Abstract
At least 10 million individuals worldwide are co-infected with immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV). These two viruses are transmitted most primarily by exposure to infected blood or blood products. Various nucleic acid assays have been developed for diagnostics and therapeutic monitoring of infections. In the present study, a multiplex real-time PCR assay for simultaneous detection of HCV and HIV-1 using molecular beacons were designed and validated. A well-conserved region in the HIV-1 pol gene and 5′NCR of HCV genome were used for primers and molecular beacon design. The analysis of scalar concentrations of the samples indicated that this multiplex procedure detects at least 1,000 copies/ml of HIV-1 and 100 copies/ml of HCV with linear reference curve (R2 > 0.94). The results demonstrate that a specificity of 100 % and sensitivity of 96 % can be achieved. The analytical sensitivity study with BLAST software demonstrated that the primers do not attach to any other sequences except for that of HIV-1 or HCV. The primers and molecular beacon probes only detected HIV-1 and all major variants of HCV. This assay may represent an alternative rapid and relatively inexpensive screening method for detection of HIV-1/HCV co-infection especially in blood screening.
Keywords: HIV-1, HCV, Multiplex real-time PCR, Molecular beacon
Introduction
In the past decade, molecular techniques and particularly PCR as a rapid and sensitive method has revolutionalized detection of a variety of infectious viruses [1, 2]. Typically NAATs1 require a significant investment in equipment, training and infrastructures. In this respect, World Health Organization (WHO) has recommended that diagnostic devices should be “ASSURED”: affordable, sensitive, specific, user-friendly, rapid and robust, equipment free, and deliverable to end users, especially in case of developing countries. Application of these molecular techniques as diagnostic tools has become increasingly important [3–7].
Multiplex assays in which two or more viruses with common transmission routes can be detected are also becoming important in particular areas, for example triplex assays in which nucleic acid of blood borne viruses HBV, HCV and HIV are detected in one assay have proven to be most useful in cutting time of screening tests and shortening of window period of these viruses, thereby improving blood safety [8–12]. Similarly, multiplex assays in which two viruses of similar transmission route that can affect treatment of co-infected patients is also becoming a useful tool [13]. For example Hepatitis C virus (HCV) infection has emerged as an important co-infection in the clinical and immunological development and treatment of HIV infection. Co-infected individuals may have an altered response to anti-retroviral treatment (ART) and are at increased risk of ART-related hepatotoxicity [14–17].
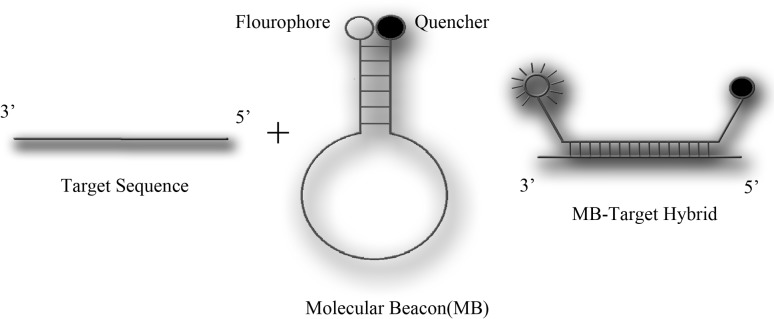
The present study was therefore designed for development of a multiplex real-time PCR assay for simultaneous detection of HCV and HIV using molecular beacons. Molecular beacons, first demonstrated by Tyagi and Kramer, are new optical tools that can be used to detect the presence of a specific oligonucleotide sequence in a mixture of targets [18]. As shown in Fig. 1, these DNA oligonucleotides contain a 5′ fluorophore, a 3′ quenching group, and 4–6 complementary bases on the 3′ and 5′ “stem” ends. When hybridized with a complementary sequence, the hairpin structure becomes linearized distancing the fluorophore and quencher resulting in fluorescence signal (Fig. 1). The hybridization signal depends on their molecular structure. Therefore, for detecting each pathogen, a separate molecular beacon with appropriate conformation in unhybridized, linearized and hybridized conformations should be designed [18–21]. This probe may provide an alternative method for detection of HIV-1 and HCV viruses with a simpler approach.
