Abstract
In eutherian mammals, dosage compensation of X-linked genes is achieved by X chromosome inactivation. X inactivation is random in embryonic and adult tissues, but imprinted X inactivation (paternal X silencing) has been identified in the extra-embryonic membranes of the mouse, rat, and cow. Few other species have been studied for this trait, and the data from studies of the human placenta have been discordant or inconclusive. Here, we quantify X inactivation using RNA sequencing of placental tissue from reciprocal hybrids of horse and donkey (mule and hinny). In placental tissue from the equid hybrids and the horse parent, the allelic expression pattern was consistent with random X inactivation, and imprinted X inactivation can clearly be excluded. We characterized horse and donkey XIST gene and demonstrated that XIST allelic expression in female hybrid placental and fetal tissues is negatively correlated with the other X-linked genes chromosome-wide, which is consistent with the XIST-mediated mechanism of X inactivation discovered previously in mice. As the most structurally and morphologically diverse organ in mammals, the placenta also appears to show diverse mechanisms for dosage compensation that may result in differences in conceptus development across species.
In placental mammals, including humans, dosage compensation in females is achieved by random X chromosome inactivation (Lyon 1961; Monk and Harper 1979; Tan et al. 1993). X inactivation is regulated by the X inactivation center (Xic). The silencing of the inactive X (Xi) spreads outwards along the chromosome from Xic. Xist is a noncoding RNA in mouse, which is transcribed only from the Xi. It coats the Xi in cis and represses X-linked gene expression. All eutherian mammals appear to perform Xist-dependent random X inactivation to achieve dosage compensation in fetal and adult tissues (Ideraabdullah et al. 2008; Payer and Lee 2008).
There is another form of X inactivation in mammals termed imprinted X inactivation. Imprinted X inactivation was discovered in both embryonic and extra-embryonic tissues in marsupials (Cooper et al. 1990; Al Nadaf et al. 2010) and in extra-embryonic tissue (which gives rise to placenta) in eutherian mammals. Imprinted X inactivation has been reported in the mouse (Huynh and Lee 2001, 2005; Heard and Disteche 2006), rat (Wake et al. 1976), and cow (Xue et al. 2002). The situation in humans is still unresolved (Goto et al. 1997; van den Berg et al. 2009; Moreira de Mello et al. 2010), and little is known about the X inactivation status in the placentae of other mammals.
In the mouse germline, the paternal X chromosome (Xp) is silenced by a process called meiotic sex chromosome inactivation (MSCI). After fertilization, Xp is activated and biallelic expression is observed for X-linked genes at the two-cell stage. Subsequently, at the eight-cell stage, only the Xp is inactivated through imprinted X inactivation (Huynh and Lee 2001, 2005; Heard and Disteche 2006). The extra-embryonic tissues retain imprinted X inactivation, and the X-linked genes (except Xist) are transcribed only from the maternal X chromosome. At implantation (E3.5–E4.5), the paternal X is reactivated, and the X-linked genes are transcribed from both parental X chromosomes in the inner cell mass (Mak et al. 2004; Okamoto et al. 2004). Then at the blastocyst stage embryonic day 6.5 (E6.5), random X inactivation takes place, and one of the two parental X chromosomes is randomly inactivated (Cheng and Disteche 2004).
Hybrids of the horse, donkey, and other equids provide an ideal system for investigating X inactivation in the placenta. In 1964, Mukherjee and Sinha (1964) found cytological evidence for random X inactivation in leukocytes of a single female mule, supporting the Lyon hypothesis. In 1969, Hamerton et al. (1969, 1971) reported nonrandom X inactivation in female mules, suggesting that the X inactivation might be aberrant in mules. However, subsequent studies revealed that this could be due to selection during cell culture and/or the sampling effect of random X inactivation (Mukherjee and Mukherjee 1970; Mukherjee and Milet 1972; Serov et al. 1978). The whole-genome level tools now available for equids and the availability of highly purified placental tissues have enabled us to address the question of X inactivation in extra-embryonic membranes in the genus Equus.
Results
Profile of transcriptome-wide allelic expression ratios is consistent with random X inactivation in mule placenta
RNA-seq was performed on placental tissue (chorionic girdle trophoblast isolated from day 33–34 conceptuses) from six horses, four donkeys, seven mules, and two hinnies (Fig. 1A). Sixty percent to 70% of the sequence reads were uniquely mapped to the horse reference genome (Supplemental Table S1). Relative allelic expression ratios in the mule and hinny conceptuses were established by directly counting the allele-specific sequence reads at SNP positions within expressed transcripts (Wang et al. 2008). These data covered more than 7000 genes with sufficient read depth at informative SNP positions to infer allele-specific expression ratios.
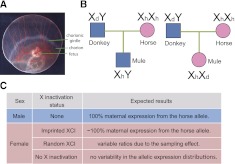
Figure 1.
Results under different forms of X inactivation in mule placenta samples. (A) Image of day 33 horse conceptus, showing fetal and placental tissues, including the invasive trophoblasts known as the chorionic girdle. (B) Transmission of X chromosome in male and female mules. (C) Expected results of allelic expression ratios for male mule and female mule under different forms of X inactivation.
