Abstract
The mid-distal region of mouse chromosome 4 (Chr 4) is homologous with human Chr 1p36. Previously, we reported that mouse Chr 4 carries a QTL with strong regulatory effect on vBMD. The intent of this report is to utilize nested congenic strains to decompose the genetic complexity of this gene rich region. Adult females and males from eighteen nested congenic strains carrying discrete C3H sequences were phenotyped for femoral mineral and volume by pQCT and for trabecular BV, TV, Trab.no and Trab.thk by MicroCT40. Our data show that the mouse Chr 4 region consists of at least 10 regulatory QTL regions that affected either or both pQCT and Micro CT40 phenotypes. The pQCT phenotypes were typically similar between sexes, whereas the microCT40 phenotypes were divergent. Individual congenic strains contained one to seven QTL regions. These regions conferred large positive or negative effects in some congenic strains, depending on the particular bone phenotype. The QTL regions, II to X, are syntenic with human 1p36, contained from 1 to 102 known genes. We identified 13 candidate genes that can be linked to bone within these regions. Six of these genes were linked to osteoblasts, three linked to osteoclasts, and two linked to skeletal development. Three of these genes have been identified in GWAS studies linked to 1p36. In region III, there is only one gene, Lck, which conferred negative pQCT and Micro CT40 phenotypes in both sexes. This gene is important to development and functioning of T cells, has been associated with osteoclast activity, and represents a novel bone regulatory gene that merits further experimental evaluation. In summary, congenic strains are powerful tools for identifying regulatory regions that influence bone biology, and offer models for testing hypotheses about gene-gene and gene-environment interactions that are not available to experimental work in humans.
Keywords: Chr 4 congenic strains, human Chr 1p36 homology, cortical and trabecular bone phenotypes, mapping of candidate genes
Introduction
Identifying the genetic basis of osteoporosis has been a major objective for many investigators in the scientific community over the past decade. Genes on most chromosomes have been implicated as bone genetics has advanced. Measurements of bone mineral density (BMD) from the hip, lumbar spine, and femoral neck, together with genotyping of alleles for Single Nucleotide Polymorphisms (SNPs) have provided most of the available genomic linkage evidence. Thus far, it is apparent that osteoporosis is a complex quantitative trait. Furthermore, the number of genes involved with skeletal regulation is extensive and the effect size of any one gene appears very modest. Among factors that have confounded progress towards understanding genetic regulation of the human skeleton are: 1) heterogeneity of the human genome, 2) phenotypic heterogeneity, 3) environmental background, 4) population sample sizes, 5) sex-based differences, 6) age-related differences, and 7) ethnicity.
Investigative approaches to the genetic basis of osteoporosis have capitalized on molecular genetics and statistical methodologies that facilitate whole genome analyses, as well as increasing knowledge of candidate genes judged critical to bone formation and resorption. Meta-analyses of Genome Wide Association Studies (GWAS; (1)) with large comparable populations have demonstrated that numerous loci associated with bone can be simultaneously addressed and some loci are common to different populations (see (2–5). An important chromosomal region in each of these meta-analyses is that associated with human Chromosome (Chr) 1p36. This region also has been reported in early GWAS investigations associated with BMD of the hip, lumbar spine, femoral neck, or wrist (6–9). Given the consistency of finding Chr 1p36 as a region important to bone, several studies have focused on potential candidate genes within this region utilizing extended families and ethnic groups. For example, Bab and Zimmer (10) reported that in a Caucasian sample of 98 osteoporotic versus 220 age-matched controls, several SNPs in CNR2, but not CNR1, were significantly associated with BMD. Huang et al. (11) reported on a study of 1243 Chinese case-controls (women and men) with significant association of hip, femoral neck and lumbar spine BMD with SNPs for PLOD and MTHFR, and suggestive association with SNPs for CNR2 and TNFRSF1b. Xiong et al. examined 1873 women and men for BMD versus 20 candidate osteoporosis genes and also found suggestive association of distal radius BMD with SNPs in TNFR2 (now TNFRSF1b) but not MTHFR (12). Zhang et al. (13) utilized lumbar spine, femoral neck, and trochanter BMD from 39 osteoporosis pedigrees and reported significant association of SNPs in 4 genes (RERE, G1P2, SSA72, and CCDC27) within an 11 Mb region of Chr 1p36. Ermakov et al. (14) reported significant association of SNPs in ALPL with bone size and hand BMD in 310 Caucasian families. Similarly, Agueda et al. (Agueda et al 2010) found that SNPs in MTHFR were significantly associated with vertebral fractures in 944 post-menopausal Spanish women. Other investigations of skeletal phenotypes for association with genomic regions have not reported Chr 1p36. Although, the evidence for bone regulatory genes in Chr 1p36 region is excellent, consensus for candidates remains a work-in-progress.
Inbred strains of mice have well defined genetic linkage homology between human and mouse chromosomal regions (see http://www.informatics.jax.org/reports/homologymap/mouse_human.shtml). Furthermore, mouse strains vary markedly in both whole body areal (a)BMD (15) as well as femoral and vertebral volumetric (v)BMD (16). These characteristics led us conduct a cross between low BMD C57BL/6J (B6) and high BMD C3H/HeJ (C3H) mice that showed significant association between BMD and several chromosomes (17). Subsequently, several of these BMD quantitative trait loci (QTL) were confirmed by development of congenic strains by repeated backcrossing of defined mice carrying chromosomal segments from C3H on to the B6 genetic background (18). One congenic strain designated B6.C3H-4T showed higher vBMD compared to the B6 progenitor strain, and carried a large C3H sequence, 24 Mb of which is homologous with human Chr 1p36. In this report, we present nested congenic strain mapping data of this 24 Mb region with associated femoral and cancellous bone phenotypes. The resultant decomposition of the Chr 4 BMD QTL (termed Bmd5) revealed a complex assembly of at least 8 regions with regulatory elements related to bone compartments as well as positive and negative effects dependent on sex.
Materials and Methods
Mice
The inbred mouse strains utilized for the studies reported herein were obtained from our research colonies at The Jackson Laboratory, Bar Harbor, Maine. The C57BL/6BmJ (B6/Bm) and B6.C3H-4 congenic sublines of mice were produced by pair matings, with progeny weaned at 22–25 days of age and housed in groups of 2–5 of the same sex in polycarbonate cages (324 cm2) with sterilized White Pine shavings. Colony environmental conditions included 14:10 hour light:dark cycles, with free access to acidified water (pH 2.5 with HCl to retard bacterial growth plus 0.4 mg/ml of vitamin K (menadione Na bisulfite)), and irradiated NIH 31 diet containing 6% fat, 19% protein, Ca:P of 1.15:0.85, plus vitamin and mineral fortification (Purina Mills International, Brentwood, MO). Animal studies were approved by the Institutional Animal Care and Use Committee of The Jackson Laboratory.
Congenic sublines and genotyping
We previously reported that a congenic mouse strain, designated B6.C3H-4T, carried femoral vBMD regulation in female and male mice (18). Based on analyses of incipient N6F2 congenic mice (18) and of B6C3F2 (17) mice, the genetic segment of interest was large and exerted a substantial regulatory effect on vBMD. To begin mapping of bone regulation associated with distal Chr 4, congenic male mice of the generation N10F4 were backcrossed to B6/Bm (B6) progenitor females, and the N11F1 offspring were intercrossed to obtain N11F2 progeny. These mice were genotyped to identify mice that were homozygous for identical but smaller C3H segments. Eighteen additional congenic strains were developed, each homozygous for a unique genomic segment of different physical size derived from distal portion of Chr 4 in B6.C3H-4T mice. These 18 nested congenic strains represent a set of overlapping sequences of the C3H genome from distal Chr 4. These congenic strains are available from The Jackson Laboratory (http://jaxmice.jax.org/findmice/index.html).
