDGFB receptor pathway. |
| (4) Transcription factors | |
| HRAS/KRAS/NRAS (Gly12Val, Gln61Lys, (Gly12Cys, Gly13Asp, Gly13Arg, Gln61Arg) | Mutation-activating RAS family members, including h-RAS, k-RAS, and n-RAS, were found in various cancers, including lung cancer, gastric cancer, acute myelogenous leukemia, and melanomas. By activating Myc, activated RAS promotes numerous down-stream genes that contribute to tumorigenesis. |
| MYB (the rearrangement of chromosome 6) | Reactivation of MYB caused by rearrangement of chromosome could be associated with the progression of melanoma, leukemia, and lymphomas. |
| Myc (amplification, translocation, and PRO57SER) | Mutations in Myc gene lead to constitutive Myc activity, which promotes oncogenesis in a variety of cancers by activating the transcription of numerous down-stream genes. |
| MYCN (amplification) | In neuroblastoma tumors, MYCN amplification promotes a rapid cell division and regulates the expression of FAK, a key tyrosine kinase involved in the survival and metastasis of human tumors. |
| (5) Metabolism | |
| GLDC | High expression of GLDC drives aerobic glycolysis and tumorigenesis |
Oncogenic Cell Plasma Membrane Receptors
Mutations of several key cell plasma membrane receptors can produce oncogenes (Table 1). Epithelial growth factor receptor (EGFR) mutants cause a constant activation of EGFR tyrosine kinase, even in the absence of growth factors; the high kinase activity increases multiple signal transduction pathways, including PI3K-AKT, JAK-STAT, Notch, TGF-β1, and VEGF signal transduction, resulting in the over-expression of various growth factors and niche factors responsible for the genesis of the TME and tumorigenesis [15–17]. Mutations of ERBB2 can cause tumorigenesis and the generation of the TME by protein kinase-mediated activation of ERBB2 down-stream signal pathways [18]. In addition, point mutations of FGFR1 (ASN546LYS and ARG576TRP) result in constitutive high FGFR1 kinase activity in glioblastomas, and FGFR1 chromosome translocations t(8;13)(p11;q11-12), t(8;22)(p11;q11), and t(12;8)(p11 p11p22) result in oncogenic fusion proteins (ZNF198-FGFR1, BCR-FGFR1, and FGFR1-OP2) with a constitutive high FGFR1 kinase activity in lymphoma and leukemia cells [18–20]. Mutations in KIT (CD117) result in constitutive activation of KIT ligand-independent tyrosine kinase activity in various malignant tumors and the genesis of the TME [21, 22]. Mutants of MET result in constitutively activated MET protein, which causes tumorgenesis in papillary renal cell carcinoma and hepatocellular carcinoma, and ROS1-FIG fusion gene results in a constitutively activated tyrosine kinase in glioblastoma cell lines [23].
Oncogenic Nonreceptor Protein Kinases
The mutations of nonreceptor protein kinases can result in production of oncogenes responsible for tumorigenesis and genesis of the TME. The BCR-Abl and Abl-NUP24 fusion proteins derived from chromosomal translocation have constant tyrosine kinase activity in the absence of growth factors, and these oncogenic proteins constitutively activate Abl down-stream signal cascades, resulting in leukemia and genesis of the TME [24–26].
Oncogenic Intracellular Signal Proteins
Mutations of intracellular signal proteins cause tumorigenesis and genesis of the TME. Both AKT1 mutant (E17K) and over-expression of AKT1 contribute to oncogenesis and genesis of the TME in various cancers [27, 28]. The BRAF mutants (V600E) have an elevated kinase activity that activates various cell surface receptors, increases production of the extracellular matrix, and triggers initiation of the TME [29, 30]. Several SRC mutants (c-SRC position 95, 117, and 527 mutations, v-SRC) display high kinase activity and contribute to the malignant progression of human colon and other cancers [31].
Oncogenic proteins in the RAS family including k-RAS, h-RAS, and n-RAS are important signal proteins in the genesis of TME. Mutations in several special positions of RAS genes can lead to the constant activation of the RAS signal transduction pathway in various cancers [32]. Activated RAS increases Myc activity that contributes to the generation of the TME [33]. Expression of oncogenic k-RAS mutant (G12D) results in NF-kappa-B activation, essential for tumor cell survival [34]. The RAF1-SRGAP3 fusion protein increases tumor angiogenesis and generation of the TME [35]. The PDGFB-COL1A1 fusion protein derived from chromosome translocation t(17;22)(q22;q13) induces tumorigenesis by activating the PDGFB pathway that contributes to genesis of the TME [36].
Oncogenic Transcription Factors
Several oncogenic transcription factors, such as c-Myc, FOS-JUN, MYB, and Fra1, play pivotal roles in the generation of the TME and tumorigenesis. Mutations of Myc, caused by genetic alterations including point mutations, abnormal amplification, and chromosome translocation, result in cancers with a distinct TME. In addition, inactivation of tumor suppressor genes such as adenomatous polyposis coli gene (APC) also leads to high constitutive Myc activity. Activated Myc promotes global gene transcription mediated by master transcription factors E2F1, E2F2, and E2F3, and increases cell proliferation, transformation, angiogenesis, abnormal cancer metabolism, the genesis of the TME, and development of cancers [37, 38]. The Myc gene is important in generating tumor stroma and blood vessels, the inflammatory response, and cancer anaerobic metabolism [39]. Inhibiting endogenous Myc triggers collapse of tumor vasculature and microenvironment, resulting in regression of pancreatic islet tumors stimulated by simian virus 40 (SV40) [40]. Therefore, Myc is important in coordinating diverse intracellular oncogenic pathways and genesis of the malignant TME. In addition to Myc, the MYB chromosome 6 translocation [41] and proto-oncogene Fra-1 remodel the TME and result in genesis and metastasis of tumors [42].
Tumor Suppressor Genes
Several tumor suppressor genes contribute to genesis of the TME. The loss of PTEN results in a malignant TME by transcriptional reprogramming of stromal cells, down-regulation of miR-320, and up-regulation of ETS2 (v-Ets erythroblastosis virus E26 oncogene homolog 2) [43]. Deficiency of p53 gene causes a regenerative microenvironment that strongly induces embryonic rhabdomyosarcoma in mice [44], and p53 mutations promote genesis of the TME by regulating cancer metabolism.
