Abstract
Mangrove wetland is a unique ecosystem and has rich bioresources. In this article, the roots, stems, branches, leaves, barks, fruits, and flowers from 12 species of the mangrove plants and six species of the accompanying mangrove plants, seawater and sediments in mangrove ecosystems in China were used as sources for isolation of yeasts. A total of 269 yeasts strains were obtained from the samples. The results of routine identification and phylogenetic analysis showed that they belonged to 22 genera and 45 species. Of all the 269 strains, Candida spp. was predominant with the proportion of 44.61%, followed by Kluyveromyces spp. (8.55%), Pichia spp. (7.44%), Kodamaea ohmeri (5.58%), Issatchenkia spp. (4.83%) and Debaryomyces hansenii (4.46%). We also found that strains N02-2.3 and ST3-1Y3 belonged to the undescribed species of Pichia sp. and Trichosporon sp. respectively while strain HN-12 was not related to any known yeast strains. This means that different yeast strains of Candida spp. especially C. tropicalis were widely distributed in the mangrove ecosystems and may have an important role in the mangrove ecosystems. The results also showed that some of them may have potential applications.
Keywords: Yeast diversity, Mangrove ecosystems, Diversity, Candida spp., Undescribed yeasts
Introduction
Mangrove ecosystems are typical intertidal wetland systems of the tropics and subtropics [1]. In recent years, microbial community in mangrove ecosystem has been intensively exploited in China and it has been found that it is mainly composed of bacteria, fungi and actinomyces [2]. At the same time, it has been found that yeasts are prevalent in salt marshes or mangrove ecosystems where the yeasts play an important role in the detrital food web [3], and they might be a food source for some marine invertebrates and zooplanktons. Yeasts also can be involved in marine habitats in decomposition and nutrient cycling, biodegradation of xenobiotics such as petroleum and its derivatives, and as parasites. Mangroves offer numerous microhabitats with the potential of harboring yeast communities [4, 5]. Some human-associated species and the prevalent species in polluted water were common in the mangrove ecosystem. Further extensive research could reveal the ecological roles of these yeasts and their interaction with the other organisms of the salt marsh. For example, the species Kluyveromyces aestuarii predominated the yeast communities of two detritus feeding crabs and Candida spp. were frequently isolated [6]. However, little has been known about yeast community in the mangrove ecosystems in China.
In this article, we dealt with all of the yeast strains isolated from the mangrove areas in South China, and tried to give a general understanding of the yeasts biodiversity of the Chinese mangrove.
Materials and Methods
Sampling
The samples (Table 1) which were collected from the mangrove ecosystem and used as the sources for yeast isolation in this study were described by Buzdar et al. [7]. Latitude and longitude of the sampling sites are shown in Table 2.
Table 1.
The tested mangrove species, their typical accompanying plants and the sites where they lived
| Tested mangrove species | DZG | DHI | NSI | HCD | LYB | SAB |
|---|---|---|---|---|---|---|
| Acanthus ilicifolius | + | + | + | + | + | |
| Aegiceras corniculata | + | + | + | + | + | |
| Avicennia mariana | + | + | + | |||
| Bruguiera gymnorrhiza | + | + | + | + | ||
| Bruguiera sexangula (Lour.) Poir. | + | |||||
| Clerodendrum inerme | + | |||||
| Ceriops tagal | + | + | ||||
| Excoecaria agallocha | + | + | + | + | ||
| Heritiera littoralis | ||||||
| Kadelia candel | + | + | + | |||
| Rhizophora stylosa | + | + | + | + | ||
| Sonneratia apetala | + | |||||
| Acrostichum aureuma | + | + | ||||
| Thespesia populneaa | + | + | ||||
| Cassytha filiformis L.a | + | + | ||||
| Phragmites communis Trin.a | + | |||||
| Pandanus tectoriusa | + | + | ||||
| Pluchea indica Less.a | + | + |
“+” Means the corresponding mangrove species from where the samples were collected, DZG Dongzhai habour, Haikou, Hainan Province; DHI Donghai island, Zhanjiang, Guangdong Province; NSI Nansan island, Zhanjiang, Guangdong Province; HCD Haicang dock, Xiamen, Fujian Province; LYB Luoyang bridge, Quanzhou, Fujian Province; SAB Su’ai bay, Shantou, Guangdong Province
aTypical accompanying plants
Table 2.