Fig. 1.
Schematic mechanism of molecular beacon
Materials and Methods
Probe and Primer Design
We designed and validated two sets of primers and molecular beacon probes on the most conserved region of pol gene in HIV-1 virus and 5′ non coding region (5′NCR) in HCV virus by aligning sequences in nucleotide database of NCBI (www.ncbi.nlm.nih.gov/nucleotide) containing full genomes of HIV-1 and HCV. All the alignments were carried out using Mega4 software. Pol gene in HIV-1 genome and 5′NCR of HCV genome were selected due to better sequence conservation in comparison with other genes.
The design of primers and probes was carried out using Beacon Designer 7 Software (Palo Alto, CA). The primer set chosen for HIV-1 virus amplified a 179 bp fragment in the HIV-1 pol gene. The primer sequences were 5′-GTACAGTGCAGGGGAAAG-3′ (forward) and 5′-CCAGAGTAGTTTTGCTGGTC-3′ (reverse). HCV primers were 5′-CATGGCGTTAGTAYGAGTG-3′ (forward) and 5′-CTATCAGGCAGTACCACAAG-3′ (reverse). The primer set selected for HCV virus amplified a 241-base pair fragment in the 5′NCR of HCV. To allow distinction between the fluorescence signal of HIV-1 and HCV, sequence-specific probes with two different reporter dyes were used.
The specific probe for HIV-1 was 5′-CCCGTGGTTTATTACAGGG ACAGCAGAACGGG-3′, labeled with the fluorescent reporter dye Tet (tetrachloro-6-carboxyfluorescein) at the 5′ end and the quencher dye BHQ-1 (Black Hole Quencher-1) at the 3′ end. The probe specific for the HCV was 5′-CCGATCAGCCATAGTGGTCTGCGGAAGAT CGG-3′ labeled with the fluorescent reporter dye FAM (6-carboxyfluorescein) at the 5′ end and the quencher dye BHQ-1 at the 3′ end. Stem sequences are indicated with underline.
Sample Preparation
Whole blood samples were collected from 70 adult individuals (All the received clinical specimens were untagged and re-labeled by numbers as sample identifier) including 30 HIV-1/HCV co-infected patients (group A), 10 HIV-1 (group B) and 10 HCV (group C) infected patients. These samples were previously screened by serological experiments and quantified by Artus HCV RG RT-PCR and Artus HIV-1 RG RT-PCR Kits (Qiagen, Hilden, Germany). In order to assess the specificity of the assay, 20 HIV-1 and HCV seronegative patients (group D) were tested. Twenty samples were manually prepared by spiking normal plasma with HIV-1 and HCV (group E).
Blood samples were collected in EDTA-containing tubes and were centrifuged at 2,500 rpm for 20 min and stored at −80 °C. Nucleic acids were extracted by QIAamp Viral RNA Mini kit (Qiagen, Hilden, Germany), eluted using 50 μl nuclease-free water, and stored at −80 °C until use.
Reverse Transcription Reaction
Following RNA extraction, the samples were reverse transcribed using expand reverse transcriptase (Roche Diagnostics GmbH Mannheim, Germany). 5 μl extracted RNA was added to 2 μl HCVR and 2 μl HIV-1R (2 mM), 2 μl dNTP (10 mM) and 1 μl distilled water and incubated at 65 °C for 10 min. The tubes were held in ice and 4 μl 5× RT-Buffer (Roche), 2 μl DTT (10 mM), 0.5 μl RNase inhibitor (20 U), 0.5 μl distilled water and 1 μl expand reverse transcriptase (50 U) was added. The cDNA synthesis was carried out at 43 °C for 60 min and then at 95 °C for 2 min for inactivation of the enzyme. The cDNA was stored at −20 °C until use.
