Abstract
Abnormal replication timing has been observed in cancer but no study has comprehensively evaluated this misregulation. We generated genome-wide replication-timing profiles for pediatric leukemias from 17 patients and three cell lines, as well as normal B and T cells. Nonleukemic EBV-transformed lymphoblastoid cell lines displayed highly stable replication-timing profiles that were more similar to normal T cells than to leukemias. Leukemias were more similar to each other than to B and T cells but were considerably more heterogeneous than nonleukemic controls. Some differences were patient specific, while others were found in all leukemic samples, potentially representing early epigenetic events. Differences encompassed large segments of chromosomes and included genes implicated in other types of cancer. Remarkably, differences that distinguished leukemias aligned in register to the boundaries of developmentally regulated replication-timing domains that distinguish normal cell types. Most changes did not coincide with copy-number variation or translocations. However, many of the changes that were associated with translocations in some leukemias were also shared between all leukemic samples independent of the genetic lesion, suggesting that they precede and possibly predispose chromosomes to the translocation. Altogether, our results identify sites of abnormal developmental control of DNA replication in cancer that reveal the significance of replication-timing boundaries to chromosome structure and function and support the replication domain model of replication-timing regulation. They also open new avenues of investigation into the chromosomal basis of cancer and provide a potential novel source of epigenetic cancer biomarkers.
DNA replication in human cells proceeds according to a defined temporal order (Hiratani et al. 2009). Several studies have identified abnormal temporal control of replication in many cancers (Amiel et al. 2001, 2002; Smith et al. 2001; Sun et al. 2001; Korenstein-Ilan et al. 2002). For example, specific chromosome translocations result in a chromosome-wide delay in replication timing (Breger et al. 2005; Chang et al. 2007) that is found frequently in cancer cells (Smith et al. 2001). Some cancer-specific replication-timing changes appear to be epigenetic in that, similar to developmental changes, they are mitotically stable but do not involve detectable genetic lesions (Eul et al. 1988; Adolph et al. 1992). A far-reaching aspect of epigenetic abnormalities is that they are potentially reversible. In fact, in a mouse lymphoma model showing aberrant replication timing, fusion of affected cells with normal mouse fibroblasts restored the normal pattern of replication timing and reversed the malignant phenotype (Eul et al. 1988; Adolph et al. 1992). Despite these observations, there has not been a comprehensive study to evaluate the extent of replication-timing abnormalities in cancer.
We recently generated genome-wide replication-timing profiles for a wide collection of human and mouse cell lines and embryonic stem cell (ESC) differentiation intermediates, revealing developmentally regulated changes in replication timing that encompass at least half of the genome (ReplicationDomain.org). Developmentally regulated changes take place in units of 400–800 kb and are associated with changes in subnuclear 3D organization of the affected domains (Hiratani et al. 2008, 2010). This replication-timing program is a highly stable epigenetic characteristic of a given cell type that is indistinguishable between the same cell types from different individuals (Pope et al. 2011). This stability has allowed for the development of tools to unambiguously determine cellular identity using their specific “replication fingerprints” (Ryba et al. 2011b). Intriguingly, replication-timing profiles correlate more strongly with genome-wide maps of the sites and frequencies of chromatin interactions (Hi-C) (Lieberman-Aiden et al. 2009) than with any other chromosomal property identified to date (Ryba et al. 2010), indicating that replication domains reflect the structural architecture of chromosomes and support the model of replication-timing domains as structural and functional large-scale units (the replication domain model). In summary, replication-timing profiles are unique to specific cell types and define an unexplored level of chromosome domain organization with intriguing potential for epigenetic fingerprinting.
We reasoned that just as specific cell types display unique replication-timing fingerprints, specific cancers may also be definable by their replication-timing fingerprints. Acute lymphoblastic leukemia (ALL) is an excellent model cancer to investigate this hypothesis due to the availability of relatively homogeneous cancer tissue from affected patients and several well-characterized genetic subtypes linked to prognosis. Current clinical risk stratification for pediatric ALL includes factors such as age, leukocyte count at time of diagnosis, and recurrent chromosomal abnormalities detected in malignant lymphoblasts (Yeoh et al. 2002; Jeha and Pui 2009; Luo et al. 2009). Chromosomal abnormalities with prognostic significance include aneuploidies, such as hypodiploidy (<44 chromosomes) and hyperdiploidy (with trisomies 4, 10, and 17), translocations, and deletions (Pui et al. 2011). However, only a minority of these abnormalities such as t(9;22) show a direct activation of an oncogene, and the underlying mechanisms of tumorigenesis for the majority of ALL subtypes remain elusive. Furthermore, ∼20% of ALL and 50% of AML cases present with a normal karyotype (Bienz et al. 2005; Kearney and Horsley 2005; Usvasalo et al. 2009; Collins-Underwood and Mullighan 2010; Pui et al. 2011) but have widely varying clinical outcomes, underscoring the need for additional epigenetic markers. Here we query the replication program genome-wide in 17 primary childhood leukemias and three ALL cell lines and report widespread instability, with some changes in common to all leukemias and others unique to specific patients. The differences that distinguish different cancers also align with the boundaries of normal developmentally regulated replication domains, supporting the replication domain model. In addition, the timing changes that distinguish cancers from normal cells do not resemble any particular tissue, extending a model derived from DNA methylation studies that cancers are characterized by widespread epigenetic instability (Hansen et al. 2011; Pujadas and Feinberg 2012).
Results
Replication timing is conserved between diverse nonleukemic lymphoblasts
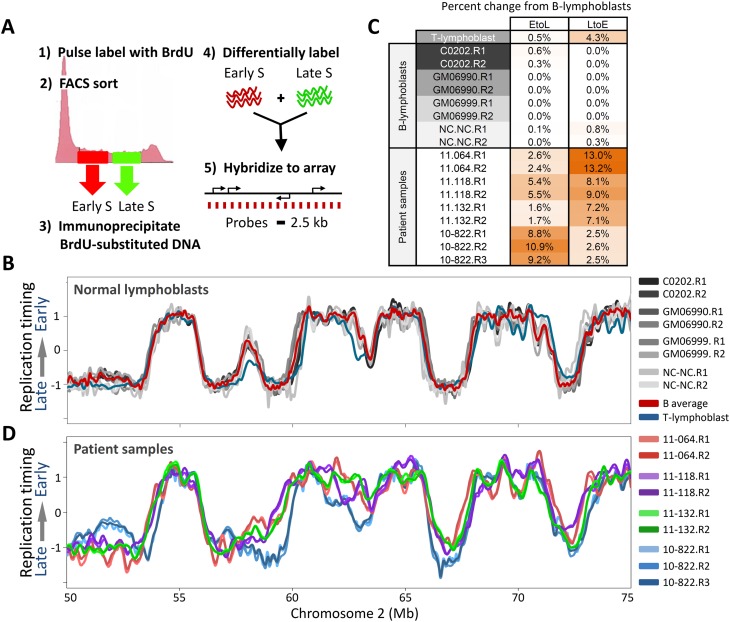
The majority of our patients during this study presented with pre-B ALL. Hence, we first evaluated the stability of replication-timing profiles between nonleukemic human B cells. Since proliferating immature B cells derived directly from patients are not available (immature B cells [hematogones] make up <5% of cells from the bone marrow of normal individuals and must be stimulated to proliferate ex vivo) (McKenna et al. 2001), we analyzed four established nonleukemic EBV-transformed mature human B lymphoblastoid cell lines: C0202, NC-NC, GM06990, and GM06999. The protocol for generating genome-wide replication-timing profiles is summarized in Figure 1A, and has been described in detail (Ryba et al. 2011a). Figure 1B shows loess-smoothed replication profiles for an exemplary 25-Mb chromosomal segment, while Figure 1C summarizes the percentage of the genome with significant timing changes between biological replicates of the four lymphoblastoid cell lines. The high degree of conservation between these lines demonstrates that their replication-timing profiles are a stable characteristic of mature human B cells, even when comparing established cell lines from different sources and histories (Supplemental Fig. 1). This extends previous results demonstrating the robust stability of replication profiles between common cell types (Hiratani et al. 2008, 2010; Ryba et al. 2010; Pope et al. 2011). The average of all replicates from these four cell lines provides a single B-cell-derived replication-timing profile that will herein be called “control” in comparisons with leukemia profiles, with the given caveat that leukemic samples are arrested at various stages in lymphoblast development from immature to more mature pre-B stages (Ferrando et al. 2002; Nemazee 2006; Mullighan et al. 2007) as indicated by their immunophenotypes where available (Supplemental Table 1). To derive an approximation of the extent to which different types of lymphoblasts vary in replication timing, and since two patients presented with T-cell ALL, we also profiled a mature CD4+ peripheral T-cell sample from a normal individual (Fig. 1B,C). These results revealed that replication timing in mature B and T cells differs by only 4.5% genome wide.