The sampling sites and their Latitude and longitude
| Sampling sites | Latitude and longitude |
|---|---|
| DZG, Haikou, Hainan Province | N19°53′ E110°19′ |
| DHI, Zhanjiang, Guangdong Province | N21°00′ E110°23′ |
| NSI, Zhanjiang, Guangdong Province | N21°08′ E110°30′ |
| HCD, Xiamen, Fujian Province | N24°26′ E118°02′ |
| LYB, Quanzhou, Fujian Province | N24°57′ E118°40′ |
| SAB, Shantou, Guangdong Province | N23°19′ E116°43′ |
Yeast Isolation
The yeast isolation from the samples was carried out by using the methods described by Buzdar et al. [7]. Colony and cell morphology observation and fermentation tests of the yeasts were performed using the methods described by Kurtzman and Robnett [8].
Metabolic Characterization of the Yeasts
The yeast strains were also identified by using BIOLOG system TM (Biolog MicroStation with Microlog System, Release 4.20, Biolog, Hayward, CA, USA), Biolog Universal Yeast Agar (Biolog) and Biolog Yeast microplate (Biolog) according to the procedures offered by the manufacturer [9].
DNA Extraction and PCR
The total genomic DNA of the yeast strains was isolated and purified by using the methods as described by Sambrook et al. [10]. Amplification and sequencing of D1/D2 26S rDNA sequences from the yeasts were performed according to the methods described by [11]. The common primers for amplification of D1/D2 26S rDNA in yeasts were used, the forward primer was NL-1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and the reverse primer was NL-4 (5′-GGTCCGTGTTTCAAGACGG-3′).
Phylogenetic Analysis and Identification of the Yeasts
The sequences obtained above were aligned by using BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST). The sequences which shared over 98% similarity with currently available sequences are considered to be the same species and multiple alignments was performed by using Clustal X 1.83 and phylogenetic trees were constructed by MEGA 4.0 [12].
Screening of Killer Toxin-Producing Yeasts
Screening of killer toxin-producing yeasts was performed according to the methods described by Wang et al. [14].
Screening of Single Cell Protein-Producing Yeasts
Screening of single cell protein-producing yeasts was conducted based on the methods described by Gao et al. [22].
Results and Discussion
Description of the Samples
The yeast strains were isolated from the roots, stems, branches, leaves, barks, fruits, and flowers from 12 species of the mangrove plants, six species of the typical accompanying plants, seawater and sediments in the mangrove ecosystems in Hainan Province, Guangdong Province and Fujian Province (Table 1). Therefore, the yeast strains isolated can represent the main yeast community in the mangrove ecosystems in China. From Table 2, it can be seen that all the trees involved in this study live in tropical and subtropical areas in China. Table 3 shows the chemical parameters at the sampling sites. The results in Table 3 indicated that the sampling sites had the characteristics of tropical and subtropical areas as well as marine environments, such as the high temperature, pH and salt.
Table 3.