HIV-1 and HCV Standards
Limit of detection of this method was calculated using, PCR products cloned in T/A cloning vector. In vitro transcription was performed in a final volume of 50 μl using the recombinant plasmid and T7 RNA Polymerase (Fermentas, Germany); in presence of 2 mM NTP mix; 10 μl transcription buffer; 50 U RNase inhibitor and 30 U T7 RNA Polymxerase in a 50 μl reaction. After transcription 5 U (2 U per 1 μg of DNA used) of RNAse free DNAse (Fermentas, Germany) was added for 30 min at 37 °C to degrade the template DNA. The transcribed RNA was purified by Trizol extraction. The integrity of RNA was checked on a 2 % formaldehyde agarose gel by electrophoresis and quantitated by measuring the optical density (OD) at 260 nm. The diluted RNA was tested for DNA contamination by PCR. Before extraction, the in vitro transcribed RNAs were added to HIV-1 and HCV negative plasma samples and mixed to achieve HIV-1 or HCV scalar dilutions of 102–106 copies/ml.
Multiplex Real-Time RT-PCR Using Molecular Beacon
HIV-1/HCV multiplex real-time PCR assay was performed in 25 μl PCR mixture volume composed of 10 μl 2× Quantifast Multiplex Probe PCR Master Mix (Qiagen), 0.4 μM HIV-1 and HCV oligonucleotide primers, 0.2 μM HIV-1 and HCV probes and 5 μl cDNA. Amplification was performed in the following conditions: activation step at 95 °C for 5 min and 45 cycles of three thermal amplification steps : 94 °C for 10 s, 56 °C for 30 s and 72 °C for 30 s. Single fluorescence detection was performed in each cycle at 56 °C to reveal the positive samples. Amplification, data acquisition and analysis were performed on Rotor-Gene 3000 (Corbett Research, Australia) using Rotor-Gene 6.1 software. All samples were run in duplicate.
Results
Probe and Primer Design
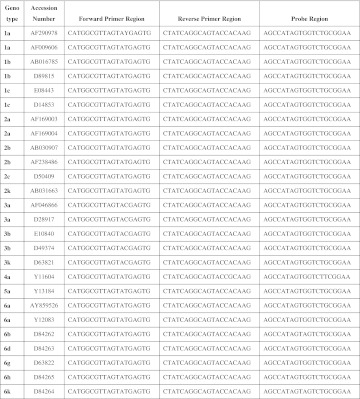
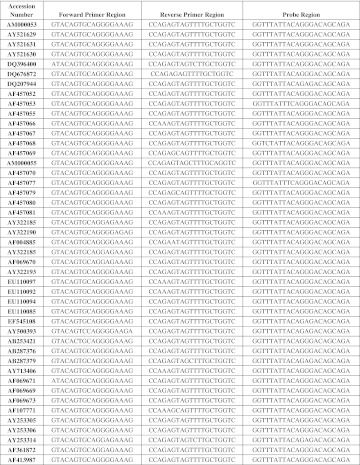
Genotype independent detection of viruses such as HIV-1 and HCV relies on finding the most conserved region of viruses by using a complete databank of aligned sequences for genome of both viruses. Comparison between HCV sequences of several genotypes and also between HIV-1 sequences of three genotypes and several subtypes are represented in (Figs. 2, 3), respectively. Two sets of primers and molecular beacon probes were designed and validated.
Fig. 2.
Conserved genomic sequences were applied for HCV primers and probe design
Fig. 3.