Because a mule is an offspring from a horse mother and a donkey father, male mules inherit their X chromosome from a horse and their Y chromosome from a donkey (Fig. 1B). This implies that 100% expression of the horse allele for X-linked genes is expected. (Fig. 1C). In female mules, each parent transmits an X chromosome (Fig. 1B), giving rise to three possible patterns of allelic expression depending on the form of X inactivation:
Imprinted or leaky imprinted X inactivation
If there is imprinted X inactivation in the placentae of equids, as in the mouse, the Xp will be inactivated. In this case, we would expect to see 100% expression from the horse (maternal) allele for X-linked genes (except XIST) (Fig. 1C). If the imprinted X inactivation is leaky, as in marsupial X inactivation in placenta, we would expect preferential expression from the maternal (horse) allele.
Random X inactivation
If X inactivation is random, expression from both parental alleles is expected in the placental tissue, with an expression ratio that is variable across individuals, due to the sampling of random X inactivation among cells (Fig. 1C). X inactivation happens early during development, when there are a limited number of placental progenitor cells. Once the X inactivation status is determined, it is fixed in the daughter cells, giving rise to a somatic clone of cells with the same X inactivated. Under random X inactivation, each cell has a 50% chance of having the maternal (or paternal) X chromosome remain active, and each cell could be viewed as an independent Bernoulli trial. In the tissues as a whole, the probability of having a certain number of cells with active maternal X would be expected to follow the binomial distribution. When the sample size of cells is small at the initiation of X inactivation, the among-individual variability will be large (Fig. 1C). The magnitude of interindividual variability due to this sampling effect has been quantified in humans and the mouse (Amos-Landgraf et al. 2006; Wang et al. 2010).
No X inactivation
The third possibility is to observe homogeneous biallelic expression, which would imply that the mule placenta has no X inactivation. In this case, the among-individual variability of allele-specific expression ratios would be comparable to that of autosomal genes.
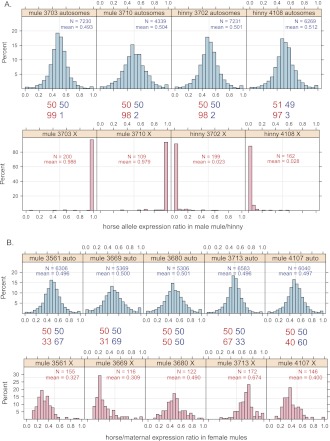
To investigate which form of X inactivation is consistent with our observed data in the chorionic girdle, we plotted the distribution of allelic expression proportions in two male mules (mule-3703 and mule-3710) and two male hinnies (hinny-3702 and hinny-4108) for the autosomal genes and X linked genes, respectively (Fig. 2A). RNA-seq data from male mule and hinny conceptuses showed a distribution of allele-specific expression for autosomal genes centered on 50:50 (Fig. 2A). These males also displayed 97%–99% expression of the maternal X allele and 1%–3% leakage due to the biallelic expression of the genes in the pseudoautosomal region (PAR) (Supplemental Fig. S1; Raudsepp et al. 2004). The allelic expression results obtained from hybrid males served as a control to demonstrate the lack of bias in our RNA-seq methods, SNP calling, and allelic expression analyses.
Figure 2.
Transcriptome-wide distribution of allele-specific expression ratios in male and female mules and hinnies for autosomal and X-linked genes. (A) Histograms of the allelic expression ratios from the horse allele in chorionic girdle samples of two male mules (mule-3703 and mule-3710) and two male hinnies (hinny-3702 and hinny-4108). (B) Histograms of the allelic expression ratios from the horse (maternal) allele in chorionic girdle samples of five female mules (mule-3561, mule-3669, mule-3680, mule-3713, and mule-4107). In these plots, the y-axis shows the percentage of genes in each category of allele-specific expression ratio. The x-axis is the proportion of allelic expression from the horse in the mules and hinnies, ranging from 0 to 1. The average allelic expression proportion and number of informative genes with sufficient expression are labeled for each individual. (Top) Plots for autosomal genes in blue; (bottom) X-linked genes plotted in pink; (middle) comparison of mean maternal vs. paternal expression ratios between autosomal and X-linked genes for each sample.
We then examined the distributions of allele-specific expression in five female mule placenta samples (mule-3561, mule-3669, mule-3680, mule-3713, and mule-4107). For autosomal genes, the distributions were centered on 50:50 without any parental bias, similar to the results in the male mules and hinnies (Fig. 2B). For X-linked genes, we also observed biallelic expression, but there was much greater among-individual variance, with the percentage paternal allele ranging from 33%–69% (Fig. 2B). The coverage of over 170 X-linked genes (990 informative X-linked SNPs) with relatively high expression level in each individual suggests that the variability is a chromosome-wide effect. This among-individual variability is specific to X-linked genes and is expected due to the sampling effect of random X inactivation. These results are consistent with random X inactivation in the mule placenta.