Chemical parameters of the sampling sites
| Sampling sites | pH | Temperature (°C) | Salt (%) |
|---|---|---|---|
| DZG | 8.1 | 35 | 2.02 |
| DHD | 8.0 | 35 | 1.70 |
| NSD | 7.9 | 35 | 1.71 |
| HCD | 8.1 | 35 | 2.30 |
| LYB | 7.8 | 34 | 1.69 |
| SAB | 7.9 | 36 | 1.57 |
Isolation and Routine Identification of the Yeast Strains
After isolation and purification, a total of 269 yeasts strains were obtained from the samples. According to the results of their colony and cell morphology, fermentation tests and metabolic characteristics, it was found that they belonged to 22 genera and 45 species. One hundreds and twenty isolates (44.61%) were identified as 14 species in the genera Candida spp. (Table 4). It was worthy to notice that 74 isolates (27.51%) were identified as Candida tropicalis and it could be found in seawater and sediments in all the mangrove ecosystems, and on fruits, root, barks, leaves and branches of most of the mangrove trees. This meant that Candida spp. especially C. tropicalis were widely distributed in the mangrove ecosystems in China. This may be related to high temperature and salts in the sampling sites (Table 3). However, C. phangngensis (0.37%), C. aaseri (0.74%), C. boidinii (0.38%), C. Hollandica (0.37%) and C. maltosa (0.37%) could not be isolated from any samples from the mangrove trees. It also has been reported that Candida spp. are the most frequently isolated genus in Brazilian mangrove ecosystem [6] and at Sepetiba Bay [13]. Among the Candida spp., C. parapsilosis, C. tropicalis, C. valida-like, C. krusei, C. sorbosa, C. colliculosa-like, C. famata-like, C. guilliermondii, C. albicans, C. silvae and C. boidinii have been found to occur in other mangrove ecosystems [1, 3, 4, 6]. However, C. valida-like, C. krusei, C. sorbosa, C. colliculosa-like, C. famata-like, C. guilliermondii, C. albicans were not found in this study (Table 4). In contrast, it can be seen from Table 4 that C. maltosa, C. orthopsilosis, C. tenuis, C. asseri, C. butyric, C. thaimueangensis were obtained in the mangrove ecosystems in China. The results in Table 4 showed that C. tropicalis outnumbered other Candida species greatly and was considered to be a highly dominant species. It has been well documented that C. tropicalis could degrade different kinds of pollutants [14, 16]. Other Candida spp. such as C. parapsilosis [15] and C. intermedia [16] also have ability to degrade oil pollutants.
Table 4.
Yeasts of Candida spp. and the amount, proportion of every species and sources
| Yeasts species | Amount of isolates | Proportion (%) | Sources |
|---|---|---|---|
| C. aaseri | 2 | 0.74 | Sediments |
| C. boidinii | 1 | 0.37 | Sediments |
| C. butyric | 9 | 3.35 | Branches, barks, flowers, fruits, roots |
| C. catenulata | 6 | 2.24 | Branches, flowers, fruits, sediments |
| C. hollandica | 1 | 0.37 | Seawater |
| C. intermedia | 11 | 4.09 | Leaves, waters, barks, fruits, flowers, branches, roots |
| C. maltosa | 1 | 0.37 | Sediments |
| C. parapsilosis | 6 | 2.23 | Leaves, fruits, barks, branches |
| C. orthopsilosis | 1 | 0.37 | Fruits |
| C. phangngensis | 1 | 0.37 | Sediments |
| C. silvae | 3 | 1.12 | Branches |
| C. tenuis | 1 | 0.37 | Branches |
| C. thaimueangensis | 3 | 1.12 | Roots, branches, leaves |
| C. tropicalis | 74 | 27.51 | Sediments, fruits, root, barks, leaves, branches, water |
| Total | 120 | 44.61 |
Other yeasts (146 strains) obtained from the mangrove ecosystems included Aureobasidium pullulans (1.12%), Clavispora lusitaniae (2.6%), Debaryomyces hansenii (4.46%), Galactomyces geotrichum (1.49%), Geotrichum sp. (3.35%), Issatchenkia occidentalis (0.37%), I. orientalis (2.97%), I. siamensis (1.49%), Kazachstania exigua (2.6%), K. aestuarii (4.46%), K. nonfermentans (0.37%), K. siamensis (3.72%), Metschnikowia koreensis (0.74%), Kodamaea ohmeri (5.58%), Pichia anomala (3.35%), P. guilliermondii (1.12%), P. mexicana (2.23%), P. spartinae (0.74%), Rhodotorula mucilaginosa (0.74%), Rhodosporidium paludigenum (0.37%), Saccharomyces exiguous (0.37%), Saccharomycete sp. (1.47%), Saturnispora mendoncae (1.47%), Trichosporon asahii (2.6%), Williopsis saturnus (0.74%), Yarrowi lipolytica (2.24%) and Zygoascus steatolyticus (0.74%) (Table 5). Among Kluyveromyces spp., K. nonfermentans have been obtained from deep-sea samples [1]. However, we found that it also appeared in the mangrove ecosystems. It has been reported that K. aestuarii was frequently isolated from marine environments and could become the feed for invertebrates in mangrove ecosystem [6]. Consistent with the finding, 4.46% of the yeast strains isolated from the mangrove ecosystems in this study were K. aestuarii. This meant that the mangrove ecosystems indeed had some characteristics of marine environments (Table 3).
