Abstract
The hierarchical heterogeneous architecture of bone imposes significant challenges to structural and dynamic studies conducted by traditional biophysical techniques. High-resolution solid-state nuclear magnetic resonance (SSNMR) spectroscopy is capable of providing detailed atomic-level structural insights into such traditionally challenging materials. However, the relatively long data-collection time necessary to achieve a reliable signal-to-noise ratio (S/N) remains a major limitation for the widespread application of SSNMR on bone and related biomaterials. In this study, we attempt to overcome this limitation by employing the paramagnetic relaxation properties of copper(II) ions to shorten the 1H intrinsic spin-lattice (T1) relaxation times measured in natural-abundance 13C cross-polarization (CP) magic-angle-spinning (MAS) NMR experiments on bone tissues for the purpose of accelerating the data acquisition time in SSNMR. To this end, high-resolution solid-state 13C CPMAS experiments were conducted on type I collagen (bovine tendon), bovine cortical bone, and demineralized bovine cortical bone, each in powdered form, to measure the 1H T1 values in the absence and in the presence of 30 mM Cu(II)(NH4)2EDTA. Our results show that the 1H T1 values were successfully reduced by a factor of 2.2, 2.9, and 3.2 for bovine cortical bone, type I collagen, and demineralized bone, respectively, without reducing the spectral resolution and thus enabling faster data acquisition. In addition, paramagnetic quenching of particular 13C NMR resonances on exposure to Cu2+ ions in the absence of mineral was also observed, potentially suggesting the relative proximity of three of the main amino acids in the protein backbone (glycine, proline, and alanine) to the bone mineral surface.
INTRODUCTION
The complex hierarchical structure of bone serves to provide the strength, toughness and stiffness required of the material.1–10 Atomic-level structure and dynamics are necessary for understanding the intimate molecular interactions between the organic matrix (primarily type I collagen) and the mineral surface (primarily poorly crystalline calcium-rich carbonated hydroxyapatite) in bone tissues that provide strength and toughness with a minimal amount of mass. Understanding molecular interactions is also a prerequisite for the development of therapeutic approaches to treat bone diseases and regenerate bone defects. However, this is a challenging task for traditionally used characterization techniques11–18 due to the heterogeneous and amorphous nature of bone tissues. Recent studies have demonstrated that high-resolution solid-state nuclear magnetic resonance (SSNMR) spectroscopy represents an invaluable method for the characterization of bone tissues and related biological materials, as it has a much higher potential to elucidate structure and dynamics in a quantitative manner on the molecular level. These NMR studies range from one- (e.g., 1H, 13C, 31P, 43Ca) to two-dimensional (e.g., 31P/1H and 13C/1H Heteronuclear Correlation) magic-angle spinning (MAS) measurements,19–43 and have markedly enriched our knowledge and perception about the structure and function of bone.
Notwithstanding the recent developments in NMR instrumentation and methodology, one of the key challenges that still hinders the widespread application of SSNMR spectroscopy for bone and related biomaterials is the long data-collection time required to achieve a reasonable signal-to-noise ratio (S/N) of the NMR spectrum. It is well known that acquiring an NMR spectrum with excellent sensitivity is crucial for the accurate analysis and interpretation of the NMR line shapes, especially for macromolecules with a large number of amino acid residues. In 13C CP MAS experiments, for example, the inter-scan recycle delay depends exclusively on the proton spin-lattice (T1) relaxation times and consumes more than 90% of the measurement time in the solid state. Therefore, the development of new approaches and methodologies to shorten these recycle delays and thereby accelerate SSNMR measurements is timely and essential.
Doping with paramagnetic reagents such as Cu2+ can effectively enhance the longitudinal 1H relaxation rates and accelerate data acquisition in solution as well as in SSNMR on peptides, proteins and model membranes.44–57 While the previous SSNMR studies have successfully investigated the use of paramagnetic doping to speed up the longitudinal relaxation process in MAS NMR measurements of crystalline peptides/proteins and hydrated lipid bilayers, the effects of paramagnetic dopants on spin–lattice (T1) relaxation times in heterogeneous and amorphous biomolecular systems have not been extensively explored, particularly for bone. Therefore, in an effort to acquire further understanding of the organic–mineral interface in bone, we present here our high-field SSNMR investigation of the utility of paramagnetic ion doping to enhance 1H longitudinal relaxation rates in one-dimensional 13C CP MAS for powdered samples of three bone specimens: type I collagen (bovine tendon), fresh bovine cortical bone, and demineralized bovine cortical bone. To this end, we have employed the paramagnetic properties of Cu2+ ions using the Cu(II)(NH4)2EDTA complex (henceforth referred to as Cu–EDTA) as a relaxation enhancement agent. Additionally, the application of one of the highest available magnetic fields (21.14 T / 900 MHz 1H Larmor frequency) has enabled us to detect the various 13C NMR resonances in native bone components with high sensitivity at natural abundance, which in turn has facilitated the investigation of the effect of Cu–EDTA doping on both the line widths and line positions of the 13C resonances in the CP MAS spectra for these samples.
EXPERIMENTAL SECTION
Sample Preparation
Bovine femora were collected from freshly slaughtered animals (2–4 years old) at a local abattoir. After cleaning of all soft tissues, slices of cortical bone were dissected from mid-diaphyseal femoral regions and machined into rectangular specimens with a band saw under continuous irrigation with calcium-buffered saline solution to avoid excessive heating of the bone pieces. Rectangular specimens, randomly selected from a collection of 10 femora with respect to longitudinal and transverse orientation, were ground into powder in a cryogenic mill under liquid nitrogen. Type I collagen (bovine tendon) was purchased from Sigma Aldrich (St. Louis, MO, USA). For consistency, a sample of type I collagen was also milled into fine powder while cryogenically cooled with liquid nitrogen. Finally, the obtained powdered samples were soaked with standard phosphate–buffered saline (PBS) solution and stored at −20 °C prior to further treatment and NMR measurements.