Conserved genomic sequences were applied for HIV-1 primers and probe design
Multiplex Real-Time RT-PCR Optimization
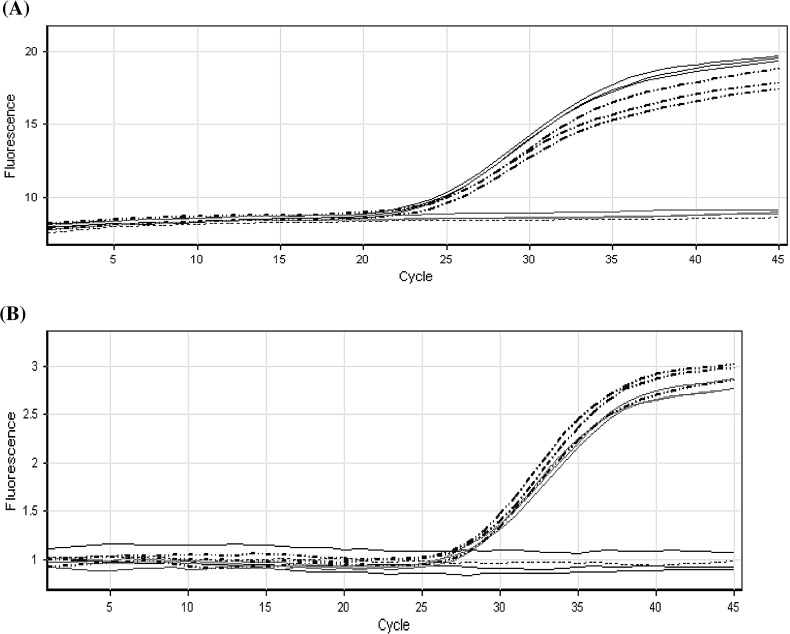
A Multiplex real-time PCR was evaluated for simultaneous detection of HIV-1 and HCV viruses in plasma samples. HIV-1 and HCV specific oligonucleotides pairs which were able to detect all significant HIV-1 subtypes and HCV genotypes were selected (Fig. 4). HIV-1 primers and probe specific oligonucleotides capable of recognizing a well-conserved region in the HIV-1 pol gene amplifying a 179 bp fragment were used. This oligonucleotide pair is known to be effective in the amplification of major HIV-1 subtypes and has been successfully employed for real-time RT-PCR technique. On the other hand, the HCV specific oligonucleotides were designed to amplify a 241 bp fragment within the 5′NCR of HCV genome. These primers and probes comply with the well-conserved sequences among different HCVs. The oligonucleotide pairs selected were checked by BLAST analysis. These sequences did not show any relevant homology with other viral or human sequences while revealing all representative HIV-1 subtypes and HCV genotypes.
Fig. 4.
Real-time detection of two different viral genomes in a multiplex format. Each reaction contained two sets of PCR primers specific for unique HIV-1 and HCV nucleotide sequences and two molecular beacons, each specific for one of the two amplicons and labeled with a differently colored fluorophore. HCV is plotted in solid black, multiplex format plotted in dotted–dashed black, HIV is plotted in solid gary and Non template control is plotted in dotted gary line. a FAM/Syber window for HCV detection b JOE/Tet window for HIV-1 detection
Sensitivity and Specificity of the Assay
The assay was evaluated on transcribed RNA dilutions of HIV-1 or/and HCV. Invitro-transcribed RNA scalar dilutions were added to uninfected plasma samples (Table 1). After extraction and real-time RT-PCR, the assay sensitivity limit, determined as the dilution in which 100 % of samples were revealed by our technique, was 1,000 copies/ml for HIV-1 and 100 copies/ml for HCV. The Ct values showed linear reference curves for both viruses (R2 > 0.94). The analytical sensitivity of Multiplex PCR could be affected by high concentration of each target sequence, a serial dilution of each viral standard were tested against a high copy number/ml (106) standard of the other one. As shown in the (Table 2) high viral load of one target sequence does not influence the analytical sensitivity of the other virus with low concentration. In order to determine the specificity, first a BLAST was carried out using NCBI Nucleotide BLAST software (http://blast.ncbi.nlm.nih.gov/). It was demonstrated that the primers do not attach to any other sequences except for that of HIV-1 or HCV. Then, some blood transmitted viruses such as HBV, HTLV-1, TTV, B19, HSV-1, HSV-2, HHV-6, HHV-8, HCMV and EBV were evaluated by the designed primers and probes. Only HIV-1 and HCV (and no other irrelevant genomes) were detected.
Table 1.
Sensitivity of multiplex real-time RT-PCR
| aHIV RNA copies/ml | Number detected | Percent detected |
| 106 | 8/8 | 100 |
| 105 | 8/8 | 100 |
| 104 | 8/8 | 100 |
| 103 | 8/8 | 100 |
| 102 | 5/8 | 62.2 |
| bHCV RNA copies/ml | Number detected | Percent detected |
| 106 | 8/8 | 100 |
| 105 | 8/8 | 100 |
| 104 | 8/8 | 100 |
| 103 | 8/8 | 100 |
| 102 | 8/8 | 100 |
Four experiments in duplicate were performed. The replicates were considered positive when HIV-1 or HCV were revealed
aSensitivity of assay for HIV-1 virus
bSensitivity of assay for HCV virus
Table 2.