Confirmation of random X inactivation in mule placenta tissues using allele-specific pyrosequencing
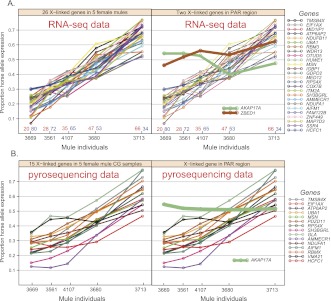
The allelic expression ratios for 26 selected X-linked genes outside the PAR in the RNA-seq data covary among individuals in a concerted way (Fig. 3A), with a variance of 0.355 and mean coefficient of variation (CV) of 0.431. Two PAR genes, AKAP17A and ZBED1, showed much less variability, with an average variance of 0.0037 and a CV of only 0.109 (Fig. 3A), significantly less than that for the non-PAR genes (P = 0.002, bootstrap test for comparing the variance for two independent groups). The remarkably tight covariation across genes for non-PAR genes is consistent with the presence of random X inactivation in mule placenta, and the low interindividual variability of PAR genes demonstrates the expected biallelic expression for these genes.
Figure 3.
Plot of allelic expression proportions in five female mules for 28 X-linked genes in RNA-seq data and 16 X-linked genes in pyrosequencing verification data. (A, left) Allelic expression profiling of 26 X-linked genes in five female mules from the RNA-seq data. The x-axis simply arrays the five individuals. Plotted on the y-axis is the proportion of expression from the horse allele. Two genes (AKAP17A and ZBED1) in the pseudoautosomal region are highlighted in the right panel. (B, left) Allelic expression profiling of 15 selected X-linked genes in five female mules from pyrosequencing verification. The x-axis arrays the five individuals as in panel A. Plotted on the y-axis is the proportion of expression from the horse allele. The PAR gene AKAP17A is highlighted in the right panel.
To confirm our finding in the RNA-seq data, we quantified the allelic-specific expression in 16 selected X-linked genes uniformly distributed along the X chromosome in the same five female mule chorionic girdle samples using pyrosequencing (Marsh 2007), as an independent verification (Supplemental Table S2; Supplemental Data S1). We plotted the proportion of the horse (maternal) expression in the five mules for the 15 non-PAR genes and observed a pattern similar to the RNA-seq data (Fig. 3B, left panel). The substantial covariation across multiple genes seen in the pyrosequencing results confirms that the mule placenta displays random X inactivation.
Profiling X inactivation status in two other tissues in the mule
The equid day 33 placenta consists of several extra-embryonic tissues: the invasive trophoblast cells of the chorionic girdle, the noninvasive trophoblast (chorion and allantochorion), and the yolk sac. Among these placental tissues, the chorionic girdle, which we examined, provides the progenitor cells of the endometrial cups that secrete equine chorionic gonadotrophin (eCG). To exclude the possibility that random X inactivation is only present in a specific placental tissue in mule, we examined the X inactivation status in another placental tissue of the same mule individuals. We performed pyrosequencing experiments on 16 X-linked genes in chorion samples of three mules (mule-3669, mule-3680, and mule-3713). Although only three individual samples were tested, substantial variability of allelic expression is captured for 15 genes outside the PAR, with the proportion of horse allelic expression ranging from 0.1–0.8 (Supplemental Fig. S2A). The allelic expression pattern is consistent with random X inactivation in the chorion samples as well. As expected from the chorionic girdle samples, in the chorion the PAR gene AKAP17A also had a 50:50 expression ratio, and no variation was observed across the three individuals (Supplemental Fig. S2A).
In the mouse embryonic tissue during early development, there is imprinted X inactivation at the eight-cell to morula stage, which is then replaced by random X inactivation in the inner cell mass of the blastocyst stage. If the fetal X inactivation process in mule is the same as in the mouse and humans, we would expect random X inactivation in fetal tissue. To test this hypothesis, we performed pyrosequencing assays to quantify allelic expression of 13 X-linked genes in fetal samples of the same five female mules. We observed covariation of the allelic expression ratios among five individuals, consistent with the sampling effect due to random X inactivation (Supplemental Fig. S2B). Therefore, the patterns of the allele-specific expression for X-linked genes in the extra-embryonic tissues (chorionic girdle and chorion) resemble the fetal tissue, suggesting that random X inactivation happens in all three tissues in day 33 mule conceptuses.
Random X inactivation in the horse placenta
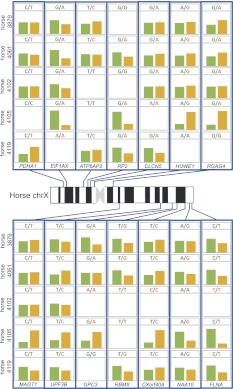
The random X inactivation discovered in the mule placenta might be a hybrid-specific artifact due to potential dysregulation of X inactivation in interspecific hybrids. To check this possibility, we selected 14 widely dispersed genes along the X chromosome with informative SNPs in five individual female horses (Supplemental Table S3; Supplemental Data S2). Allele-specific expression in horse day 33 chorionic girdle samples was quantified by pyrosequencing in all five female horses and a control male horse. In every informative (heterozygous) conceptus and for every gene tested, expression from both parental alleles was observed, demonstrating lack of imprinted X inactivation in horse placenta (Fig. 4). The allelic expression levels in the five chorionic girdle samples covary across the 14 genes, a pattern that is consistent with the previously described sampling effect due to random X inactivation (Fig. 4).
Figure 4.
Pyrosequencing results for 14 X-linked genes in female horse chorionic girdle trophoblast samples. Plotted are allele-specific expression percentages in horse day 33 chorionic girdle samples quantified by pyrosequencing in five female horse samples. (Green) Horse reference allele; (orange) alternative allele. Uninformative homozygous loci are not shown. We observed variable biallelic expression for all 14 genes rather than preferential maternal expression, suggesting random X inactivation in horse placenta. Within genes, we also observed the cis-eQTL effect (EIF1AX, RGAG4, RBMX, and FLNA). The horse X chromosome ideogram was drawn from the work by Bowling et al. (1997).