Table 5.
Other yeasts, the numbers of the strains, proportion of every species and sources
| Yeasts species | Numbers of strains | Proportion (%) | Sources |
|---|---|---|---|
| Aureobasidium pullulans | 3 | 1.12 | Roots, fruits, leaves |
| Clavispora lusitaniae | 7 | 2.60 | Barks, leaves, branches, sediments |
| Debaryomyces hansenii | 12 | 4.46 | Leaves, branches, roots, barks, sediments |
| Galactomyces geotrichum | 4 | 1.49 | Branches, leaves |
| Geotrichum sp. | 9 | 3.35 | Waters, roots, fruits, sediments |
| Issatchenkia occidentalis | 1 | 0.37 | Branches of Amcerana marma |
| Issatchenkia orientalis | 8 | 2.97 | Sediments, branches, fruits, |
| Issatchenkia siamensis | 4 | 1.49 | Sediments, leaves, fruits |
| Kazachstania exigua | 7 | 2.60 | Sediments, branches, fruits, leaves |
| Kluyveromyces aestuarii | 12 | 4.46 | Branches, sediments, leaves, fruits |
| Kluyveromyces nonfermentans | 1 | 0.37 | Leaves |
| Kluyveromyces siamensis | 10 | 3.72 | Branches, flowers, fruits, barks, roots |
| Metschnikowia koreensis | 2 | 0.74 | Leaves |
| Kodamaea ohmeri | 15 | 5.58 | Roots, leaves, fruits, flowers, barks |
| Pichia anomala | 9 | 3.35 | Flowers, sediments, leaves, roots |
| Pichia guilliermondii | 3 | 1.12 | Roots, branches, fruits |
| Pichia mexicana | 6 | 2.23 | Leaves, fruits, roots, sediments |
| Pichia spartinae | 2 | 0.74 | Flowers, sediments |
| Rhodotorula mucilaginosa | 2 | 0.74 | Leaves |
| Rhodosporidium paludigenum | 2 | 0.74 | Fruits and leaves of Aegiceras corniculatum |
| Rhodosporidium sphaerocarpum | 1 | 0.37 | Fruits of Ceriops tagal |
| Saccharomyces exiguous | 1 | 0.37 | Leaves of Aegiceras corniculatum |
| Saccharomycete sp. | 4 | 1.47 | Sediments |
| Saturnispora mendoncae | 4 | 1.47 | Leaves and roots |
| Trichosporon asahii | 7 | 2.60 | Barks, fruits, branches, sediments |
| Williopsis saturnus | 2 | 0.74 | Sediments |
| Yarrowi lipolytica | 6 | 2.24 | Roots, fruits, leaves, branches |
| Zygoascus steatolyticus | 2 | 0.74 | Leaves, fruits |
| Pichia sp. N02-2.3 (undescribed species) | 1 | 0.37 | Barks of Bruguiera gymnorrhiza |
| Trichosporon sp. ST3-1Y3 (undescribed species) | 1 | 0.37 | Leaves of Pluchea indica |
| Strain HN1-2 (undescribed genus) | 1 | 0.37 | Leaves of Kadelia candel |
| Total | 149 | 55.3 |
Pichia spp. is also frequently obtained from marine environments. For example, P. guilliermondii, P. ohmeri, P. fermentans, P. burtonii and P.chia anomala were obtained from surface of Sargassum pallidum, gut of Hexagrammos otakii, gut of yellow-fin tuna, gut of Lepidorigla microptera and gut of sea squirt [17]. In this study, 7.44% of the yeast strains obtained from the mangrove ecosystems were Pichia spp. (Table 4). P. anomala and P. membranaefaciens, also have been reported to occur in mangrove ecosystems [1, 4–6].
In our previous study, we found that K. ohmerii is widely distributed in different marine environments and the marine-derived yeast K. ohmerii BG3 can produce a large amount extracellular phytase [18]. In this study, 5.58% of the yeast strains isolated from the mangrove ecosystems were found to be K. ohmerii (Table 4). This revealed that the mangrove ecosystems are also the natural habitat for growth of K. ohmerii.