Multiplex real time RT-PCR sensitivity evaluated in different ratios of standard plasma dilution conditions
| HCV copies/ml | HCV positive replicates | HIV-1 copies/ml | HIV-1 positive replicates |
|---|---|---|---|
| 106 | 4/4 | 106 | 4/4 |
| 106 | 4/4 | 105 | 4/4 |
| 106 | 4/4 | 104 | 4/4 |
| 106 | 4/4 | 103 | 4/4 |
| 106 | 4/4 | 102 | 1/4 |
| 106 | 4/4 | 106 | 4/4 |
| 105 | 4/4 | 106 | 4/4 |
| 104 | 4/4 | 106 | 4/4 |
| 103 | 4/4 | 106 | 4/4 |
| 102 | 4/4 | 106 | 4/4 |
Two experiments in duplicate were performed. The replicates were considered positive when both HIV-1 and HCV were revealed
Analysis of Patients’ Plasma by Multiplex Real-Time RT-PCR Using Molecular Beacon
These HIV-1/HCV positive samples were previously screened by serological experiments and quantified by Artus HCV RG RT-PCR and Artus HIV-1 RG RT-PCR Kits (Qiagen, Hilden, Germany). Samples quantified between 75 IU/ml and 6,700 IU/ml were chosen. Multiplex real-time RT-PCR using molecular beacon on co-infected patients, we used group A and E. 48 out of 50 positive samples were positive for both viruses and two samples were positive only for HCV. Storage conditions of the extracted sample and repeated freeze and thawing may have caused the negative results. We did not have access to the patients for resampling. Moreover, all samples in group B (HIV-1 positive) and C (HCV positive) with viral load over 1,000 copies/ml and 100 copies/ml, respectively were detected. Finally, analysis of group D (seronegative plasma samples) did not show any positive signal, confirming the specificity of the assay.
Discussion
The paper describes an approach for simultaneous detection of HIV-1 and HCV in plasma samples. The method uses a molecular beacon to run multiplex PCR in real-time. Application of advanced immunological method is limited during the seronegative window period as well as in cases of delitescence and non-immunogenic forms of HCV and HIV-1 infections. The most advanced techniques for detection of transfusion transmitted viruses such as HIV-1 and HCV are base on real-time detection, as a NAT method, which essentially shorten the duration of analysis, increase the sensitivity and specificity, and provide simultaneous quantification as well. In contrast to a single-target format, the multiplex format of the HIV-1/HCV assay developed in the present study has advantages with respect to both time and cost [21, 22]. The time needed to complete initial screening is decreased due to the ability to amplify multiple targets simultaneously in one tube. Multiplex assays using molecular beacon probes save cost of testing not only by reducing consumption of reagents but also by enabling its use in systems such as NASBA2 [1, 22], strand displacement amplification, and rolling-circle amplification which avoids necessity of expensive equipment such as thermal cycler [21].
The use of nonspecific DNA binding probes such as SYBR Green I is limited to detection of total amount of all amplification products synthesized in on reaction tube and melting curve analysis is required for multiplex detection [13]. So amplification detection base on specific probes is desired. Although several commercial techniques such as COBAS TaqMan HIV-1 or HCV (Roche Molecular Systems, Inc.) are FDA approved real-time PCR but they are monoplex and more expensive than in house designed multiplex real-time PCR. In this study, the use of molecular beacons was chosen as the probe of choice due to their advantages such as quenching efficiency over other probes such as Taqman and Hyprobe. In molecular beacon, the close distance between the dye and the quencher decreases the likelihood of their detachment, leading to lower background fluorescence resulting in enhanced detection and higher sensitivity [21]. Molecular beacons are especially suitable for identifying point mutations since they can distinguish targets with only a single nucleotide difference in equivalent length [18, 20, 21]. Molecular beacons have also been used for quantification of pathogens, virus replication, and gender detection in embryos. With the use of molecular beacons in real-time RT-PCR, no post-amplification handling is required and the chance of contamination of samples with amplicons which leads to false-positive results decrease. The hairpin structure of molecular beacons in comparison to Taqman made it more specific than corresponding conventional linear probes [19].