Powdered bovine bone was demineralized in 0.2 N hydrochloric acid solution for 5 hours at room temperature,58 then washed in excess of PBS solution and filtered. 30 mM Cu–EDTA PBS solution was prepared by dissolving Cu(II)(NH4)2EDTA powder (Sigma Aldrich, St. Louis, MO, USA) in standard PBS buffer. Bone and collagen samples were suspended and mixed with 30 mM Cu–EDTA PBS solution for about 10 minutes, and filtered for each NMR experiment.
NMR Spectroscopy
Preliminary NMR experiments were performed on a Varian VNMRJ 600 MHz solid-state NMR spectrometer using a 4 mm double-resonance MAS probe. To acquire high-resolution 13C NMR spectra with better sensitivity, all solid-state NMR experiments reported herein were performed at 21.14 T on a Bruker AVANCE 900 MHz spectrometer operating at a resonance frequency of 226.3 MHz for 13C and 899.9 MHz for 1H, and equipped with a 4-mm triple-resonance magic angle spinning (MAS) probe. The 13C MAS spectra were acquired under 10 kHz MAS conditions at 25 °C using a ramped-amplitude cross-polarization (Ramp-CP) pulse sequence during an acquisition period of 5 ms with a 2 ms cross-polarization contact time. A 75 kHz two-pulse phase-modulation (TPPM)59 scheme was applied to decouple protons during signal acquisition. 13C NMR chemical shifts were referenced with respect to TMS using adamantane as a secondary external standard. The 1H T1 values were calculated from a series of data collected by 1H spin-inversion recovery experiments detected in 13C Ramp-CP MAS, in which a 1H π-pulse followed by an inversion recovery delay were introduced prior to the Ramp-CP pulse sequence and TPPM decoupling scheme. Six data points with increasing inversion recovery delays were recorded for each sample. The recycle delays were set to 7 s for samples containing no Cu–EDTA and 3 s for those with 30 mM Cu–EDTA, and 2000 scans were accumulated for each inversion recovery experiment. The signal intensities were measured for peaks in the aliphatic (10–70 ppm) and the carboxyl/carbonyl (165–185 ppm) regions to determine the corresponding 1H T1 values. Only those residues whose signals are distinctly resolved in the 13C MAS spectrum and have adequate S/N have been considered for data analysis. The average of the 1H T1 values determined for these residues was adopted as the 1H T1 value of the sample in question.
RESULTS AND DISCUSSION
Prior to commencing the discussion on the SSNMR results, a comment should be made on the bone demineralization procedure used in this study. A common bone demineralization procedure is treatment with EDTA.60 Therefore, it is reasonable to question whether the use of Cu(II)(NH4)2EDTA as the paramagnetic relaxation agent would cause the exchange of Cu2+ for Ca2+ ions in the hydrated surface layer of bone or within the inorganic crystal lattice. For comparison, the log formation constant for the copper–EDTA complex is 18.8, while the log formation constant for the calcium–EDTA complex is 10.70.61 Thus, the copper complex is favored by eight orders of magnitude relative to the calcium complex. Conditional formation constants, which describe the effects of pH and other ligands, would be expected to differ by about eight orders of magnitude as well, and therefore the possibility of exchanging copper for calcium in the copper-EDTA complex is vanishingly low. To experimentally verify this assertion, we used Inductively Coupled Plasma – Optical Emission Spectroscopy (ICP – OES) to measure the concentration of Ca2+ ions in fresh powdered bovine cortical bone soaked with standard PBS solution in the absence and in the presence of 30 mM Cu–EDTA. Our results show the Ca2+ concentration in bone to be, within experimental error, essentially the same with and without 30 mM Cu–EDTA ([Ca2+] = 155 ± 16 µg/L without Cu–EDTA and 148 ± 25 µg/L with 30 mM Cu–EDTA). Therefore, one can unambiguously conclude that the addition of the Cu–EDTA complex to bone does not cause the exchange of copper for calcium in the bone mineral phase.