Debaryomyces hansenii has been found to be the common member in marine environment as it can tolerate high concentration of NaCl [19]. In this study, 4.46% of the yeast strains obtained from the mangrove ecosystems were D. hansenii. This again demonstrated that the mangrove ecosystems had some characteristics of marine environments (Table 3).
Aureobasidium pullulans is popularly known as black yeast and widely distributed in the phyllosphere of many crop plants, on various tropical fruits, in fresh water, estuarine, marine sediments, hypersaline habitats, seawater and deep sea [20]. In this study, the mangrove swamp was proven to be another natural habitat that was suitable for A. pullulans though it was not the major species.
The results in Table 4 indicated that Saccharomycete sp. could not be isolated from any samples of the mangrove trees, either. We found that strains N02-2.3 and ST3-1Y3 were identified as the undescribed species of Pichia sp. and Trichosporon sp. respectively while strain HN-12 was assigned to the undescribed genus (Table 4). This suggested that there were still unknown yeast species or genera in the mangrove ecosystems in China and more new genera or species can be found if further studies are carried on.
The results in Tables 4 and 5 also showed that the diversity of the yeast species was very rich and they belonged to 22 genera and 45 species.
Phylogenetic Analysis of Partial Sequences of the D1/D2 26S rDNA
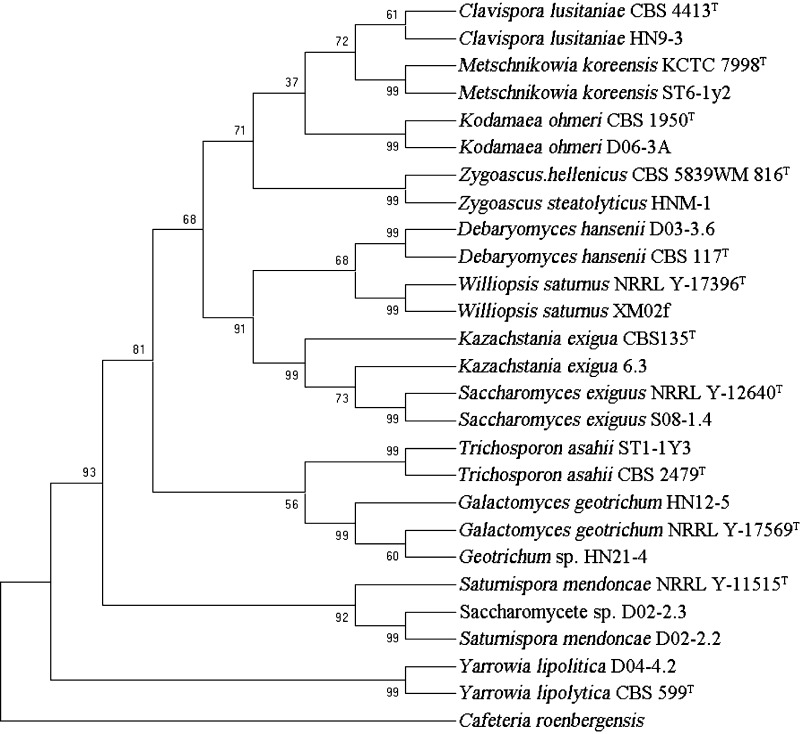
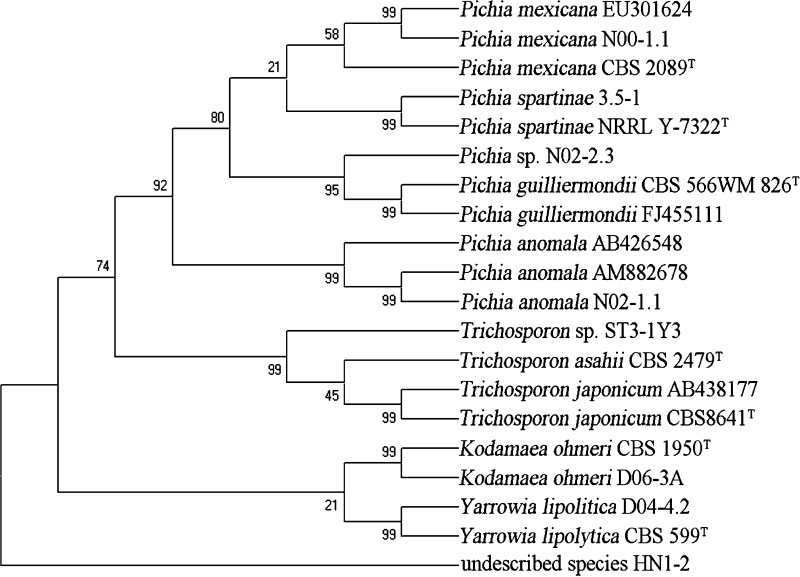
It has been confirmed that most yeast species can be easily identified at species level from sequence divergence in the D1/D2 domain [21]. In this study, in order to certify the taxonomic status of the isolates and the inter- and intra-specific relationships, 26S domain D1/D2 of each isolate obtained was sequenced, the sequences obtained were aligned. The phylogenetic trees were constructed when Cafeteria roenbergensis and Ricciocarpos natans were used as out-groups, respectively. The topology of the phylogram in Figs. 1, 2 and 3 demonstrated that the 266 isolates obtained in this study were closely related to the known different type strains, respectively. However, the results in Fig. 4 indicated that strains N02-2.3 and ST3-1Y3 belonged to the undescribed species of Pichia sp. and Trichosporon sp. respectively while strain HN-12 was not related to any known yeast strains. This meant that bioresource and diversity of yeasts in the mangrove ecosystems needed to be further exploited.
Fig. 1.