Niche-driving oncogenes in tumor cells interact with each other and integrate with a variety of cell signal proteins. This causes a complex signaling network (Fig. 2) that controls intracellular transcriptome and secretome to produce and release various niche factors, including growth factors, cytokines, chemokines, metalloproteinases (MMPs), and ECMs. This results in tumorigenesis and initiation of a malignant TME (Figs. 1 and 2). Therefore, these niche-driving oncogenes are an important factor in developing the malignant TME.
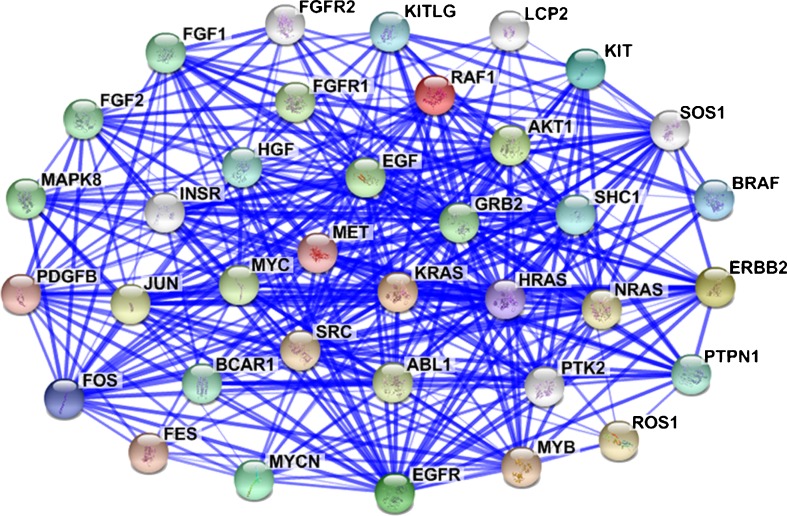
Fig. 2.
Protein-protein interactions among oncogenes result in a molecular network to initiate the genesis of the TME. Protein-protein interaction among oncogenes forms a stable/strong network, as shown by STRING analysis of niche-driving oncogenes. When an oncogene is activated, it sequentially induces the expression of target genes by itself or other nodes in the network. As a result, the activated oncogenic network stimulates the initiation and development of TME by multiple reactions. ( , protein-protein interaction previously reported; the thickness of the blue line is qualitatively proportional to the reliability of the data)
, protein-protein interaction previously reported; the thickness of the blue line is qualitatively proportional to the reliability of the data)
Abnormal Cancer Metabolism and Tumor Microenvironment
In malignant tumor cells, anaerobic metabolism (Warburg effect) occurs in an oxygen-independent manner, and cancer cells maintain glycolytic phenotype in areas of an adequate oxygen supply [45]. Glycolysis and glutaminolysis are boosted in cancers to meet the high ATP and NADPH demands of tumor cells. At the initial stage of tumorigenesis, a small tumor (< 2 mm) usually does not have a distinct TME, but the tumor has a robust cancer anaerobic metabolism even in a normoxic environment. The anaerobic metabolism generates lactic acid and results in an acidic TME [46]. Lactic acidosis and an acidic environment in the tumor tissue result in over-expression of various genes, including glycine decarboxylase (GLDC) [13], glucose transporters, SIRT1, COX2, GAC, PDHK1, PFK1, ANT2, and HIF1A. However, acidic environment may cause genetic instability and activation of oncogenes, resulting in expression of hypoxia-induced factor-A (HIF1A). Over-expression of these cancer-metabolism–related genes, oncogenes, and HIF1A triggers genesis of the TME [13, 47–50].
The metabolic enzyme glycine decarboxylase (GLDC) is an oncogenic protein that induces generation of cancer stem cells and tumorigenesis of non-small cell lung cancer [13]. This protein causes an increase in glycolysis and the metabolism of glycine, serine, and pyrimidine, promoting stem cell proliferation in several types of cancers. In addition, IDH1 mutants and HMGCR have potential oncogenic activities [51, 52]. The mTOR up-regulation of PKM2 through HIF1A-mediated transcriptional activation is important for aerobic glycolysis and tumor growth [53]. Oncogenic k-RAS modulates mitochondrial metabolism in human colon cancer cells by activation of HIF1A and HIF2A, which promotes over-expression of hundreds of HIF-target genes including those involved in cancer anaerobic metabolism, tumor angiogenesis, metastasis, and genesis of the malignant TME [53, 54]. Furthermore, over-expression of several cancer metabolic genes in cancer-associated fibroblasts and epithelial cells, including PKM1, PKM2, cathepsin B, MCT4, BNIP3L, TOMM20, MT-CO1, and SDH-B, drives stromal nutrient production and tumor growth [55, 56]. In contrast, loss of caveolin-1 expression in cancer-associated fibroblasts results in the generation of the TME in breast and prostate cancers [8], and caveolin-1 is a useful marker of oxidative stress, hypoxia, and autophagy in the TME [57]. Therefore, these data suggest that anaerobic metabolism is an important initiator and feature of TME.
Cancer Stem Cells and Tumor Microenvironment
Cancer stem cells (CSCs) are important in tumor initiation, progression, and drug resistance [58]. At the initial stage of tumorigenesis, intrinsic and extrinsic factors cause intracellular genetic mutations and epigenetic alterations, resulting in generation of oncogenes that induce the production of CSCs and tumorigenesis [6]. The CSCs can be produced from precancerous stem cells [59–64], cell de-differentiation [65], or an epithelial-mesenchymal transition [66–68]. Malignant mesenchymal stem cells have been found in the niche of cancers [66, 67], and an epithelial-mesenchymal transition may be an early key step in the initiation of TME and tumorigenesis [68]. The CSCs may expand by symmetric division, produce progenitor cells by asymmetric division, and differentiate to multiple lineages of tumor cells, resulting in a rapid increase in tumor mass (Figs. 3 and 4).
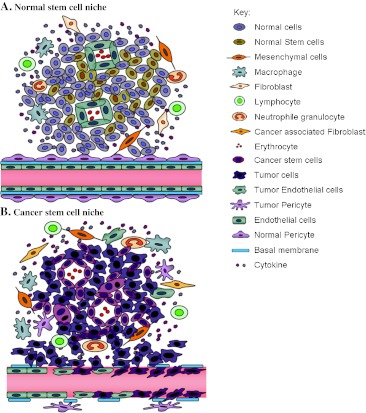
Fig. 3.