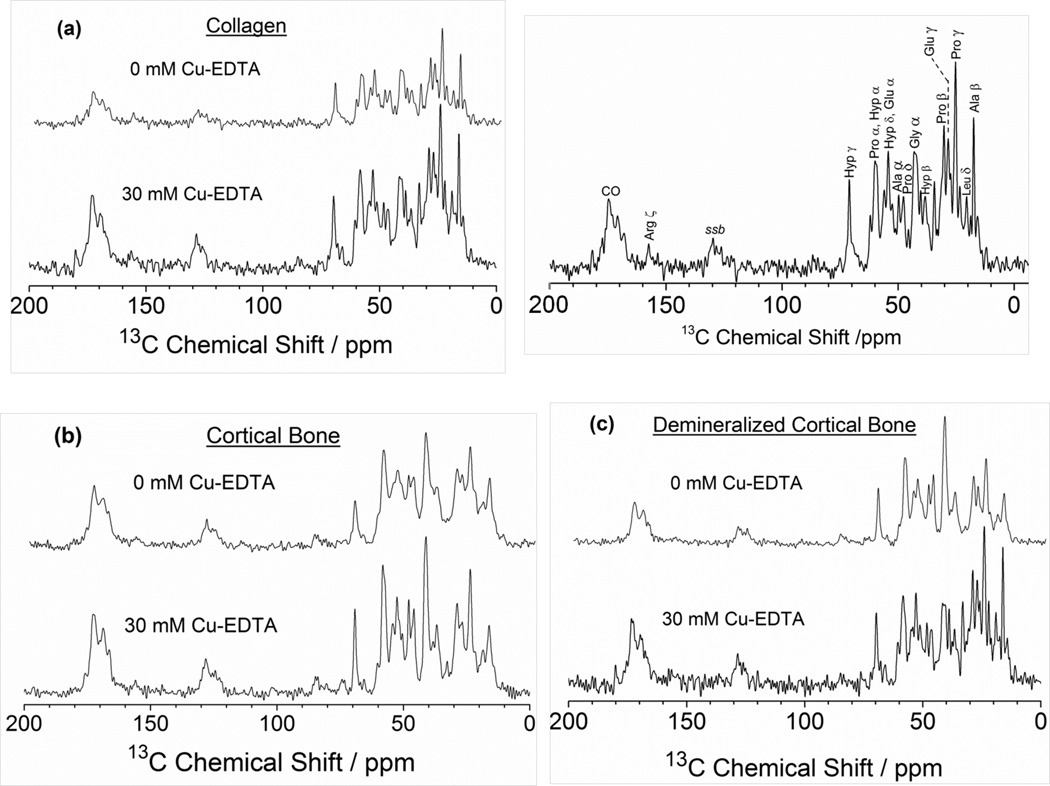
Since it has been well established that copper chelates have the ability to produce an overall relaxation enhancement in the solid state,48–52 we chose to investigate the paramagnetic relaxation effect of Cu2+ ions on the 1H intrinsic spin-lattice (T1) relaxation times measured in 13C CP MAS NMR experiments on bone tissues for the purpose of accelerating the data acquisition time in SSNMR. The natural abundance 13C Ramp-CP MAS NMR experiments were performed on powdered samples of type I collagen, fresh bovine cortical bone, and demineralized bone. The 13C chemical shift NMR spectra corresponding to these samples are shown in Figure 1 using an optimized recycle delay after determination of the 1H T1 values for type I collagen (Figure 1a), fresh bovine cortical bone (Figure 1b), and demineralized bone (Figure 1c). These spectra were collected without Cu–EDTA (top) and with 30 mM Cu–EDTA (bottom), for which the recycle delays were set to 3 s for the undoped samples and 2 s for the doped ones. It is evident that using the shorter 1H spin-lattice relaxation times in the presence of 30 mM Cu–EDTA resulted in faster signal acquisition with a noticeable sensitivity enhancement. As expected, the 13C NMR resonances in all these spectra are dominated by amino acid signals originating from type I collagen. Most of these resonances could be easily recognized and assigned,19,31,40 as shown for the collagen 13C MAS spectrum in Figure 1. It is also obvious that no significant variations in the 13C NMR chemical shifts were observed upon addition of 30 mM Cu–EDTA. This indicates that no major structural variations are likely to occur in the investigated bone samples due to the presence of Cu–EDTA.
Figure 1.
Carbon-13 proton-decoupled Ramp-CP MAS NMR spectra of bone samples recorded at a 13C NMR frequency of 226.3 MHz under 10 kHz MAS conditions for (a) type I collagen, (b) powdered bovine cortical bone, and (c) demineralized cortical bone. Samples were soaked in PBS buffer without Cu–EDTA (top) and with 30 mM Cu–EDTA (bottom). Also shown in (a) the 13C NMR chemical shift assignments of various amino acids constituting type I collagen in the 13C Ramp-CP MAS spectrum of collagen soaked in PBS buffer. All spectra were recorded at 25 °C on a Bruker AVANCE 900 MHz solid-state NMR spectrometer equipped with a 4-mm triple-resonance MAS probe. Other experimental parameters include a 2 ms ramp-cross-polarization time, a 75 kHz TPPM proton decoupling during acquisition. Ala, alanine; Leu, leucine; Pro, proline; Glu, glutamic acid; Hyp, hydroxyproline; Gly, glycine; Arg, arginine; CO, carbonyl; ssb, spinning sidebands.