Figure 1.
RT profiles are stable in nonleukemic lymphoblasts, but diverge in leukemic samples. (A) Method for generating genome-wide replication-timing profiles. Dividing cells are pulse labeled with BrdU and FACS sorted into early and late S-phase fractions, and nascent BrdU-substituted DNA is differentially labeled and hybridized on a tiling CGH microarray with even probe spacing. (B) Overlaid replication-timing profiles of a segment of human chromosome 2 for four nonleukemic EBV transformed human B-cell lines: C0202, GM06990, GM06999, and NC-NC. Each cell line is represented by loess-smoothed curves of two high-quality biological replicates (denoted R1 and R2; see Methods). The red profile is the average of the four B-cell lines, and in blue is a corresponding primary T-cell line. (C) Percentage of the genome with significant (>1 RT unit) timing changes toward earlier (L to E) or later (E to L) replication from the average normal B-cell profile, for each of the individual replicate profiles in B and C. (D) Profiles of four arbitrary patient samples, which diverge from each other and from lymphoblastoid B cells in a chromosome that did not harbor karyotypic rearrangements.
Heterogeneous replication timing in leukemia cells
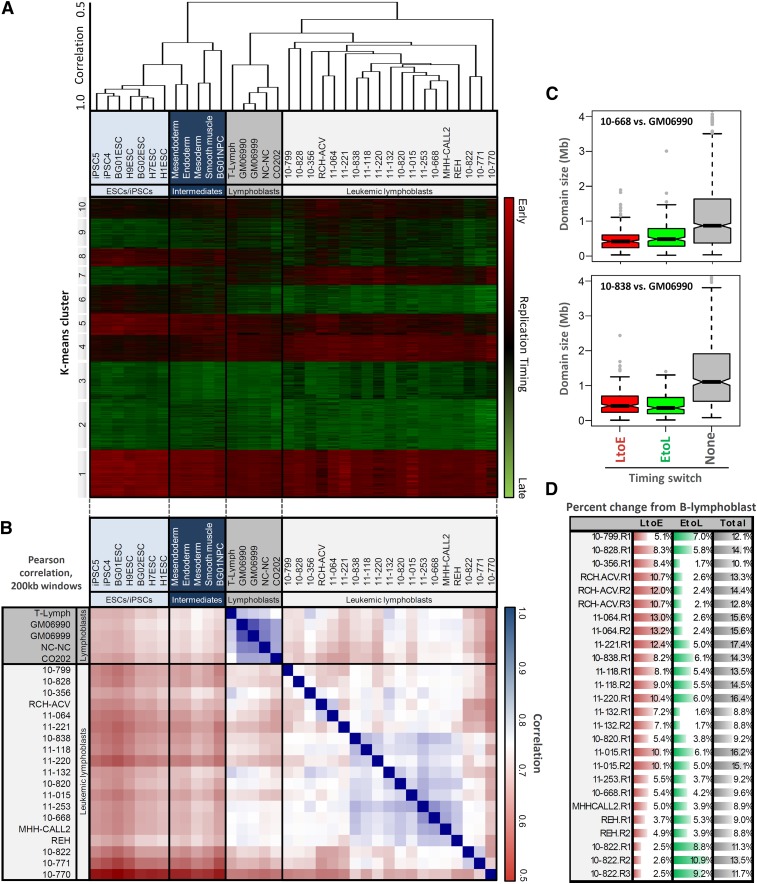
Figure 1D shows profiles from four exemplary pre-B ALL patient samples across the same segment of chromosome 2 as shown in Figure 1B. In contrast to control mature B-cell lines, these cells show numerous differences in replication timing, even more than seen between mature B and T cells, while replicates of each patient sample are virtually indistinguishable. Altogether, we profiled three B-ALL cell lines, and 13 B-ALL, two T-ALL, and one AML leukemic patient samples. The properties of all 20 leukemic and five normal samples are summarized in Supplemental Figure 1, and cell cycle analyses for all samples are shown in Supplemental Figure 2. As an initial comparison, the genome was divided into 12,625 nonoverlapping 200-kb windows, and replication profiles were hierarchically clustered to create a dendrogram expressing relatedness between the various cell samples (Fig. 2A). This, along with genome-wide correlations between control and leukemic lymphoblasts and cell types previously profiled (Fig. 2B) confirmed that replication profiles of individual leukemic samples were widely divergent and easily distinguished from control lymphoblasts, other human cell types, and each other. However, many differences from control cells were shared between leukemia samples despite their various stages of developmental arrest, suggesting that there are replication abnormalities in common between many types of leukemia. Control mature B- and T-cell profiles were distinct, but were more similar to each other than to leukemias of any origin. Nonetheless, T-ALL patient samples (10-828, 10-799) clustered separately from B-ALL, and samples with TCF3/PBX1 translocations (11-064, RCH-ACV) as well as those with mostly normal karyotypes (10-838, 11-118) formed their own clusters, suggesting conservation of features among developmentally related subgroups.
Figure 2.
Leukemic cells show global changes in replication profiles. (A) Hierarchical clustering of genome-wide replication-timing patterns for the four lymphoblastoid B-cell lines and those of other human cell types, showing relatively stable profiles between mature B and T lymphoblasts, and clustering of samples with similar genetic makeup, including T-cell leukemias (10-799/10-828), those with TCF3/PBX1 translocations (11-064/RCH-ACV), and those with mostly normal karyotype (10-838/11-118). (B) Genome-wide correlations between replication-timing data sets used in this study. Correlations between divergent B cells are consistently above 0.9, while those between leukemic samples generally range from 0.60 to 0.85. (C) Domain-wide switches to earlier (L to E) or later (E to L) replication timing occur in units of ∼400–800 kb, smaller than static early or late domains and consistent with developmentally regulated changes in timing. (D) Percentage of the genome with significant timing changes toward earlier (L to E) or later (E to L) replication from the normal B-cell profiles in indicated cell types.
Timing changes between leukemic and control cells occurred in a tight size distribution consistent with the 400–800-kb unit size of natural developmentally regulated changes in replication timing (Fig. 2C), and 9%–18% of domains detectably deviated from the controls in each leukemic cell line or patient sample (Fig. 2D), with consistent changes between replicates. With the notable exception of patient 10-822, most profiles had a significantly higher fraction of the genome replicating earlier than the controls (LtoE), rather than later (average 7.8% LtoE differences; 4.7% EtoL). The amount of change was generally not as great as the 20% of domains that differ between most cell types, but significantly higher than the 2%–4% of domains that deviate between cells of the same type, the 4.5% between B and T cells, or the 6.0% between human ESC-derived early endoderm vs. mesoderm tissues (Figs. 1C, 2; Hiratani et al. 2010; Ryba et al. 2010, 2011a). Replication-timing changes were distributed throughout the genome on all chromosomes (Supplemental Fig. 3) more evenly than the breakpoints present in patient samples, and unlike the phenomenon of chromothripsis, where multiple breaks are clustered on a single chromosome (Liu et al. 2011). All replication profiles reported here are freely available to view or download at www.ReplicationDomain.org (Weddington et al. 2008).