Mice were genotyped by preparing genomic DNA from digestion of 1 mm tail tips in 0.5 ml of 50 mM NaOH for 10 min at 95°C, then pH was adjusted to 8.0 with 1M Tris-HCl. Genotyping of individual mouse DNAs was accomplished by polymerase chain reaction (PCR) using oligonucleotide primer pairs from several sources (Research Genetics, Birmingham, AL; Invitrogen, Carlsbad, CA; IDT, Coralville, IA; Qiagen, Valencia, CA). These primer pairs amplify sequence length polymorphisms (SSLP) consisting of CA repeat sequences wherein the length repeat is strain specific (Mit markers, www-genome.wi.mit.edu/cgi-bin/mouse/index). Details of standard PCR reaction conditions have been described previously (19). PCR products from B6/Bm, C3H and F1 hybrids were used as electrophoretic standards in every gel to identify the genotypes of mice (i.e., b6/b6, b6/c3h, c3h/c3h). In addition to the SSLP markers described above, two additional markers were developed in our laboratory to further define the congenic regions. Specifically, “novel” SSLP were identified in the region of interest and primers were designed to amplify these regions by PCR. Primers for flanking sequences of CA repeats were designed with the aid of Frodo.wi.mit.edu/cgi-bin/primer3/ and MacVector. Custom primer pairs were obtained from Invitrogen and tested against B6/Bm and C3H genomic DNA to confirm polymorphisms. In addition, 15 Single Nucleotide Polymorphisms (SNPs, NCBI Ensembl Build 37, Release 59) that were assayed for the sublines are listed in this report (listed in Table 1).
Table 1.
Primers for genotyping the Chr 4 congenic strains.
| Primer Name | Forward primer | Reverse primer |
|---|---|---|
| rs27558236 (Phc2) | GCTTCCGATTCATTTCCTGTAGT | TTGTAGGCAAAGTCCACCCGTC |
| rs3726101 (Lck) | AGGCTCCAGGCTGTTTGCTTTC | TGGGCTTTGAGAAGGGTGAAC |
| rs13477977 | AGGTGGGAAATGATGGACAGC | TAGGTCTATGGCAGCAGGAAGC |
| rs27492041 (Col161a) | AGGTAAGACTGACTGGGAGGTAAGG | GGCACAAATGAATGATGAGGCTG |
| rs27559177 | GCAGAGTGGCTCCAATGGTAAAC | CCAAGTAAGAGACAGGCAGATGTGC |
| D4kls1-3 | ACTACCCGGCACAAACTGTC | TCACCATCTTTGTGGCAGAG |
| rs3678225 | GCGGAGAGATGATTATGCCCAG | TGTTTGAGTCAGAGCAGCCCTAAG |
| D4kls2-1 | CAGCAAATCCGTGTCACCTA | GCAGAGTCATTGTCCATTCG |
| rs27573590 (Zbtb40) | AAAAGCGTGACCCTCAGAACG | ATGGAACACCCAAGAGATAGCAAG |
| rs32784466 | TGACTGGTTGGTGGCTTGAGAG | AAGGCTGAGGAAGTGTGGAGAAGG |
| rs27559554 | TCAAGCCTCCATCAGTTCCTCTC | TGACACGCTCATCTGCATTCAG |
| rs27560689 (Wnt4) | GGAGTTGAAGGACAGCAGAGATAGG | CTTGTATGCCTGAAGTCACCGTC |
| rs27626795 | ACCTGAACCCTCACCTTCCTTG | TCCCTGCCCAATGTCCTTTG |
| rs27603541 (Plod1) | GGAAAGATTCACAAGAGGCACAGTC | ATCCAGGGTGGCTACGAAAACG |
| rs27617493 (Mthfr) | GGAGTGGCTCTGTCTTGCTGTTAC | CCAGGTTGACCAGGAAATAGTTGTC |
Necropsy
At 16 weeks of age, when mice have acquired their adult femoral mass (16), females from the B6/Bm progenitors and the newly developed distal Chr 4 sublines were necropsied and whole body weights recorded. Skeletal preparations (lumbar vertebral columns, pelvis and attached hind limbs) were placed in 95% ethanol (EtOH) for a period of 3 or more weeks. Lumbar vertebral columns, tibias, and femurs were dissected free of remaining muscle and connective tissue, and placed in 95% EtOH for storage. Femoral volumetric BMD (vBMD) and distal trabecular bone were assessed by peripheral quantitative computed tomography (pQCT) and by MicroCT (μCT) as described below.
pQCT densitometry for vBMD
Femoral mineral and volume were measured on left femurs from groups of female and male B6/Bm and congenic subline mice as previously described (18). Briefly, isolated femur lengths were measured with digital calipers (Stoelting, Wood Dale, IL). Femurs were measured for density using the SA Plus densitometer (Orthometrics, White Plains, NY). Calibration of the SA Plus instrument was accomplished daily with a manufacturer supplied phantom. The bone scans were analyzed with Orthometrics software vers. 5.50. Femur mineral content and volume were determined with thresholds such that mineral from most partial voxels (0.07 mm) would be included in the analysis. Isolated femurs were scanned at 7 locations at 2 mm intervals, beginning 0.8 mm from the distal ends of the epiphyseal condyles. Due to variation in femur lengths, the femoral head could not be scanned at the same location for each bone, and thus was not included in final data. Total vBMD values were calculated by dividing the total mineral content (mineral, primarily cortical bone) by the total bone volume (volume, within periosteal envelope) and expressed as mg/mm3.
MicroCT40 for distal trabecular bone
Trabecular bone components of the distal femur were measured by Micro-computed Tomography 40 (μCT40, Scanco Medical AG, Bassersdorf, Switzerland) as previously described (20). The μCT40 unit is calibrated weekly with a phantom standard provided by Scanco. The femurs were scanned at the energy level of 55 KeV, and intensity of 177 μA, followed by analysis of the trabecular bone volume (BV), tissue volume (TV), trabecular number (Trab.no), and trabecular thickness (Trab.thk) with the Scanco software version 5.0. Approximately 150 cross-sectional slices were made at 12 μ interval at the distal end. One hundred contiguous slices were selected for analysis, beginning at the proximal edge of the growth plate and extending toward the femoral head.
Statistical assessment
Statistical evaluation of all data was undertaken using JMP version 7.0 software (SAS, Cary, NC). For the pQCT and μCT40 data, effects of body size differences between strains were accounted for by using body weight and femur length as covariates when they contributed significantly to an overall ANCOVA model (20). B6 progenitor controls were collected in small groups coincidental with the period of time that congenic strains were developed and tested for bone phenotypes. Data from these small groups of B6 mice were pooled for statistical purposes. Data are expressed as adjusted least squares mean ± SEM in all tables and figures. Differences between means for B6/Bm and each congenic subline were tested by Student’s t-test. Significance was declared when a p ≤ 0.003 (‘p’ of 0.05/18 comparisons) was observed. Those means with ‘p’ values between 0.01 – 0.003 were declared as suggestively significant.
Results
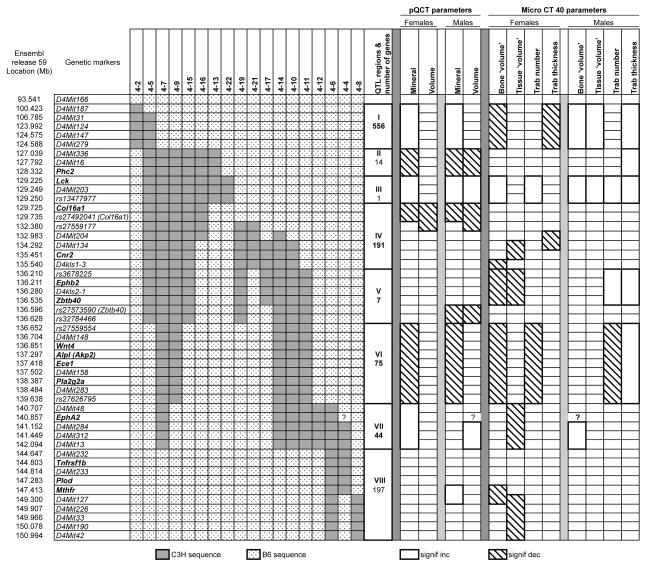
Eighteen congenic strains were developed to fine map bone phenotypes and locate QTL for volumetric density and trabecular parameters. These strains are formally designated as B6.C3H (B6 background.donor strain sequence) followed by the number 4 for Chr 4 and by a strain number. For simplicity of presentation, Figure 1 presents congenic strains designated by Chr number followed by the strain number (i.e., 4-7). The light gray segments represent C3H genetic sequences and the stippled segments represent the B6 genetic sequences. Physical distances (Mb) and genetic markers are presented along the left margin. Fourteen genes in bold print are inserted at correct positions among the genetic markers. The overlapping regions of C3H sequences within the nested congenic strains indicate where to search for the correlation of phenotype with genetic region. Figure 1 also presents a graphical summary of combined findings from congenic subsets for the six femoral bone phenotypes partitioned by measuring instrumentation and sex. QTL regions are presented and described below.