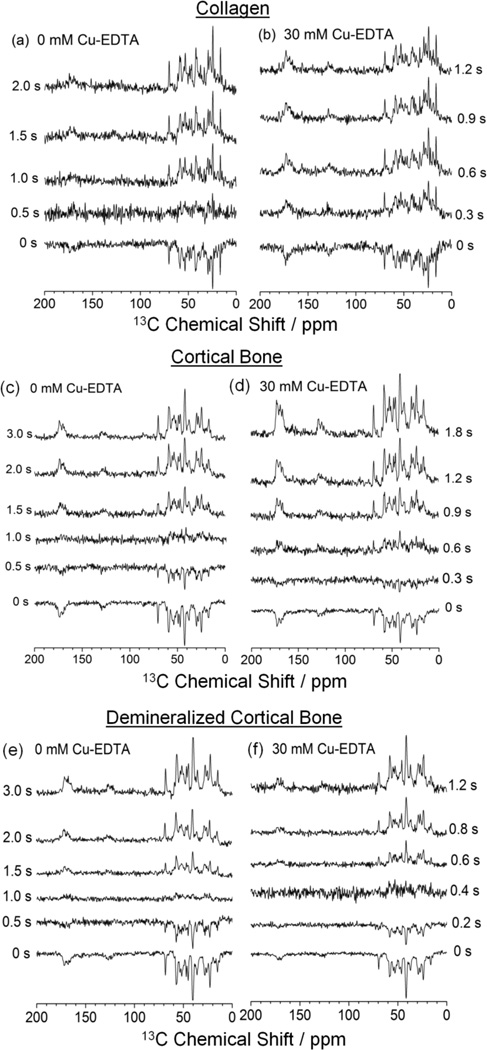
To obtain a detailed quantitative insight into the paramagnetic effect of copper-chelates on relaxation, the 1H T1 values for various amino acid residues were calculated from the data collected by 1H inversion recovery experiments detected by 13C Ramp-CP MAS. Figure 2 shows the 13C Ramp-CP MAS spectra of the three bone samples acquired at different inversion recovery delays without (a, c, e) and with (b, d, f) 30 mM Cu–EDTA. The 1H T1 values for different amino acid residues in the investigated samples are significantly reduced by the addition of Cu–EDTA. To illustrate, the series of 13C MAS spectra of type I collagen in Figure 2(b) show a null signal between 0 and 0.3 s in the presence of Cu–EDTA, while the spectra without Cu–EDTA (Figure 2a) display an almost null signal after 0.5 s; the average 1H T1 values determined for collagen from these experiments are (a) 632 ± 49 ms and (b) 218 ± 38 ms. Similarly, the 13C MAS spectra of cortical bone display a null signal between 0.3 s and 0.6 s in the presence of Cu–EDTA in Figure 2(d), while the spectra in the absence of Cu–EDTA show a signal close to null only after 1.0 s (Figure 2c); the average 1H T1 values obtained from these experiments for cortical bone are (c) 1174 ± 71 ms and (d) 540 ± 48 ms. For demineralized bone, a near null signal is observed after 0.4 s in presence of Cu–EDTA (Figure 2f) and after 1.0 s without Cu–EDTA (Figure 2e), thus yielding a calculated average 1H T1 values of (e) 1261 ± 63 ms and (f) 394 ± 28 ms for this sample. These numerical results indicate that 11H T1 values were successfully reduced by a factor of 2.2, 2.9, and 3.2 for bovine cortical bone, type I collagen, and demineralized bone, respectively. By virtue of the paramagnetic Cu–EDTA doping method, these SSNMR measurements can therefore be accelerated up to three times using the shorter 1H T1 values, given that the sample characteristics (quantity, stability, etc.) and the duty cycle of the NMR probe are not the impeding elements.
Figure 2.
1H inversion recovery spectra of 13C Ramp-CP MAS NMR experiments of (a, b) type I collagen, (c, d) cortical bone, and (e, f) demineralized bone. Samples were soaked in PBS buffer (a, c, e) without Cu–EDTA and (b, d, f) with 30 mM Cu–EDTA. All spectra were recorded under 10 kHz MAS at 25 °C. Other experimental and data processing details are mentioned in Figure 1 caption. Inversion recovery delays are indicated for each spectrum.
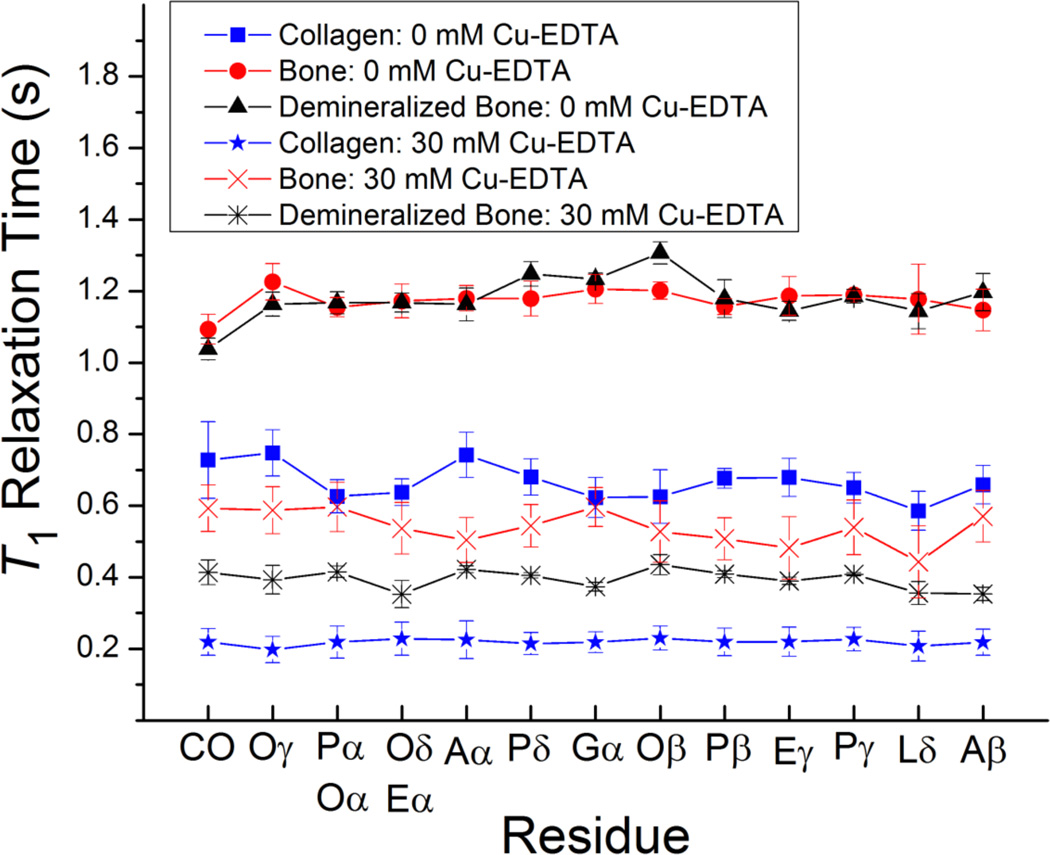
Shown in Figure 3 are the calculated site-specific 1H T1 relaxation times for the different amino acids in each of the three materials in the presence of 30 mM Cu–EDTA in comparison with the undoped samples. In the absence of Cu–EDTA, the 1H T1 relaxation times are different among protons of a variety of amino acid residues at different positions; these variations of the 1H T1 relaxation times are most likely due to internal structural flexibility and relative molecular mobility. In the presence of Cu–EDTA, the average 1H T1 relaxation times are considerably shortened while the overall trend for relaxation times as a function of the amino acid residues is essentially preserved. However, a closer examination of the measured 1H T1 delays reveals that the shortening in T1 times is not consistent among all the amino acid residues.
