Abstract
Hypermethylated in cancer 1(HIC1) was identified as a strong suppressor gene in chromosome region 17p13.3 telomeric to TP53. This gene encodes a transcriptional repressor and is ubiquitously expressed in normal tissues but downexpressed in different tumor tissues where it is hypermethylated. The hypermethylation of this chromosomal region leads to epigenetic inactivation of HIC1, which would prompt cancer cells to alter survival and signaling pathways or specific transcription factors during the period of tumorigenesis. In vitro, HIC1 function is mainly a sequence-specific transcriptional repressor interacting with a still growing range of histone deacetylase(HDAC)-dependent and HDAC-independent corepressor complexes. Furthermore, a role for HIC1 in tumor development is firmly supported by Hic1 deficient mouse model and two double heterozygote models cooperate with p53 and Ptch1. Notably, our findings suggest that potential factors derived from tumor microenviroment may play a role in modulating HIC1 expression in tumor cells by epigenetic modification, which is responsible for tumor progression. In this review, we will describe genomic and proteinic structure of HIC1, and summary the potential role of HIC1 in human various solid tumors and leukemia, and explore the influence of tumor microenviroment on inducing HIC1 expression in tumor cells.
Keywords: HIC1, Epigenetic modification, Tumor microenviroment, Tumor progression
Introduction
HIC1 (Hypermethylated in Cancer 1) was originally identified as a new candidate tumor suppressor gene located at 17p13.3 region telomeric to TP53 [1]. It is more clear that epigenetic changes, located in the chromosomal region 17p13.3, often show loss of heterozygosity or DNA hypermethylation that resides in CpG islands in area of promoter in various types of human solid tumors and leukemia. In the majority of cases, the hypermethylation of this chromosomal region leads to epigenetic inactivation of HIC1 [2]. This inactivation of HIC1 might impel cancer cells to alter survival and signaling pathways or lineage-specific transcription factors during the early stages of tumorigenesis. In addition, exogenously delivered HIC1 leads to a significant decrease in clonogenic survival in cancer cell lines[3]. Furthermore, an epigenetic inactivation role for HIC1 in tumor development is further supported by Hic1 deficient mice model [4] and two double heterozygote models that Hic1 can respectively cooperate with p53 or Ptch1 [5, 6]. Apart from HIC1 hypermethylation modification, several posttranslational regulatory mechanisms have been described for affecting its function. The first is glycosylation of the HIC1 protein that preferentially occurs in the DNA-binding domain but does not affect its specific DNA-binding activity. In addition, SUMOylation of the conserved lysine K314 in the central region of HIC1 reduces its repressive activity. The third is acetylation of the same K314 residue, which also can affect HIC1 transcriptional activity [2, 3].
HIC1 gene encodes a transcriptional repressor comprising three known functional domain, an N-terminal BTB/POZ domain, a C-terminal DNA binding domain containing five Krüppel-like C2H2 zinc fingers and the central region which may recruit the C-terminal binding proteins (CtBPs) for repression. This transcriptional repressor is in the presence of a p53 binding site in the 5’ flanking region and its identified target genes are involved in proliferation, tumor growth, angiogenesis and invasion, etc. Currently, many researchers have demonstrated some HIC1 target genes and some putative HIC1 tumor suppressor pathways as well as one target that is inactivated by HIC1 via so-called “HIC1 bodies” (HIC1 sequestered CtBPs to nuclear dot-like structures) [3, 7–9], which will be described below.
HIC1 Genomic Structure and Function Domain
HIC1 Gene Structure
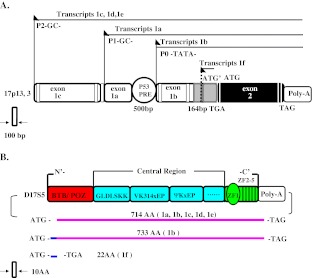
Currently, It is clearer that the exon-intron structures of the human and murine HIC1 genes appeared very similar [10, 11]. As show in Fig. 1A, the human HIC1 transcription can initiate at three separate promoters called as P0, P1 and P2, which give rise to three alternative first exons 1a, 1b and 1c, followed by a second coding exon contained the 3’ untranslated region. Exons 1a and 1c are associated with the major GC-rich promoters P1 and P2 respectively and both noncoding, whereas exon 1b is associated with the P0 TATA box promoter and partially coding. Exon 1b contains an ATG’ codon, which is in frame with the ATG initiation codon located in exon 2. Two new transcripts named 1d and 1e are rooted from additional alternative splice events in exon 1c, and both transcribed from promoter P2 [12]. The unspliced transcript 1f is initially translated from ATG’ within exon 1b, but ended at a TGA stop codon in the unspliced intron sequence. In brief, six HIC1 transcripts (1a–f) are generated from three different promoters through alternative splicing events. The HIC1 transcripts 1a, 1b and 1c were detected in various normal tissues with strong predominance of the exon 1a transcript and are upregulated by p53 and other p53 family members [1, 11]. The p53 responsive element in HIC1 is highly conserved in the intron position between exons 1a and 1b and in sequence among various species [13]. Unspliced HIC1 transcript 1f was discovered in human leukocytes, which may modulate HIC1 protein levels in cancer cells [14].
Fig. 1.