Normal and cancer stem cell (CSC) niches. a The normal stem cell niche includes endothelial cells, stem cells, and stromal cells, with only few inflammatory cells; the stem cells are located near blood vessels, and the inner surface of blood vessels is lined by endothelial cells covered by pericytes. b Vascular CSC niche includes vasculogenic tumor cells, cancer-associated stromal cells, endothelial cells, and other cells. The vasculogenic tumor cells include precancerous stem cells, CSCs, and differentiated tumor cells; vasculogenic CSCs in the tumor blood vessel wall can migrate to the niche and undergo proliferation and differentiation
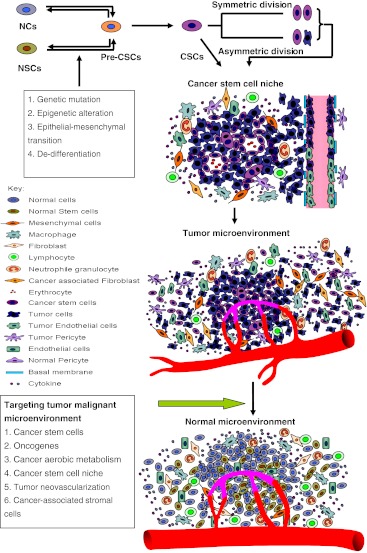
Fig. 4.
Genesis of the TME and strategy for normalization of malignant niche. Normal cells (NCs) and normal stem cells (NSCs) can become precancerous stem cells (Pre-CSCs) and cancer stem cells (CSCs) by genetic mutation, epigenetic alteration, epithelial-mesenchymal transition (EMT), and de-differentiation. The CSCs can self-renew and expand by symmetric division, and also undergo asymmetric division to produce various differentiated tumor cells. The tumor cells and stromal cells form a CSC niche, and then recruit bone marrow and blood cells to form a TME. Strategies for normalization of the TME may include targeting niche-driving oncogenes, cancer aerobic metabolism, CSCs, CSC niche, tumor blood vessels, tumor-associated macrophages (TAMs), and cancer-associated stromal cells
There are several possible ways for CSCs to initiate and stimulate the genesis of TME. First, CSCs actively participate in the development of the CSC niche [11]. Second, CSCs may trans-differentiate into cancer-associated stromal cells [58–62, 65–69], such as cancer-associated fibroblasts and tumor endothelial cells [70, 71]. The CSC-differentiated tumor endothelial cells are important in tumor neovascularization and the genesis of TME [72–75]. Third, CSCs release various stem cell factors that induce normal stromal cells to become cancer-associated stromal cells, such as cancer-associated fibroblasts (CAFs) [1] and tumor endothelial cells (TEMs) [76], and to recruit immune cells from bone marrow and blood to the tumor tissue. Normal macrophages are converted into tumor-associated macrophages (TAMs) in the TME [77]. Therefore, CSCs and derived factors work with stromal cells to initiate and develop a malignant TME that facilitates tumor growth, progression, metastasis, and drug resistance (Figs. 3 and 4). The CSCs have different characteristics than normal cells, suggesting that the CSCs are an important feature of malignant TME.
Tumor Neovascularization and Cancer Stem Cell Niche
The expansion, invasion, and metastasis of malignant tumors is intimately linked to the ability of the tumor to establish an adequate vascular supply [78]. A small tumor (< 2 mm diameter) can grow without blood vessels. When a tumor grows > 2 mm, the central zone of the tumor becomes hypoxic and acidic because of the aerobic metabolism of the tumor. For further growing up, tumor cells need blood vessels to supply oxygen and nutrients and to discharge waste metabolic materials [58, 59, 69]. After a tumor generates its own vasculature, the tumor grows rapidly. Preclinical and clinical studies have shown that aggressive and metastatic tumors usually are rich in tumor vasculature [70, 78]. Tumor blood vessels provide the tumor with oxygen, nutrients, cells from outside tissues, a route for metastasis, and an important component of the vascular stem cell niche. Generation of tumor blood vessels is an important initial step for genesis of the TME.
Malignant tumors can generate blood vessels in 6 distinct ways including angiogenesis, vasculogenesis, intussusception, vessel co-option, vasculogenic mimicry, and CSC-tumor endothelial cell trans-differentiation [70, 71]. Hypoxia is a stimulator of tumor angiogenesis and genesis of the TME. In addition, tumor cells and CSCs use host tissue as “soil” to expand rapidly and also actively build up their own unique microenvironment.
The mechanisms of tumor cell-dominant vasculogenic mimicry and tumor cell-vascular endothelial cell trans-differentiation are unknown. We recently studied tumor cell-dominant vasculogenic mimicry and examined the vasculogenic capability of 62 human tumor cell lines using an in vitro tumor cell-mediated tube formation system. We found that 25 of 62 tumor cell lines formed capillary tubes in vitro, and most of the aggressive tumor cells actively participated in tumor vascularity in vivo [79]. The vasculogenic human ovarian cancer cell line Hey1B had marked vasculogenic capability. When transplanted in vivo, Hey1B cells induced development of a tumor infiltrated vasculature, with the Hey1B cells directly assembling as an integral component of the newly formed vasculature. In addition, Hey1B cells expressed 18 known vasculogenic and angiogenic genes. Most of these genes are highly expressed at an early stage of embryonic development and are important in the vasculogenesis required to create the immature, primitive vasculature [79]. Furthermore, VE-cadherin plays an essential role in tumor neovascularization, and the VE-cadherin-binding protein T2-tryptophanyl tRNA synthetase (T2-TrpRS) effectively inhibits tumor angiogenesis in vivo [80, 81].
The CSCs are not randomly distributed in tumor tissue, but the cells live in a specific CSC niche. There are 2 types of CSC niches, osteoblast CSC niche and vascular CSC niche, that regulate CSC self-renewal and differentiation [3, 10]. The niches consist of CSCs, stromal cells, and blood vessels. The vascular CSC niche is at the unique location that directly contacts with both local tissue and the circulating blood, and is important in CSC generation, expansion, and the genesis of TME [82–86]. In normal tissue, the vascular stem cell niche consists of endothelial cells, stem cells, and stromal cells, with few inflammatory cells; the inner layer of blood vessels is lined by endothelial cells covered by pericytes (Fig. 3). In contrast, the vascular CSC niche is composed of CSCs, endothelial cells, and various cancer-associated stromal cells (Fig. 3). The blood vessels of the vascular CSC niche had been thought to consist of endothelial cells, but recent data have suggested that tumor cells and CSCs actively participate in generation of the blood vessels in the vascular CSC niche (Fig. 3). These tumor cells include precancerous stem cells [59–61], CSCs [62–64], and vasculogenic tumor cells [71–75].