Figure 3.
Spin-lattice (T1) 1H relaxation times for the various residues in collagen, powdered cortical bone, and demineralized bone in the absence and in the presence of Cu–EDTA (30 mM). The T1 values were determined from 1H-spin-inversion recovery experiments detected in 13C Ramp-CP MAS, and the reported errors were estimated from the best-fitting of experimental data. All measurements were performed on a 900 MHz Bruker AVANCE solid-state NMR spectrometer. Other experimental and data processing details are mentioned in Figure 1 caption. A, alanine; L, leucine; P, proline; E, glutamic acid; O, hydroxyproline; G, glycine; CO, carbonyl. The signals from (Pα, Oα) and (Oδ, Eα) overlap in the 13C NMR spectrum.
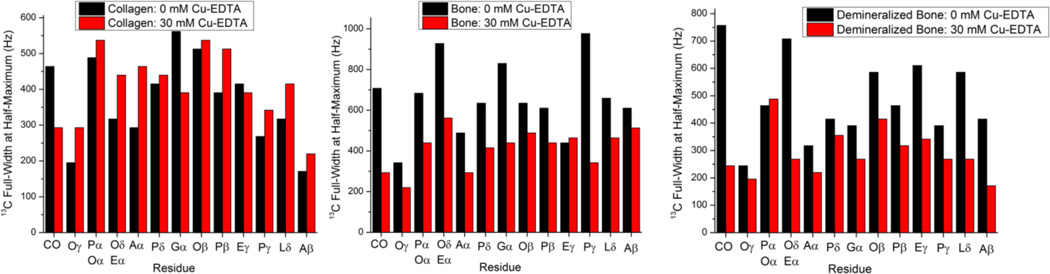
A more detailed analysis of the effect of Cu–EDTA on the 13C line width is given in Figure 4 which shows the 13C line width measured as the full width at half-maximum (FWHM) of the signal from each of the amino acid residues with and without Cu–EDTA. Interestingly, we find that while the presence of 30 mM Cu–EDTA in type I collagen induces a modest increase in the average 13C line width from (370 ± 121) Hz to (409 ± 98) Hz, it has an opposite effect on the average 13C line width in the other two specimens; the average carbon line width in bovine bone decreases from (657 ± 180) Hz to (413 ± 99) Hz while that of demineralized bone decreases from (488 ± 151) Hz to (294 ± 88) Hz upon doping in 30 mM Cu–EDTA. It has been proven that Cu2+ has the ability to reduce the T1 of protons while minimally affecting the 13C line shapes in CP MAS experiments on microcrystalline proteins in the absence of close contact.48–50 Even though line-narrowing rather than line-broadening due to Cu–EDTA doping was observed in our case for cortical and demineralized bone samples, further systematic studies of the paramagnetic effect of Cu2+ on the 13C spin-spin relaxation times (T2) are needed to provide more reliable information in that aspect.
Figure 4.
Full-width at half-maximum (FWHM) values obtained from 13C Ramp-CP MAS NMR spectra of bone samples recorded at a 13C NMR frequency of 226.3 MHz under 10 kHz MAS conditions for type I collagen, powdered bovine cortical bone, and demineralized cortical bone without Cu–EDTA (black) and with 30 mM Cu–EDTA (red). Other experimental and data processing details are mentioned in Figure 1 caption. A, alanine; L, leucine; P, proline; E, glutamic acid; O, hydroxyproline; G, glycine; CO, carbonyl. The signals from (Pα, Oα) and (Oδ, Eα) overlap in the 13C NMR spectrum.