HIC1 genomic structure and function domain. A. Gene structure schema of human HIC1. HIC1 transcription can separately initiate at P0, P1 and P2 promoters that generates three alternative first exons 1a, 1b and 1c, followed by a second coding exon containing the 3’ untranslated region. The scale is 100 bp. Black color represents translated exons. p53 PRE represents p53 responsive element. B. Functional domains of HIC1 protein and possible isoforms. Splicing variants leading to the respective protein isoforms are indicated. The scale is 10 AA (amino acid)
HIC1 Protein Structure
As show in Fig. 1B, HIC1 gene splicing variants can lead to the respective protein isoform. The major 1a-type transcripts directly synthesize a 714 amino acids protein encoded by the ORF in exon 2. The 1b-type transcripts can encode an alternative putative protein containing 19 additional N-terminal amino acids (12 derived from exon 1b and 7 derived from 5’ sequences in exon 2) [11]. The unspliced transcript 1f might result in a 22 amino acid polypeptide due to a premature stop codon in the intron [14].
The N-terminal BTB/POZ of HIC1 protein stands for Broad complex, Tramtrack and Bric à brac/Poxviruses and Zinc finger domain of about 120 aminoacid, which is a dimerization domain known to play direct or indirect roles in protein-protein interactions through conformational effects. The region also contains an autonomous transcriptional repression domain [15–17]. The interaction domain is necessary for its binding on HIC1 responsive elements located in the promoter of its target genes. Recently, Pinte et al. have identified the sequence 5′-C/G NG C/G GGGCA C/A CC-3′ as an optimal HIC1 binding site (HiRE for HIC1-responsive element, GGCA consensus) [18]. It is reported that the B cell lymphoma 6(BCL6) BTB/POZ domain directly recruits nuclear corepressors SMRT, N-CoR or B-CoR/HDAC complexes in an exclusive manner [19, 20], but it is insensitive to trichostatin A (TSA), a specific inhibitor of class I and class II HDACs [21]. The HIC1 BTB/POZ domain is able to directly bind the class III HDAC(SIRT1) promoter to form a transcriptional repression complex to repress SIRT1’s transcription [22].
The C-terminal end region contains a cluster of four conserved C2H2 zinc fingers (ZF2-5). The region can bind a defined DNA sequence termed the HiRE and a more distant and isolated upstream Zinc finger motif (ZF1), which is conserved but is unlikely to contribute to DNA-binding [23]. The ZF2-5 are separated by the typical 7–8 amino acid conserved H/C links found in Krüppel-like Zinc fingers, likely to be involved in sequence-specific DNA binding [18].
The central region is a second autonomous transcriptional repression domain of the HIC1 [24] including 4 peptidic motifs perfectly conserved from human to zebrafish [25]. One of them, GLDLSKK motif, is found in proteins interacting with the co-repressor CtBP, thus extending the CtBP binding site [26, 27]. However, this repression domain exhibits both CtBP-dependent and CtBP-independent repression mechanisms both sensitive to TSA. The second conserved motif is an YRWM/VK314xEP motif on which it contains a potential SUMOylation consensus site of ψKxE. SUMOylation of K314 does not affect HIC1 subnuclear localization and interaction between HIC1 with CtBP, HDAC4 and SIRT1, but down-regulate the transcription. The third ψKxEP motif with the proline residue conserved from human to zebrafish is relevant to the G/SKxxP consensus motif for acetylation by CBP/P300. It is shown that HIC1 is acetylated on various lysine residues including K314. The ψKxEP motif is an acetylation/SUMOylation switch, which is related to the fourth conserved ψKxEPxxSP. Phosphorylation-regulated SUMOylation-acetylation switch motif (SAS) is found in the major myocyte enhancer factor2 (MEF2) isoforms[24, 28, 29].
HIC1 by Epigenetic Modification Modulates Tumor Progression
HIC1 Promoter is Hypermethylated Frequently in Cancers
It is well known that cancer initiation and progression are modulated by both genetic and epigenetic events. The epigenetic mechanisms that alter gene expression, but don’t alter the primary DNA sequence include changes in DNA methylation, histone modifications and small noncoding microRNAs (miRNA) et al. Disruption of epigenetic processes can lead to altered gene function and malignant cellular transformation. Actually, aberrant epigenetic modifications are widely described as the essential players in cancer progression [30].
It has been widely suggested that HIC1 is a tumor suppressor gene epigenetically silenced in primary tumors and some hematological malignancies, for example in prostate cancer (PCa) [31], non-small cell lung cancer [32, 33], breast cancer [34], gastric and liver cancer [35, 36], esophageal cancer [37, 38], human male non-seminomatous germ cell tumor [39] and the most common malignant brain tumor of childhood medulloblastoma [40–42] and the glial malignancy, ependymoma [43]. After being used HIC1 probe as well as more sensitive and informative assays such as methylation-specific PCR (MSP) and bisulfite sequencing, HIC1 has been frequently observed to be hyper-methylated in human various solid tumors and leukemia. It is generally assumed that hypermethylation of the HIC1 promoter region leads to silencing of HIC1 gene expression [1, 34, 36, 44] and overall HIC1 expression levels were decreased during the development of cancer.
However, HIC1 promoter hypermethylation was also found in normal brain tissue of children [40], in adult brain [45] and in prostate epithelium [31]. Moreover, HIC1 is only rarely methylated in acute leukemia at diagnosis (10 %) and in the chronic-phase of chronic myelogenous leukemia CML (50 %)[46, 47]. But HIC1 is found methylated in all recurrent acute lymphocytic leukemia and in the blast-crisis of all CML. Thus, HIC1 hypermethylation has been considered to be a late event in hematopoietic neoplasm and suggests that other inhibitory mechanism may exist to lower HIC1 expression.