Figure 1. Haplotype map of 18 congenic strains used for mapping bone phenotypes.
The grey segments represent C3H sequence and white segments represent B6 sequence. Physical map distances (Mb) and genetic markers are listed on the left side. Fourteen bone candidate genes in bold text are located at appropriate positions among the markers. Ten (I-X) proposed QTL regions and the number of genes therein are shown. The pQCT and Micro CT40 phenotypes for regions that differed significantly from B6 controls are indicated by boxes that have dense stippling (increase) or black in color (decrease). The genotype for EphA2 could not be determined because there is no genetic variation between B6 and C3H (represented by a ‘?’).
Results of pQCT phenotyping of whole femurs from females and males of each congenic strain are presented in Tables 2 and 3. Phenotypes chosen for mapping the QTL regions were mineral and volume. These are primary measures considered as likely to indicate gene action, whereas the more commonly utilized BMD is a composite measure utilizing mineral and volume. Inspection of data in Table 2 shows that the C3H sequences present in the congenic strain females led to 9 significant increases in mineral and volume, whereas 3 significant and 2 suggestive decreases were recorded. In more than half of the strains with significant or suggestive change, mineral and volume were independent. With respect to the size of the change, the greatest increase for mineral was observed for 4-10 (+8.36%), while the greatest decrease was observed in 4-4 (−6.62%). Similarly, the greatest increase in volume was observed in 4-9 (+4.27%), while the greatest decrease was observed in 4-4 (−2.90%).
Table 2.
Femoral pQCT data from 4 month old congenic females. Data presented as mean ± SEM.
| Congenic strain (n) | Mineral (mg) | ‘p’ for mineral | Volume (mm3) | ‘p’ for volume | vBMD (mg/mm3) | ‘p’ for vBMD |
|---|---|---|---|---|---|---|
| B6 (77) | 11.48 ± 0.08 | 19.68 ± 0.09 | 0.583 ± 0.003 | |||
| 4-2 (23) | 10.99 ± 0.15d | 0.0094 | 19.13 ± 0.16 | - - - | 0.574 ± 0.005 | - - - |
| 4-5 (22) | 10.87 ± 0.15c | 0.0006 | 20.03 ± 0.16 | - - - | 0.543 ± 0.005c | 0.0001 |
| 4-7 (26) | 11.91 ± 0.12 | - - - | 19.95 ± 0.13 | - - - | 0.597 ± 0.004 | - - - |
| 4-9 (19) | 12.37 ± 0.14a | 0.0001 | 20.52 ± 0.15a | 0.0001 | 0.602 ± 0.005a | 0.0002 |
| 4-15 (26) | 12.20 ± 0.14a | 0.0011 | 20.35 ± 0.15a | 0.0020 | 0.599 ± 0.005 | - - - |
| 4-16 (29) | 11.90 ± 0.13 | - - - | 20.13 ± 0.14a | 0.0020 | 0.591 ± 0.004 | - - - |
| 4-13 (27) | 11.75 ± 0.13 | - - - | 19.95 ± 0.15 | - - - | 0.588 ± 0.004 | - - - |
| 4-22 (27) | 11.07 ± 0.13d | 0.0078 | 19.68 ± 0.15 | - - - | 0.562 ± 0.004c | 0.0001 |
| 4-19 (28) | 11.58 ± 0.13 | - - - | 20.32 ± 0.15a | 0.0008 | 0.569 ± 0.004 | - - - |
| 4-21 (27) | 11.64 ± 0.13 | - - - | 20.04 ± 0.15 | - - - | 0.580 ± 0.004 | - - - |
| 4-17 (25) | 11.51 ± 0.14 | - - - | 19.79 ± 0.15 | - - - | 0.581 ± 0.005 | - - - |
| 4-14 (27) | 11.95 ± 0.13 | - - - | 19.98 ± 0.15 | - - - | 0.597 ± 0.004 | - - - |
| 4-10 (23) | 12.44 ± 0.14a | 0.0001 | 20.38 ± 0.16a | 0.0002 | 0.610 ± 0.005a | 0.0001 |
| 4-11 (26) | 12.34 ± 0.14a | 0.0001 | 19.96 ± 0.15 | - - - | 0.621 ± 0.005a | 0.0001 |
| 4-12 (31) | 11.28 ± 0.12 | - - - | 19.60 ± 0.14 | - - - | 0.575 ± 0.004d | 0.0052 |
| 4-6 (26) | 11.31 ± 0.14 | - - - | 19.70 ± 0.15 | - - - | 0.573 ± 0.005 | - - - |
| 4-4 (20) | 10.72 ± 0.16c | 0.0001 | 19.11 ± 0.17c | 0.0011 | 0.561 ± 0.005 | - - - |
| 4-8 (29) | 11.26 ± 0.13 | - - - | 19.65 ± 0.14 | - - - | 0.573 ± 0.004d | 0.0099 |
= significantly (p < 0.003) or
= suggestively (p < 0.01) increased from B6 controls;
= significantly (p < 0.003) or
= suggestively (p < 0.01) decreased from B6 controls
Table 3.
Femoral pQCT data from 4 month old congenic males. Data presented as mean ± SEM.
| Congenic strain (n) | Mineral (mg) | ‘p’ for mineral | Volume (mm3) | ‘p’ for volume | vBMD (mg/mm3) | ‘p’ for vBMD |
|---|---|---|---|---|---|---|
| B6 (75) | 13.69 ± 0.12 | 22.90 ± 0.15 | 0.598 ± 0.003 | |||
| 4-2 (22) | 12.55 ± 0.23c | 0.0001 | 21.39 ± 0.27d | 0.0068 | 0.587 ± 0.006 | - - - |
| 4-5 (21) | 13.59 ± 0.25 | - - - | 23.15 ± 0.29 | - - - | 0.589 ± 0.006 | - - - |
| 4-7 (28) | 14.36 ± 0.20b | 0.0104 | 23.21 ± 0.24 | - - - | 0.619 ± 0.005a | 0.0028 |
| 4-9 (26) | 15.11 ± 0.21a | 0.0001 | 24.40 ± 0.25a | 0.0001 | 0.618 ± 0.005a | 0.0009 |
| 4-15 (26) | 14.67 ± 0.21a | 0.0001 | 23.88 ± 0.25a | 0.0001 | 0.613 ± 0.005 | - - - |
| 4-16 (25) | 14.44 ± 0.21a | 0.0020 | 23.28 ± 0.25 | - - - | 0.620 ± 0.005a | 0.0014 |
| 4-13 (30) | 14.56 ± 0.20a | 0.0005 | 23.62 ± 0.23 | - - - | 0.616 ± 0.005a | 0.0027 |
| 4-22 (26) | 13.39 ± 0.21 | - - - | 22.03 ± 0.25c | 0.0001 | 0.607 ± 0.005 | - - - |
| 4-19 (19) | 14.52 ± 0.20a | 0.0030 | 24.13 ± 0.24a | 0.0001 | 0.599 ± 0.006 | - - - |
| 4-21 (28) | 14.17 ± 0.20 | - - - | 23.70 ± 0.24 | - - - | 0.597 ± 0.005 | - - - |
| 4-17 (25) | 13.54 ± 0.21 | - - - | 22.56 ± 0.25 | - - - | 0.599 ± 0.005 | - - - |
| 4-14 (27) | 15.25 ± 0.21a | 0.0001 | 24.18 ± 0.24a | 0.0001 | 0.631 ± 0.005a | 0.0001 |
| 4-10 (23) | 14.60 ± 0.22a | 0.0002 | 23.64 ± 0.26b | 0.0039 | 0.616 ± 0.006b | 0.0098 |
| 4-11 (27) | 15.08 ± 0.20a | 0.0001 | 23.68 ± 0.24b | 0.0061 | 0.638 ± 0.005a | 0.0001 |
| 4-12 (27) | 13.31 ± 0.21 | - - - | 22.33 ± 0.24 | - - - | 0.595 ± 0.005 | - - - |
| 4-6 (25) | 12.68 ± 0.21c | 0.0005 | 21.85 ± 0.25c | 0.0012 | 0.578± 0.005 | - - - |
| 4-4 (17) | 12.32 ± 0.27c | 0.0001 | 21.53 ± 0.31c | 0.0020 | 0.569 ± 0.007c | 0.0008 |
| 4-8 (22) | 13.14 ± 0.23 | - - - | 22.42 ± 0.27 | - - - | 0.584 ± 0.006 | - - - |
= significantly (p < 0.003) or
= suggestively (p < 0.01) increased from B6 controls;
= significantly (p < 0.003) or
= suggestively (p < 0.01) decreased from B6 controls
The mineral and volume phenotyping data for males in Table 3 show that presence of C3H sequences were associated with 13 significant and 2 suggestive increases and 5 significant and 1 suggestive decreases when compared with the B6 controls. In the male data, more than half of strains with significant or suggestive changes in both mineral and volume were in the same direction. The greatest increase in mineral was observed in 4-14 (+11.40%), whereas the greatest decrease was observed in 4-4 (−10.01%). The greatest increase in volume was observed in 4-9 (+6.55%), while the greatest decrease was observed in 4-2 (−6.59%).