Replication profiles detect karyotypic abnormalities and copy-number variation
Although >90% of mononucleate cells from bone-marrow aspirates are leukemic, only 5%–10% of cells were in S phase (Supplemental Fig. 2). Hence, it was important to validate that our replication profiles were indeed derived from leukemic cells rather than proliferating contaminants. Most leukemias contain karyotypic abnormalities that distinguish them from contaminating cells (Supplemental Fig. 1). We reasoned that many of these lesions should be detectable in replication-timing data, serving as internal validation for the leukemic source of the replication-timing profiles. For example, aneuploidies were readily detectable as copy-number variation (CNV) derived from the sum of raw signal values (Cy3 + Cy5) for probes encompassing those chromosomes (Supplemental Fig. 4), providing validation for many samples.
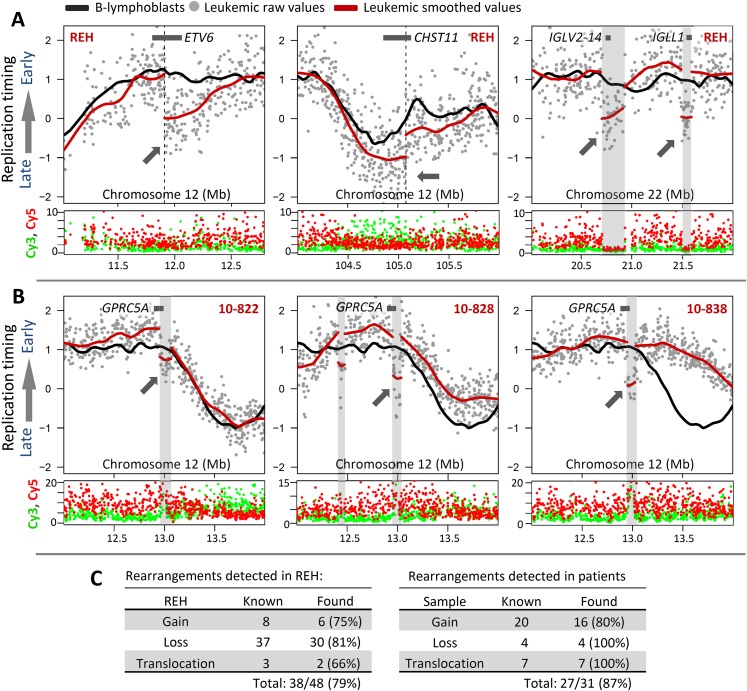
We also reasoned that translocation breakpoints that juxtapose early and late-replication domains should be detectable as unnaturally sharp transitions in replication timing at the breakpoint where sequences are no longer in their original genomic position. As proof of principle we examined a translocation in cell line REH for which the breakpoint junctions of both translocation partners have been precisely mapped (Wiemels et al. 2000), a translocation that fuses the ETV6 (formerly TEL1) gene at 12p13 with RUNX1 (formerly AML1) at 21q22. As shown in Figure 3A, this breakpoint was readily detected within ETV6 as an abrupt shift toward later replication timing that coincides with the molecularly mapped position of the breakpoint in REH. Downstream from the normally late-replicating RUNX1 partner, a shift to earlier replication also localized to the molecularly mapped breakpoint position (data not shown). Using this principle, we were able to more precisely map an additional REH translocation that mapped cytogenetically between 94.8 and 107.5 Mb (Horsley et al. 2006) of 12q23 and by replication timing to 105.08 Mb (Fig. 3A, middle). This locus is within the CHST11 gene that was found to be aberrant in other subtypes of leukemia (e.g., CLL) (Hiraoka et al. 2000; Okuda et al. 2000; Schmidt et al. 2004). Hence, this method was able to provide further validation of sample source (Fig. 3C) and demonstrates that replication-timing data can facilitate the localization of translocation breakpoints. It should be noted that translocations that fuse loci with similar replication timing are not expected to produce abrupt shifts. This is consistent with results in patient 10-668, in which a Philadelphia chromosome translocation t(9;22)(q34;q11) fuses two regions that are normally late replicating, and remain late after translocation (data not shown).
Figure 3.
Abrupt shifts in replication-timing localize a subset of rearrangements. (A, left) Abrupt timing changes in REH at 12p13 map within ETV6 (TEL1) at 11.95 Mb, consistent with the molecularly mapped translocation site. (Middle) A breakpoint at 12q23 (94.8–107.5 Mb) can be mapped more precisely within CHST11 by an abrupt shift in timing values at 105.08 Mb. (Right) Abrupt timing changes in regions not included in published karyotypes of REH represent deletions of IGL loci involved in B-cell maturation, evidenced by a sharp drop in overall Cy5 and Cy3 signal intensity. (B) Examples of abrupt timing changes in patients 10-822, 10-828, and 10-838 undetected by karyotype analysis, but suggesting a shared amplification of ∼12.95–13.05 Mb in these three patients, which overlaps suspected tumor suppressor GPRC5A. (C) Summary of rearrangements detected in REH and patient samples (Supplemental Figs. S4–S8) by either CNV in raw replication-timing data or abrupt timing shifts lacking CNV (translocations).
We also found abrupt timing shifts not represented by translocations in published karyotypes of REH (Fig. 3A, right). Abrupt shifts in replication timing could also result from localized CNV. Amplifications could delay the time for replication forks to arrive to normally adjacent sequences and would appear as shifts toward later replication, while homozygous deletions would result in background levels of hybridization that would average to a log ratio of zero. Hence, we computationally identified abrupt shifts in replication-timing ratio data and determined whether each corresponded to significant CNV determined from raw array values (Fig. 3A; Supplemental Figs. 5–8). A CNV was considered significant if it encompassed ≥2 probes within 10 kb with overall intensity outside of the 99.9th/0.1st percentiles. This analysis revealed that abrupt shifts in replication timing coinciding with sites of karyotypically defined translocations did not accompany significant CNV (Fig. 3A left, middle), whereas abrupt shifts that were not at known translocation sites represented either deletions or amplifications (Fig. 3A, right, Fig. 3B). For example, abrupt replication-timing changes at 20.70–20.92 Mb and 21.49–21.59 Mb of Chr. 22 (Fig. 3A, right), which include the IGLV2-14 and IGLL1 loci involved in B-cell maturation and often rearranged in leukemia (Tang et al. 1991; Brauninger et al. 2001), are clearly due to large deletions encompassing those sequences that suddenly bring the replication-timing ratio to zero. Importantly, using our algorithms and comparing our data to known CGH data for REH, we were able to identify 79% of known gains (6/8), losses (30/37), and translocations (2/3) (Fig. 3C, left). We believe that this is an underestimate; since REH is an established cell line, it is possible that additional genetic changes exist between our cells and those that were analyzed for CGH.