Consensus tree of 120 isolates based on 26S rRNA gene sequences obtained in this study. The type strains were selected from CBS database, and the previously published sequences of the type strains were obtained from GenBank. The outgroup we used was R. natans. The numbers above the branches are bootstrap. T stands for type strains
Fig. 2.
Consensus tree of 64 isolates based on 26S rRNA gene sequences obtained in this study. The type strains were selected from CBS database and the previously published 26S rRNA gene sequences of the type strains were obtained from GenBank. The outgroup we used was R. natan. The numbers above the branches are bootstrap. NT means neotype and T stands for type strains
Fig. 3.
Consensus tree of 82 isolates based on 26S rRNA gene sequences obtained in this study. The type strains were selected from CBS database while M. koreensis KCTC 7998T was from GenBank taxonomy database and the previously published 26S rRNA gene sequences of the type strains were obtained from GenBank. The outgroup we used was C. roenbergensis. The numbers above the branches are bootstrap. T stands for type strains
Fig. 4.
Consensus tree of 3 isolates based on 26S rRNA gene sequences obtained in this study. The type strains were selected from CBS database and the previously published 26S rRNA gene sequences of the type strains were obtained from GenBank. The numbers above the branches are bootstrap. T stands for type strains
We also found that the 26S rRNA gene sequence of each species isolated from different niches in the mangrove ecosystems was the same (Figs. 1, 2, 3, 4).
All the results in Figs. 1, 2, 3 and 4 were in agreement with those of fermentation texts and metabolic identification by Biolog (Tables 4, 5).
Potential Applications of the Yeasts Isolated from the Mangrove Systems
The dried cells of some of the 269 yeasts strains obtained above were found to contain more than 30% (w/w) of crude proteins, especially the dried cells of P. anomala No2-1.1 had more than 44.8% (w/w) protein (Table 6). It have been confirmed that single-cell proteins have many uses in food and feed industries as they have high content of protein, high percentage of essential amino acids and other nutrients [22]. In addition, after the single cell proteins were hydrolyzed by alkaline protease, the hydrolysates obtained have angiotensin converting enzyme inhibitory activity and antioxidant activity [23]. In general, the protein content in the single cells for protein production should be between 29 and 73 g per 100 g of cell dry weight [24]. This meant that many yeast strains obtained from the mangrove systems have many potential uses.
Table 6.