Micro CT40 phenotyping results from distal femoral diaphyses of females and males are compared with B6 controls and presented in Tables 4 and 5. The phenotypes of BV, TV, Trab.no, and Trab.thk were chosen for mapping as they are considered to be primary measures likely to indicate gene action, whereas the more commonly utilized BV/TV is a composite measure BV and TV. In female data in Table 4, the BV means show 8 significant increases and 1 significant decrease. In the TV data, there were 6 significant and 2 suggestive increases, but no decreases. On the other hand, the Trab.no data show 4 significant and 2 suggestive increases, plus 4 significant decreases. For the Trab.thk, there were 5 significant and 1 suggestive increase, plus 2 significant decreases. Thus, the C3H sequences in the congenic strain females resulted in positive effects on femoral BV and TV, but showed more negative effects on Trab.no and Trab.thk. The greatest increases or decreases, respectively, in these 4 female trabecular parameters were: a) BV in 4-17 (+59.9%) and in 4-22 (−36.9%); b) TV in 4-21 (+16.3%) and no observed decreases; c) Trab.no in 4-11 (+9.5%) and in 4-22 (−10.5%); and d) Trab.thk in 4-2 (+86.2%) and in 4-22 (−6.8%).
Table 4.
Distal femoral microCT40 data from 4 month old congenic females. Data presented as mean ± SEM.
| Congenic strain | BV (mm2) | ‘p’ for BV | TV (mm2) | ‘p’ for TV | BV/TV (%) | ‘p’ for BV/TV | Trab.no (1/μm) | ‘p’ for No | Trab.thk (μm) | ‘p’ for Thk |
|---|---|---|---|---|---|---|---|---|---|---|
| B6 (64) | 0.157 ± 0.004 | 1.960 ± 0.013 | 8.0 ± 0.2 | 3.89 ± 0.02 | 47.1 ± 0.4 | |||||
| 4-2 (15) | 0.184 ± 0.008a | .0020 | 1.995 ± 0.028 | - - - | 8.8 ± 0.4 | - - - | 3.84 ± 0.05 | - - - | 87.7 ± 2.1a | .0001 |
| 4-5 (17) | 0.184 ± 0.007a | .0033 | 2.081 ± 0.029a | .0009 | 8.9 ± 0.3 | - - - | 3.85 ± 0.05 | - - - | 49.8 ± 0.8b | .0042 |
| 4-7 (25) | 0.191 ± 0.007a | .0001 | 2.038 ± 0.027b | .0090 | 9.3 ± 0.3a | .0006 | 4.04 ± 0.04a | .0033 | 48.0 ± 0.7 | - - - |
| 4-9 (20) | 0.176 ± 0.007 | - - - | 1.975 ± 0.025 | - - - | 9.0 ± 0.3b | .0053 | 4.00 ± 0.04b | .0128 | 48.3 ± 0.7 | - - - |
| 4-15(20) | 0.148 ± 0.006 | - - - | 1.976 ± 0.023 | - - - | 7.4 ± 0.3 | - - - | 3.77 ± 0.04b | .0138 | 48.8 ± 0.7 | - - - |
| 4-16 (20) | 0.152 ± 0.006 | - - - | 1.988 ± 0.024 | - - - | 7.6 ± 0.3 | - - - | 3.76 ± 0.04a | .0032 | 48.9 ± 0.7 | - - - |
| 4-13 (17) | 0.144 ± 0.007 | - - - | 1.973 ± 0.026 | - - - | 7.3 ± 0.3 | - - - | 3.79 ± 0.04 | - - - | 45.6 ± 0.7 | - - - |
| 4-22 (20) | 0.099 ± 0.007c | .0001 | 2.020 ± 0.028 | - - - | 4.7 ± 0.3c | .0001 | 3.48 ± 0.05c | .0001 | 43.9 ± 0.8c | .0011 |
| 4-19 (27) | 0.164 ± 0.008 | - - - | 2.145 ± 0.008a | .0001 | 7.6 ± 0.4 | - - - | 3.59 ± 0.04c | .0001 | 49.2 ± 0.8 | - - - |
| 4-21 (22) | 0.234 ± 0.008a | .0001 | 2.279 ± 0.026a | .0001 | 10.2 ± 0.3a | .0001 | 3.49 ± 0.04 c | .0001 | 53.8 ± 0.8a | .0001 |
| 4-17 (22) | 0.251 ± 0.007a | .0001 | 2.271 ± 0.031a | .0001 | 11.0 ± 0.3a | .0001 | 3.56 ± 0.04c | .0001 | 54.4 ± 0.7a | .0001 |
| 4-14 (20) | 0.175 ± 0.007 | - - - | 2.025 ± 0.023 | - - - | 8.6 ± 0.3 | - - - | 3.97 ± 0.05 | - - - | 47.6 ± 0.8 | - - - |
| 4-10 (20) | 0.206 ± 0.006a | .0001 | 2.046 ± 0.024b | .0044 | 10.2 ± 0.3a | .0001 | 4.23 ± 0.04a | .0001 | 48.0 ± 0.7 | - - - |
| 4-11 (23) | 0.241 ± 0.010a | .0001 | 1.923 ± 0.022 | - - - | 12.6 ± 0.5a | .0001 | 4.26 ± 0.04a | .0001 | 50.4 ± 0.8a | .0002 |
| 4-12 (19) | 0.143 ± 0.007 | - - - | 2.009 ± 0.024 | - - - | 7.1 ± 0.3 | - - - | 3.86 ± 0.04 | - - - | 44.0 ± 0.7c | .0005 |
| 4-6 (19) | 0.186 ± 0.007a | .0004 | 2.158 ± 0.025a | .0001 | 8.5 ± 0.3 | - - - | 3.87 ± 0.04 | - - - | 48.1 ± 0.7 | - - - |
| 4-4 (21) | 0.175 ± 0.007 | - - - | 1.994 ± 0.026 | - - - | 8.7 ± 0.3 | - - - | 3.96 ± 0.04 | - - - | 76.5 ± 2.2a | .0001 |
| 4-8 (20) | 0.179 ± 0.009 | - - - | 2.152 ± 0.040a | .0001 | 8.3 ± 0.4 | - - - | 3.85 ±0.05 | - - - | 47.5 ± 0.9 | - - - |
= significantly (p < 0.003) or
= suggestively (p < 0.01) increased from B6 controls;
= significantly (p < 0.003) or
= suggestively (p < 0.01) decreased from B6 controls
Table 5.