The Potential Targets by HIC1
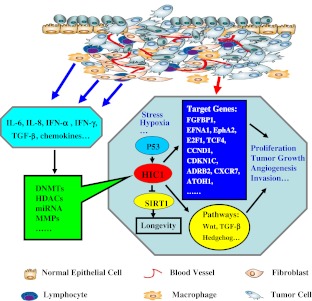
Interestingly, some HIC1 critical target genes and pathways have been identified as shown in Fig. 2. An important transcriptional target of HIC1 identified is the silent mating type information regulation 2 homolog 1 (SIRT1) deacetylase[22, 48]. SIRT1 can act on p53 through deacetylation as a result of attenuating its ability to activate downstream targets involved in regulation of apoptosis and/or proliferation. It has been reported that under normal physiological conditions HIC1 represses SIRT1 transcription and therefore inhibits p53 deacetylation, but in tumor cells HIC1 is inactivated by epigenetic modification, which induces the increased SIRT1 levels. It leads to deacetylation and therefore inactivation of p53 activity and allows cells to bypass apoptosis and survive avoiding from DNA damage. Moreover, the PRE (p53 responsive element) identified in the promoter region of HIC1 suggest that HIC1 is a direct p53 target gene [49] and p53 can activate the transcription of HIC1, independently of its methylation status. Therefore, this HIC1/SIRT1/p53 regulatory loop is an essential pathway through HIC1 may function as a tumor suppressor gene and cooperate with p53 [50].
Fig. 2.
Modulating HIC1 expression in tumor cell. Responding to some outer stress such as DNA damage factors and hypoxia, putative HIC1 critical target genes and pathways have been identified. Notably, some potential stromal cells and inflammatory cells within tumor microenvironment secrete a large amount factors, including chemokines, cytokines, et al., which are capable of silencing certain tumor suppressor genes including HIC1 and reshaping the phenotypes of tumor cells by modulating its activity of DNMTs, HDACs or miRNA, et al. It is also responsible for proliferation, tumor growth, angiogenesis and invasion, etc. FGFBP1, fibroblast growth factor binging protein; EFNA1, ephrin-A1; EphA2, tyrosine kinase receptor; E2F1, cell cycle and apoptosis regulator gene; TCF4, T-cell-specific transcription factor 4; CCND1, Cyclin D1; CDKN1C, cell cycle inhibitor P57KIP2; ADRB2, β-2 adrenergic receptor; CXCR7, chemokine (CXC motif) receptor 7; ATOH1, atonal homolog1
Briones et al. found that the core binding sequence for HIC1 is GGCA, and this sequence is present in the fibroblast growth factor binging protein (FGF-BP1) promoter located in the region from −785 to −782 bp (GGCA). The authors showed that mutation of this HIC1 core binding site in the FGF-BP1 promoter markedly impaired the transcriptional repression mediated by transforming growth factor-β (TGF-β). TGF-β is the prototypical member of a family of growth factors that play important roles in normal vascular development and human diseases. The data implicated that HIC1 is involved in the transcriptional repression of FGF-BP1 by TGF-β signaling regulation of angiogenesis in cancer [51]. In brief, inactivation of HIC1 in tumour cells may increase expression of FGF-BP1, resulting in an increase in angiogenesis and/or proliferation.
Zhang W. et al. found that HIC1 could regulate a direct transcriptional repressor of ephrin-A1(EFNA1) gene, a cell surface ligand for Eph receptors implicated in the pathogenesis of epithelial cancers [52]. The data showed that mouse embryos lacking both Hic1 alleles manifested developmental defects spatially relative to mis-expression of ephrin-A1. Overexpression of ephrin-A1 is a feature of tumors arising in Hic1 heterozygous. Restoration of HIC1 function in breast cancer cells could lead to a reduction in tumor growth in vivo, and the effect could be partially rescued by co-overexpression of ephrin-A1. It was concluded that the epigenetic silencing of HIC1 in cancer cells can result in upregulated expression of ephrin-A1 in favour of tumor growth in vivo. As a newest direct target gene of HIC1, the tyrosine kinase receptor EphA2 was identified and whose ligand ephrin-A1 is also a HIC1 target gene [53]. The ectopic expression of HIC1 in the highly malignant MDA-MB-231 breast cancer cell line severely impaired cell proliferation, migration and invasion. Inactivation of endogenous HIC1 through RNA interference in normal breast epithelial cells resulted in upregulating EphA2 and increasing cellular migration. Therefore, loss of the regulation of this Eph pathway through HIC1 epigenetic silencing may be an important mechanism in the pathogenesis of epithelial cancers [53].
Recently, Zhang B. et al. identified the E2F1 gene encodes an HIC1-binding consensus sites (GGCA) in its promoters and HIC1 targets E2F-responsive genes for transcriptional regulation and growth suppression. Brg1 is as a central component of the SWI/SNF chromatin-remodeling family and is required for the transcriptional regulation of multiple cell cycle control-related genes, including E2F-responsive promoters. They found that HIC1 can recruit Brg1 to E2F-responsive promoters and its transcriptional repression of these genes is dependent upon Brg1. These data indicated that HIC1 is a central molecule in a novel mechanism controlling cell growth. The disruption of this HIC1-mediated pathway may lead to abnormal cell proliferation and cancer ultimately. Thus inactivation of HIC1 leads to undue activation of E2F signaling, favoring cell cycle progression and tumor growth [54].