Leukemia stemlike cells, precancerous stem cells, and glioma stem cells could form tumor vasculature by tumor vasculogenesis [59–64, 71, 72]. Glioma stem cells contribute to tumor neovascularization by trans-differentiation of the cells to tumor endothelial cells, and > 60 % vascular cells are tumor endothelial cells derived from the trans-differentiation of glioma stem cells [74, 75].
Based on these data, we propose a new model of vascular CSC niche (Fig. 4). In this model, vasculogenic CSCs directly form tumor blood vessels, and the tumor cells in the vascular wall can interact with blood cells, plasma factors, and various stromal cells near the vessels. The CSCs on the vascular wall can migrate to the middle of the niche, proliferate, differentiate, and metastasize to other organs using the blood circulation. Therefore, CSC-dominant neovascularization and the vascular CSC niche are distinct features of malignant TME.
Chronic Inflammation and Tumor Microenvironment
Chronic inflammation is important in the development of cancer, and the amount of various inflammatory cells is much greater in tumor tissue than normal tissues. Tumor-associated inflammatory cells, such as tumor-associated macrophages (TAMs) [77, 87] and Tie-2 expressing monocytes [88], are important in the genesis of TME. During tumor initiation and progression, tumor cells can produce and release chemokines that induce migration of monocytes and macrophages to tumor tissue. Tumor cells may cause TAMs to lose anti-tumor immunologic function and gain tumor assistant functions. The activated TAMs can release various niche factors, such as growth factors, chemokines, and proteolytic enzymes to promote TME formation. Bone marrow-derived monocytes and macrophages contribute to vasculogenesis in multiple myeloma, and bone marrow macrophages from multiple myeloma patients can form a capillarylike network and contribute to the malignancy of multiple myeloma [89, 90]. In addition, Tie2-expressing monocytes have nonredundant function to promote tumor angiogenesis and genesis of TME in mice [88]. Therefore, these tumor-associated inflammatory cells in TME, such as TAMs and Tie2-expressing monocytes, have distinct features and contribute to the development of the TME.
Tumor-Associated Stromal Cells and Tumor Microenvironment
The TME consists of 2 discrete but interactive compartments: tumor cells and stroma [1–3]. The stroma is a mixture of nonmalignant cells and stromal elements, including vascular endothelial cells, fibroblasts, mesenchymal cells, inflammatory cells, and extracellular macromolecules such as collagen, fibronectin, laminin, fibrin, proteoglycans, and hyaluronan [91]. During the development of TME, the tumor cells and CSCs are important as “seeds”; however, tumor cells alone are not enough to develop a malignant TME without the stroma as “soil” to provide a favorable environment. Tumor-associated stromal cells, such as CAFs, TAMs, and TECs, trigger the genesis of the TME [1–3, 76, 77]. The CAFs can release growth factors, cytokines, and chemokines, and are important tumor-associated stromal cells in the genesis of the TME [1]. The TAMs contribute to the generation of the TME [77], and TECs also are important in tumor neovascularization and the formation of the vascular stem cell niche [76].
Tumor-associated stromal cells can interact with tumor cells to facilitate the development of TME. The cells release various niche factors, including growth factors, cytokines, chemokines, MMPs, ECMs, and cell adhesion molecules to promote the genesis of the TME [91]. Various growth factors promote TME formation, including AREG, BDNF, CTGF, CSF2, CSF3, EGF, FGF1, HGF, IGF1, LEF, VEGF, PDGF, SDF-1, SDF4, SCF, TCF, TF, and TGFβ1. Various cytokines increase generation of the TME, including IL-1, IL-10, IL-12, IL-15, IL-21, IL-3, IL-6, IL-7, IL-17, IL-8, interferons, and TNFs [60, 62] . The C-C motif chemokines and their receptors trigger the formation of TME in cancers, including CCL2/MCP-1, CCL8/MCP-2, CCL7/MCP-3, CCL21, CCL22, CCL25, CCR1, CCR2, CCR5, CCR9, CX3CL1, CX3CR1, CXCL10, CXCL12, CXCR3, and CXCR4. Various MMPs, including MMP2, MMP3, MMP9, MMP10, MMP13, MMP14, MMP15, and ADAM1, are essential for the genesis of the TME. The ECM proteins, such as collagen, elastin, fibronectin, and laminin, are essential for the TME. Cell adhesion molecules, such as cadherins, catenins, and integrins, are critical for the formation of the TME. Collectively, these proteins and factors promote genesis of the TME and nurture tumor growth and metastasis.
Although stromal elements, including stromal cells and derived factors, are important in the genesis of the TME, the microenvironment may be a required, but not sufficient “facilitator or promoter” for malignant transformation [1–3]. However, stromal cells with genetic and epigenetic abnormalities primarily induce the generation of CSCs and the development of malignant tumors [13]. Targeted deletion of Dicer1, the “landscape-modifying” miRNA-processing endonuclease in osteoprogenitor cells, causes a myelodysplastic syndrome (MDS) in knockout mice, and the Dicer1-deleted mice develop acute myelogenous leukemia [13, 92]. These abnormalities are dependent on the type of stromal cells in the microenvironment, because MDS was not observed when Dicer1 was deleted from mature osteoblasts. Therefore, the phenotypic changes were microenvironment-dependent, and the dysfunction of stromal cells can initiate neoplastic disease and microenvironment abnormality because stromal cell malfunction can stimulate oncogenesis. Thus, cancer-associated stromal cells may be a key factor of malignant TME.