Distal femoral microCT40 data from 4 month old congenic males. Data presented as mean ± SEM
| Congenic strain | BV (mm2) | ‘p’ for BV | TV (mm2) | ‘p’ for TV | BV/TV (%) | ‘p’ for BV/TV | Trab.no (1/μm) | ‘p’ for No | Trab.thk (μm) | ‘p’ for Thk |
|---|---|---|---|---|---|---|---|---|---|---|
| B6 (44) | 0.656 ± 0.023 | 2.676 ± 0.045 | 24.2 ± 0.5 | 5.44 ± 0.04 | 64.6 ± 0.5 | |||||
| 4-2 (15) | 0.555 ± 0.037 d | .0135 | 2.374 ± 0.069 c | .0002 | 22.9 ± 0.9 | - - - | 5.53 ± 0.06 | - - - | 56. 6± 1.0 c | .0001 |
| 4-5 (15) | 0.961 ± 0.058 a | .0001 | 3.051 ± 0.094 a | .0010 | 30.9 ± 1.2 a | .0001 | 5.72 ± 0.07 a | .0006 | 62.3 ± 1.1 | - - - |
| 4-7 (23) | 0.669 ± 0.030 | - - - | 2.633 ± 0.061 | - - - | 24.8 ± 0.8 | - - - | 5.67 ± 0.05 a | .0003 | 58.3 ± 0.8 c | .0001 |
| 4-9 (21) | 0.672 ± 0.030 | - - - | 2.742 ± 0.062 | - - - | 24.6 ± 0.7 | - - - | 5.41 ± 0.05 | - - - | 59.5 ± 0.7 c | .0001 |
| 4-15 (20) | 0.638 ± 0.033 | - - - | 2.766 ± 0.068 | - - - | 22.9 ± 0.8 | - - - | 5.23 ± 0.06 | - - - | 59.4 ± 0.8 c | .0001 |
| 4-16 (19) | 0.592 ± 0.033 | - - - | 2.640 ± 0.066 | - - - | 22.4 ± 0.9 | - - - | 5.38 ± 0.06 | - - - | 57.1 ± 0.9 c | .0001 |
| 4-13 (20) | 0.624 ± 0.032 | - - - | 2.707 ± 0.067 | - - - | 23.0 ± 0.7 | - - - | 5.29 ± 0.07 | - - - | 59.4 ± 0.7 c | .0001 |
| 4-22 (20) | 0.467 ± 0.032 c | .0001 | 2.320 ± 0.061 c | .0001 | 20.5 ± 0.8 c | .0001 | 5.01 ± 0.06 c | .0001 | 59.7 ±1.0 c | .0001 |
| 4-19 (23) | 0.707 ± 0.033 | - - - | 2.988 ± 0.060 a | .0001 | 23.0 ± 0.8 | - - - | 5.04 ± 0.06 c | .0001 | 60.9 ± 1.0 d | .0059 |
| 4-21 (22) | 0.735 ± 0.033 | - - - | 2.996 ± 0.062 a | .0002 | 24.4 ± 0.8 | - - - | 4.88 ± 0.07 c | .0001 | 64.5 ± 0.8 | - - - |
| 4-17 (22) | 0.640 ± 0.031 | - - - | 2.774 ± 0.062 | - - - | 23.0 ± 0.8 | - - - | 5.04 ± 0.05 c | .0001 | 62.0 ± 0.9 | - - - |
| 4-14 (20) | 0.764 ± 0.032 b | .0040 | 2.745 ± 0.064 | - - - | 27.8 ± 0.7 a | .0001 | 5.94 ± 0.05 a | .0001 | 57.1 ± 0.7 c | .0001 |
| 4-10 (18) | 0.588 ± 0.035 | - - - | 2.594 ± 0.069 | - - - | 22.5 ± 0.8 | - - - | 5.73 ± 0.05 a | .0001 | 52.3 ± 0.9 c | .0001 |
| 4-11 (20) | 0.673 ± 0.031 | - - - | 2.593 ± 0.063 | - - - | 26.0 ± 0.7 | - - - | 5.74 ± 0.05 a | .0001 | 56.7 ± 0.8 c | .0001 |
| 4-12 (19) | 0.506 ± 0.033 c | .0009 | 2.567 ± 0.065 | - - - | 19.4 ± 0.8 c | .0001 | 5.37 ± 0.05 | - - - | 52.3 ± 0.9 c | .0001 |
| 4-6 (21) | 0.468 ± 0.033 c | .0001 | 2.636 ± 0.063 | - - - | 18.1 ± 0.9 c | .0001 | 5.20 ± 0.08 | - - - | 50.7 ± 0.8 c | .0001 |
| 4-4 (20) | 0.573 ± 0.037 | - - - | 2.562 ± 0.039 | - - - | 21.9 ± 0.9 | - - - | 5.68 ± 0.05 a | .0005 | 52.9 ± 0.9 c | .0001 |
| 4-8 (16) | 0.592 ± 0.038 | - - - | 2.734 ± 0.069 | - - - | 21.6 ± 0.9 | - - - | 5.32 ± 0.07 | - - - | 54.7 ± 0.9 c | .0001 |
= significantly (p < 0.003) or
= suggestively (p < 0.01) increased from B6 controls;
= significantly (p < 0.003) or
= suggestively (p < 0.01) decreased from B6 controls
Lastly, the Micro CT40 phenotyping data for congenic strain males compared with B6 controls are listed in Table 5. Fewer significant changes in BV or TV associated with C3H sequences were observed in males compared with females. In the BV data, there are 1 significant and 1 suggestive increase, plus 3 significant and 1 suggestive decreases. In the TV data, there are 3 significant increases and 2 significant decreases. In the Trab.no data, there are 6 significant increases and 4 significant decreases. For Trab.thk data, there areno increases, whereas there are 14 significant and 1 suggestive decreases. Thus, the C3H sequences do not show a clear pattern of effects on male BV, TV, or Trab.no, whereas there was a predominantly negative effect on Trab.thk. The greatest increases or decreases, respectively, in these 4 trabecular parameters were: a) BV in 4-5 (+46.5%) and in 4-22 (−28.8%); b) TV in 4-5 (+14.0%) and in 4-22 (−13.3%); c) Trab.no in 4-14 (+9.2%) and in 4-21 (−10.3%); and d) Trab.thk (no increase) and in 4-6 (−21.5%).
The combined genetic haplotype and phenotype data summary are present in Figure 1. The left hand side of the figure shows the summary of the genotyping data for each congenic strain, while the right hand side shows the phenotype data partitioned by sex. Based upon the segregation of measured phenotypes (Tables 2 – 5) among the 18 congenic strains, we propose 10 distinct QTL regions in the mid-distal portion of Chr 4 as presented in Figure 1. The 10 regions (Roman numerals, I to X) are located between the haplotype map and the associated phenotypes. The numbers of genes by QTL region were determined from genetic databases and ranged from 1 to 556. A given phenotype was assigned to the QTL region by examination of data from congenic strains that shared a genomic sequence. A phenotype was assigned as positive or negative when the congenic data yielded a net significant and suggestive effect. Not more than one non-conforming strain was allowed per QTL assignment. The QTL region boundaries were determined by the proximal and distal genetic markers of a congenic subset sharing the phenotypic change.
Addressing the femoral pQCT data first, we observed that in females, the femoral mineral in females decreased in region I, while the femoral volume increased volume only in region IV. In males, we found that femoral mineral increased in regions II, VI, and VII, while mineral decreased in region IX. For femoral volume, we observed increases in regions VI and VII, while decreases were observed in region IX.
The trabecular bone assessed by Micro CT40 showed remarkable complexity. Assessment of female data showed region I increased BV and region X increased TV. Region VII increased Trab.no, while QTL regions III and V decreased trab.no and region I increased Trab.thk. The male data did not reveal QTL regions for BV or TV that were associated with congenic strains sharing a common genomic sequence. On the other hand, region VII increased Trab.no., while QTL regions III, VI, VIII, and X decreased Trab,thk.
An inspection of Figure 1 by QTL region across bone phenotypes was undertaken to look for similarities and differences in allele effects on bone compartments by sex. The only QTL region that is associated with the same response in females and males appears to be region VII for Trab.no. The remaining QTL appear to be sex specific and with variety of phenotypic effects.
Table 6 presents the QTL regions derived from our data, the physical segment encompassed by each region, candidate bone genes identified within each region, selected references, and SNP differences between B6 and C3H alleles that have potential regulatory effects. We searched databases and original literature to find possible candidate genes that could be credibly linked to bone. The SNP database found in The Center for Genome Dynamics maintained at The Jackson Laboratory (http://cgd.jax.org/) was used to determine the number of SNP differences between B6 and C3H for each gene. Table 6 lists the number of SNPs that are non-synonymous, or located in the 3′ or 5′UTRs.
