Abstract
X chromosome inactivation (XCI) achieves dosage balance in mammals by repressing one of two X chromosomes in females. During XCI, the long noncoding Xist RNA and Polycomb proteins spread along the inactive X (Xi) to initiate chromosome-wide silencing. Although inactivation is known to commence at the X-inactivation center (Xic), how it propagates remains unknown. Here, we examine allele-specific binding of Polycomb repressive complex 2 (PRC2) and chromatin composition during XCI and generate a chromosome-wide profile of Xi and Xa (active X) at nucleosome-resolution. Initially, Polycomb proteins are localized to ∼150 strong sites along the X and concentrated predominantly within bivalent domains coinciding with CpG islands (“canonical sites”). As XCI proceeds, ∼4000 noncanonical sites are recruited, most of which are intergenic, nonbivalent, and lack CpG islands. Polycomb sites are depleted of LINE repeats but enriched for SINEs and simple repeats. Noncanonical sites cluster around the ∼150 strong sites, and their H3K27me3 levels reflect a graded concentration originating from strong sites. This suggests that PRC2 and H3K27 methylation spread along a gradient unique to XCI. We propose that XCI is governed by a hierarchy of defined Polycomb stations that spread H3K27 methylation in cis.
X chromosome inactivation (XCI) provides an excellent model by which to study Polycomb regulation and the role of long noncoding RNAs (lncRNAs) in inducing facultative heterochromatin (Lyon 1999; Wutz and Gribnau 2007; Payer and Lee 2008; Lee 2011). XCI is controlled by the X-inactivation center (Xic), an X-linked region that controls the counting of X chromosomes, the mutually exclusive choice of Xa and Xi, and the recruitment and propagation of silencing complexes. The 17-kb Xist RNA initiates the silencing step as it accumulates on the X (Brockdorff et al. 1992; Brown et al. 1992; Clemson et al. 1996). Although recent studies have shown that Xist RNA directly recruits Polycomb repressive complex 2 (PRC2) to the Xi (Zhao et al. 2008) and that loading of the Xist-PRC2 complex occurs first at a YY1-bound nucleation center located within the Xic (Jeon and Lee 2011), how the silencing complexes spread throughout the X after this obligatory nucleation step remains a major unsolved problem.
Because autosomes with ectopic Xic sequences are subject to long-range silencing (Wutz and Gribnau 2007; Payer and Lee 2008), it is thought that spreading elements cannot be unique to the X. One hypothesis suggests that repetitive elements of the LINE1 class facilitate spreading (Lyon 2000). However, this hypothesis has been difficult to test, as linking repeats to locus-specific function has been complicated by their repetitive nature. Some studies have provided correlative evidence (Bailey et al. 2000; Wang et al. 2006; Chow et al. 2010), whereas others find that species lacking active LINE1s nonetheless possess XCI (Cantrell et al. 2009). Other classes of repeats may be more enriched on the X (Chow et al. 2005). Matrix-associated proteins, such as HNRNPU (also known as SAF-A), have also been proposed to facilitate spreading (Helbig and Fackelmayer 2003; Hasegawa et al. 2010; Pullirsch et al. 2010), but a direct link has also not been demonstrated.
In general, the identification of spreading elements has been thwarted by the lack of high-throughput approaches that distinguish Xi and Xa at sufficient resolution. Epigenomic studies have primarily focused on male cells (Bernstein et al. 2006; Boyer et al. 2006; Barski et al. 2007; Mikkelsen et al. 2007; Ku et al. 2008), though one recent ChIP-seq analysis with partial allele-specific coverage used female mouse embryonic stem (ES) cells but without addressing PRC2 binding. The reported 1.2-fold enrichment of H3K27me3 on Xi (Marks et al. 2009) is unexpectedly low and at odds with intense cytological H3K27me3 immunostaining (Plath et al. 2003; Silva et al. 2003)—likely caused by low-density polymorphisms between Xi and Xa. As a result, the quest for an Xi chromatin state map and spreading elements has remained unrealized.