Normalization of Tumor Microenvironment for Anti-cancer Therapy
At the early stage of tumorigenesis, tumor cells initiate and develop a favorable tumor microenvironment by using intrinsic and extrinsic tumorigenic factors to overcome anti-tumorigenic factors [93]. In traditional Chinese medicine, diseases including cancers are described as having been induced by loss of the balance between yin and yang [94]. Evaluating the TME in cancers shows 6 features that may result in the imbalance of yin and yang in TME. The yin represents the TME-stimulating and promoting forces, and yang represents the TME-inhibiting forces. In tumor tissues, the yin is escalated but the yang is diminished. The imbalance of yin and yang results in genesis of malignant the TME and tumorigenesis [93, 94]. Therefore, blocking of yin and increase in yang to improve yin-yang balance may result in the reversal of the malignant TME. We propose to target the 6 distinct features in the TME to normalize the malignant TME for effective anti-cancer therapy (Fig. 4).
Targeting Niche-Driving Oncogenes
There are 18 oncogenes that contribute to the initiation of TME and tumorigenesis (Table 1). However, the understanding of the mechanism of malignant TME generation by niche-driving oncogenes is limited. Further research may identify the new oncogenes responsible for genesis of the TME and elucidate the mechanisms of oncogene-drived formation of TME. We may target key oncogenes for the normalization of malignant TME by zinc finger protein (ZFP) technology or specific inhibitory therapeutics.
Correction of Abnormal Cancer Metabolism
Malignant tumor cells take up much more glucose than normal cells and use aerobic glycolysis to produce ATP for syntheses of proteins, DNA, and fatty acids. Furthermore, tumor cells produce large quantities of secreted lactate, which causes an acidic environment to stimulate reprogramming gene expression and the genesis of TME [8, 45–57]. Key genes responsible for abnormal cancer metabolism, such as DGLC and several other aerobic metabolic enzymes, can be targeted using their pharmacological inhibitors in glucose, glutamine, and fatty acid metabolic pathways. Correction of the abnormal cancer metabolism may provide a new approach for the normalization of malignant TME.
Targeting Cancer Stem Cells in the Tumor Microenvironment
The CSCs are important in the initiation of the TME and generation of malignant tumors, and these cells are potential targets in attempts to normalize the TME. Several options potentially may target CSCs in the TME. First, precancerous cells are reversible and may be induced to regress to normal cells, depending on the environmental cue [59–61]. Second, oncogenic mesenchymal stem cells could be reversed to normal epithelial cells using the approach of mesenchymal-epithelial transition, because epithelial-mesenchymal transition may be reversible [65–68]. Furthermore, differentiation of CSCs and inhibition of the proliferation of these cells by changes in environmental cues may be additional options to normalize malignant TME.
Targeting Cancer Stem Cell Niche and Disorganized Tumor Vasculature
The CSCs live in a favorable CSC niche that supports stem cell proliferation and differentiation and protects stem cells from being killed by cytotoxic drugs. Therefore, disruption of the CSC niche, or inhibition of CSC niche formation, may result in correction of malignant the TME. Options include elimination or suppression of the CSCs in the niche; blocking the interaction between the CSCs and stromal cells; impeding the generation of the niche factors by both tumor and stromal cells; and using new approaches to hinder tumor neovascularization and to normalize the disorganized tumor blood vessels as we have recently addressed [95]. Solid tumors require blood vessels for generation of the CSC niche, growth, and metastasis; therefore, the tumor vasculature has been targeted using anti-angiogenic drugs to decrease the tumor vascular supply, but the success of anti-angiogenesis at the early stage is limited by insufficient efficacy and development of drug resistance [96–98]. Normalization of the vascular abnormalities is a complementary therapeutic approach for cancer [99–101]. Furthermore, new approaches to normalize the disorganized tumor blood vessels may be considered, by targeting vasculogenic genes and cancer cells, combining anti-angiogenic therapeutics with vascular normalization drugs, and integrating Western medicine and traditional Chinese medicine [95]. The correction of the disorganized tumor vasculature may limit the genesis of the TME and may normalize the malignant TME.
Targeting Cancer-Associated Stromal Cells and Tumor-Associated Inflammatory Cells
During tumor development, the microenvironment promotes the genesis of cancer-associated stromal cells. Targeting these cancer-associated stromal cells, such as CAFs and malignant mesenchymal stem cells, may normalize the TME. Epithelial-mesenchymal transition is reversible, and the TME potentially may be normalized in various epithelial cancers by inducing the reversal of malignant mesenchymal stem cells to normal epithelial cells by mesenchymal-epithelial transition. Targeting TAMs and other tumor-associated inflammatory cells in the TME may result in the inhibition of the malignant TME.
Combination Therapy
The TME consists of cancer cells and various nonmalignant stromal cells, and the genesis of TME is a dynamic process that involves multiple genes, cells, and both intrinsic and extrinsic factors. Therefore, targeting a single gene, cell type, or signal pathway may yield low efficacy in anti-cancer therapy. Instead, targeting multiple key niche-driving and promoting genes, cells, signal pathways, stromal cells, and derived niche factors in the TME may cause a synergistic effect in the normalization of malignant TME, and may enable the development of novel anti-tumor drugs for effective anti-cancer therapy.
Acknowledgments
This study was supported by grants from National Natural Science Foundation of China (Grant No. 30971138), Chinese Academy of Science Special National Strategic Leader Project (No. XDA01040200), Suzhou City Scientific Research Funds (No. SWG0904, SS201004, and SS201138), and a project funded by the priority academic program development (PAPD) of Jiangsu Higher Education Institutions.