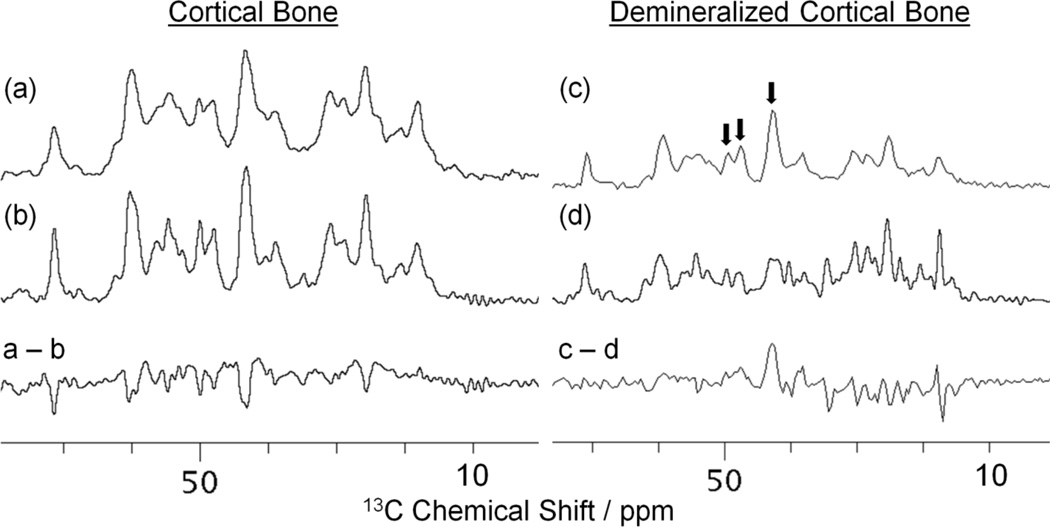
Finally, the resolution of observed spectral lines in the 13C CP MAS spectra for cortical bone and demineralized bone was investigated with and without 30 mM Cu–EDTA. Figure 5 shows the aliphatic regions of the 13C CP MAS spectra for cortical bone (a, b) and demineralized bone (c, d) along with the difference between the spectra acquired without and with 30 mM Cu–EDTA for both samples. As mentioned previously, no major changes in the 13C resonance positions were observed by the addition of 30 mM Cu–EDTA. Although line narrowing instead of line broadening due to Cu–EDTA was detected, the intensities of few resonances (denoted by arrows in Figure 5) were significantly reduced in the spectrum of the demineralized bone. In the absence of mineral, the quenching of these signals (namely Gly Cα, Pro Cδ, and Ala Cα), in which the 13C spins undergo faster spin-spin (T2) relaxation due to paramagnetic Cu2+ proximity, indicates that these residues are more exposed to the bone mineral surface than other residues in the bone organic matrix. Theoretically, the observed signal quenching can be attributed to the fact that the paramagnetic T2 relaxation rates are inversely proportional to r6, where r is the distance between the paramagnetic ion and the observed spin. 53,62 Thus, these residues must correspond to locations of close contact between the mineral and the organic matrix in bone such that in the absence of mineral, Cu2+ can come into close proximity of the newly exposed residues. It is therefore plausible that signals for amino acid residues in closer proximity to the bone mineral surface are significantly quenched, while the signals for the majority of other residues remain unaffected by paramagnetic T2 relaxation enhancement. Further investigations and analyses are required in order to assess the validity of using such enhancements to obtain quantitative structural information from bone samples.
Figure 5.
Carbon-13 Ramp-CP MAS NMR spectra of (a, b) powdered cortical bone and (c, d) demineralized bone samples recorded at a 13C NMR frequency of 226.3 MHz under 10 kHz MAS without Cu–EDTA (a, c) and with 30 mM Cu–EDTA (b, d). For each sample, the difference between the spectra obtained without and with Cu–EDTA is also shown. Other experimental and data processing details are mentioned in Figure 1 caption.
CONCLUSIONS
In conclusion, we have demonstrated that the doping of powdered bone tissues in a copper-EDTA complex results in a two- to three-fold enhancement in the intrinsic spin-lattice relaxation rates of protons, which leads to faster data acquisition without affecting the resolution of their 13C CP MAS NMR spectra. Although this enhancement is not as drastic as those reported for microcrystalline proteins, it is still significant and will be of ample applicability in structural and dynamic studies of traditionally complex materials like bone. It is worth noting that while previous solid-state NMR reports have discussed the use of paramagnetic doping to shorten the proton T1 values in MAS NMR measurements of crystalline proteins and peptides, the effects of paramagnetic ions on T1 relaxation times in heterogeneous biomolecular systems have not been tackled in depth. In this study, this approach has been explored in one-dimensional 13C Ramp-CP MAS for powdered bones, and it is possible that the same approach can be implemented in CP MAS or stationary NMR experiments on intact bone, as intact bone would give more useful results than the cryogenically milled bone powders used in the present experiments. The successful enhancement of the longitudinal relaxation rates of protons in this study opens up new avenues for sensitivity enhancements for the majority of signals in complex biological systems in more complicated multi-dimensional SSNMR experiments. Moreover, the combined application of paramagnetic doping and radio frequency pulse sequences tailored specifically for this purpose under much faster MAS rates would also provide further signal enhancements. Finally, a comprehensive study involving comparison of the paramagnetic relaxation effects of various metal-chelates like Cu2+, Mn2+, Ni2+, and lanthanide ions (e.g., Gd3+ and Dy3+) on bone samples is currently underway in our laboratory. We strongly believe that such a study can provide additional high-resolution insights on the structure and dynamics in bone and related biomaterials.
ACKNOWLEDGMENT
This research was supported by NIH grants AR056657 and AR052010, and RR023597.