Using these methods, we were able to identify 87% of known karyotypic and genetic abnormalities from patient samples, providing important validation for the ability of replication timing to query proliferating leukemic cells directly from bone-marrow samples (Fig. 3C, right; Supplemental Figs. 4–8). However, as with REH, we found several examples of abrupt timing changes in patient samples at sites not detected as lesions in karyotypic data, some of which were conserved among multiple samples of different ALL subtypes. For instance, samples 10-822, 10-828, and 10-838 all displayed a sharp shift to later replication at the same location near 12.95 Mb on chromosome 12, but did not show karyotypic abnormalities at this site (in fact, 10-838 showed a normal karyotype). In all three cases, this replication-timing change was associated with a gain in copy number (Fig. 3B). Intriguingly, this locus is within suspected tumor-suppressor gene GPRC5A (Tao et al. 2007; Acquafreda et al. 2009), suggesting the possibility that persistent STAT3 activation due to mutation of GPRC5A (Chen et al. 2010) may be a contributing factor in these patients. Interestingly, both samples 10-822 [carrying the t(17;19)] and 10-838 were from relapsed patients who eventually died of their disease, suggesting that GPRC5A disruption should be investigated for potential prognostic significance. The break region also contains a binding site for the B-cell CLL/lymphoma 11A (BCL11A) protein, which mediates gamma-globin expression and blood-cell maturation through long-range chromatin interactions (Xu et al. 2010).
Taken together, we conclude that genome-wide replication-timing profiles generated from bone-marrow samples of leukemia patients accurately reflect the replication program of their leukemic cells. Moreover, they simultaneously report on both replication-timing abnormalities and CNV. We estimate that, depending on the timing difference between the regions and the proportion of cells with the translocation, at least 30%–50% of translocations and 75% of CNVs will be detectable from replication-timing profiles (Fig. 3C; Supplemental Figs. 4–8). Moreover, replication-timing analyses reveal long-range influences of a breakpoint on replication timing, which may propagate hundreds of kilobases from the break site, and such changes would not be detected by conventional CGH or genome sequencing. Such distal changes could be very important. For example, deregulation of genes hundreds of kilobases from a common breakpoint in anaplastic large cell lymphoma (ALCL) has been shown to play a causal role in ALCL (Mathas et al. 2009).
Patient-specific epigenetic replication-timing fingerprints
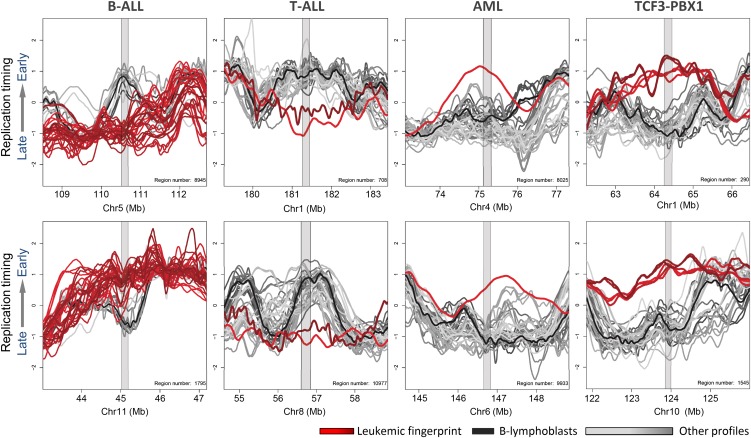
The premise for this study was to test the hypothesis that, just as specific cell types display unique epigenetically regulated replication-timing fingerprints, cancers might display their own class of epigenetic replication-timing differences. These replication-timing “fingerprints” can be identified using a previously described replication-fingerprinting algorithm (Ryba et al. 2011b), which isolates regions of unique replication timing between any predesignated sets of samples. As shown in Figure 4, different classes of leukemia can be distinguished by their common differences in replication timing from all other samples. Replication fingerprints found only in B-ALL (n = 16), T-ALL (n = 2), AML (n = 1), or patients and cell lines with TCF3/PBX1 translocations (n = 2) were identified. A complete list of these fingerprints can be found in Supplemental Table 2. As expected from Figure 1, comparison to normal mature human CD4+ T cells verified that some T-ALL-specific fingerprints were likely due to normal developmental differences. Without access to normal human proliferating immature lymphoblasts, it is difficult to rule out the possibility that any individual feature of the fingerprint may reflect arrest at a particular stage of immature B-cell development. However, a much higher proportion of differences from B-cell controls were shared between B-ALL and T-ALL than were exclusive to either one, suggesting that many differences from controls were not due to the developmental stage of the leukemias, and at least some were common to all leukemias (discussed below).
Figure 4.
Leukemia type-specific replication-timing differences. Example fingerprint regions that depict leukemia type-specific timing differences in B-cell ALL, T-cell ALL, AML, and TCF3/PBX1 translocation-positive cell lines and patient samples. Colors correspond to the color key at right, with colors of fingerprint profiles highlighted in red, other profiles in gray, and an average of karyotypically normal B-cell controls in black. Tables of fingerprint regions and genes are given in Supplemental Table 2.
Fingerprinting identifies a small number (∼20) of the largest replication-timing differences. This contrasts with hierarchical clustering, which highlights widespread but small differences in replication timing across the genome. For example, in Figure 2, patients 10-838, 11-118, 11-220, 11-132, 10-820, 11-015, 11-253, and 10-668 clustered together, but we did not find a well-defined fingerprint able to distinguish this group of patients from all others. Importantly, the criteria for fingerprinting largely excluded changes associated with genetic lesions. In fact, using the criteria described in Figure 3, >74% of fingerprint regions did not exhibit CNV or abrupt changes in Cy3/Cy5 ratios ±1 Mb from the fingerprint region (e.g., Fig. 5A). Due to their lack of association with genetic changes detectable by karyotype or CNV analysis, we refer to these as “epigenetic replication-timing fingerprints” (albeit, genetic changes that do not affect CNV or replication time such as inversions within regions of constant replication timing would not be detected). Some epigenetic fingerprints were specific to a single cell line or patient sample (Fig. 5A). These changes may serve as unique identifiers of the different patient leukemias regardless of whether they represent arrested development or are causally linked, and should be pursued for their potential as biomarkers for risk stratification.
Figure 5.
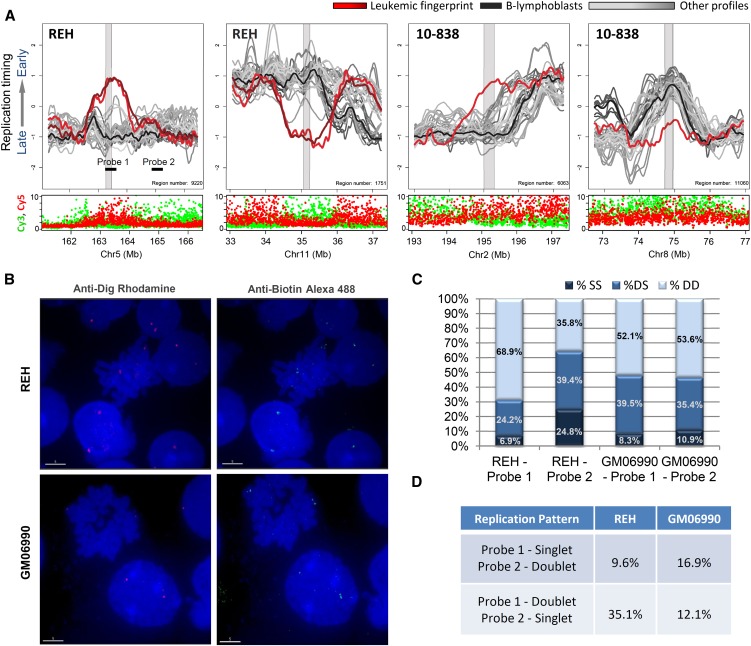
Replication-timing differences in karyotypically normal regions. (A) Shown are sample-specific fingerprint regions in cell line REH and patient 10-838 that lack genetic lesions under karyotypic analysis (both samples, with 10-838 being karyotypically normal), total Cy3 + Cy5 intensity (both samples) or Sanger CGH (REH), and therefore represent apparently epigenetic timing changes. Such regions may be explained by changes in long-range interactions or by subkaryotypic or CGH-resolution rearrangements. As in Figure 4, fingerprint profiles (REH or 10-838) are highlighted (red) against a background of other leukemic samples (gray) and B-lymphoblastoid cells (black). (B) Fluorescent in situ hybridization images of cell lines REH and GM06990 showing region-specific binding in metaphase nuclei and doublet/singlet hybridization patterns in interphase nuclei. (C) Quantification of observed singlet/singlet (SS), doublet/singlet (DS), and doublet/doublet (DD) configuration of allelic homologs for each probe shown in A. Only nuclei displaying at least one doublet allele (189 GM06990 and 296 REH) in either probe were scored, which may exaggerate the percentage of nuclei that appear to have replicated the regions asynchronously (single-doublets). (D) Quantification of the frequency with which one probe appeared to replicate prior to the other as a percentage of total chromosomes scored for which cis-linked probes 1 and 2 show a singlet–doublet configuration (378 GM06990 and 592 REH). In REH, probe 1 appears to replicate prior to probe 2 nearly 75% of the time, whereas in GM06990 either probe may replicate first.