Interestingly, Mathias et al. found that HIC1 is a new transcriptional target of the cell cycle and apoptosis regulator E2F1. E2F1 induces HIC1 via two E2F DNA binding sites within the TATA-box containing HIC1 P0 promoter. E2F1 binds to this HIC1 promoter region in vivo and induces endogenous HIC1 mRNA expression. Furthermore, HIC1 expression is induced by E2F1 regardless of HIC1 P0 promoter hypermethylation [55]. These findings imply that there is a regulation loop between E2F1 and HIC1. Potentially, endogenous E2F1 protein directly combines with the HIC1 P0 promoter to activate HIC1 expression in DNA damage responses to etoposide treatment; in return, HIC1 binds to E2F1 promoter gene that contains HIC1-binding consensus sites, which suppress E2F1 transcription reducing cell growth.
Notably, it has been reported that HIC1 can directly target T-cell-specific transcription factor 4 (TCF4), a positive cell cycle regulator frequently amplified in tumors called Cyclin D1 (CCND1) and the cell cycle inhibitor P57KIP2 (CDKN1C) [26, 56]. TCF4 interacts with β-catenin in active Wnt signaling and co-activates downstream target genes. This activity is important during normal development, but its deregulation plays a pivotal role in cancer progression. HIC1 also sequestered TCF4 as well as TCF4 bound β-catenin via CtBP to“HIC1-bodies”and thereby attenuated Wnt signaling. This may suppress tumor formation, since TCF4 and β-catenin are prevented from activating TCF-responsive genes possibly involved in tumor development, such as c-Myc (MYC) or Cyclin D1[57]. This apparent contradiction that HIC1 represses a cell cycle accelerator Cyclin D1 and inhibitor P57KIP2, may be explained by the observation that low levels of P57KIP2 are able to promote cyclin/CDK complex formation and thus cell cycle progression[58].
More recently, through promoter luciferase activity, ChIP and sequential ChIP experiments, β-2 adrenergic receptor (ADRB2) is demonstrated as a direct target gene of HIC1[59]. ADRB2 encodes a G-protein-coupled-receptor (GPCR) activated by adrenaline/noradrenaline. ADRB2 promoter was shown to be presented in many putative HiRE, particularly 600 bp upstream of the translation start site. Overexpression of HIC1 in WI-38 normal lung embryonic fibroblasts by retroviral infection induced a marked decrease of ADRB2 mRNA and a slight decrease of protein levels. Conversely, inhibition of endogenous HIC1 expression in WI-38 cells by siRNA resulted in a concomitant increase in ADRB2 transcripts and proteins. In MDA-MB-231, a metastatic breast cancer cell line expressing high levels of ADRB2, HIC1 re-expression strongly repressed ADRB2 expression and prevented its activation of migration and invasion. These data suggest that loss of HIC1 in tumorigenesis may favor metastasis through upregulation of ADRB2 in breast epithelial cells.
Recently, Capucine et al. have found scavenger chemokine receptor 7 (CXCR7) was downregulated in HIC1-deficient U2OS osteosarcoma cells transduced with an adenoviral HIC1-expressing vector. In WI38 cells, some assays showed that endogenous HIC1 binds to HIC1-responsive elements in the CXCR7 promoters to repress its expression [60]. Our initial findings suggest that HIC1 were abundantly methylated in plasma and tissues of PCa patient compared with those of normal control patient. Similar results were observed in PCa cell lines. In vitro assays, restoring HIC1 expression in PCa cells markedly inhibited proliferation, migration, and invasion and induced the apoptosis in these cells. Moreover, mice bearing PCa cells with overexpression of HIC1 had a significant effect on reducing tumor growth and osseous metastases. Notably, we also identified chemokine receptor CXCR7 is a potential downstream target gene of HIC1 in PCa cells by microarray and ChIP assays. Therefore, CXCR7 promoter in PCa cells is reversely regulated by HIC1, which may contribute to PCa progression (our unpublished data).
The Function of HIC1 in Mice Models
The findings from the constructed Hic1 deficient mice demonstrated that some interesting clues to HIC1 function as a tumor suppressor [4]. Two double heterozygote models have shown that Hic1 can respectively cooperate with p53 or Ptch1 in tumorigenesis [5, 6]. Homozygous disruption of Hic1 impairs development and results in embryonic and perinatal lethality [10], while heterozygous Hic1+/− mice develop an age- and gender-dependent spectrum of malignant tumors with 44 % of epithelial cancers [4]. By bisulfite genomic sequencing of human, mice tumor cell lines and normal tissues, Chen et al. found that the reason of function loss of the remaining Hic1 allele in heterozygous mice is not gross chromosomal deletions but hypermethylation of major alternative promoters of the gene in human cancers.
Actually, Hic1 and p53 are located on the same chromosome in humans (chromosome 17) and mice (chromosome 11). The remaining p53 and Hic1 wild-type alleles are simultaneously deleted due to loss of the entire chromosome in these cis tumors. But the trans Hic1+/− p53+/− mice have no acceleration of tumorigenesis as compared to p53+/− mice. The remaining Hic1 allele is retained and epigenetically silenced by hypermethylation in the trans tumors, while the p53 allele undergoes interstitial deletion [5].