Mechanism of X inactivation in equids: Characterizing the XIST transcript in the horse, donkey, and mule
To study the mechanism of X inactivation in equids, we identified and annotated the XIST transcript in the horse and donkey. The noncoding XIST transcript is present in the horse reference genome, but its gene model is not annotated. From our RNA-seq data in the female horse, donkey, and mule, we found that the horse/donkey XIST is expressed in day 33 chorionic girdle. We discovered seven transcribed exons in the RNA-seq data, corresponding to the seven exons in the mouse Xist RefSeq gene model NR_001463 (Supplemental Fig. S3; Supplemental Table S4). The horse XIST is on the minus strand of the X chromosome (55,226,251–55,258,045 bp, UCSC equCab2 assembly), the same orientation as in the mouse and humans. We aligned the horse XIST sequence to the mouse sequence using MUSCLE, version 3.8 (Edgar 2004), and found that the average sequence identity between the horse and mouse is 63%. The horse exon 1 is separated into three fragments by two gaps in the genome assembly. The exon structure is conserved between the horse and mouse, except for some exon length differences (Supplemental Table S4).
The sequence identity between the horse and human XIST is 77%. Although, the human XIST has higher sequence similarity to the horse than the mouse transcript, there are differences in the exon models. The human RefSeq gene has six exons (Supplemental Fig. S3), five of which correspond to mouse and horse exons 1, 3, 4, 6, and 7. Human exon 2, which is also present in other primate species (Horvath et al. 2011), could not be found in the mouse or horse. From the sequence alignment, mouse and horse exon 2 and 5 could be identified in humans by homology, but they are not expressed in the human XIST transcript (A Clark, M Ho, and M Roberson, unpubl.). From the horse and donkey RNA-seq data, the XIST exon model in the horse and donkey is the same, without any major exon length or splicing difference detected. At the sequence level, the horse and donkey XIST have high sequence similarity, with only 23-nucleotide differences between the horse and donkey in the transcripts studied.
Mechanism of X inactivation in equids: XIST expression in male and female equids
In somatic cells, the expression of the noncoding XIST transcript has been shown to be critical for X inactivation in the mouse and humans. It is expressed only from the inactive X chromosome (Xi), and spreads in cis to coat the inactivated X. XIST is not expressed in males because males have only one X chromosome and it is always active. If the random X inactivation we observed in female horses and mules is regulated by the horse XIST transcript, we would expect that it is expressed only in females. In fact, we were only able to detect XIST expression in female horse, donkey, mule, and hinny chorionic girdle samples (Supplemental Figs. S4, S5) and in female mule and hinny fetus samples (Supplemental Fig. S5). This finding suggests that equine XIST likely plays the homologous role in X inactivation in equids.
Mechanism of X inactivation in equids: Allele-specific expression of XIST is associated with the allelic expression of other X-linked genes chromosome-wide
Because XIST is only expressed from the inactive X in individual cells, the proportion of cells in a given tissue expressing XIST from one parent could determine the proportion of cells with inactive X from that parent. Therefore, we could use the allelic expression ratio from paternal XIST allele to estimate the maternal expression ratio of other X-linked genes, if the two parental XIST alleles have similar expression levels in a single cell (Supplemental Fig. S6A).
However, in F1 mice from crosses of inbred strains, the allelic expression for X-linked genes cannot be estimated using Xist allelic expression, because the expression levels from the two parental Xist alleles are different. In hybrid mouse F1 animals, the relative Xist expression from the two parental alleles varied substantially depending on the cross, due to the Xce effect. Xce is the X chromosome control element that maps to a region on the X chromosome containing Rr18 (formerly known as Xite) and the promoter of Tsix (Cattanach and Isaacson 1967; Simmler et al. 1993; Courtier et al. 1995; Chadwick et al. 2006; Valley and Willard 2006). In F1 mice, the X chromosome with a stronger Xce allele has lower expression from the Xist allele. In any given tissue, the relative number of cells with an active Xm (maternally inherited X) and Xp (paternally inherited X) cannot be estimated by the allelic expression ratio of the two parental Xist alleles (Wang et al. 2010) because the expression level from the two Xist alleles is dramatically different, due to different strengths of Xce (X controlling element) alleles. From our AKR–PWD reciprocal crosses, we observed ∼90% of Xist is expressed from the AKR allele, and 80% of Xist expression is from the B6 allele in B6-CAST reciprocal crosses (Wang et al. 2010). This strong cis-eQTL (cis-regulating expression quantitative trait locus) effect is due to the different strength of the Xce alleles among mouse inbred lines (Supplemental Figs. S6B, S7).