References
- 1.Allen M, Louise Jones J. Jekyll and Hyde: the role of the microenvironment on the progression of cancer. J Pathol. 2011;223:162–176. doi: 10.1002/path.2803. [DOI] [PubMed] [Google Scholar]
- 2.Weber CE, Kuo PC (2011) The tumor microenvironment. Surg Oncol Sep 29 [DOI] [PubMed]
- 3.Borovski T, Sousa E, Melo F, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer Res. 2011;71:634–639. doi: 10.1158/0008-5472.CAN-10-3220. [DOI] [PubMed] [Google Scholar]
- 4.Paget S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev. 1889;8:98–101. [PubMed] [Google Scholar]
- 5.Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 7.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 8.Sotgia F, Martinez-Outschoorn UE, Howell A, Pestell RG, Pavlides S,Lisanti MP (2012) Caveolin-1 and Cancer Metabolism in the Tumor Microenvironment: Markers, Models, and Mechanisms. Annu Rev Pathol 7:423–467 [DOI] [PubMed]
- 9.Sengupta A, Cancelas JA. Cancer stem cells: a stride towards cancer cure? J Cell Physiol. 2010;225:7–14. doi: 10.1002/jcp.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabarcas SM, Mathews LA,Farrar WL (2011) The cancer stem cell niche—there goes the neighborhood? Int J Cancer 129:2315–27 LID, doi:10.1002/ijc.26312 [DOI] [PMC free article] [PubMed]
- 11.Zhang LZ, Zhang CQ, Yan ZY, Yang QC, Jiang Y,Zeng BF (2011) Tumor-initiating cells and tumor vascularization. Pediatr Blood Cancer 56:335–40 LID—doi:10.1002/pbc.22886 [DOI] [PubMed]
- 12.Medema JP, Vermeulen L (2011) Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature 474:318–26 LID—doi:10.1038/nature10212 [DOI] [PubMed]
- 13.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, Bhakoo KK, Jayapal SR, Nichane M, Yu Q, Ahmed DA, Tan C, Sing WP, Tam J, Thirugananam A, Noghabi MS, Huei Pang Y, Ang HS, Robson P, Kaldis P, Soo RA, Swarup S, Lim EH, Lim B (2012) Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148:259–272 [DOI] [PubMed]
- 14.Zhang G, Shang B, Yang P, Cao Z, Pan Y, Zhou Q (2012) Induced pluripotent stem cell consensus genes: implication for the risk of tumorigenesis and cancers in induced pluripotent stem cell therapy. Stem Cells Dev 21(6):955–964 [DOI] [PubMed]
- 15.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science (New York, N.Y.) 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 16.Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323–327. doi: 10.1038/nature09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang SE, Yu Y, Criswell TL, Debusk LM, Lin PC, Zent R, Johnson DH, Ren X, Arteaga CL. Oncogenic mutations regulate tumor microenvironment through induction of growth factors and angiogenic mediators. Oncogene. 2010;29:3335–3348. doi: 10.1038/onc.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rand V, Huang J, Stockwell T, Ferriera S, Buzko O, Levy S, Busam D, Li K, Edwards JB, Eberhart C, Murphy KM, Tsiamouri A, Beeson K, Simpson AJ, Venter JC, Riggins GJ, Strausberg RL. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci U S A. 2005;102:14344–14349. doi: 10.1073/pnas.0507200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, Stone R, Weissman SM, Hudson TJ, Fletcher JA. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 20.Popovici C, Adelaide J, Ollendorff V, Chaffanet M, Guasch G, Jacrot M, Leroux D, Birnbaum D, Pebusque MJ. Fibroblast growth factor receptor 1 is fused to FIM in stem-cell myeloproliferative disorder with t(8;13) Proc Natl Acad Sci U S A. 1998;95:5712–5717. doi: 10.1073/pnas.95.10.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad Tunio G, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science (New York, N.Y.) 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 22.Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, Fletcher JA, Dewell S, Maki RG, Zheng D, Antonescu CR, Allis CD, Sawyers CL. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–853. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charest A, Lane K, McMahon K, Park J, Preisinger E, Conroy H, Housman D. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21) Genes Chromosomes Cancer. 2003;37:58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 24.Heisterkamp N, Stephenson JR, Groffen J, Hansen PF, Klein A, Bartram CR, Grosveld G. Localization of the c-ab1 oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature. 1983;306:239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- 25.Clark SS, McLaughlin J, Crist WM, Champlin R, Witte ON. Unique forms of the abl tyrosine kinase distinguish Ph1-positive CML from Ph1-positive ALL. Science (New York, N.Y.) 1987;235:85–88. doi: 10.1126/science.3541203. [DOI] [PubMed] [Google Scholar]
- 26.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, Vermeesch JR, Stul M, Dutta B, Boeckx N, Bosly A, Heimann P, Uyttebroeck A, Mentens N, Somers R, MacLeod RA, Drexler HG, Look AT, Gilliland DG, Michaux L, Vandenberghe P, Wlodarska I, Marynen P, Hagemeijer A. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–1089. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 27.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 28.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science (New York, N.Y.) 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 29.Nucera C, Lawler J, Parangi S. BRAF(V600E) and microenvironment in thyroid cancer: a functional link to drive cancer progression. Cancer Res. 2011;71:2417–2422. doi: 10.1158/0008-5472.CAN-10-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 31.Naoki K, Chen TH, Richards WG, Sugarbaker DJ, Meyerson M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62:7001–7003. [PubMed] [Google Scholar]
- 32.Santos E, Martin-Zanca D, Reddy EP, Pierotti MA, Della Porta G, Barbacid M. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science (New York, N.Y.) 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 33.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/S1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 34.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Frohling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alavi A, Hood JD, Frausto R, Stupack DG, Cheresh DA. Role of Raf in vascular protection from distinct apoptotic stimuli. Science (New York, N.Y.) 2003;301:94–96. doi: 10.1126/science.1082015. [DOI] [PubMed] [Google Scholar]
- 36.Muchemwa FC, Jinnin M, Wakasugi S, Sakamoto M, Inoue Y, Ishihara T, Ihn H. A novel COL1A1 exon 14/PDGFB fusion gene in dermatofibrosarcoma protuberans. Eur J Dermatol. 2010;20:390–391. doi: 10.1684/ejd.2010.0906. [DOI] [PubMed] [Google Scholar]