To verify these findings by an independent method, one of the replication-timing changes specific to the cell line REH was analyzed by the singlet-doublet DNA replication assay (Fig. 5B–D). After cellular fixation methods that separate sister chromatids, fluorescence in situ hybridization (FISH) reveals replicated homologs as doublet signals in the nucleus, while unreplicated homologs appear as singlets. These results confirmed that the REH-specific fingerprint region displayed a substantially higher frequency of doublets than the same region in nonleukemic GM06990 or than an adjacent region that replicates late in both normal and leukemic cells. This result was further confirmed through replication timing in situ hybridization (ReTiSH) (Schlesinger et al. 2009; Supplemental Fig. 9).
A “pan-leukemia” replication-timing fingerprint
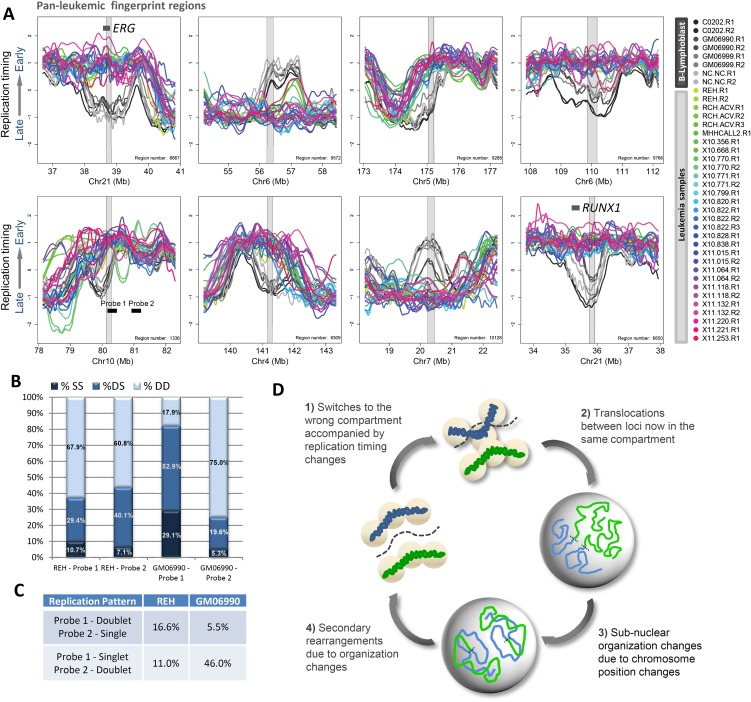
Many replication-timing changes were found to be in common between all leukemia cells, referred to as “pan-leukemic fingerprints” (Fig. 6A–C). These changes are unlikely to be the result of differences in the developmental stage of the ALL samples vs. the B-cell baseline, as they were also found in the few T-ALL and AML samples that we profiled, but may represent early events in leukemogenesis (Fig. 6D). Since each type of leukemia has a different genetic constitution, pan-leukemic changes are likely to be epigenetic in origin in most leukemias. One such aberration is located at the RUNX1 locus (Fig. 6A), at the site of the ETV6/RUNX1 translocation that in our study was found in the cell line REH and several patient samples, and which causes a large shift from late to early replication over several hundred kilobases. Intriguingly, every leukemic cell type profiled had this same extensive replication-timing fingerprint, terminating at the same boundaries, independent of the breakpoint. This finding indicates that the replication-timing change reflects an epigenetic misregulation that precedes (possibly predisposes) breakage at this site and that the replication-timing domain boundaries—rather than the site of translocation—determine the range of influence (Fig. 6D). The RUNX1 gene is thought to be involved in normal hematopoiesis and is one of the most frequently disrupted genes in leukemia (Niebuhr et al. 2008). These observations support the hypothesis that RUNX1 serves a gatekeeping function for leukemia (Niebuhr et al. 2008) and suggest that the pan-leukemic replication-timing fingerprint may be related to the disease state due to an epigenetic phenomenon rather than a mutation or translocation.
Figure 6.
Pan-leukemic replication-timing changes suggest common early events in leukemogenesis. (A) Example regions from a pan-leukemic fingerprint between all leukemic cells versus B-cell controls. Fingerprint regions are highlighted in gray overlay. (B) Percentages of SS, DS, and DD configurations for each of the FISH probes indicated in A, as probes 1 and 2 were scored as in Figure 5C (192 GM06990 and 516 REH nuclei scored). (C) Quantification of the frequency with which one probe appeared to replicate prior to the other as in Figure 5D (384 GM06990 and 1032 REH chromosomes scored). (D) A prospective model for common early events in leukemogenesis: (1) Loci in late-replicating compartments on the periphery undergo a switch to earlier replication together with a switch to the wrong nuclear compartment, which may be precipitated by loss of anchorage on the periphery or incorporation of accessibility-promoting chromatin factors in early-S phase. (2) Translocations occur between loci that now occupy the same compartment. (3) Large rearrangements between chromosomes disrupt the normal distribution of chromatin in the nucleus, leading to further subnuclear organization changes. (4) Subnuclear organization changes bring together additional loci that would normally not be in contact or share the same compartment, leading to accumulation of additional secondary rearrangements and genome instability.
Other interesting examples in the pan-leukemic fingerprint include a region downstream from the RUNX1 gene at ∼39 Mb on chromosome 21 in all leukemic cell types profiled (Fig. 6A). Like the RUNX1 region, this region is much earlier replicating than control B cells in all leukemic cell types. Interestingly, this region contains the ERG gene, which is involved in hematopoietic regulation and chromosomal translocations in other types of cancer, including AML (Marcucci et al. 2005). Another example is a region of the extended MHC (Major Histocompatibility Complex) that harbors two gene clusters: the BTN (butyrophilin) and the major histone gene cluster, HIST1. The BTN cluster contains a total of seven genes, including the BTN1A1 gene, and the two subfamilies BTN2 and BTN3, each of which contain three genes (Rhodes et al. 2001). The precise role of these genes in immune response is unknown, but BTN1A1 and the BTN2 genes have been implicated as negative regulators of T-cell activation (Smith et al. 2010). BTN3 mRNA is widely expressed in immune cells such as T cells, B cells, macrophages, dendritic cells, and monocytesm with most protein expression occurring at the cell surface (Rhodes et al. 2001; Compte et al. 2004; Smith et al. 2010). Additionally, most members of the BTN family contain a 30.2 protein domain, which is a 170-amino acid globular domain found at the C terminus of proteins for which there is evidence of involvement in inflammatory response (Compte et al. 2004). The histone gene cluster contains a total of six genes, HIST1H4H, HIST1H2BI, HIST1H3G, HIST1H2BH, HIST1H3F, and HIST1H4G. Interestingly, this gene cluster is also present in a group of replication-timing fingerprints specific to pluripotent cell types (Ryba et al. 2011b). Finally, to confirm predictive ability of the pan-leukemic fingerprint for new samples, we applied leave-one-out cross-validation (LOOCV) as described (Ryba et al. 2011b) to predict the identity of each sample using regions selected in the absence of that sample. In this test, leukemic/nonleukemic identity was accurately assessed in 40/40 test cases, and 87% of fingerprint regions were conserved throughout cross-validation. A complete list of the pan-leukemic fingerprint regions can be found in Supplemental Table 2.