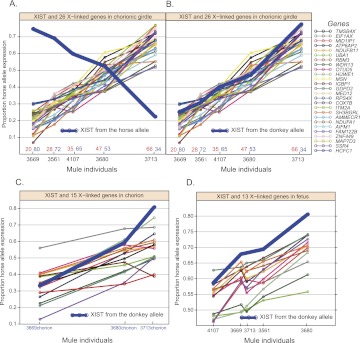
To further understand the mechanism of X inactivation in equids, we performed pyrosequencing experiments to quantify the allele-specific expression for XIST in the female mule chorionic girdle trophoblast. When the allelic expression ratios for XIST and other X-linked genes were compared in five female mules, we observed expression of XIST from both the horse and donkey alleles (Fig. 5A). This is unlike the situation in the mouse placenta in which Xist is imprinted with 100% paternal expression, another observation consistent with a lack of imprinted X inactivation in mule placenta. In addition, a negative correlation in expression between XIST and the alleles of other genes on the cis chromosome was observed (Fig. 5A). Since we assume that XIST is expressed from the inactive X, we plotted the XIST allelic expression ratios from the donkey allele (Fig. 5B), and it overlapped perfectly with other X-linked genes (average correlation coefficient, 0.969), suggesting equine XIST functions by repressing the expression of the Xi. The same pattern holds in mule chorion and fetal samples (Fig. 5C,D). This relationship was not observed in crosses of mouse inbred strains, because of the strong Xce effect (Supplemental Fig. S6B). Our finding is consistent with a mechanism in which XIST regulates random X inactivation by coating the inactive X in cis in the mule placenta and fetus, as has been described in the mouse.
Figure 5.
Allelic expression of XIST transcripts for five female mules in chorionic girdle, chorion, and fetus samples. (A) The horse/maternal expression of XIST transcripts in five female mule chorionic girdle samples (thick blue line). The allelic expression proportions of 26 X-linked genes from the RNA-seq data are plotted in the background. On the x-axis are the five individuals. Plotted on the y-axis is the proportion of expression from the horse allele. (B) The donkey/paternal expression proportion (1-horse) of XIST transcript in five female mule chorionic girdle samples (thick blue line). (C) The donkey/paternal expression proportion (1-horse) of XIST transcript in three female mule chorion samples (thick blue line). The allelic expression proportions of 15 X-linked genes from the pyrosequencing results are plotted in the background. On the x-axis are the three individuals. Plotted on the y-axis is the proportion of expression from the horse allele. (D) The donkey/paternal expression proportion (1-horse) of XIST transcript in five female mule fetus samples (thick blue line). The allelic expression ratios of 13 X-linked genes from the pyrosequencing results are plotted in the background. On the x-axis are the five individuals. Plotted on the y-axis is the proportion of expression from the horse allele.
From the mule XIST pyrosequencing data, we also conclude that the Xce alleles have similar strength in the horse and donkey, at least across the alleles sampled in this study. Interestingly, we should be able to use the allelic expression ratio for the XIST gene to predict the allelic expression ratio for all X-linked genes chromosome-wide in any given tissue of any given female, which is not possible in crosses of mouse inbred strains because of the strong Xce effect.
Degree of sampling effect due to random X inactivation varies from tissue to tissue
A key characteristic of random X inactivation is the sampling effect of relative allelic expression that is seen across different individuals. If the X inactivation is random and the determination of X inactivation status in each tissue is independent, the sampling effect of X inactivation is predicted to vary across tissues. In tissues that have a small, homogeneous population of cells at the time of X inactivation, such as the chorionic girdle and chorion, the among-individual variability in allelic expression ratio will be high, due to the smaller number of trials. In contrast, for tissues with a large number of founder cells at the time of X inactivation, there will be lower among-individual variability for allelic expression ratios of X-linked genes. For samples of mixed tissues or cell types, the distribution of allelic expression ratios is no longer binomially distributed but instead is a mixture distribution usually with lower variance.
We observed significant variation in allelic expression for the X-linked genes among multiple female mule individuals in three different tissues (Fig. 6), consistent with X inactivation establishment at a time in development when each tissue is composed of a limited number of cells.
Figure 6.
Plot of allelic expression of 13 X-linked genes in female mule chorionic girdle, chorion, and fetus samples. Plotted in this figure is the allelic expression profiling of 13 X-linked genes in five female mules from the RNA-seq data. The x-axis simply arrays the five fetus samples, three chorion samples, and five chorionic girdle samples. Plotted on the y-axis is the proportion of expression from the horse allele.
We predict that tissues with greater heterogeneity of cell types at the initiation of X inactivation will have smaller variance. Indeed, for chorionic girdle and chorion trophoblast samples, which are derived from a pure population of a single cell type, the variability was larger (range, 0.1–0.8; mean variances, 0.025 and 0.018); whereas in the fetal sample, which consists of a mixture of different tissues and cell types, the variation was lower (range, 0.45–0.75; mean variances, 0.004). If each individual tissue is an independent trial of random X inactivation, we would also predict that there will be no correlation in different tissues within the same individual. As expected, we found no significant correlation (r = 0.147, P > 0.05) across tissues (Supplemental Fig. S8). Both results provided further evidence that the X inactivation in mule placenta is truly random.