- 37.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 38.Morse B, Rotherg PG, South VJ, Spandorfer JM, Astrin SM. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988;333:87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- 39.Whitfield JR, Soucek L (2012) Tumor microenvironment: becoming sick of Myc. Cell Mol Life Sci 69:931–934 [DOI] [PMC free article] [PubMed]
- 40.Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25:907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassago A, Ferreira AP, Ferreira IM, Fornezari C, Gomes ER, Greene KS, Pereira HM, Garratt RC, Dias SM, Ambrosio AL (2012) Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc Natl Acad Sci U S A 109:1092–1097 [DOI] [PMC free article] [PubMed]
- 42.Luo YP, Zhou H, Krueger J, Kaplan C, Liao D, Markowitz D, Liu C, Chen T, Chuang TH, Xiang R, Reisfeld RA. The role of proto-oncogene Fra-1 in remodeling the tumor microenvironment in support of breast tumor cell invasion and progression. Oncogene. 2010;29:662–673. doi: 10.1038/onc.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, Srinivasan R, Trimboli AJ, Martin CK, Li F, Yu L, Fernandez SA, Pecot T, Rosol TJ, Cory S, Hallett M, Park M, Piper MG, Marsh CB, Yee LD, Jimenez RE, Nuovo G, Lawler SE, Chiocca EA, Leone G, Ostrowski MC (2012) Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol 14:159–67 LID—doi:10.1038/ncb2396 [DOI] [PMC free article] [PubMed]
- 44.Camboni M, Hammond S, Martin LT, Martin PT (2012) Induction of a regenerative microenvironment in skeletal muscle is sufficient to induce embryonal rhabdomyosarcoma in p53-deficient mice. J Pathol 226:40–9 LID—doi:10.1002/path.2996 [DOI] [PMC free article] [PubMed]
- 45.Kareva I. Prisoner’s dilemma in cancer metabolism. PLoS One. 2011;6:e28576. doi: 10.1371/journal.pone.0028576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero-Garcia S, Lopez-Gonzalez JS, Báez-Ez-Viveros JL, Aguilar-Cazares D, Prado-Garcia H. Tumor cell metabolism: an integral view. Cancer Biol Ther. 2011;12:939–948. doi: 10.4161/cbt.12.11.18140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 49.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science (New York, N.Y.) 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 50.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clendening JW, Pandyra A, Boutros PC, El Ghamrasni S, Khosravi F, Trentin GA, Martirosyan A, Hakem A, Hakem R, Jurisica I, Penn LZ. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci U S A. 2010;107:15051–15056. doi: 10.1073/pnas.0910258107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, Wang Y, Jing Y, Yang H, Chen R, Chang L, Zhang Y, Goto J, Onda H, Chen T, Wang MR, Lu Y, You H, Kwiatkowski D, Zhang H. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for anaerobic glycolysis and tumor growth. Proc Natl Acad Sci U S A. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Israelsen WJ, Vander Heiden MG. ATP consumption promotes cancer metabolism. Cell. 2010;143:669–671. doi: 10.1016/j.cell.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 54.Chun SY, Johnson C, Washburn JG, Cruz-Correa MR, Dang DT, Dang LH. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1alpha and HIF-2alpha target genes. Mol Cancer. 2010;9:293. doi: 10.1186/1476-4598-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldberg MS, Sharp PA (2012) Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J Exp Med 209:217–224 [DOI] [PMC free article] [PubMed]
- 56.Chiavarina B, Whitaker-Menezes D, Martinez-Outschoorn UE, Witkiewicz AK, Birbe RC, Howell A, Pestell RG, Smith J, Daniel R, Sotgia F, Lisanti MP (2011) Pyruvate kinase expression (PKM1 and PKM2) in cancer-associated fibroblasts drives stromal nutrient production and tumor growth. Cancer Biol Ther 12 [DOI] [PMC free article] [PubMed]
- 57.Sotgia F, Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Lisanti MP. Understanding the Warburg effect and the prognostic value of stromal caveolin-1 as a marker of a lethal tumor microenvironment. Breast Cancer Res. 2011;13:213. doi: 10.1186/bcr2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen SY, Huang YC, Liu SP, Tsai FJ, Shyu WC, Lin SZ. An overview of concepts for cancer stem cells. Cell Transplant. 2011;20:113–120. doi: 10.3727/096368910X532837. [DOI] [PubMed] [Google Scholar]
- 59.D’Angelo RC, Wicha MS. Stem cells in normal development and cancer. Prog Mol Biol Transl Sci. 2010;95:113–158. doi: 10.1016/B978-0-12-385071-3.00006-X. [DOI] [PubMed] [Google Scholar]
- 60.Gao JX. Cancer stem cells: the lessons from pre-cancerous stem cells. J Cell Mol Med. 2008;12:67–96. doi: 10.1111/j.1582-4934.2007.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menakuru SR, Brown NJ, Staton CA, Reed MW. Angiogenesis in pre-malignant conditions. Br J Cancer. 2008;99:1961–1966. doi: 10.1038/sj.bjc.6604733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dieter SM, Ball CR, Hoffmann CM, Nowrouzi A, Herbst F, Zavidij O, Abel U, Arens A, Weichert W, Brand K, Koch M, Weitz J, Schmidt M, von Kalle C, Glimm H (2011) Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell 9:357–65 LID—doi:10.1016/j.stem.2011.08.010 [DOI] [PubMed]
- 63.Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, Green J, Colman S, Piacibello W, Buckle V, Tsuzuki S, Greaves M, Enver T. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science (New York, N. Y.) 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 64.Davies EJ, Marsh V, Clarke AR (2011) Origin and maintenance of the intestinal cancer stem cell. Mol Carcinog 50:254–63 LID—doi:10.1002/mc.20631 [DOI] [PubMed]
- 65.Floor S, van Staveren WC, Larsimont D, Dumont JE, Maenhaut C (2011) Cancer cells in epithelial-to-mesenchymal transition and tumor-propagating-cancer stem cells: distinct, overlapping or same populations. Oncogene 30:4609–21 LID—doi:10.1038/onc.2011.184 [DOI] [PubMed]
- 66.Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, Tessema M, Leng S, Belinsky SA. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011;71:3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hotz HG, Hotz B, Buhr HJ. Genes associated with epithelial-mesenchymal transition: possible therapeutic targets in ductal pancreatic adenocarcinoma? Anticancer Agents Med Chem. 2011;11:448–454. doi: 10.2174/187152011795677436. [DOI] [PubMed] [Google Scholar]
- 68.Said NA, Williams ED. Growth factors in induction of epithelial-mesenchymal transition and metastasis. Cells Tissues Organs. 2011;193:85–97. doi: 10.1159/000320360. [DOI] [PubMed] [Google Scholar]