Protein contents of some yeast strains isolated from the mangrove systems
| Species | Protein content (%) |
|---|---|
| Pichia anomala No2-1.1 | 44.8 ± 1.4 |
| Candida silvae ST3-1R1 | 35.9 ± 1.1 |
| Candida butyri ST2-4R2 | 37.6 ± 0.4 |
| Kluyveromyces siamensis HN12-1 | 30.0 ± 1.8 |
| Trichosporon asahii ST3-1Y1 | 30.6 ± 0.9 |
| Metschnikowia koreensis ST6-1y2 | 33.3 ± 1.3 |
| Rhodosporidium paludigenum ST2-7G | 35.0 ± 1.1 |
| Rhodosporidium sphaerocarpum n04-2.1 | 34.1 ± 0.7 |
Data are given as means ± SD, n = 3
It was also found that K. siamensis D04-1.4 among them could actively produce killer toxin against M. bicuspidate WCY, a pathogenic yeast in crab (Fig. 5). The killer toxins produced by marine yeasts can be applied to biocontrol, fermentation and pharmaceutical industries [25].
Fig. 5.
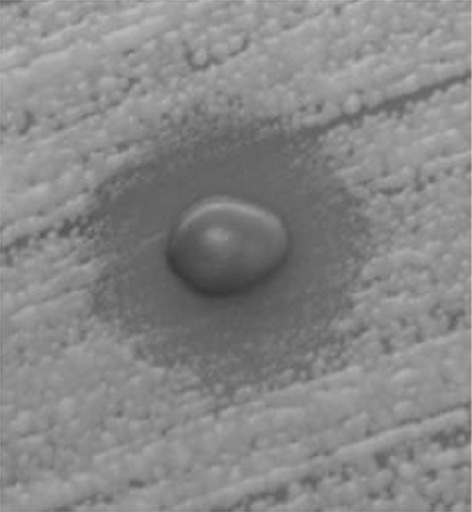
The clear zone formed by the yeast K. siamensis D04-1.4 grown in the plate seeded with the pathogenic yeast M. bicuspidata WCY at 28°C for 4 days
Acknowledgment
This work was supported by the National Infrastructure of Natural Resources for Science and Technology Program of China (the Grant No. 2005DKA21209).
References
- 1.Nagahama T. Yeast biodiversity in freshwater, marine and deep-sea environments. The yeast handbook biodiversity and ecophysiology of yeasts. Berlin: Springer; 2006. pp. 241–262. [Google Scholar]
- 2.Cao QM, Zheng KZ, Chen G, Chen GZ. A review of studies on microbiology of mangrove ecosystems. Ecol Environ. 2008;17:839–845. [Google Scholar]
- 3.Meyers SP, Ahearn DG, Alexander SK, Cook WL. Pichia spartinae, a dominant yeast of the Spartina salt marsh. Dev Ind Microbiol. 1975;16:262–267. [Google Scholar]
- 4.Naumova ES, Sukhotina NN, Naumov GI. Molecular-genetic differentiation of the dairy yeast Kluyveromyces lactis and its closest wild relatives. FEMS Yeast Res. 2004;5:263–269. doi: 10.1016/j.femsyr.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Meyers SP, Ahearn DG, Miles P. Characterization of yeasts in Baratara Bay. La St Univ Coastal Stud Bull. 1971;6:7–15. [Google Scholar]
- 6.Araujo EV, Soares CAG, Hagler AN, Mendonqa-Hagler LC. Ascomycetous yeast communities of marine invertebrates in a Southeast Brazilian mangrove ecosystem. Antonie van Leeuwenhoek. 1995;68:91–99. doi: 10.1007/BF00873096. [DOI] [PubMed] [Google Scholar]
- 7.Buzdar MA, Chi Z, Wang Q, Hua MX, Chi ZM. Production, purification, and characterization of a novel killer toxin from Kluyveromyces siamensis against a pathogenic yeast in crab. Appl Microbiol Biotechnol. 2011 doi: 10.1007/s00253-011-3220-8. [DOI] [PubMed] [Google Scholar]
- 8.Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/A:1001761008817. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Yue L, Chi ZM, Wang X. Marine killer yeasts active against a yeast strain pathogenic to crab Portunus trituberculatus. Dis Aquat Organ. 2008;80:211–218. doi: 10.3354/dao01943. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch EF, Maniatis T (1989) Preparation and analysis of eukaryotic genomic DNA. In: Molecular cloning: a laboratory manual, 2nd edn (Chinese translating edn.), Cold Spring Harbor Laboratory Press, Beijing, pp. 367–370