Discussion
Random (not imprinted) X inactivation in the equid placenta
The vast majority of studies of X inactivation have analyzed human or mouse samples; investigations of X inactivation in other mammals are rare and limited in scope, relying on a single or a few X-linked gene(s). This study represents the first systematic analysis of global X chromosome inactivation outside of humans and mice. We studied reciprocal hybrid placental tissue of horse and donkey crosses, because their level of genome divergence made it possible to distinguish maternal and paternal alleles of nearly every gene. We analyzed all X-linked genes with detectable expression levels, using RNA-seq and allele-specific pyrosequencing to accurately quantify allelic expression with a CV of 2%. Seven thousand autosomal genes and X-linked genes in the PAR were used as controls. Three different tissues were examined in five female mule conceptuses, a sample sufficiently large to assess interindividual variation in allelic expression ratios. The XIST expression profile in the mule trophoblast was consistent with the mechanism for X inactivation demonstrated earlier in mice. Finally, we tested 14 X-linked genes in horse placental trophoblast cells to provide intraspecies verification of our results. These aspects of our design and data set make us confident that X chromosome inactivation in the placentae of mules and horses is random and not imprinted.
Is there dysregulation of X inactivation in interspecific hybrids?
Aberrant X inactivation profiles have been reported in cloned animals (Xue et al. 2002), and there was some evidence of dysregulation of X inactivation in mules and hinnies (Hamerton et al. 1969, 1971). Subsequent studies found that the nonrandom X inactivation in the mule and hinny may be an artifact due to the cell selection effect (Mukherjee and Mukherjee 1970; Mukherjee and Milet 1972). In our RNA-seq study, we did not find evidence for hybrid dysregulation of X inactivation in mule tissues. Whether there is hybrid dysregulation of X inactivation in other hybrid systems remains an open question.
Evolutionary considerations of X inactivation in the mammalian placenta
Our findings provide evidence that imprinted X inactivation in the placenta might not be the general case in eutherian mammals. These results beg the question, why are there different forms of X inactivation in the eutherian placenta but not in other tissues? From an evolutionary perspective, monoallelic maternal expression in imprinted X inactivation has a clear disadvantage. By giving up diploidy, every X-linked gene acquires an expression pattern that is effectively hemizygous. When the maternal copy of the X harbors a recessive deleterious allele, the effect could be lethal for organisms with imprinted X inactivation, whereas organisms with random X inactivation will still have 50% of cells expressing the normal paternal allele. However, the mammalian placenta is a specialized organ that is critical for maternal–fetal nutrient, gas, and waste exchange. Important cell signaling, cell–cell interaction, and immunological functions are performed in the placenta, where direct contact of the maternal and fetal tissues occurs. In imprinted X inactivation, expressing only the maternal X-linked genes could potentially reduce the possibility of female immune response to paternal antigens and reduce the maternal–fetal inconsistencies by matching the maternal genotype for X-linked genes.
If we assume that imprinted X inactivation is the ancestral state because of its presence in marsupials, eutherian mammals evolved different strategies in the placenta: mice, rats, and cattle retain imprinted X inactivation, whereas the horse (and most probably the donkey) has developed random X inactivation. With the powerful tools of RNA sequencing now available, it should be possible to identify the type of X inactivation employed in the placentae of various mammals and to determine the evolutionary branchpoints that led to divergence for this trait.
Methods
Horse, donkey, and mule conceptus recovery and dissection
Adult male and female horses and donkeys were maintained at facilities at Cornell's Baker Institute for Animal Health. Timed intra-and interspecies pregnancies were established using standard methods in equine reproduction (collection of semen and artificial insemination, transrectal ultrasound determination of ovulation and pregnancy). Equine conceptuses from pregnant horse mares and jenny donkeys were collected on days 33–35 after ovulation using standard nonsurgical techniques (Antczak et al. 1987). After collection, the conceptuses were micro-dissected into distinct tissues (chorionic girdle, allantochorion, fetus, etc.) with the aid of a dissection microscope and ophthalmic instruments. Tissues were snap frozen in liquid nitrogen and stored at −80°C until use. Animal care was performed in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee of Cornell University under protocol 1986-0216.
RNA sample extractions and QC
Total RNA samples were extracted from snap-frozen placental tissues using the RNeasy or RNeasy Plus mini kit (Qiagen) following the manufacturer's protocol. Samples were eluted in sterile MilliQ water and stored at −80°C. RNA quality and concentration was assayed on an Agilent BioAnalyzer 2100 (Agilent) for a RNA Integrity Number (RIN) score. All samples tested had RIN scores above 9.0, and most had scores from 9.8–10.0.
Illumina sequencing of the transcriptome and allele-specific expression analysis
Our initial mRNA-seq experiments were performed on total RNA samples from one horse (horse-3879, 20 lanes), one donkey (donkey-3689, 10 lanes), one mule (mule-3703, eight lanes), and one hinny (hinny-3702, eight lanes) chorionic girdle sample (day 33–34) using an Illumina Genome Analyzer (Illumina). The mRNA-seq libraries were made with 3 μg of starting total RNA samples using the mRNA-seq 8-Sample Prep Kit (Illumina), following the Illumina protocol for mRNA sequencing sample preparation. To quantify the variability of allelic expression ratios for X-linked genes in female samples, we also did RNA-seq on additional samples from five horses, three donkeys, and five mules (Supplemental Table 1), with the Illumina Genome Analyzer IIx (GAIIx). Image analysis and base calling were performed with the Illumina pipeline (version 1.3). Data analysis and statistical methods used for estimation of total expression level and allelic expression ratios from RNA-seq data are available in the work of Wang et al. (2011).