In principle, silencing complexes could initially load at the Xic and spread serially from nucleosome to nucleosome. Alternatively, they could spread outwardly via “way stations” located at defined sites along the X that would anchor and relay silencing complexes (Gartler and Riggs 1983). To test these models, we herein devise an allele-specific ChIP-seq strategy that enables the generation of chromosome-wide developmental profiles at unprecedented allelic resolution. We report a high-density Xi chromatin state map and identification of discrete Polycomb stations.
Results
Allele-specific ChIP-seq
Mammalian PRC2 contains four core subunits: EED, SUZ12, RBAP48 (RBBP4 in mouse), and EZH2, the subunit responsible for trimethylating H3K27. Because Polycomb recruitment is a central feature of XCI (Plath et al. 2003; Silva et al. 2003; Zhao et al. 2008), we obtained allele-specific ChIP-seq profiles for EZH2 and H3K27me3 and compared them to those for activating marks, including RNA polymerase II holoenzyme RNAPII-S5P (active RNAPII), H3K4me3 (transcriptional initiation), and H3K36me3 (transcriptional elongation). To distinguish Xi from Xa, we used female cell lines carrying one X of Mus castaneus origin (XCast) and one of M. musculus 129 origin (X129) and analyzed three developmental stages. First, we examined undifferentiated female ES cells (d0), which carry two Xa but recapitulate XCI during differentiation. Second, we examined differentiating ES cells on day 7 (d7), a time point corresponding to a mid-XCI state where ∼40% of cells are establishing XCI (Supplemental Fig. S1A). Due to this heterogeneity the actual level of H3K27me3 and EZH2 deposition may be somewhat higher than determined in this analysis. Disabling the Tsix allele on X129 (TsixTST/+) (Ogawa et al. 2008) ensured inactivation of X129 in the ES line. Third, we examined post-XCI, clonal hybrid female mouse embryonic fibroblasts (MEF; F1, ♂XCast × ♀X129) which have inactivated X129. Approximately 0.6 million SNPs and insertions/deletions (indels) of ∼23 million genome-wide SNPs/indels between M. castaneus and M. musculus were used to distinguish XCast and X129 (Keane et al. 2011).
Using paired-end sequencing, ≥83% of all read pairs aligned uniquely, and ∼36% provided allele-specific information (Supplemental Table S1). All tracks (Cast, 129, Composite) were first normalized to their corresponding input controls to minimize potential artifacts stemming from differential chromosome compaction, crosslinking, or sonication efficiencies. ES and MEF input data mapped proportionally to chromosome length (Supplemental Fig. S1B) and equally well to both homologs of ChrX and Chr13 (Supplemental Fig. S1C); this showed that experimental bias between Xi and Xa was negligible. The composite track (comp) represents total epitope abundance, whereas Cast and 129 tracks reflect relative allelic abundance based on local SNP densities (see Supplemental Methods). To validate our approach, we inspected loci with established mono- or biallelic expression (Supplemental Fig. S2). At the imprinted Dlk1-Meg3 and Zim1-Peg3 loci, allele-specific H3K4me3 and RNAPII-S5P profiles were consistent with known parent-of-origin effects. At the Xist promoter, H3K4me3 and RNAPII-S5P were specifically enriched on X129 (Xi). The opposite pattern was observed at Iqsec2, a gene subject to XCI. In contrast, biallelic H3K4me3 and RNAPII-S5P enrichment was seen at the neighboring Kdm5c (formerly Jarid1c), a gene known to escape XCI (Greenfield et al. 1998; Carrel and Willard 2005). These results demonstrate the allele-specific nature of our ChIP-seq.