Specificity and sensitivity of 100 % for oligonucleotide microarray for real-time PCR detection of HIV-1, HBV and HCV was reported by Khodakov et al. [23]. The results obtained using the developed assay in the present study demonstrates that a specificity of 100 % and sensitivity of 96 % can be achieved. This sensitivity was due to two negative clinical samples in 50 samples. Actually, we could not determine the reason of this negative result as we did not have access to the patient for resampling.
The detection limit for HCV-RNA was demonstrated as 100 copies/ml and for HIV-1 it is 1,000 copies/ml that is better than results obtained by De Crignis et al. That were used SYBR Green I [8]. Our data was obtained using mono-infected as well as co-infected samples. These results are in concomitance with previously reported results [9].
All subtypes detection of any virus is reliant on finding the most conserved region of the viral genome by the use of a complete databank of aligned sequences of the virus genomes. The design of primers set was carried out to cover every known polymorphism in the genomes of viral subtypes. Furthermore, all the primer and probe sets were tested against a panel of anonymous samples previously found positive for viruses such as HBV, HTLV-1, TTV, B19, HSV-1, HSV-2, HHV-6, HHV-8, HCMV and EBV. Using these primers set and probes, no other genomes were detected.
It can therefore be concluded that application of molecular beacons to enable multiple target detection can improve reliability, speed, cost, and ease of diagnostic clinical assays [1]. The assay introduced here is demonstrated to be suitable for concomitant detection of HCV and HIV-1 in plasma or serum samples for clinical diagnosis with a further possible use in blood screening.
Acknowledgments
We would like to extend the gratitude to the management of Day General Hospital Laboratory, Tehran, for providing funding of this project.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Nucleic acid amplification techniques.
nucleic acid sequence-based amplification.
References
- 1.Lau LT, Feng XY, Lam TY, Hui HK, Yu AC. Development of multiplex nucleic acid sequence-based amplification for detection of human respiratory tract viruses. J Virol Methods. 2010;168(1–2):251–254. doi: 10.1016/j.jviromet.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 2.Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42(4):1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaBarre P, Hawkins KR, Gerlach J, Wilmoth J, Beddoe A, Singleton J, Boyle D, Weigl B. A simple, inexpensive device for nucleic acid amplification without electricity-toward instrument-free molecular diagnostics in low-resource settings. PLoS One. 2011;6(5):e19738. doi: 10.1371/journal.pone.0019738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mabey D, Peeling RW, Ustianowski A, Perkins MD. Diagnostics for the developing world. Nat Rev Microbiol. 2004;2(3):231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 5.Ngom B, Guo Y, Wang X, Bi D. Development and application of lateral flow test strip technology for detection of infectious agents and chemical contaminants: a review. Anal Bioanal Chem. 2010;397(3):1113–1135. doi: 10.1007/s00216-010-3661-4. [DOI] [PubMed] [Google Scholar]
- 6.Peeling RW, Mabey D, Herring A, Hook EW., 3rd Why do we need quality-assured diagnostic tests for sexually transmitted infections? Nat Rev Microbiol. 2006;4(12):909–921. doi: 10.1038/nrmicro1555. [DOI] [PubMed] [Google Scholar]
- 7.Tucker JD, Bu J, Brown LB, Yin YP, Chen XS, Cohen MS. Accelerating worldwide syphilis screening through rapid testing: a systematic review. Lancet Infect Dis. 2010;10(6):381–386. doi: 10.1016/S1473-3099(10)70092-X. [DOI] [PubMed] [Google Scholar]