Replication-timing fingerprints align with developmentally regulated replication domains
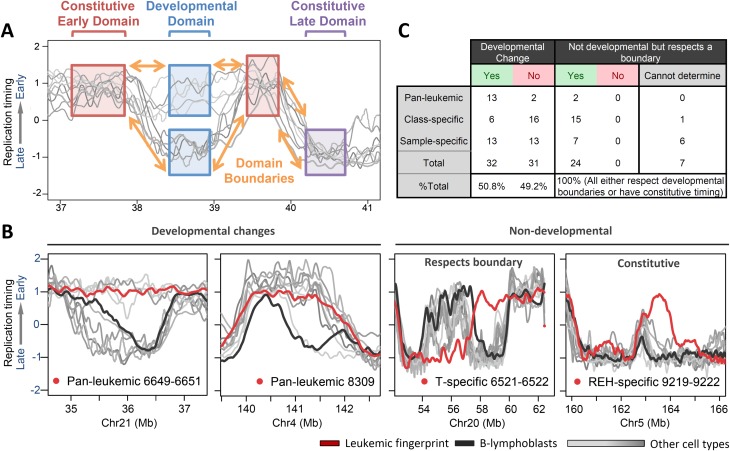
Replication-timing fingerprints are of the same size range as normal developmentally regulated replication domains (Fig. 2C). This raised the possibility that they represent misregulation of developmental control over replication timing, as has been described for DNA methylation changes in cancers (Hansen et al. 2011; Pujadas and Feinberg 2012). To test this hypothesis, we compared the collection of leukemia fingerprints with profiles of the same genomic regions from nine cell types that we have profiled in the past (Weddington et al. 2008; Pope et al. 2011; Ryba et al. 2011b). More than half of these aligned in register to a developmentally regulated replication domain boundary (Fig. 7; Supplemental Fig. 10). The remaining half of the fingerprints shared boundaries present in all queried cell types, or in rare cases appeared to create a new boundary within a large early- or late-replicating region. These latter cases could be developmental replication-timing boundaries in cell types that we have not queried. Importantly, in no case did a leukemia fingerprint boundary pass over a developmental boundary or appear in a novel position out of register with a known boundary. These results indicate that the replication-timing differences that distinguish leukemias from each other and from normal cell types are the same units of chromosomes that distinguish cell types from each other. Interestingly, however, the cohort of fingerprints for any particular leukemia correlated poorly (R = 0.43–0.61) with that of any profiled human cell type (Ryba et al. 2011b), indicating that leukemias do not take on the identity of any particular cell type, but acquire misregulated features of many different cell types.
Figure 7.
Replication changes in leukemia respect normal developmental boundaries. (A) Diagram of normal boundaries of replication in development, defining regions of constitutively early or late replication timing, conserved boundaries between these regions, and developmentally regulated domains. (B). Examples of each class of domains in leukemic fingerprints that switch replication timing (red) against a background of other human cell types (gray) and normal B-cell controls (black). Here, two pan-leukemic fingerprint regions align to both developmental boundaries and timing values. T-cell profiles switch timing in opposite directions from others, but at the same transition region as is used in B cells and other cell types, while an REH-specific fingerprint region aligns to one boundary but switches earlier than other cell types profiled. Additional examples of domain boundary alignment are shown in Supplemental Figure S10. (C) A summary of the number of domains in leukemic fingerprints that align to developmental boundaries, either with or without acquiring the timing of other cell types.
Discussion
Here we show that genome-wide replication-timing analyses detect widespread deregulation of replication timing in leukemias. While control cell lines show remarkably stable and cell-type-specific replication profiles, leukemic samples deviate substantially from controls and from each other, demonstrating a high degree of instability in the replication program. These differences occurred largely in units of 400–800 kb and align with developmentally programmed changes in replication timing, supporting the concept of the replication domain as a unit of chromosome structure and function and suggest that mechanisms acting at the level of these units are misregulated in cancer. Despite their heterogeneity, leukemic cells all share certain replication-timing aberrations, indicating common early events in leukemogenesis that appear to be conserved. Some of these commonalities occur at sites of translocations but are remarkably identical independent of the translocation, suggesting that the changes precede the translocation and that the distance over which replication timing is influenced is determined by misregulation of replication-timing domains rather than by the site of translocation. Our results provide the first comprehensive assessment of replication misregulation in cancer, identify novel epigenetic events occurring early in leukemogenesis, and suggest the possibility that specific subtypes of leukemia may be linked to specific replication-timing fingerprints that should be pursued for their potential as a novel genre of cancer biomarkers.
Genome-wide assessments of transcription, sites of DNA methylation, and the distribution of chromatin proteins and their modifications have received a lot of recent attention and offer great promise (Bibikova et al. 2006). However, each is informative for only the fraction of the genome affected by each property and some of these methods are expensive and laborious. Replication profiles comprehensively and reliably assess epigenetic state genome wide, and are considerably less expensive to generate and easier to interpret than these other markers. Full genome-scale profiling can be performed with less than a million cells (Gilbert 2010; Hiratani et al. 2010). Analysis of replication timing is fundamentally different from other common genome-wide methodologies in that it queries large-scale organization of the genome, which has otherwise been assessed only through challenging chromatin-conformation capture methods (Lieberman-Aiden et al. 2009). In fact, the uncanny alignment of replication timing to existing chromatin interaction maps (Ryba et al. 2010) implies that replication-timing profiles predict megabase-level spatial organization of chromosomes. Hence, replication-timing changes likely reflect novel spatial relationships (e.g., unusual juxtapositions of chromosome segments) that may predispose cells to particular translocation events. Consistently, a recent analysis found significant linkages between cancer rearrangements and replication timing (De and Michor 2011). Hence, replication-timing abnormalities have the potential to inform early cancer diagnosis.
In translocations between temporally distinct replication domains, replication timing will necessarily change across hundreds of kilobases. Since different types of chromatin are assembled at different times, this may transmit chromatin changes long distances from the break site. In fact, attempts to implicate the gene loci near translocation breakpoints in the etiology of the associated cancer have met with limited success (Hunger et al. 1998; Strefford et al. 2009) Hence, replication profiling has the potential to detect long-range effects of a chromosome break. In addition, complex genome rearrangements smaller than ∼1 Mb that do not alter copy number, such as inversions, will escape detection by both spectral karyotyping and comparative genomic hybridization (CGH) methods, but may replicate at a distinctly different time from their native location. Finally, most of the replication-timing differences we identified between leukemias are unlikely to be associated with any genetic lesion and would not be detected by any other current method. At present, we do not know the significance of replication-timing changes to the cancer phenotype. There is a general correlation between replication timing and gene expression, but it is promoter specific and appears to reflect transcriptional competence rather than transcription per se (Hiratani et al. 2009). The similarity in sizes of replication domains and regions of long-range epigenetic silencing (LRES) that has been observed in many cancers (Coolen et al. 2010; Hsu et al. 2010; Dallosso et al. 2012) and the observation that LRES consolidates the cancer genome to reduce transcriptional plasticity while replication timing consolidates during differentiation (Hiratani et al. 2008), suggest a potential relationship between these mechanisms. Taken together, replication profiling can identify novel genetic lesions and their associated long-range effects, as well as epigenetic changes that escape detection by other diagnostic methods.