- 11.Chi Z, Ma C, Wang P, Li HF. Optimization of medium and cultivation conditions for alkaline protease production by the marine yeast Aureobasidium pullulans. Biores Technol. 2007;98:534–538. doi: 10.1016/j.biortech.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 13.Pagnocca FG, Mendonça-Hagler LC, Hagler AN. Yeasts associated with the white shrimp Penaeus schmitti, sediment, and water of Sepetiba Bay, Rio de Janeiro, Brasil. Yeast. 1989;15:479–483. [PubMed] [Google Scholar]
- 14.Wang XH, Chi ZM, Yue LX, Li J, Li MJ, Wu LF. A marine killer yeast against the pathogenic yeast strain in crab (Portunus trituberculatus) and an optimization of the toxin production. Microbiol Res. 2007;162:77–85. doi: 10.1016/j.micres.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Omar SH, Rehm HJ. Degradation of n-alkanes by Candida parapsilosis and Penicillium frequentans immobilized on granular clay and aquifer sand. Appl Microbiol Biotechnol. 1988;28:103–108. doi: 10.1007/BF00250507. [DOI] [Google Scholar]
- 16.Fialova A, Boschke E, Bley T. Rapid monitoring of the biodegradation of phenol-like compounds by the yeast Candida maltosa using BOD measurements. Int Biodeterior Biodegrad. 2004;54:69–76. doi: 10.1016/j.ibiod.2004.02.004. [DOI] [Google Scholar]
- 17.Li J, Chi ZM, Wang XH, Wand L, Sheng J, Gong F. Occurrence and diversity of Pichia spp. in marine environments. J Ocean Univ Chin. 2008;7:281–286. doi: 10.1007/s11802-008-0281-0. [DOI] [Google Scholar]
- 18.Li XY, Liu ZQ, Chi ZM. Production of phytase by a marine yeast Kodamaea ohmeri BG3 in an oats medium: optimization by response surface methodology. Biores Technol. 2008;99:6386–6390. doi: 10.1016/j.biortech.2007.11.065. [DOI] [PubMed] [Google Scholar]
- 19.Prista C, Loureiro-Dias MC, Garcı MVR, Ramos J. Mechanisms underlying the halotolerant way of Debaryomyces hansenii. FEMS Yeast Res. 2005;5:693–701. doi: 10.1016/j.femsyr.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Chi ZM, Wang F, Chi Z, Yue LX, Liu GL, Zhang T. Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl Microbiol Biotechnol. 2009;82:793–804. doi: 10.1007/s00253-009-1882-2. [DOI] [PubMed] [Google Scholar]
- 21.Kurtzman CP, Fell JW (2000) The yeasts. A taxonomic study, 4th revised and enlarged edn., Elsevier, Amsterdam, Lausanne, New York, Oxford, Shannon, Singapore, Tokyo. pp 222–360
- 22.Gao LM, Chi ZM, Sheng J, Ni XM. Single-cell protein production from Jerusalem artichoke extract by a recently isolated marine yeast Cryptococcus aureus G7a and its nutritive analysis. Appl Microbiol Biotechnol. 2007;77:825–832. doi: 10.1007/s00253-007-1210-7. [DOI] [PubMed] [Google Scholar]
- 23.Ni XM, Yue L, Chi ZM, Li J, Wang XH, Madzak C. Alkaline protease gene cloning from the marine yeast Aureobasidium pullulans HN2-3 and the protease surface display on Yarrowia lipolytica for bioactive peptide production. Mar Biotechnol. 2009;11:81–89. doi: 10.1007/s10126-008-9122-9. [DOI] [PubMed] [Google Scholar]
- 24.Patil RS, Ghormade V, Deshpande MV. Chitinolytic enzymes: an exploration. Enzyme Microb Technol. 2000;26:473–483. doi: 10.1016/S0141-0229(00)00134-4. [DOI] [PubMed] [Google Scholar]
- 25.Chi ZM, Liu GL, Zhao SF, Li J, Peng Y. Marine yeasts as biocontrol agents and producers of bio-products. Appl Microbiol Biotechnol. 2010;86:1227–1241. doi: 10.1007/s00253-010-2483-9. [DOI] [PubMed] [Google Scholar]