- 8.Crignis E, Re MC, Cimatti L, Zecchi L, Gibellini D. HIV-1 and HCV detection in dried blood spots by SYBR Green multiplex real-time RT-PCR. J Virol Methods. 2010;165(1):51–56. doi: 10.1016/j.jviromet.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 9.Giachetti C, Linnen JM, Kolk DP, Dockter J, Gillotte-Taylor K, Park M, Ho-Sing-Loy M, McCormick MK, Mimms LT, McDonough SH. Highly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA. J Clin Microbiol. 2002;40(7):2408–2419. doi: 10.1128/JCM.40.7.2408-2419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis L, Becker J, Tender A, Cleland A, Queiros L, Aquiar A, Azevedo J, Aprili G, Bressan F, Torres P, Nieto S, Ursitti A, Montoro J, Vila E, Ramada C, Saldanha J. Evaluation of the Roche cobas s 201 system and cobas TaqScreen multiplex test for blood screening: a European multicenter study. Transfusion. 2008;48(9):1853–1861. doi: 10.1111/j.1537-2995.2008.01779.x. [DOI] [PubMed] [Google Scholar]
- 11.Meng Q, Wong C, Rangachari A, Tamatsukuri S, Sasaki M, Fiss E, Cheng L, Ramankutty T, Clarke D, Yawata H, Sakakura Y, Hirose T, Impraim C. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J Clin Microbiol. 2001;39(8):2937–2945. doi: 10.1128/JCM.39.8.2937-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt M, Pichl L, Jork C, Hourfar MK, Schottstedt V, Wagner FF, Seifried E, Muller TH, Bux J, Saldanha J. Blood donor screening with cobas s 201/cobas TaqScreen MPX under routine conditions at German Red Cross institutes. Vox Sang. 2010;98(1):37–46. doi: 10.1111/j.1423-0410.2009.01219.x. [DOI] [PubMed] [Google Scholar]
- 13.Gibellini D, Gardini F, Vitone F, Schiavone P, Furlini G, Re MC. Simultaneous detection of HCV and HIV-1 by SYBR Green real time multiplex RT-PCR technique in plasma samples. Mol Cell Probes. 2006;20(3–4):223–229. doi: 10.1016/j.mcp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Busch MP, Kleinman SH, Jackson B, Stramer SL, Hewlett I, Preston S. Committee report. Nucleic acid amplification testing of blood donors for transfusion-transmitted infectious diseases: report of the Interorganizational Task Force on Nucleic Acid Amplification Testing of Blood Donors. Transfusion. 2000;40(2):143–159. doi: 10.1046/j.1537-2995.2000.40020143.x. [DOI] [PubMed] [Google Scholar]
- 15.Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 16.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C virus prevalence among patients infected with human immunodeficiency virus: a cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34(6):831–837. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 17.Sulkowski MS, Mast EE, Seeff LB, Thomas DL. Hepatitis C virus infection as an opportunistic disease in persons infected with human immunodeficiency virus. Clin Infect Dis. 2000;30(Suppl 1):S77–S84. doi: 10.1086/313842. [DOI] [PubMed] [Google Scholar]
- 18.Goel G, Kumar A, Puniya AK, Chen W, Singh K. Molecular beacon: a multitask probe. J Appl Microbiol. 2005;99(3):435–442. doi: 10.1111/j.1365-2672.2005.02663.x. [DOI] [PubMed] [Google Scholar]
- 19.Marras SA, Tyagi S, Kramer FR. Real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes. Clin Chim Acta. 2006;363(1–2):48–60. doi: 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 20.Monroe WT, Haselton FR. Molecular beacon sequence design algorithm. Biotechniques. 2003;34(1):68–70. doi: 10.2144/03341st02. [DOI] [PubMed] [Google Scholar]
- 21.Vet JA, Majithia AR, Marras SA, Tyagi S, Dube S, Poiesz BJ, Kramer FR. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc Natl Acad Sci USA. 1999;96(11):6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong Y, Lemieux B, Kong H. Multiple strategies to improve sensitivity, speed and robustness of isothermal nucleic acid amplification for rapid pathogen detection. BMC Biotechnol. 2011;11:50. doi: 10.1186/1472-6750-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khodakov DA, Zakharova NV, Gryadunov DA, Filatov FP, Zasedatelev AS, Mikhailovich VM. An oligonucleotide microarray for multiplex real-time PCR identification of HIV-1, HBV, and HCV. Biotechniques. 2008;44(2):241–246. doi: 10.2144/000112628. [DOI] [PubMed] [Google Scholar]