Verification of allele-specific expression in mules by pyrosequencing
To verify the allelic expression ratios inferred from the RNA-seq data in the mule chorionic girdle samples using an independent method, we selected 16 X-linked genes with detectable expression level and informative SNPs in the RNA-seq data (TMSB4X, EIF1AX, ATP6AP2, DDX3X, UBA1, MSN, PDZD11, RPS4X, SH3BGRL, GLA, AMMECR1, NDUFA1, AIFM1, RBMX, VMA21, and HCFC1). The selected genes are evenly distributed along the X chromosome to assess whether effects were chromosome-wide (Supplemental Table S2). Unlike other X-linked genes with only one copy in males, genes in the PAR have two alleles in both male and females. To check the allelic expression ratios for PAR genes, we included two of them, AKAP17A and ZBED1 (Supplemental Table S2). To study the mechanism of X inactivation, we annotated the horse and donkey XIST transcripts and did SNP discovery using this data resource. We selected an informative SNP between the horse and donkey in XIST, which does not overlap the predicted TSIX transcript region (Supplemental Table S2).
We quantified the allelic expression for all 19 selected X-linked genes in total RNA samples of five female and one male mule chorionic girdles by allele-specific pyrosequencing. Pyrosequencing PCR and sequencing primers were designed with PyroMark Assay Design Software, version 2.0.1.15 (Qiagen). To guarantee that there are no SNPs within primers, SNP positions between and within the horse and donkey were labeled and excluded for the primer design. The PCR amplification of cDNA templates was carried out in 40 μL volumes using Ampli-Taq Gold polymerase (Life Technologies) under the following cycling conditions: one cycle of for 5 min at 95°C; 45 cycles 45 sec at 95°C, 30 sec at 50°C–57°C, and 20 sec at 72°C; followed by one cycle of 10 min at 72°C. PCR products were prepared immediately according to the manufacturer's protocol and then loaded on the PSQ 96MA Pyrosequencer (Qiagen) with the PyroMark Gold Reagents (Qiagen) using the Allele Quantification method (AQ). Two technical replicates were done for each gene in each sample, and the average ratio was used for data analysis.
Allelic expression ratios were quantified in embryonic tissue; pyrosequencing assays were performed for the same 19 X-linked genes in fetus of the same five female mule individuals tested in chorionic girdle. We also carried out allele-specific pyrosequencing in another placental tissue (chorion) in three of the five female mules for which we have that tissue.
Verification of allele-specific expression in horses by pyrosequencing
To quantify the allelic expression ratios in progeny of horse × horse crosses, we needed informative SNPs within horse. DNA sequence heterozygosity within the female horse samples is much lower than horse–donkey differences, and it is especially difficult to find informative SNPs that are heterozygous across all five female horses in the RNA-seq data. To confirm our X inactivation results in horse × horse crosses, we selected three X-linked genes for pyrosequencing verification within horse. Based on the SNP calling from the horse RNA-seq data in chorionic girdle samples, 14 genes (PDHA1, EIF1AX, ATP6AP2, RP2, CLCN5, HUWE1, RGAG4, MAGT1, UPF3B, GPC3, RBMX, CXorf40A, NAA10, and FLNA) with informative heterozygous SNPs in at least three female horses were selected (Fig. 4; Supplemental Table S3). To confirm the genotypes called from RNA-seq data, we did SNP genotyping in one male and five female horse genomic DNA samples using pyrosequencing. The results matched the RNA-seq calls. Allele-specific expression ratios were quantified using pyrosequencing of cDNAs made from the chorionic girdle of the six horse samples.
Data access
IIlumina RNA-seq data in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE36352.
Acknowledgments
We thank Jenny Xiang, Ying (Diana) Shao, Amanda Manfredo, and Li (Grace) Chi for assistance with Illumina sequencing and pyrosequencing experiments. We also thank Jim Hardy, Scott Hoffay, and Emily Silvela for management of the Baker Institute equid herd. This work was supported in part by a Meinig Family Investigator Award to A.G.C. D.F.A. is an investigator of the Dorothy Russell Havemeyer Foundation, Inc. We thank the Cornell Center for Vertebrate Genomics, the Zweig Memorial Fund, and the Morris Animal Foundation for funding.
Author contributions: X.W., D.C.M., D.F.A., and A.G.C. designed the experiment; D.C.M. and D.F.A. dissected the tissues; X.W. and D.C.M. performed the experiments; X.W., D.C.M., D.F.A., and A.G.C. analyzed the data; and X.W., D.C.M., D.F.A., and A.G.C. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138487.112.