- 69.Hill RP, Marie-Egyptienne DT, Hedley DW. Cancer stem cells, hypoxia and metastasis. Semin Radiat Oncol. 2009;19:106–111. doi: 10.1016/j.semradonc.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 70.Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 71.Ping YF, Bian XW (2011) Consice review: Contribution of cancer stem cells to neovascularization Stem Cells (Dayton, Ohio) 29:888–94 LID—doi:10.1002/stem.650 [DOI] [PubMed]
- 72.Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe’er J, Trent JM, Meltzer PS, Hendrix MJ. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol. 1999;155:739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Vasculogenic mimicry and tumour-cell plasticity: lessons from melanoma. Nat Rev Cancer. 2003;3:411–421. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- 74.Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, Fligelman B, Leversha M, Brennan C, Tabar V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- 75.Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, Maria R. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 76.Hida K, Hida Y, Amin DN, Flint AF, Panigrahy D, Morton CC, Klagsbrun M. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 2004;64:8249–8255. doi: 10.1158/0008-5472.CAN-04-1567. [DOI] [PubMed] [Google Scholar]
- 77.Coffelt SB, Hughes R, Lewis CE. Tumor-associated macrophages: effectors of angiogenesis and tumor progression. Biochim Biophys Acta. 2009;1796:11–18. doi: 10.1016/j.bbcan.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 78.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Q, Cao Z, Shang B, Zhang G, Pan Y, Guo P (2011) The molecular and celluar progress of tumor-cell- mediated angiogenesis and vasculogenesis. Blood 118(21):1405–1406
- 80.Zhou Q, Kiosses WB, Liu J, Schimmel P. Tumor endothelial cell tube formation model for determining anti-angiogenic activity of a tRNA synthetase cytokine. Methods (San Diego, Calif.) 2008;44:190–195. doi: 10.1016/j.ymeth.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Q, Kapoor M, Guo M, Belani R, Xu X, Kiosses WB, Hanan M, Park C, Armour E, Do MH, Nangle LA, Schimmel P, Yang XL. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat Struct Mol Biol. 2010;17:57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Melero-Martin JM, Dudley AC (2011) Concise review: vascular stem cells and tumor angiogenesis. Stem Cells (Dayton, Ohio) 29:163–8 LID—doi:10.1002/stem.583 [DOI] [PMC free article] [PubMed]
- 83.Zhao Y, Bao Q, Renner A, Camaj P, Eichhorn M, Ischenko I, Angele M, Kleespies A, Jauch KW, Bruns C. Cancer stem cells and angiogenesis. Int J Dev Biol. 2011;55:477–482. doi: 10.1387/ijdb.103225yz. [DOI] [PubMed] [Google Scholar]
- 84.Charles N, Holland EC. The perivascular niche microenvironment in brain tumor progression. Cell Cycle (Georgetown, Tex.) 2010;9:3012–3021. doi: 10.4161/cc.9.15.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doan PL, Chute JP (2012) The vascular niche: home for normal and malignant hematopoietic stem cells. Leukemia 26:54–62 LID—doi:10.1038/leu.2011.236 [DOI] [PubMed]
- 86.Zhu TS, Costello MA, Talsma CE, Flack CG, Crowley JG, Hamm LL, He X, Hervey-Jumper SL, Heth JA, Muraszko KM, DiMeco F, Vescovi AL, Fan X. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71:6061–6072. doi: 10.1158/0008-5472.CAN-10-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Siveen KS, Kuttan G. Role of macrophages in tumour progression. Immunol Lett. 2009;123:97–102. doi: 10.1016/j.imlet.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 88.Palma M, Naldini L. Tie2-expressing monocytes (TEMs): novel targets and vehicles of anticancer therapy? Biochim Biophys Acta. 2009;1796:5–10. doi: 10.1016/j.bbcan.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 89.Ria R, Piccoli C, Cirulli T, Falzetti F, Mangialardi G, Guidolin D, Tabilio A, Renzo N, Guarini A, Ribatti D, Dammacco F, Vacca A. Endothelial differentiation of hematopoietic stem and progenitor cells from patients with multiple myeloma. Clin Cancer Res. 2008;14:1678–1685. doi: 10.1158/1078-0432.CCR-07-4071. [DOI] [PubMed] [Google Scholar]
- 90.Scavelli C, Nico B, Cirulli T, Ria R, Pietro G, Mangieri D, Bacigalupo A, Mangialardi G, Coluccia AM, Caravita T, Molica S, Ribatti D, Dammacco F, Vacca A. Vasculogenic mimicry by bone marrow macrophages in patients with multiple myeloma. Oncogene. 2008;27:663–674. doi: 10.1038/sj.onc.1210691. [DOI] [PubMed] [Google Scholar]
- 91.Raaijmakers MH. Niche contributions to oncogenesis: emerging concepts and implications for the hematopoietic system. Haematologica. 2011;96:1041–1048. doi: 10.3324/haematol.2010.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian RP, Scadden EO, Aung Z, Matza M, Merkenschlager M, Lin C, Rommens JM, Scadden DT. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu WR, Lin HS, Chen XY, Zhang Y. Yin-yang balance therapy on regulating cancer stem cells. J Tradit Chin Med. 2011;31:158–160. doi: 10.1016/S0254-6272(11)60032-0. [DOI] [PubMed] [Google Scholar]
- 95.Shang B, Cao Z, Zhou Q (2012) Progress in tumor vascular normalization for anticancer therapy: challenges and perspectives. Front Med 6(1):67–78 [DOI] [PubMed]
- 96.Ebos JM, Kerbel RS. Antiangiogenic therapy: impact on invasion, disease progression, and metastasis . Nat Rev Clin Oncol. 2011;8:210–221. doi: 10.1038/nrclinonc.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cutsem E, Lambrechts D, Prenen H, Jain RK, Carmeliet P. Lessons from the adjuvant bevacizumab trial on colon cancer: what next? J Clin Oncol. 2011;29:1–4. doi: 10.1200/JCO.2010.32.2701. [DOI] [PubMed] [Google Scholar]
- 98.Miles D, Harbeck N, Escudier B, Hurwitz H, Saltz L, Cutsem E, Cassidy J, Mueller B, Sirzen F. Disease course patterns after discontinuation of bevacizumab: pooled analysis of randomized phase III trials. J Clin Oncol. 2011;29:83–88. doi: 10.1200/JCO.2010.30.2794. [DOI] [PubMed] [Google Scholar]
- 99.Carmeliet P, Jain RK. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat Rev Drug Discov. 2011;10:417–427. doi: 10.1038/nrd3455. [DOI] [PubMed] [Google Scholar]
- 100.Goel S, Duda DG, Xu L, Munn LL, Boucher Y, Fukumura D, Jain RK. Normalization of the vasculature for treatment of cancer and other diseases. Physiol Rev. 2011;91:1071–1121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sato Y. Persistent vascular normalization as an alternative goal of anti-angiogenic cancer therapy. Cancer Sci. 2011;102:1253–1256. doi: 10.1111/j.1349-7006.2011.01929.x. [DOI] [PubMed] [Google Scholar]