Genes that escape XCI
We used allele-specific profiles to identify genes that escape XCI by scoring H3K4me3 peaks within 3 kb of annotated transcriptional start sites (TSSs) and noting allelic skew with statistical significance (P < 0.05, normal approximation of binomial). Genes with significant XCast (Xa) skewing were considered monoallelic. Genes with insignificant skewing or two-allele H3K4me3 enrichment were designated biallelic. Genes lacking H3K4me3 peaks within 3 kb of annotated promoters were considered repressed (“off”). Those lacking sufficient SNP density were excluded (not determined, “n/d”). On Chr13, ∼400 out of 843 genes were biallelic, ∼300 genes were off, ∼100 were indeterminate (n/d), and few were monoallelic (Supplemental Fig. S3A).
XCast skewing was evident for almost all MEF genes (Supplemental Fig. S3A), consistent with the genes being subject to XCI (Carrel and Willard 2005; Berletch et al. 2010). Eleven genes showed biallelic H3K4me3 marks (Supplemental Fig. S3B,C), in excellent agreement with recent expression analysis (Yang et al. 2010). Eight out of 11 genes appeared on both lists (e.g., Kdm6a [Utx], Kdm5c). Three noncoding genes, including Jpx (Enox), Ftx (B230206F22Rik), and 5530601H04Rik (Supplemental Fig. S3B), appeared only in our study, though they had been shown by others to be biallelically expressed (Johnston et al. 2002; Reinius et al. 2010; Chureau et al. 2011). These results suggest that allele-specific ChIP-seq could be an effective method for identifying monoallelically expressed genes on a genome-wide scale.
Allelic profiles of Xa and Xi
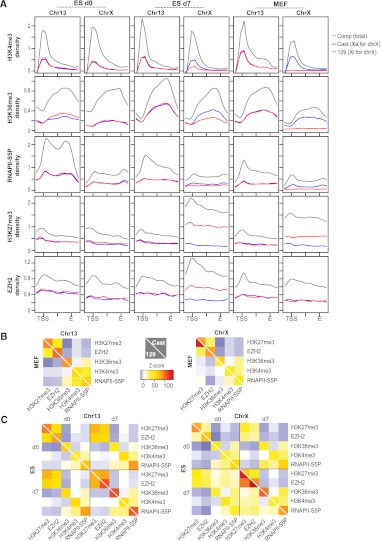
We then performed metagene analysis to examine average epitope densities within genes on chromosomes X (ChrX) and 13 (Chr13). In 16.7 ES cells, Chr13 was the only autosome that was fully M. castaneus for one homolog and fully 129 for the other in TsixTST /+ ES cells (other autosomes had meiotically recombined in the F1 germline). For Chr13 genes, the marks of gene activation remained relatively constant, and there was little allele-specific distinction before (d0 ES), during (d7 ES), and after (MEF) XCI. On both homologs, RNAPII-S5P and H3K4me3 were enriched over promoters, and H3K36me3 occurred along the gene body, as expected.
In contrast, ChrX showed dynamic changes. While XCast and X129 profiles were similar on d0 (pre-XCI), they diverged significantly at d7 (mid-XCI) and remained distinct in MEFs (post-XCI) (Fig. 1A). Active marks (RNAPII-S5P, H3K4me3, and H3K36me3) showed stereotypical enrichment on both homologs in the pre-XCI state (d0) but became substantially depleted on Xi (X129) allele in d7 cells, dropping to about half of Xa (XCast) levels, as might be expected for a mid-XCI stage. In MEFs (post-XCI), the average composite values were reduced by ∼50%, and the Xi trace flatlined, consistent with complete loss of RNAPII-S5P, H3K4me3, and H3K36me3 from Xi. Conversely, EZH2 binding increased ∼twofold on Xi between d0 and d7, and H3K27me3 levels increased ∼threefold. On Xa (XCast), EZH2 and H3K27me3 levels stayed relatively constant. These results demonstrated de novo recruitment of EZH2 and H3K27 trimethylation en masse to X-genes during XCI. In post-XCI MEFs, however, allelic differences remained for H3K27me3, but EZH2 binding was no longer significantly enriched in genes on Xi over Xa. These results indicate that massive PRC2 recruitment occurs during the establishment phase of XCI, but maintenance does not require large amounts of genic PRC2, consistent with the already trimethylated state of H3K27 in post-XCI cells. It is possible that PRC2 on Xi is concentrated in nongenic regions.