In Ptch1+/−/Hic1+/− heterozygous knockout mice, the data showed a markedly increased incidence of medulloblastoma as compared to Ptch1+/− heterozygous mice. The Patched (Ptch)1 is a tumor suppressor, capable of inhibiting Hedgehog(HH) signaling. Ptch1+/−/Hic1+/− heterozygous knockout mice showed a fourfold increased incidence of medulloblastoma as compared to Ptch1+/− heterozygous mice [6]. Moreover, it was found that the proneural transcription factor Atonal Homolog 1 (Atoh1) is directly suppressed by Hic1. Atoh1 is a putative target of Hh signaling and also essential for cerebellar growth and development. Briggs et al. concluded that Hic1 and Ptch1 tumor suppressors cooperate to silence Atoh1 expression during a critical phase in the granule cell precursors (GCPs) of the cerebellum differentiation in which malignant transformation may lead to medulloblastoma [61]. The Ptch1+/− heterozygotes spontaneously develop medulloblastomas at a frequency of 10–15 % [62]. However, deletion of chromosome 17p occurs in up to 50 % of the cases and is frequently restricted to the 17p13.1-13.3 region containing several tumor suppressor genes including p53, REN, MNT and HIC1 [63]. Loss of function of p53 significantly increases the frequency of medulloblastomas occurring in Ptch1+/− p53+/− animals as compared to Ptch1+/− heterozygote [6, 64]. Apart from this increased incidence, the time frame and the type of tumors did not very vary between these two cohorts, in striking contrast with the p53+/− Hic1+/− heterozygote where distinct tumors appear earlier than in the p53+/− cohort [5].
HIC1 is a Marker of Cancer Stem Cell
Many studies have highlighted the key roles of epigenetic signatures in stem-cell identity [65]. In pluripotent embryonic stem cells (ES), the promoters of some developmental transcription factors are in a “bivalent state” with both activating (H3K9Ac and H3K4me) and repressive (H3K27me) epigenetic marks. Thus, these promoters are “poised” in a transcription-ready state which, depending on the developmental cues, can be tipped toward activation of tissue-specific genes or reversible silencing of genes involved in other developmental pathways. Indeed, in differentiated cells, the promoters of many non-transcribed genes have lost the activating marks and are enriched with the H3K27 trimethylated mark which is deposited by the Polycomb repressor complex 2 (PRC2). Some studies in ES cells have determined that the epigenetic status of restricted lists of genes defined as tumor suppressor genes are frequently hypermethylated in colon, breast and ovarian cancer pre-marked with trimethylated H3K27 and other PRC2 components in ES cells [66, 67].
HIC1 has been defined as a “stem cell gene” because the gene can be marked with at least two out of the three components SUZ12, EED and H3K27me3 in human ES cells in colorectal cancer, ovarian cancer and CD34 positive hematopoietic progenitor cells [67]. But this exception in breast cancer may be due to its hemi-methylation in normal breast epithelium [34]. In addition, HIC1 is unmethylated in human ES cells and partially or fully methylated in two embryonal carcinomas cell lines, Tera-1 and Tera-2 respectively [68]. However, to be fully understood, the function of HIC1 in cancer stem cell is warranty investigated.
Modulating HIC1 Expression by the Tumor Microenvironment
DNA Methylation Status of Specific Gene Can Be Involved in Formation of Tumor Microenvironment
These findings suggest that DNA methylation is inherited upon somatic cell division, and methylation of a promoter CpG island silences its downstream gene. Altering methylation in cancer cells can cause inactivation of both tumor-suppressor genes (driver) and other genes (passenger), and by global hypomethylation [69]. Many studies revealed that aberrant methylation is presented even in non-cancerous tissues, but its level is associated with cancer risk in an epigenetic field for cancerization. Quantification of methylation revealed that aberrant methylation can be induced much more frequently than mutations [70], and it has already been indicated that methylation alterations are involved in epithelial-mesenchymal transition (EMT) [71, 72]. The high frequency of methylation alterations also suggested that they could involve in phenotypic changes of stromal cells, and thus formation of cancer microenvironment [73, 74]. DNA methylation alterations are likely to be important players not only in transformation of epithelial cells but also in the formation of cancer microenvironment by stromal cells, which indicated that DNA methylation of specific genes can be induced in a significant fraction of cells even in polyclonal tissue. There is a possibility that altered methylation statuses may involve in formation of tumor microenvironments. Notably, epigenetic silencing of specific genes due to changes in histone modification has been reported in tumor endothelial cells [75]. Mesenchymal cells produced by EMT of tumor cells may contribute to formation of tumor microenvironments [76].
Tumor Microenvironment May Modulate HIC1 Expression of Tumor Cell
As mentioned above, HIC1 as suppressor gene is not mutated in observed solid tumors but epigenetically mediated loss of function may help drive key stages of tumorigenesis. Despite multiple epigenetic modifications, including glycosylation, sumoylation, acetylation, HIC1 is often hypermethylated in 50 % or more of such tumors due in a large part to the enhanced DNMTs activity by multiple factors [1, 2, 7]. Xia et al. recently found that prostaglandin E2 (PGE2) silences certain tumor-suppressor and DNA-repair genes through enhancing the CGI methylation in the promoters of these genes to promote intestinal tumor growth [77]. Ng et al. indicated that microRNA-143 regulates DNA methyltransferases 3A in colorectal cancer. These findings prompted us to postulate whether some factors derived from tumor microenviroment also may modulate HIC1 expression by epigenetic modifications. Indeed, the data suggested that different tumor environmental cues influence epigenetic modification of histones or DNA and alter access of transcription factors (TFs) to the DNA sequence, thereby affecting gene expression [78, 79]. Moreover, our initial findings indicate that some inflammatory factors and microRNAs secreted by stromal cells within tumor microenviroment are capable of increasing the activity of DNA methyltransferases (DNMTs) and histone deacetylased (HDACs) of tumor cells, therefore silencing certain tumor suppressor genes including HIC1 and reshaping the phenotypes of tumor cells (Our unpublished data) as shown in Fig. 2. So far, the antitumoral properties of novel epigenetic therapies have largely been attributed to the reactivation of silenced tumor-suppressor genes in tumor cells [80, 81]. However, given their universal gene regulatory loops, it is pivotal that epigenetic therapy will also pay a more attention to the role of tumor microenviroment. It may provide more novel options in cancer treatment.