References
- Al Nadaf S, Waters PD, Koina E, Deakin JE, Jordan KS, Graves JA 2010. Activity map of the tammar X chromosome shows that marsupial X inactivation is incomplete and escape is stochastic. Genome Biol 11: R122 doi: 10.1186/gb-2010-11-12-r122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Cottle A, Plenge RM, Friez M, Schwartz CE, Longshore J, Willard HF 2006. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am J Hum Genet 79: 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antczak DF, Oriol JG, Donaldson WL, Poleman C, Stenzler L, Volsen SG, Allen WR 1987. Differentiation molecules of the equine trophoblast. J Reprod Fertil Suppl 35: 371–378 [PubMed] [Google Scholar]
- Bowling AT, Breen M, Chowdhary BP, Hirota K, Lear T, Millon LV, Ponce de Leon FA, Raudsepp T, Stranzinger G 1997. International system for cytogenetic nomenclature of the domestic horse. Report of the Third International Committee for the Standardization of the domestic horse karyotype, Davis, CA, USA, 1996. Chromosome Res 5: 433–443 [DOI] [PubMed] [Google Scholar]
- Cattanach BM, Isaacson JH 1967. Controlling elements in the mouse X chromosome. Genetics 57: 331–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick LH, Pertz LM, Broman KW, Bartolomei MS, Willard HF 2006. Genetic control of X chromosome inactivation in mice: Definition of the Xce candidate interval. Genetics 173: 2103–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MK, Disteche CM 2004. Silence of the fathers: Early X inactivation. Bioessays 26: 821–824 [DOI] [PubMed] [Google Scholar]
- Cooper DW, Johnston PG, Vandeberg JL, Robinson ES 1990. X-chromosome inactivation in marsupials. Aust J Zool 37: 411–417 [Google Scholar]
- Courtier B, Heard E, Avner P 1995. Xce haplotypes show modified methylation in a region of the active X chromosome lying 3′ to Xist. Proc Natl Acad Sci 92: 3531–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Wright E, Monk M 1997. Paternal X-chromosome inactivation in human trophoblastic cells. Mol Hum Reprod 3: 77–80 [DOI] [PubMed] [Google Scholar]
- Hamerton JL, Giannelli F, Collins F, Hallett J, Fryer A, McGuire VM, Short RV 1969. Non-random x-inactivation in the female mule. Nature 222: 1277–1278 [DOI] [PubMed] [Google Scholar]
- Hamerton JL, Richardson BJ, Gee PA, Allen WR, Short RV 1971. Non-random X chromosome expression in female mules and hinnies. Nature 232: 312–315 [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM 2006. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Horvath JE, Sheedy CB, Merrett SL, Diallo AB, Swofford DL, Nisc Comparative Sequencing P, Green ED, Willard HF 2011. Comparative analysis of the primate X-inactivation center region and reconstruction of the ancestral primate XIST locus. Genome Res 21: 850–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Lee JT 2001. Imprinted X inactivation in eutherians: A model of gametic execution and zygotic relaxation. Curr Opin Cell Biol 13: 690–697 [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT 2005. X-chromosome inactivation: A hypothesis linking ontogeny and phylogeny. Natl Rev 6: 410–418 [DOI] [PubMed] [Google Scholar]
- Ideraabdullah FY, Vigneau S, Bartolomei MS 2008. Genomic imprinting mechanisms in mammals. Mutat Res 647: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303: 666–669 [DOI] [PubMed] [Google Scholar]
- Marsh S. 2007. Pyrosequencing protocols. Humana Press, Totowa, NJ. [Google Scholar]
- Monk M, Harper MI 1979. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281: 311–313 [DOI] [PubMed] [Google Scholar]
- Moreira de Mello JC, de Araujo ES, Stabellini R, Fraga AM, de Souza JE, Sumita DR, Camargo AA, Pereira LV 2010. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS ONE 5: e10947 doi: 10.1371/journal.pone.0010947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee BB, Milet RG 1972. Nonrandom X-chromosome inactivation–an artifact of cell selection (mouse chimera). Proc Natl Acad Sci 69: 37–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee BB, Mukherjee AB 1970. Is inactivation of the X chromosome in the female mule non-random? Nature 228: 1321–1322 [DOI] [PubMed] [Google Scholar]
- Mukherjee BB, Sinha AK 1964. Single-active-X hypothesis: Cytological evidence for random inactivation of X-chromosomes in a female mule complement. Proc Natl Acad Sci 51: 252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649 [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Santani A, Wallner B, Kata SR, Ren C, Zhang HB, Womack JE, Skow LC, Chowdhary BP 2004. A detailed physical map of the horse Y chromosome. Proc Natl Acad Sci 101: 9321–9326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serov OL, Zakijan SM, Kulichkov VA 1978. Analysis of mechanisms regulating the expression of parental alleles at the GPD locus in mule erythrocytes. Biochem Genet 16: 379–386 [DOI] [PubMed] [Google Scholar]
- Simmler MC, Cattanach BM, Rasberry C, Rougeulle C, Avner P 1993. Mapping the murine Xce locus with (CA)n repeats. Mamm Genome 4: 523–530 [DOI] [PubMed] [Google Scholar]
- Tan SS, Williams EA, Tam PP 1993. X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat Genet 3: 170–174 [DOI] [PubMed] [Google Scholar]
- Valley CM, Willard HF 2006. Genomic and epigenomic approaches to the study of X chromosome inactivation. Curr Opin Genet Dev 16: 240–245 [DOI] [PubMed] [Google Scholar]
- van den Berg IM, Laven JS, Stevens M, Jonkers I, Galjaard RJ, Gribnau J, van Doorninck JH 2009. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet 84: 771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake N, Takagi N, Sasaki M 1976. Non-random inactivation of X chromosome in the rat yolk sac. Nature 262: 580–581 [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS ONE 3: e3839 doi: 10.1371/journal.pone.0003839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG 2010. Paternally biased X inactivation in mouse neonatal brain. Genome Biol 11: R79 doi: 10.1186/gb-2010-11-7-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG 2011. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics 189: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X 2002. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet 31: 216–220 [DOI] [PubMed] [Google Scholar]