Figure 1.
Allelic chromatin profiles of Xa and Xi. (A) Metagene analysis for chromatin epitopes on Chr13 and ChrX at indicated time points. Coverages were averaged over all genes (843 for Chr13, 1007 for ChrX) and scaled from the TSS to the 3′ end, E. Profiles extend 3 kb upstream of the TSS and 3 kb downstream from E. Densities were normalized to the average gene coverage over the chromosome. (B,C) Pearson correlations for pairwise comparisons between epitopes were compared to a permutation-based random model and resulting Z-scores plotted in heat maps. (B) Plots of MEF results. (C) Plots of ES d0 and d7 results. Numerical Z-scores are color-coded and scaled identically for all heat maps. (Yellow–red patches) Significant positive correlation, (blue patches) significant negative correlation. A white diagonal line separates Cast and 129 results.
We then examined whole-chromosome coverages. Allelic heat maps display positional correlations between different epitopes at different time points, with color-coded Z-scores representing the significance of the correlation estimated using a permutation-based random model (Fig. 1B,C). In MEFs (Fig. 1B), allelic profiles for Chr13 were essentially identical (colors mirrored across the diagonal) with strong positive correlations of RNAPII-S5P with H3K4me3 and H3K36me3, and good anti-correlations between RNAPII-S5P/H3K36me3 and H3K27me3/EZH2. However, allelic profiles for ChrX were significantly different for a number of epitopes (discordant colors across the diagonal), consistent with XCI having occurred in MEFs. To ask what happens during XCI establishment, we examined d0 and d7 ES cells (Fig. 1C). Chr13 showed almost no allelic differences for any chromatin epitope, but significant allelic deviations were seen on ChrX. First, while the positive correlation between H3K27me3 and EZH2 on Xi was significant on d0 (Z-score = 15), the correlation grew very strong on d7 (Z-score = 96). Second, whereas H3K27me3/EZH2 were anticorrelated with active marks on d0, they became unexpectedly positively correlated on d7. Notably, the active marks showed high correlation between d0 and d7, suggesting that their density maps did not change dramatically. This dynamic of active versus repressive marks raised the intriguing possibility that, during XCI, EZH2 and H3K27me3 appear in domains marked by active RNAPII and H3K4me3. This feature was Xi-specific and absent on Xa and Chr13.
Strong EZH2 recruiting sites on Xi are bivalent domains
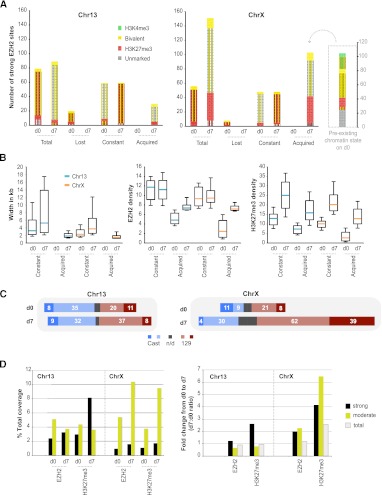
Chromatin regions marked concurrently by H3K4me3 and H3K27me3 have been termed “bivalent domains,” typically associated with transcriptionally poised developmental genes in ES cells (Bernstein et al. 2006). In undifferentiated ES cells, CpG islands associated with bivalent domains account for most EZH2 sites (Ku et al. 2008), and H3K27me3 is rarely detected in the absence of H3K4me3 (Mikkelsen et al. 2007). We asked if EZH2 localization and H3K27me3 followed similar patterns on ChrX during XCI. On d0, most “strong” EZH2 sites—defined as sites with significant EZH2 ChIP-seq coverage (P < 10−5, according to a permutation-based random model)—were often found within canonical bivalent domains (Fig. 2A), frequently coinciding with CpG islands (striped), as well as a few that were solely H3K27me3-marked. This was true for both ChrX and Chr13 and consistent with previous reports (Bernstein et al. 2006; Mikkelsen et al. 2007; Ku et al. 2008).