Conclusions and Prospects
HIC1 is a central transcriptional regulator of a few key genes controlling cell growth as well as death in response to p53-dependent apoptotic DNA damage through binding to SIRT1 promoter. By combining with PATCHED, HIC1 may play a inhibitory role in Hedgehog pathway for medulloblastomas and capable of regulating Wnt pathway involved in function of stem cell. In fact, HIC1 is frequently hypermethylated as a result of silence or low-level in a variety of solid tumors and leukemia, therefore making it a new therapeutic target for DNA methyltransferase inhibitors such as 5-Aza-2′-deoxycytidine(decitabine) [82].
However, some intriguing questions of HIC1 still remain unclear. Firstly, how does transcription modification by HIC1 protein occur in the target gene DNA-binding domain. Secondly, given many potential physiological roles of HIC1, the number of HIC1-characterized target genes appears to be a small amount. Thirdly, it is not clear how tumor microenvironment modulate HIC1 expression of tumor cells. Finally, other inhibitory mechanisms other than hypermethylation of the HIC1 promoter may exist due to the findings that low HIC1 levels contribute to cancer development.
In summary, further exploring HIC1 function and its characterized targets would not only help to understand its role as a tumor suppressor gene and provide new insights into epigenetic and tumor microenviroment in general but would also offer some clues of new therapeutic approaches to major human tumors.
Acknowledgements
We apologize to the many authors whose excellent work we could not cite owing to space limitation. Research in the authors’ laboratory is supported by National Natural funding of China (81071747), National Basic Research Program of China (973 Program, 2011CB510106, and 2011CB504300), Shanghai Education Committee Key Discipline and Specialties Foundation Project Number: J50208, Program for Professor of Special Appointment (Eastern Scholar for J. W.),Shanghai Pujiang Program (10PJ1406400), Ph.D innovation fund from Shanghai Jiao Tong University School of Medicine (BXJ201103) and Shanghai Natural Science Foundation (11ZR1419600).
References
- 1.Wales MM, et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1(6):570–577. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- 2.Fleuriel C, et al. HIC1 (hypermethylated in cancer 1) epigenetic silencing in tumors. Int J Biochem Cell Biol. 2009;41(1):26–33. doi: 10.1016/j.biocel.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenal M, et al. (2010) Inactivation of the hypermethylated in cancer 1 tumour suppressor—not just a question of promoter hypermethylation? Swiss Med Wkly 140(w13106) [DOI] [PubMed]
- 4.Chen WY, et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat Genet. 2003;33(2):197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, et al. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Canc Cell. 2004;6(4):387–398. doi: 10.1016/j.ccr.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 6.Briggs KJ, et al. Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev. 2008;22(6):770–785. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dehennaut V, Leprince D. Implication of HIC1 (hypermethylated in cancer 1) in the DNA damage response. Bull Canc. 2009;96(11):E66–E72. doi: 10.1684/bdc.2009.0959. [DOI] [PubMed] [Google Scholar]
- 8.Boulay G, et al. (2011) Loss of hypermethylated in cancer 1 (HIC1) in breast cancer cells contributes to stress induced migration and invasion through beta-2 adrenergic receptor (ADRB2) misregulation. J Biol Chem [DOI] [PMC free article] [PubMed]
- 9.Foveau B, et al. (2012) Receptor tyrosyne kinase Epha2 is a direct target-gene of Hic1 (hypermethylated in cancer 1). J Biol Chem [DOI] [PMC free article] [PubMed]
- 10.Carter MG, et al. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller-Dieker syndrome. Hum Mol Genet. 2000;9(3):413–419. doi: 10.1093/hmg/9.3.413. [DOI] [PubMed] [Google Scholar]
- 11.Guerardel C, et al. Identification in the human candidate tumor suppressor gene HIC-1 of a new major alternative TATA-less promoter positively regulated by p53. J Biol Chem. 2001;276(5):3078–3089. doi: 10.1074/jbc.M008690200. [DOI] [PubMed] [Google Scholar]
- 12.Pinte S, et al. Identification of a second G-C-rich promoter conserved in the human, murine and rat tumor suppressor genes HIC1. Oncogene. 2004;23(22):4023–4031. doi: 10.1038/sj.onc.1207504. [DOI] [PubMed] [Google Scholar]
- 13.Britschgi C, et al. Identification of the p53 family-responsive element in the promoter region of the tumor suppressor gene hypermethylated in cancer 1. Oncogene. 2006;25(14):2030–2039. doi: 10.1038/sj.onc.1209240. [DOI] [PubMed] [Google Scholar]
- 14.Mondal AM, et al. Identification and functional characterization of a novel unspliced transcript variant of HIC-1 in human cancer cells exposed to adverse growth conditions. Cancer Res. 2006;66(21):10466–10477. doi: 10.1158/0008-5472.CAN-06-0352. [DOI] [PubMed] [Google Scholar]
- 15.Albagli O, et al. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6(9):1193–1198. [PubMed] [Google Scholar]