Table 6.
Proposed Chr 4 bone QTLs, physical map positions, candidate bone genes, and biological effects. Number of SNP differences between B6 and C3H with potential regulatory effects.
| QTL | Genetic region (Mb) | Bone gene | Skeletal effects & references | -------------------SNPs------------------ | ||
|---|---|---|---|---|---|---|
| Non-synon | 3′ UTR | 5′ UTR | ||||
| I | 100.423 – 124.588 | ? | Not determined | |||
| II | 127.039 – 128.322 | Phc2 | Axial skeletal Development (30) |
0 | 2 | 0 |
| III | 129.225 – 129.250 | Lck | OC bone resorption inhibited (25, 26) | 0 | 2 | 0 |
| IV | 129.725 – 129.735 | Col16a1 | Role in chondrogenesis (32) | 7 | 1 | 9 |
| V | 134.292 – 135.540 | Cnr2 | OBs, enhanced formation (10) | 0 | 0 | 0 |
| VI | 136.535 – 136.628 | Zbtb40 | Human BMD, GWAS (3, 5) | 5 | 40 | 0 |
| VII | 136.652 – 139.638 | Wnt4 | OBs increased formaion (34) | 0 | 0 | 0 |
| Alpl | OBs, tissue non-specific, promotes bone mineralization (35) | 1 | 1 | 0 | ||
| Ece1 | OBs, bone formation (23) | 3 | 19 | 1 | ||
| Pla2g2a | OC genesis (36) | 3 | 0 | 0 | ||
| VIII | 140.707 – 142.094 | EphA2 | OC genesis enhanced, OB genesis suppressed (37) | 0 | 0 | 0 |
| IX | 144.647 – 147.413 | Tnfrsf1b | Human BMD 20-gene test (8), GWAS (11) | 4 | 6 | 0 |
| Plod1 | OBs, bone matrix collagen maturation (38) | 0 | 0 | 1 | ||
| Mthfr | Human GWAS, BMD and Fx (11) | 0 | 1 | 0 | ||
| X | 149.300 – 150.994 | ? | None identified | |||
Discussion
The distal region of mouse Chr 4 in B6C3F2 segregating progeny was originally shown to have significant regulation for femoral and vertebral vBMD (17). This was confirmed in congenic strain B6.C3H-4T carrying 70 Mb of C3H sequence on Chr 4 in a B6 background (18). Other investigators using crosses among additional inbred mouse strains have also found bone QTLs in the mid-distal region of Chr 4 (21) as reviewed by Ackert-Bicknell and colleagues (22). Mouse Chr 4 is recognized as being gene-rich, consequently it was not surprising that our data revealed substantial regulation of bone phenotypes. QTL region I extends from approximately 100.423 to 124.588 Mb physical distance and contains at least 556 functionally described genes. This region is homologous with human Chr regions 1p31 to 1p34. Bone phenotypes were readily associated with QTL region I, however, we lacked congenic strain resources needed to undertake phenotype mapping of this large segment. The region of interest homologous with human 1p36 is defined by proposed QTL regions II - X within the Chr 4 congenic strains, includes approximately 24Mb, and contains at least 716 known genes from NCBI Ensembl Mouse Build 37 release 59. The numbers of candidate genes present in the distal region of Chr 4 that participate in regulation of bone proved substantial. Nevertheless, bone has major roles in mammalian physiological systems and that implies the necessity of complex genetic support.
Congenic strain mapping of mouse bone phenotypes is advantageous for several reasons. For example, subtle effects can be detected at very high resolution in mice homozygous for a specific genetic sequence by comparison with mice of the genetic background, such as B6. Phenotypic variation arises only from the genetic segment of interest on one chromosome while other genes affecting the phenotype in the genetic background remain constant. Furthermore, it is relatively easy to obtain the number of mice needed for hypothesis testing. Finally, congenic strains carrying defined QTL provide excellent tools for biological studies to follow-up on genetic findings.
The congenic strains with the shorter C3H sequences are more informative since they contain fewer QTL regions that affect a given phenotype. For example, the congenic strain 4-5 (see Figure 1) covers as many as 6 QTL regions. The individual QTL regions within the 4-5 congenic strain have both positive and negative effects on femoral bone mineral in females and males. Thus, the net phenotype for mineral in the 4-5 congenic strain is a consequence of the allelic effects in all six regions. The same issue pertains to all congenics that include C3H sequences from more than one region.
The QTL regions within one congenic strain occasionally modify effects from neighboring regions. For example, congenic strains 4-7, 4-9, 4-10, and 4-11 span QTL region III, V, and VII and have increased female Trab.no. Based on female data from 4-15, 4-13, 4-22, 4-19, 4-21, and 4-17, we observed strong negative effects associated with regions III and V. We interpret this to mean that the positive effect in region VII is dominant to both regions III and V, resulting in the increased Trab.no observed in congenics 4-7, 4-9, 4-10, and 4-11. We did not clearly observe this interaction in the males from these congenic strains. Proof of this proposed genetic interaction requires a set of F2 progeny with both sexes analyzed for segregation of phenotypes with genetic segments.
As noted above, the QTL regions, II to X, are of greatest interest since they are homologous with human Chr 1p36. The numbers of known genes by region (Table 6) ranged from modest numbers (1 to 14) in four regions to large numbers (30 to 102) in the other five regions. Nevertheless, within the nine QTL regions, we extracted 13 candidate genes from databases and original literature reporting effects on bone cells or processes. These candidate genes included six linked to osteoblasts - Cnr2, Wnt4, Alpl, Ece1, EphA2, Plod1, three linked to osteoclasts -Lck, Pla2g2a, EphA2, and two linked to skeletal development - Phc2, Col16a1. Three genes -Zbtb40, Tnfrsf1b, and Mthfr - were reported in meta-analyses of GWAS focused on human BMD or fractures in large populations. As yet, there are no specific literature reports associating these latter three genes to actions in bone. However, QTL regions VI and IX in the Chr 4 congenic strains contain Zbtb40 and Mthfr genes, respectively, lending some support to their possible roles in bone.
Comparison of alleles of the 13 candidate genes was undertaken to determine sequence differences with functional consequences. Table 6 shows that 6 of 13 possible candidate genes (Col16a1, Zbtb40, Alpl, Ece1, Pla2g2a, Tnfrsf1b) have non-synonymous SNPs within exons, which strongly support the possibility that the C3H alleles of these genes could be coding for a functional difference in mRNA. Eight of 13 genes have SNP differences in the 3′UTR, which could be associated with changes in mRNA stability, localization, or translation. Furthermore, 2 of 13 genes have SNP differences in the 5′UTR that may also have functional implications for the regulation of these genes. Three genes, Cnr2, Wnt4 and EphA2, have no SNP differences between B6 and C3H sequences and thus may not be good candidates for direct action in our congenic strains. Nevertheless, we cannot rule out interaction with other genes that would lead to a phenotypic difference. In summary, the sequence variations among 10 of the 13 genes provide ample evidence for viable candidates that could participate in regulation of bone.
The Lck gene is the only known gene in the small 0.025 Mb segment within QTL region III. Lck protein is found in the cytoplasm and is recruited to the plasma membrane together with Src protein to form a functioning T-cell receptor (23,24). The Lck function is best seen in a gene knock-out model that has thymic atrophy and decreased peripheral T-cells resulting in the inhibition of OC bone resorption (23). Thus, we speculate that this gene could be an example of an interface between bone and the immune system as synthesized in the developing field of Osteoimmunology (see reviews in: (25–27)). The C3H mice have been shown to have reduced bone resorption and high bone mass (28). SNP databases indicate many polymorphisms exist between B6 and C3H within the Lck genetic sequence. Although there are no non-synonymous SNP differences in exons, there are 2 SNPs in the 3′UTR. Thus, a polymorphism in the Lck 3′UTR in the C3H allele may lead to altered function and reduced bone phenotype in the 4-22 congenic mice. This hypothesis was tested by real time PCR on cDNA from whole bone and spleen from B6 and C3H females at 8 weeks of age. No differences in Lck expression were observed between strains for either tissue. The LCK protein has not yet been investigated for quantitative differences.