REFERENCES
- 1.Lowenstam HA, Weiner S. On Biomineralization. New York: Oxford University Press; 1989. [Google Scholar]
- 2.Robey PG. Noncollagenous Bone Matrix Proteins. In: Bilezikian JP, Raisz LG, Martin TJ, editors. Principles of Bone Biology. Vol. 1. Burlington, MA: Academic Press; 2008. pp. 335–350. [Google Scholar]
- 3.Robey PG, Boskey AL. The Composition of Bone. In: Rosen CJ, editor. Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism. 7 ed. Washington D.C.: The American Society for Bone and Mineral Research; 2008. [Google Scholar]
- 4.Reichert D, Pascui O, deAzevedo ER, Bonagamba TJ, Arnold K, Huster D. Magn. Reson. Chem. 2004;42:276–284. doi: 10.1002/mrc.1334. [DOI] [PubMed] [Google Scholar]
- 5.LeGeros RZ. Biological and Synthetic Apatites. In: Brown PW, Constantz B, editors. Hydroxyapatite and Related Materials. Boca Raton: CRC Press; 1994. pp. 3–28. [Google Scholar]
- 6.Gupta HS, Seto J, Wagermaier W, Zaslansky P, Boesecke P, Fratzl P. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17741–17746. doi: 10.1073/pnas.0604237103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currey JD. J. Biomech. 1988;21:131–139. doi: 10.1016/0021-9290(88)90006-1. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Bank RA, TeKoppele JM, Agrawal CM. J. Orthop. Res. 2001;19:1021–1026. doi: 10.1016/S0736-0266(01)00047-X. [DOI] [PubMed] [Google Scholar]
- 9.Zioupos P. J. Biomater. Appl. 2001;15:187–229. doi: 10.1106/5JUJ-TFJ3-JVVA-3RJ0. [DOI] [PubMed] [Google Scholar]
- 10.Zioupos P, Currey JD, Hamer AJ. J. Biomed. Mater. Res. 1999;45:108–116. doi: 10.1002/(sici)1097-4636(199905)45:2<108::aid-jbm5>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 11.Boskey A, Pleshko Camacho N. Biomaterials. 2007;28:2465–2478. doi: 10.1016/j.biomaterials.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maroudas A, Wachtel E, Grushko G, Katz EP, Weinberg P. Biochim. Biophys. Acta. 1991;1073:285–294. doi: 10.1016/0304-4165(91)90133-2. [DOI] [PubMed] [Google Scholar]
- 13.Ohgushi H, Miyake J, Tateishi T. Novartis Found. Symp. 2003;249:118–127. [PubMed] [Google Scholar]
- 14.Penel G, Leroy N, Van Landuyt P, Flautre B, Hardouin P, Lemaitre J, Leroy G. Bone. 1999;25:81S–84S. doi: 10.1016/s8756-3282(99)00139-8. [DOI] [PubMed] [Google Scholar]
- 15.Stolz M, Gottardi R, Raiteri R, Miot S, Martin I, Imer R, Staufer U, Raducanu A, Duggelin M, Baschong W, Daniels AU, Friederich NF, Aszodi A, Aebi U. Nat. Nanotechnol. 2009;4:186–192. doi: 10.1038/nnano.2008.410. [DOI] [PubMed] [Google Scholar]
- 16.Takata S, Shibata A, Yonezu H, Yamada T, Takahashi M, Abbaspour A, Yasui N. J. Med. Invest. 2004;51:133–138. doi: 10.2152/jmi.51.133. [DOI] [PubMed] [Google Scholar]
- 17.Wachtel E, Maroudas A. Biochim. Biophys. Acta. 1998;1381:37–48. doi: 10.1016/s0304-4165(97)00158-x. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Chen W, Li Y, Fan S, Weng J, Zhang X. Biomaterials. 1998;19:1387–1392. doi: 10.1016/s0142-9612(98)00014-3. [DOI] [PubMed] [Google Scholar]
- 19.Aliev AE. Biopolymers. 2005;77:230–245. doi: 10.1002/bip.20217. [DOI] [PubMed] [Google Scholar]
- 20.Aue WP, Roufosse AH, Glimcher MJ, Griffin RG. Biochemistry. 1984;23:6110–6114. doi: 10.1021/bi00320a032. [DOI] [PubMed] [Google Scholar]
- 21.Beshah K, Rey C, Glimcher MJ, Schimizu M, Griffin RG. J. Solid State Chem. 1990;84:71–81. [Google Scholar]
- 22.Cho G, Wu Y, Ackerman JL. Science. 2003;300:1123–1127. doi: 10.1126/science.1078470. [DOI] [PubMed] [Google Scholar]
- 23.Duer MJ, Friscic T, Murray RC, Reid DG, Wise ER. Biophys. J. 2009;96:3372–3378. doi: 10.1016/j.bpj.2008.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu YY, Rawal A, Schmidt-Rohr K. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22425–22429. doi: 10.1073/pnas.1009219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huster D, Schiller J, Arnold K. Magn. Reson. Med. 2002;48:624–632. doi: 10.1002/mrm.10272. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger C, Groom NS, Bowe EA, Horner A, Davies ME, Murray RC, Duer MJ. Chem. Mater. 2005;17:3059–3061. [Google Scholar]
- 27.Kaflak-Hachulska A, Chmielewski D, Gorecki A, Slosarczyk A, Kolodziejski W. Solid State Nucl. Magn. Reson. 2006;29:345–348. doi: 10.1016/j.ssnmr.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Kaflak-Hachulska A, Samoson A, Kolodziejski W. Calcif. Tissue Int. 2003;73:476–486. doi: 10.1007/s00223-002-2111-5. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee S, Song Y, Oldfield E. J. Am. Chem. Soc. 2008;130:1264–1273. doi: 10.1021/ja0759949. [DOI] [PubMed] [Google Scholar]
- 30.Reid DG, Duer MJ, Murray RC, Wise ER. Chem. Mater. 2008;20:3549–3550. [Google Scholar]
- 31.Saito H, Yokoi M. J. Biochem. 1992;111:376–382. doi: 10.1093/oxfordjournals.jbchem.a123765. [DOI] [PubMed] [Google Scholar]