Methods
Cell lines and culture
Sources of control cells lines CO202, GM06990, GM06999, and NC-NC are provided in Supplemental Figure 1. Cell lines REH, RCH-ACV, and CALL-2 were purchased from ATCC. Cells were cultured in standard media of RPMI with 10% fetal bovine serum, 4 mM glutamine, 1% penicillin, and streptomycin.
Sample collection
Patient bone-marrow samples were collected as part of ongoing clinical trials at OHSU from patients who consented for enrollment in biologic studies. Currently, patients <21 yr of age with suspected leukemia are eligible for enrollment for biologic studies at the Oregon Health & Science University (OHSU) with Institutional Review Board Approval. In most cases of newly diagnosed pediatric ALL, a bone-marrow aspiration is performed to confirm the diagnosis. Samples from subjects on the biologic study were assigned a unique identifier for health information protection. Subjects include all genders, minorities, and children eligible for the study. Bone-marrow aspirate generally contains close to 1 × 106 cells /mL and is usually >80% lymphoblasts. Other fresh bone-marrow samples were obtained from the Children’s Oncology Group (COG) ALL Cell Bank for a pilot study. These fresh samples were processed to purify mononuclear cells by centrifugation through Ficoll. The mononuclear cell ring was then isolated and counted. Then, 0.5 – 1 × 107 cells were labeled with 10 μg/mL BrdU for 2 h in RPMI, 10% FBS media, fixed in 70% ethanol, and shipped to FSU.
Genome-wide replication-timing analysis
Genome-wide replication timing was analyzed as described in detail (Ryba et al. 2011a) using NimbleGen HD2 arrays (3 × 720 K format) with average interprobe spacing of 2509 bp. Probes were designed against build Hg18 (NCBI 36) of the human genome.
Computational methods
Replication-timing data were normalized within and between arrays using the limma package in R, and smoothed using loess with a span of 300 kb, as described (Ryba et al. 2011a). To quantify the relative percentage of the genome with significant changes in timing, we calculated the fraction of loess-smoothed points with RT value differences of 1 or greater. For clustering and fingerprint analysis, data were averaged into nonoverlapping 200-kb windows, and replication fingerprints were created as described (Ryba et al. 2011b) to identify regions of shared replication-timing changes in defined groups of samples. Hierarchical clustering was performed using the pvClust package in R with absolute correlation as a distance metric. Methods for CNV detection were applied using the CGHweb R package and R/Bioconductor scripts to identify regions encompassing ≥2 probes within 10 kb, with overall intensity outside of the 99.9th/0.1st percentiles.
Data access
The replication-timing data sets used in this study are available at the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/), under accession number GSE37987.
Acknowledgments
We thank R. Didier for assistance with flow cytometry, K. Roux, I. Lucas, M. Thayer, and M. LeBeau for critical comments on the manuscript, as well as T. Chandra for control T cells, and B. Chadwick for the GM06999 cell line. This work was supported by NIH GM085354 and Bankhead Coley 1BF01 to D.M.G. B.H.C. is supported in part by the Oregon Child Health Research Center (National Institute of Child Health and Development 1K12HD057588-01) and the St. Baldrick’s Foundation. COG samples were processed at the COG ALL Molecular Reference Laboratory and Bank (J. Gastier-Foster, Principal Investigator), which is supported by NIH/NCI U24 CA114766 and U10 CA98543.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138511.112.
References
- Acquafreda T, Soprano KJ, Soprano DR 2009. GPRC5A: A potential tumor suppressor and oncogene. Cancer Biol Ther 8: 963–965 [DOI] [PubMed] [Google Scholar]
- Adolph S, Hameister H, Schildkraut CL 1992. Molecular analysis of the aberrant banding pattern on chromosome 15 in murine T-cell lymphomas. Chromosoma 101: 388–398 [DOI] [PubMed] [Google Scholar]
- Amiel A, Elis A, Sherker S, Gaber E, Manor Y, Fejgin MD 2001. The influence of cytogenetic aberrations on gene replication in chronic lymphocytic leukemia patients. Cancer Genet Cytogenet 125: 81–86 [DOI] [PubMed] [Google Scholar]
- Amiel A, Elis A, Maimon O, Ellis M, Herishano Y, Gaber E, Fejgin MD, Lishner M 2002. Replication status in leukocytes of treated and untreated patients with polycythemia vera and essential thrombocytosis. Cancer Genet Cytogenet 133: 34–38 [DOI] [PubMed] [Google Scholar]
- Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, et al. 2006. Human embryonic stem cells have a unique epigenetic signature. Genome Res 16: 1075–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M, Ludwig M, Leibundgut EO, Mueller BU, Ratschiller D, Solenthaler M, Fey MF, Pabst T 2005. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res 11: 1416–1424 [DOI] [PubMed] [Google Scholar]
- Brauninger A, Goossens T, Rajewsky K, Kuppers R 2001. Regulation of immunoglobulin light chain gene rearrangements during early B cell development in the human. Eur J Immunol 31: 3631–3637 [DOI] [PubMed] [Google Scholar]
- Breger KS, Smith L, Thayer MJ 2005. Engineering translocations with delayed replication: Evidence for cis control of chromosome replication timing. Hum Mol Genet 14: 2813–2827 [DOI] [PubMed] [Google Scholar]
- Chang BH, Smith L, Huang J, Thayer M 2007. Chromosomes with delayed replication timing lead to checkpoint activation, delayed recruitment of Aurora B and chromosome instability. Oncogene 26: 1852–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Deng J, Fujimoto J, Kadara H, Men T, Lotan D, Lotan R 2010. Gprc5a deletion enhances the transformed phenotype in normal and malignant lung epithelial cells by eliciting persistent Stat3 signaling induced by autocrine leukemia inhibitory factor. Cancer Res 70: 8917–8926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-Underwood JR, Mullighan CG 2010. Genomic profiling of high-risk acute lymphoblastic leukemia. Leukemia 24: 1676–1685 [DOI] [PubMed] [Google Scholar]
- Compte E, Pontarotti P, Collette Y, Lopez M, Olive D 2004. Frontline: Characterization of BT3 molecules belonging to the B7 family expressed on immune cells. Eur J Immunol 34: 2089–2099 [DOI] [PubMed] [Google Scholar]
- Coolen MW, Stirzaker C, Song JZ, Statham AL, Kassir Z, Moreno CS, Young AN, Varma V, Speed TP, Cowley M, et al. 2010. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat Cell Biol 12: 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallosso AR, Oster B, Greenhough A, Thorsen K, Curry TJ, Owen C, Hancock AL, Szemes M, Paraskeva C, Frank M, et al. 2012. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene doi: 10.1038/onc.2011.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F 2011. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol 29: 1103–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eul J, Gronemeyer H, Adolph S, Hameister H 1988. Suppression of tumorigenicity in T-cell lymphoma hybrids is correlated with changes in myc expression and DNA replication of the myc chromosomal domain. Chromosoma 96: 248–254 [DOI] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1: 75–87 [DOI] [PubMed] [Google Scholar]