A remarkable feature of the Chr 4 congenic strains was the phenotype effect size introduced by the C3H sequence in comparison with values recorded in B6 controls. Both pQCT and μCT40 yielded differences that were readily measured with the differences found by Micro CT40 generally greater than found by pQCT. These effect sizes are large enough to support studies undertaken to evaluate treatments, such as pharmacologic agents or gonadectomy. These effects support the use of these animal models to define candidate genes in the human Chr 1p36 region.
The commonly utilized phenotypes of vBMD and BV/TV were gathered in our studies of data derived from the pQCT and Micro CT40 instruments, and these indices are presented in Tables 2 to 5. Both vBMD and BV/TV are ratios derived from calculations with measurements gained about other phenotypes. These measures are clinically important because they can be readily obtained and are accurately reflective of a patient’s skeletal status. Clearly, a ratio such as vBMD, can increase or decrease through changes in the numerator, denominator or both terms. Determination of why a ratio behaved as it did following a treatment requires information about the components. As investigators press the search for mechanistic evidence that a candidate gene regulates bone, accurate knowledge is needed about how traits such as mineralization, bone size or both, are affected in a given study. Thus, we chose the 6 described phenotypes for mapping bone regulation to mid-distal Chr 4 in congenic strains with the intent of gaining insight to specific bone changes associated with defined genomic regions. Our goal has been realized by dividing the 24 Mb region of mouse mid-distal Chr 4 into smaller QTL regions that demonstrate gender and bone compartment regulation. The information gained about bone mineral, size, and architectural QTL would be enhanced by comparisons with QTL derived from mapping phenotypes of strength such as maximum load, energy to break, and modulus. Other important bone phenotypes such as mid-shaft bone dimensions and cortical thickness, as well as functional load modeling have been demonstrated to be associated with this Chr 4 region (29). Demonstration of co-segregation of QTL for multiple bone phenotypes would contribute to prioritizing candidate gene analyses.
In summary, we found that the mid-distal region of mouse Chr 4 was a rich source of potential candidate genes regulating both cortical and trabecular bone. Several of these genes are biologically plausible factors in determining bone acquisition and/or maintenance. In addition these data are very consistent with GWAS studies that have identified polymorphisms in genes that may be adjacent to each other and confer additive effects on bone mass and/or fracture. Inbred and congenic strains are powerful tools for identifying regulatory regions that influence bone turnover. An important limitation to our congenic strain approach is that QTL regions could be better refined by phenotyping and genotyping reciprocal F2 intercross progenies from each congenic crossed with B6. Such progeny would also permit assessment of gene-gene interactions, as well as testing for epigenetic effects on phenotype expression. Nevertheless, we speculate that genetic engineering methods using a conditional approach can provide definitive proof of biologic plausibility for individual genes. The challenge remains in both mice and humans, to understand gene-gene and gene by environment interactions.
Acknowledgments
This work was supported by NIAMS grants AR043618 and AR56404-ARRA. We thank Drs. Ken Johnson and Greg Cox for critical review and suggestions for the ms.
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
Author’s contributions to manuscript:
WGB, KLS, LRD, and CJR collectively designed the research; KLS, HFC, LGH managed record systems, matings, necropsies, and phenotyping of congenic mice; WGB, KLS, CJR analyzed data and wrote the manuscript; WGB had final responsibility for manuscript and data. All authors read and approved the final manuscript.
Contributor Information
Wesley G. Beamer, Email: wesley.beamer@jax.org.
Kathryn L. Shultz, Email: kathy.shultz@jax.org.
Harold F. Coombs, III, Email: harold.coombs@jax.org.
Lindsay G. Horton, Email: lhorton85@gmail.com.
Leah Rae Donahue, Email: leahrae.donahue@jax.org.
Clifford J. Rosen, Email: crofen@gmail.com.
References
- 1.Hardy J, Singleton A. Genomewide association studies and human disease. NEJM. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Rho Y, Choi S, Ji J, Song G. Meta-analysis of genome-wide linkage studies for bone mineral density. J Human Genetics. 2006;51:480–486. doi: 10.1007/s10038-006-0390-9. [DOI] [PubMed] [Google Scholar]
- 3.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, Bagger Y, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. Multiple genetic loci for bone mineral density and fractures. NEJM. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 4.Rivadeneira F, Styrkarsdottir U, SEstrada K, Halldorsson VH, Hsu Y-H, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Grundberg E, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra B, Pastinen T, Pols HAP, Sigurdsson G, Soranzo N, Thorleifsson G, Thorsteinsdottir U, Williams FMK, Wilson SG, Zhou Y, Ralston SH, van Duijn CM, Spector T, Kiel Dp, Stefansson K, Ionnidis JPA Uitterlinden AG Consortium GFFOG. Twenty bone mineral deisity loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Styrkarsdottir U, Halldorsson BV, Gudbjartsson DF, Tang NLS, Koh J-M, Xiao S-M, Kwok TCY, Kim GS, Chan JCN, Cherny S, Lee SH, Kwok A, Ho S, Gretarsdottir S, Kostic JP, Palsson ST, Sigurdsson G, Sham PC, Kim B-J, Kung AWC, Kim S-Y, Leung P-C, Kong A, Thorsteinsdottir U, Stefansson K. European bone mineral density loci are also associated with BMD in East-Asian populations. PLos ONE. 2010;5(10):e13217. doi: 10.1371/journal.pone.0013217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karasik D, Cupples LA, Hannan MT, Kiel DP. Genome screen for a combined bone phenotype using principle component analysis: The Framingham study. Bone. 2004;34:547–556. doi: 10.1016/j.bone.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Devoto M, Spotilla LD, Stabley DL, Wharton GN, Rydbeck H, Korkko J, Kosich R, Prockop D, Tenenhouse A, Sol-Church K. Univariate and bivariate variance compnent linkage analysis of a whole-genome scan for loci contributing to bone mineral density. European J Human Genetics. 2005;13:781–788. doi: 10.1038/sj.ejhg.5201411. [DOI] [PubMed] [Google Scholar]
- 8.Xiao P, Shen H, Guo YF, Xiong DH, Liu YZ, Zhao LJ, Long JR, Guo Y, Reker RR, Deng HW. Genomic regions identified for BMD in a large sample including epistatic interactions and gender-specific effects. J Bone Miner Res. 2006;21:1536–1544. doi: 10.1359/jbmr.060717. [DOI] [PubMed] [Google Scholar]
- 9.Streeten E, MacBride D, Pollin T, Rayan K, Shapiro J, Ott S, Mitchell B, Shuldiner A, O’Connell J. Quantitative trait loci for BMD identified by autosome-wide linkage scan to Chromosome 7Q and 21Q in men from the Amish family osteoporosis study. J Bone Min Res. 2006;21:1433–1442. doi: 10.1359/jbmr.060602. [DOI] [PubMed] [Google Scholar]
- 10.Bab I, Zimmer A. Cannabinoid receptors and the regulation of bone mass. Br J Pharmacol. 2008;153:182–188. doi: 10.1038/sj.bjp.0707593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Q, Li GH, Kung AW. Multiple osteoporosis susceptibility genes on chromosome 1p36 in Chinese. Bone. 2009;44:984–988. doi: 10.1016/j.bone.2009.01.368. [DOI] [PubMed] [Google Scholar]