- 16.Stogios PJ, et al. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6(10):R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly KF, Daniel JM. POZ for effect–POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16(11):578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Pinte S, et al. The tumor suppressor gene HIC1 (hypermethylated in cancer 1) is a sequence-specific transcriptional repressor: definition of its consensus binding sequence and analysis of its DNA binding and repressive properties. J Biol Chem. 2004;279(37):38313–38324. doi: 10.1074/jbc.M401610200. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad KF, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12(6):1551–1564. doi: 10.1016/S1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 20.Ghetu AF, et al. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell. 2008;29(3):384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deltour S, Guerardel C, Leprince D. Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: the case of HIC-1 and gammaFBP-B. Proc Natl Acad Sci U S A. 1999;96(26):14831–14836. doi: 10.1073/pnas.96.26.14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen WY, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123(3):437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Deltour S, et al. The carboxy-terminal end of the candidate tumor suppressor gene HIC-1 is phylogenetically conserved. Biochim Biophys Acta. 1998;1443(1–2):230–232. doi: 10.1016/s0167-4781(98)00219-x. [DOI] [PubMed] [Google Scholar]
- 24.Stankovic-Valentin N, et al. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27(7):2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertrand S, et al. Identification and developmental expression of the zebrafish orthologue of the tumor suppressor gene HIC1. Biochim Biophys Acta. 2004;1678(1):57–66. doi: 10.1016/j.bbaexp.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Deltour S, et al. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK motif. Mol Cell Biol. 2002;22(13):4890–4901. doi: 10.1128/MCB.22.13.4890-4901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39(9):1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 28.Shalizi A, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311(5763):1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 29.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8(12):947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 30.Kanwal R, Gupta S (2011) Epigenetic modifications in cancer. Clin Genet [DOI] [PMC free article] [PubMed]
- 31.Morton RA, Jr, et al. Hypermethylation of chromosome 17P locus D17S5 in human prostate tissue. J Urol. 1996;156(2 Pt 1):512–516. doi: 10.1097/00005392-199608000-00073. [DOI] [PubMed] [Google Scholar]
- 32.Eguchi K, et al. DNA hypermethylation at the D17S5 locus in non-small cell lung cancers: its association with smoking history. Cancer Res. 1997;57(21):4913–4915. [PubMed] [Google Scholar]
- 33.Hayashi M, et al. Reduced HIC-1 gene expression in non-small cell lung cancer and its clinical significance. Anticancer Res. 2001;21(1B):535–540. [PubMed] [Google Scholar]
- 34.Fujii H, et al. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 1998;16(16):2159–2164. doi: 10.1038/sj.onc.1201976. [DOI] [PubMed] [Google Scholar]
- 35.Kanai Y, et al. DNA hypermethylation at the D17S5 locus is associated with gastric carcinogenesis. Cancer Lett. 1998;122(1–2):135–141. doi: 10.1016/S0304-3835(97)00380-7. [DOI] [PubMed] [Google Scholar]
- 36.Kanai Y, et al. DNA hypermethylation at the D17S5 locus and reduced HIC-1 mRNA expression are associated with hepatocarcinogenesis. Hepatology. 1999;29(3):703–709. doi: 10.1002/hep.510290338. [DOI] [PubMed] [Google Scholar]
- 37.Huang J, et al. High frequency allelic loss on chromosome 17p13.3-p11.1 in esophageal squamous cell carcinomas from a high incidence area in northern China. Carcinogenesis. 2000;21(11):2019–2026. doi: 10.1093/carcin/21.11.2019. [DOI] [PubMed] [Google Scholar]
- 38.Eads CA, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61(8):3410–3418. [PubMed] [Google Scholar]
- 39.Koul S, et al. (2002) Characteristic promoter hypermethylation signatures in male germ cell tumors. Mol Cancer 1(8) [DOI] [PMC free article] [PubMed]
- 40.Rood BR, et al. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62(13):3794–3797. [PubMed] [Google Scholar]
- 41.Rathi A, et al. Aberrant methylation of the HIC1 promoter is a frequent event in specific pediatric neoplasms. Clin Cancer Res. 2003;9(10 Pt 1):3674–3678. [PubMed] [Google Scholar]
- 42.Waha A, et al. Epigenetic silencing of the HIC-1 gene in human medulloblastomas. J Neuropathol Exp Neurol. 2003;62(11):1192–1201. doi: 10.1093/jnen/62.11.1192. [DOI] [PubMed] [Google Scholar]
- 43.Waha A, et al. Analysis of HIC-1 methylation and transcription in human ependymomas. Int J Cancer. 2004;110(4):542–549. doi: 10.1002/ijc.20165. [DOI] [PubMed] [Google Scholar]
- 44.Nishida N, et al. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47(3):908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlmann K, et al. Distinct methylation profiles of glioma subtypes. Int J Cancer. 2003;106(1):52–59. doi: 10.1002/ijc.11175. [DOI] [PubMed] [Google Scholar]
- 46.Issa JP, Baylin SB, Herman JG. DNA methylation changes in hematologic malignancies: biologic and clinical implications. Leukemia. 1997;11(Suppl 1):S7–S11. [PubMed] [Google Scholar]
- 47.Issa JP, et al. HIC1 hypermethylation is a late event in hematopoietic neoplasms. Cancer Res. 1997;57(9):1678–1681. [PubMed] [Google Scholar]
- 48.Huffman DM, et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67(14):6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 49.Stocklein H, et al. Detailed mapping of chromosome 17p deletions reveals HIC1 as a novel tumor suppressor gene candidate telomeric to TP53 in diffuse large B-cell lymphoma. Oncogene. 2008;27(18):2613–2625. doi: 10.1038/sj.onc.1210901. [DOI] [PubMed] [Google Scholar]