- Gilbert DM 2010. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka N, Nakagawa H, Ong E, Akama TO, Fukuda MN, Fukuda M 2000. Molecular cloning and expression of two distinct human chondroitin 4-O-sulfotransferases that belong to the HNK-1 sulfotransferase gene family. J Biol Chem 275: 20188–20196 [DOI] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245 doi: 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi S, Lu J, Gilbert DM 2009. Replication timing and transcriptional control: Beyond cause and effect–part II. Curr Opin Genet Dev 19: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley SW, Mackay A, Iravani M, Fenwick K, Valgeirsson H, Dexter T, Ashworth A, Kearney L 2006. Array CGH of fusion gene-positive leukemia-derived cell lines reveals cryptic regions of genomic gain and loss. Genes Chromosomes Cancer 45: 554–564 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, Weng YI, Deatherage DE, Chen Z, Pereira JS, et al. 2010. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res 20: 733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger SP, Fall MZ, Camitta BM, Carroll AJ, Link MP, Lauer SJ, Mahoney DH, Pullen DJ, Shuster JJ, Steuber CP, et al. 1998. E2A-PBX1 chimeric transcript status at end of consolidation is not predictive of treatment outcome in childhood acute lymphoblastic leukemias with a t(1;19)(q23;p13): A Pediatric Oncology Group study. Blood 91: 1021–1028 [PubMed] [Google Scholar]
- Jeha S, Pui CH 2009. Risk-adapted treatment of pediatric acute lymphoblastic leukemia. Hematol Oncol Clin North Am 23: 973–990 [DOI] [PubMed] [Google Scholar]
- Kearney L, Horsley SW 2005. Molecular cytogenetics in haematological malignancy: current technology and future prospects. Chromosoma 114: 286–294 [DOI] [PubMed] [Google Scholar]
- Korenstein-Ilan A, Amiel A, Lalezari S, Lishner M, Avivi L 2002. Allele-specific replication associated with aneuploidy in blood cells of patients with hematologic malignancies. Cancer Genet Cytogenet 139: 97–103 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XQ, Ke ZY, Huang LB, Guan XQ, Zhang YC, Zhang XL 2009. High-risk childhood acute lymphoblastic leukemia in China: Factors influencing the treatment and outcome. Pediatr Blood Cancer 52: 191–195 [DOI] [PubMed] [Google Scholar]
- Marcucci G, Baldus CD, Ruppert AS, Radmacher MD, Mrozek K, Whitman SP, Kolitz J, Edwards CG, Vardiman JW, Powell BL, et al. 2005. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: A cancer and leukemia group B study. J Clin Oncol 23: 9234–9242 [DOI] [PubMed] [Google Scholar]
- Mathas S, Kreher S, Meaburn KJ, Johrens K, Lamprecht B, Assaf C, Sterry W, Kadin ME, Daibata M, Joos S, et al. 2009. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc Natl Acad Sci 106: 5831–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna RW, Washington LT, Aquino DB, Picker LJ, Kroft SH 2001. Immunophenotypic analysis of hematogones (B-lymphocyte precursors) in 662 consecutive bone marrow specimens by 4-color flow cytometry. Blood 98: 2498–2507 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764 [DOI] [PubMed] [Google Scholar]
- Nemazee D 2006. Receptor editing in lymphocyte development and central tolerance. Nat Rev Immunol 6: 728–740 [DOI] [PubMed] [Google Scholar]
- Niebuhr B, Fischer M, Tager M, Cammenga J, Stocking C 2008. Gatekeeper function of the RUNX1 transcription factor in acute leukemia. Blood Cells Mol Dis 40: 211–218 [DOI] [PubMed] [Google Scholar]
- Okuda T, Mita S, Yamauchi S, Matsubara T, Yagi F, Yamamori D, Fukuta M, Kuroiwa A, Matsuda Y, Habuchi O 2000. Molecular cloning, expression, and chromosomal mapping of human chondroitin 4-sulfotransferase, whose expression pattern in human tissues is different from that of chondroitin 6-sulfotransferase. J Biochem 128: 763–770 [DOI] [PubMed] [Google Scholar]
- Pope BD, Tsumagari K, Battaglia D, Ryba T, Hiratani I, Ehrlich M, Gilbert DM 2011. DNA replication timing is maintained genome-wide in primary human myoblasts independent of D4Z4 contraction in FSH muscular dystrophy. PLoS ONE 6: e27413 doi: 10.1371/journal.pone.0027413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Carroll WL, Meshinchi S, Arceci RJ 2011. Biology, risk stratification, and therapy of pediatric acute leukemias: An update. J Clin Oncol 29: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E, Feinberg AP 2012. Regulated noise in the epigenetic landscape of development and disease. Cell 148: 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DA, Stammers M, Malcherek G, Beck S, Trowsdale J 2001. The cluster of BTN genes in the extended major histocompatibility complex. Genomics 71: 351–362 [DOI] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM 2011a. Genome-scale analysis of replication timing: From bench to bioinformatics. Nat Protoc 6: 870–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, Dalton S, Gilbert DM 2011b. Replication timing: A fingerprint for cell identity and pluripotency. PLoS Comput Biol 7: e1002225 doi: 10.1371/journal.pcbi.1002225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S, Selig S, Bergman Y, Cedar H 2009. Allelic inactivation of rDNA loci. Genes Dev 23: 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HH, Dyomin VG, Palanisamy N, Itoyama T, Nanjangud G, Pirc-Danoewinata H, Haas OA, Chaganti RS 2004. Deregulation of the carbohydrate (chondroitin 4) sulfotransferase 11 (CHST11) gene in a B-cell chronic lymphocytic leukemia with a t(12;14)(q23;q32). Oncogene 23: 6991–6996 [DOI] [PubMed] [Google Scholar]
- Smith L, Plug A, Thayer M 2001. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc Natl Acad Sci 98: 13300–13305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IA, Knezevic BR, Ammann JU, Rhodes DA, Aw D, Palmer DB, Mather IH, Trowsdale J 2010. BTN1A1, the mammary gland butyrophilin, and BTN2A2 are both inhibitors of T cell activation. J Immunol 184: 3514–3525 [DOI] [PubMed] [Google Scholar]
- Strefford JC, An Q, Harrison CJ 2009. Modeling the molecular consequences of unbalanced translocations in cancer: Lessons from acute lymphoblastic leukemia. Cell Cycle 8: 2175–2184 [DOI] [PubMed] [Google Scholar]
- Sun Y, Wyatt RT, Bigley A, Krontiris TG 2001. Expression and replication timing patterns of wildtype and translocated BCL2 genes. Genomics 73: 161–170 [DOI] [PubMed] [Google Scholar]
- Tang JQ, Bene MC, Faure GC 1991. Alternative rearrangements of immunoglobulin light chain genes in human leukemia. Leukemia 5: 651–656 [PubMed] [Google Scholar]
- Tao Q, Fujimoto J, Men T, Ye X, Deng J, Lacroix L, Clifford JL, Mao L, Van Pelt CS, Lee JJ, et al. 2007. Identification of the retinoic acid-inducible Gprc5a as a new lung tumor suppressor gene. J Natl Cancer Inst 99: 1668–1682 [DOI] [PubMed] [Google Scholar]
- Usvasalo A, Raty R, Harila-Saari A, Koistinen P, Savolainen ER, Vettenranta K, Knuutila S, Elonen E, Saarinen-Pihkala UM 2009. Acute lymphoblastic leukemias with normal karyotypes are not without genomic aberrations. Cancer Genet Cytogenet 192: 10–17 [DOI] [PubMed] [Google Scholar]
- Weddington N, Stuy A, Hiratani I, Ryba T, Yokochi T, Gilbert DM 2008. ReplicationDomain: A visualization tool and comparative database for genome-wide replication timing data. BMC Bioinformatics 9: 530 doi: 10.1186/1471-2105-9-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemels JL, Alexander FE, Cazzaniga G, Biondi A, Mayer SP, Greaves M 2000. Microclustering of TEL-AML1 translocation breakpoints in childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 29: 219–228 [DOI] [PubMed] [Google Scholar]
- Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH 2010. Transcriptional silencing of γ-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev 24: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, et al. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1: 133–143 [DOI] [PubMed] [Google Scholar]