- 12.Xiong D-H, Shen H, Zhao L-J, Xiao P, Yang T-L, Guo Y, Wang W, Guo Y-F, Liu Y-J, Recker RR, Deng H-W. Robust and comprehensive analysis of 20 osteoporosis candidate genes by very high-density single-nucleotide polymorphism screen among 405 white nuclear families identified significant association and gene-gene interaction. J Bone Miner Res. 2006;21:1678–1695. doi: 10.1359/JBMR.060808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Sol-Church K, Rydbeck H, Stabley DL, Spotilla LD, Devoto M. High resolution linkage and linkage disequilibrium analyses of chromosome 1p36 SNPs identify new positional candidate genes for low bone mineral density. Osteoporosis Int. 2009;20:341–346. doi: 10.1007/s00198-008-0668-1. [DOI] [PubMed] [Google Scholar]
- 14.Ermakov S, Toliat MR, Cohen Z, Malkin I, Altmuller J, Livshits J, Nurnberg P. Association of ALPL and ENPP1 gene polymorphisms with bone strength related traits in a Chuvashian population. Bone. 2010;46:1244–1250. doi: 10.1016/j.bone.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Donahue L. Mouse Phenome Database web site. The Jackson Laboratory; Bar Harbor, Maine USA: 2011. Bone mineral density, body composition, and craniofacial characterization in 30 inbred strains of mice. MPD:11506. http://phenome.jax.org. [Google Scholar]
- 16.Beamer WG, Donahue LR, Rosen CJ, Baylink DJ. Genetic variability in adult bone density among inbred strains of mice. Bone. 1996;18:397–403. doi: 10.1016/8756-3282(96)00047-6. [DOI] [PubMed] [Google Scholar]
- 17.Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JE, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16:1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 18.Shultz KL, Donahue LR, Bouxsein ML, Baylink DJ, Rosen CJ, Beamer WG. Congenic strains of mice for verification and genetic decomposition of quantitative trait loci for femoral bone mineral density. J Bone Miner Res. 2003;18:175–185. doi: 10.1359/jbmr.2003.18.2.175. [DOI] [PubMed] [Google Scholar]
- 19.Svenson KL, Cheah Y-C, Shultz KL, Mu J-L, Paigen B, Beamer WG. Strain distribution pattern for the SSLP markers in the SWXJ recombinant inbred strain set: Chromosomes 1 to 6. Mam Genome. 1995;6:867–872. doi: 10.1007/BF00292437. [DOI] [PubMed] [Google Scholar]
- 20.Lang DH, Sharkey NA, Lionikas A, Mack HA, Larsson L, Vogler GP, Vandenbergh DJ, Blizzard DA, Stout JT, Stitt JP, McClearn GE. Adjusting data to body size: A comparison of methods as applied to Quantitative Trait Loci analysis of musculoskeletal phenotypes. J Bone Miner Res. 2005;20:748–757. doi: 10.1359/JBMR.041224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saless N, Litscher SJ, Lopez Franco GE, Houlihan MJ, Sudhakaran S, Raheem KA, O’Neil TK, Vanderby R, Demant P, Blank RD. Quantitative trait loci for biomechanical performance and femoral deometry in an intercross of recombinant congenic mice: restriction of the Bmd7 candidate interval. FASEB J. 2009;23:2142–2154. doi: 10.1096/fj.08-118679. [DOI] [PubMed] [Google Scholar]
- 22.Ackert-Bicknell CL, Karasik D, Li Q, Smith RV, SHsu Y-H, Churchill GA, Paigen BJ, Tsaih S-W. Mouse BMD Quantitative Trait Loci show improved concordance with human genome-wide association loci when recalculated on a new, common mouse genetic map. J Bone Miner Res. 2010;25:1808–1820. doi: 10.1002/jbmr.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 24.Shakespeare W, Yang M, Bohacek R, Cerasoli F, Stebbins K, Sundaramoorthi R, Azimioara M, Vu M, Pradeepan S, Metcalf C, II, Haraldson C, Merry T, Delgarno D, Narula S, Hatada M, Lu X, van Schravendijk MR, Adams S, Violette S, Smith J, Guan E, Bartlett C, Herson J, Liliucci J, Weigele M, Sawyer T. Structure-based design of an osteoclast-selective, nonpeptide Src homology 2 inhibitor with in vivo antiresorptive activity. PNAS. 2000;97:9373–9378. doi: 10.1073/pnas.97.17.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horowitz MC, Bothwell ALM, Hesslein DGT, Pflugh DL, Schatz DG. B cells and osteoblast and osteoclast developent. Immunological Reviews. 2005;208:141–153. doi: 10.1111/j.0105-2896.2005.00328.x. [DOI] [PubMed] [Google Scholar]
- 26.Ishimori N, Li R, Walsh K, Korstanje R, Rollins J, Petkov P, Pletcher M, Wiltshire T, Donahue L, Rosen C, Beamer W, Churchill G, Paigen B. Quantitaive trait loci that determine BMD in C57BL/6J and 129S1/SvImJ inbred mice. J Bone Miner Res. 2006;21:105–112. doi: 10.1359/JBMR.050902. [DOI] [PubMed] [Google Scholar]
- 27.Kacena MA, Nelson T, Cloough ME, Lee S-K, Lorenzo JA, Gundberg CM, Horowitz MC. Megakaryocyte inhibition of osteoclast development. Bone. 2006;39:991–999. doi: 10.1016/j.bone.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Linkhart TA, Linkhart SG, Kodama Y, Farley JR, PDH, Wright KR, Wergedal JE, Sheng MA, Beamer WG, Donahue LR, Rosen CJ, Baylink D. Osteoclast formation in bone marrow cultures from two inbred strains of mice with different bone densities. J Bone Mineral Res. 1999;14:39–46. doi: 10.1359/jbmr.1999.14.1.39. [DOI] [PubMed] [Google Scholar]
- 29.Robling AG, Li J, Shultz KL, Turner CH. Evidence for a skeletal mechanosensitivity gene on mouse chromosome 4. FASEB Journal. 2003;17:324–326. doi: 10.1096/fj.02-0393fje. [DOI] [PubMed] [Google Scholar]
- 30.Isono K, Fujimura Y, Shinga J, Yamaki Y, O-Wang J, Takihara Y, Murahashi Y, Takada Y, Mizutani-koseki Y, Koseki H. Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mole Cell Biol. 2005;25:6694–6706. doi: 10.1128/MCB.25.15.6694-6706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiya I, Vuoristo JT, Larson BL, Procop DJ. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. PNAS. 2002;99:4397–4402. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao C, Irie N, Takada Y, Shimoda K, Miyamoto T, Nishiwaki T, Suda T, Matsuo K. Bidirectional ephrinB2-EphB4 signaling controls bone homeostasis. Cell Metab. 2006;4:111–121. doi: 10.1016/j.cmet.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Krause C, de Gorter DJJ, Karperien M, ten Dijke P. Signal transduction cascades controlling osteoblast differentiation. In: Rosen CJ, Compston JE, Lian JB, editors. Primer on the metabolic bone diseases and disorders of mineral metabolism. 7. American Society for Bone and Mineral Research; Washington DC: 2008. pp. 10–16. [Google Scholar]
- 34.Tesch W, Vandenbos T, Roschgr P, Fratzl-Zelman N, Klaushofer K, Beertsen W, Fratzl P. Orientation of mineral crystallites and mineral density during skeletal development in mice deficient in tissue nonspecific alkaline phosphatase. J Bone Miner Res. 2003;18:117–125. doi: 10.1359/jbmr.2003.18.1.117. [DOI] [PubMed] [Google Scholar]
- 35.Mintz MB, Sowers R, Brown KM, Hilmer SC, Mazza B-A, Huvos AG, Meyers PA, LaFleur B, McDonough WS, Henry MM, Ramsey KE, Antonescu CR, Chen W, Healey JH, Daluski A, Berens ME, MacDonald TJ, Gorlick R, Stephan DA. An expression signature classifies chemotherapy-resistant pediatric osteocarcoma. Cancer Res. 2005;65:1748–1754. doi: 10.1158/0008-5472.CAN-04-2463. [DOI] [PubMed] [Google Scholar]
- 36.Irie N, Takada Y, Watanabe Y, Matsuzaki Y, Naruse C, Asano M, Iwakura T, Matsuo K. Bidirectinal signaling through EphrinA2-EphA2 enhances osteoclastogenesis and suppresses osteoblastogenesis. J Biol Chem. 2009;284:14637–14644. doi: 10.1074/jbc.M807598200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takaluoma K, Hyry M, Lantto J, Sormunen R, Bank RA, Kivirikko KI, Myllyharju J, Soininen R. Tissue-specific changes in the hydroxylysine content and cross-links of collagens and alterations in fibrile morphology in lysyl hydroxylase 1 knock-out mice. J Biol Chem. 2007;282:6588–6596. doi: 10.1074/jbc.M608830200. [DOI] [PubMed] [Google Scholar]