- 50.Tseng RC, et al. Distinct HIC1-SIRT1-p53 loop deregulation in lung squamous carcinoma and adenocarcinoma patients. Neoplasia. 2009;11(8):763–770. doi: 10.1593/neo.09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Briones VR, et al. Mechanism of fibroblast growth factor-binding protein 1 repression by TGF-beta. Biochem Biophys Res Commun. 2006;345(2):595–601. doi: 10.1016/j.bbrc.2006.04.052. [DOI] [PubMed] [Google Scholar]
- 52.Zhang W, et al. A potential tumor suppressor role for Hic1 in breast cancer through transcriptional repression of ephrin-A1. Oncogene. 2010;29(17):2467–2476. doi: 10.1038/onc.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foveau B, et al. The receptor tyrosine kinase EphA2 is a direct target gene of hypermethylated in cancer 1 (HIC1) J Biol Chem. 2012;287(8):5366–5378. doi: 10.1074/jbc.M111.329466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B, et al. Requirement for chromatin-remodeling complex in novel tumor suppressor HIC1-mediated transcriptional repression and growth control. Oncogene. 2009;28(5):651–661. doi: 10.1038/onc.2008.419. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Jenal M, et al. The tumor suppressor gene hypermethylated in cancer 1 is transcriptionally regulated by E2F1. Mol Canc Res. 2009;7(6):916–922. doi: 10.1158/1541-7786.MCR-08-0359. [DOI] [PubMed] [Google Scholar]
- 56.Valenta T, et al. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. EMBO J. 2006;25(11):2326–2337. doi: 10.1038/sj.emboj.7601147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rechem C, et al. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol Cell Biol. 2010;30(16):4045–4059. doi: 10.1128/MCB.00582-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pateras IS, et al. p57KIP2: Kipping the cell under control. Mol Canc Res. 2009;7(12):1902–1919. doi: 10.1158/1541-7786.MCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 59.Boulay G, et al. Loss of hypermethylated in cancer 1 (HIC1) in breast cancer cells contributes to stress-induced migration and invasion through beta-2 adrenergic receptor (ADRB2) misregulation. J Biol Chem. 2012;287(8):5379–5389. doi: 10.1074/jbc.M111.304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rechem C, et al. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target gene of HIC1 (hypermethylated in cancer 1) J Biol Chem. 2009;284(31):20927–20935. doi: 10.1074/jbc.M109.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marcotullio L, et al. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci U S A. 2004;101(29):10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodrich LV, et al. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277(5329):1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 63.Ferretti E, et al. Hedgehog checkpoints in medulloblastoma: the chromosome 17p deletion paradigm. Trends Mol Med. 2005;11(12):537–545. doi: 10.1016/j.molmed.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 64.Wetmore C, Eberhart DE, Curran T. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res. 2001;61(2):513–516. [PubMed] [Google Scholar]
- 65.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8(4):263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 66.Lee TI, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 68.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39(2):237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalari S, Pfeifer GP (2010) Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv Genet 70(277–308) [DOI] [PMC free article] [PubMed]
- 70.Saito K, et al. Aberrant methylation status of known methylation-sensitive CpG islands in gastrointestinal stromal tumors without any correlation to the state of c-kit and PDGFRA gene mutations and their malignancy. Canc Sci. 2008;99(2):253–259. doi: 10.1111/j.1349-7006.2007.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dumont N, et al. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci U S A. 2008;105(39):14867–14872. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 73.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 74.Qiu W, et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40(5):650–655. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hellebrekers DM, et al. Identification of epigenetically silenced genes in tumor endothelial cells. Cancer Res. 2007;67(9):4138–4148. doi: 10.1158/0008-5472.CAN-06-3032. [DOI] [PubMed] [Google Scholar]
- 76.Jing Y, et al. (2011) Epithelial-mesenchymal transition in tumor microenvironment. Cell Biosci 1(29) [DOI] [PMC free article] [PubMed]
- 77.Xia D, et al. Prostaglandin E(2) promotes intestinal tumor growth via DNA methylation. Nat Med. 2012;18(2):224–226. doi: 10.1038/nm.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng EK, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101(4):699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Claes B, Buysschaert I, Lambrechts D. Pharmaco-epigenomics: discovering therapeutic approaches and biomarkers for cancer therapy. Heredity (Edinb) 2010;105(1):152–160. doi: 10.1038/hdy.2010.42. [DOI] [PubMed] [Google Scholar]
- 80.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Canc Inst. 2005;97(20):1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 81.Zhou P, Lu Y, Sun XH (2012) Effects of a novel DNA methyltransferase inhibitor zebularine on human lens epithelial cells. Mol Vis 18(22–28) [PMC free article] [PubMed]
- 82.Christman JK. 5-azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]