- 32.Santos RA, Wind RA, Bronnimann CE. J. Magn. Reson. B. 1994;105:183–187. doi: 10.1006/jmrb.1994.1120. [DOI] [PubMed] [Google Scholar]
- 33.Scheidt HA, Schibur S, Magalhaes A, de Azevedo ER, Bonagamba TJ, Pascui O, Schulz R, Reichert D, Huster D. Biopolymers. 2010;93:520–532. doi: 10.1002/bip.21386. [DOI] [PubMed] [Google Scholar]
- 34.Schulz J, Pretzsch M, Khalaf I, Deiwick A, Scheidt HA, Salis-Soglio G, Bader A, Huster D. Calcif. Tissue Int. 2007;80:275–285. doi: 10.1007/s00223-007-9007-3. [DOI] [PubMed] [Google Scholar]
- 35.Tseng YH, Mou CY, Chan JC. J. Am. Chem. Soc. 2006;128:6909–6918. doi: 10.1021/ja060336u. [DOI] [PubMed] [Google Scholar]
- 36.Weber F, Bohme J, Scheidt HA, Grunder W, Rammelt S, Hacker M, Schulz-Siegmund M, Huster D. NMR Biomed. 2012;25:464–475. doi: 10.1002/nbm.1649. [DOI] [PubMed] [Google Scholar]
- 37.Wilson EE, Awonusi A, Morris MD, Kohn DH, Tecklenburg MM, Beck LW. Biophys. J. 2006;90:3722–3731. doi: 10.1529/biophysj.105.070243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wise ER, Maltsev S, Davies ME, Duer MJ, Jaeger C, Loveridge N, Murray RC, Reid DG. Chem. Mater. 2007;19:5055–5057. [Google Scholar]
- 39.Wu Y, Ackerman JL, Kim HM, Rey C, Barroug A, Glimcher MJ. J. Bone Miner. Res. 2002;17:472–480. doi: 10.1359/jbmr.2002.17.3.472. [DOI] [PubMed] [Google Scholar]
- 40.Zhu P, Xu J, Sahar N, Morris MD, Kohn DH, Ramamoorthy A. J. Am. Chem. Soc. 2009;131:17064–17065. doi: 10.1021/ja9081028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, Zhu P, Morris MD, Ramamoorthy A. J. Phys. Chem. B. 2011;115:9948–9954. doi: 10.1021/jp205663z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J, Zhu P, Gan Z, Sahar N, Tecklenburg M, Morris MD, Kohn DH, Ramamoorthy A. J. Am. Chem. Soc. 2010;132:11504–11509. doi: 10.1021/ja101961x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yesinowski JP, Eckert H. J. Am. Chem. Soc. 1987;109:6274–6282. [Google Scholar]
- 44.Laage S, Sachleben JR, Steuernagel S, Pierattelli R, Pintacuda G, Emsley L. J. Magn. Reson. 2009;196:133–141. doi: 10.1016/j.jmr.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Bertini I, Emsley L, Lelli M, Luchinat C, Mao J, Pintacuda G. J. Am. Chem. Soc. 2010;132:5558–5559. doi: 10.1021/ja100398q. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto K, Vivekanandan S, Ramamoorthy A. J. Phys. Chem. B. 2011;115:12448–12455. doi: 10.1021/jp2076098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto K, Xu J, Kawulka KE, Vederas JC, Ramamoorthy A. J. Am. Chem. Soc. 2010;132:6929–6931. doi: 10.1021/ja102103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickramasinghe NP, Kotecha M, Samoson A, Past J, Ishii Y. J. Magn. Reson. 2007;184:350–356. doi: 10.1016/j.jmr.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickramasinghe NP, Parthasarathy S, Jones CR, Bhardwaj C, Long F, Kotecha M, Mehboob S, Fung LW, Past J, Samoson A, Ishii Y. Nat. Methods. 2009;6:215–218. doi: 10.1038/nmeth.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickramasinghe NP, Shaibat M, Ishii Y. J. Am. Chem. Soc. 2005;127:5796–5797. doi: 10.1021/ja042188i. [DOI] [PubMed] [Google Scholar]
- 51.Linser R, Fink U, Reif B. J. Am. Chem. Soc. 2009;131:13703–13708. doi: 10.1021/ja903892j. [DOI] [PubMed] [Google Scholar]
- 52.Linser R, Chevelkov V, Diehl A, Reif B. J. Magn. Reson. 2007;189:209–216. doi: 10.1016/j.jmr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 53.Cai S, Seu C, Kovacs Z, Sherry AD, Chen Y. J. Am. Chem. Soc. 2006;128:13474–13478. doi: 10.1021/ja0634526. [DOI] [PubMed] [Google Scholar]
- 54.Nadaud PS, Helmus JJ, Sengupta I, Jaroniec CP. J. Am. Chem. Soc. 2010;132:9561–9563. doi: 10.1021/ja103545e. [DOI] [PubMed] [Google Scholar]
- 55.Eletsky A, Moreira O, Kovacs H, Pervushin K. J. Biomol. NMR. 2003;26:167–179. doi: 10.1023/a:1023572320699. [DOI] [PubMed] [Google Scholar]
- 56.Hiller S, Wider G, Etezady-Esfarjani T, Horst R, Wuthrich K. J. Biomol. NMR. 2005;32:61–70. doi: 10.1007/s10858-005-3070-8. [DOI] [PubMed] [Google Scholar]
- 57.Bertini I, Luchinat C, Rosato A. Prog. Biophys. Mol. Biol. 1996;66:43–80. doi: 10.1016/s0079-6107(96)00016-8. [DOI] [PubMed] [Google Scholar]
- 58.Castro-Cesena AB, Novitskaya EE, Chen PY, Hirata GA, McKittrick J. Mater. Sci. Eng. C. 2011;31:523–530. doi: 10.1016/j.msec.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 59.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 60.Skinner RA. Decalcification of Bone Tissue. In: An YH, Martin KL, editors. Handbook of Histology Methods for Bone and Cartilage. Totawa, NJ: Humana Press; 2003. pp. 167–184. [Google Scholar]
- 61.NIST Standard Reference Database 46. Gaithersburg, MD: National Institute of Standards and Technology (NIST); 2010. NIST Critically Selected Stability Constants of Metal Complexes: Version 8.0. [Google Scholar]
- 62.Solomon I. Phys. Rev. 1955;99:559–565. [Google Scholar]