Figure 2.
Many strong EZH2 sites are defined bivalent domains. (A) Characteristics of constant, acquired, and lost strong EZH2 sites on d0 and d7. The portion of sites overlapping CpG islands is indicated by stripes. (B) Changes in peak widths and densities (coverage per bp) for EZH2 and H3K27me3 between d0 and d7. Medians (turquoise, Chr13; orange, ChrX), 25–75 percentile (box) and 10–90 percentile (error bars) are shown. (C) Allelic skewing of strong EZH2 sites. Numbers of sites skewing to Cast (blue) or 129 (red) are shown. Those with significant skewing (P < 0.05, norm. approx. of binomial) are shown in darker blue and red. (Gray bars) Nonpolymorphic sites; (n/d), not determined. (D) Summed coverage of EZH2 and H3K27me3 at strong and moderate sites as a percentage of total chromosomal coverage on d0 and d7 (left) and as fold-change (d7/d0, right).
However, during cell differentiation and XCI, the chromatin composition and number of strong EZH2 sites diverged dramatically between ChrX and Chr13. On Chr13, 20 out of 79 d0 EZH2 sites were lost and few new EZH2 sites were acquired (30). Almost all d7 sites were still bivalent domains associated with CpG islands (Fig. 2A). On the other hand, ChrX lost very few sites (8/48), and a large number (>100) of new EZH2 sites was acquired on d7. In contrast to constant sites, many acquired sites on ChrX were neither bivalent nor CpG islands. Among acquired sites, about half were already marked weakly by H3K27me3 on d0 but were below the significance cutoff for EZH2; the rest were previously marked solely by H3K4me3 or unmarked by either epitope (Fig. 2A, sidebar). Interestingly, strong EZH2 sites on both Chr13 and ChrX showed changes in widths of the sites, EZH2 densities, and H3K27me3 densities (Fig. 2B), which suggests a genome-wide increase in PRC2 activity from d0 and d7. Acquired sites on ChrX, however, may be key regulators of XCI, because they appear primarily on Xi during differentiation, as shown by allelic skewing of coverage (Fig. 2C).
Therefore, ChrX and Chr13 differed in several respects during XCI. First, ChrX gained a large number of EZH2 sites. Second, EZH2 binding on ChrX was allelically skewed (to future Xi). Third, whereas acquired and constant Chr13 sites were mostly bivalent, only about half of acquired X-linked sites were bivalent before XCI. Fourth, acquired sites on ChrX experienced larger increases in EZH2 and H3K27me3 densities than on Chr13. Given that many new EZH2 sites did not conform to the established paradigm of being bivalent, we suspected that noncanonical EZH2-binding sites may be central to the spread of XCI.
Noncanonical EZH2 sites on Xi for spreading
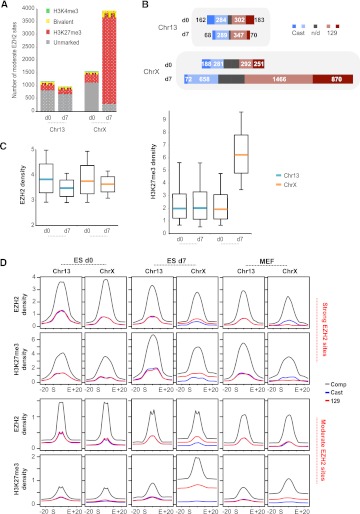
In light of de novo recruitment of noncanonical sites and the finding that many such sites were already weakly H3K27me3-positive (but below cutoff) on d0, we asked whether we might find additional functional EZH2 sites by relaxing the EZH2 enrichment threshold (Supplemental Fig. S4A). We also noted that, although chromosome-wide coverages of EZH2 and H3K27me3 were greater for ChrX than Chr13 (d7), the cumulative coverages over strong sites accounted for a small fraction of the total and were actually less for ChrX than for Chr13 (Fig. 2D).
By relaxing the cutoff to a density of >3/bp over a 1-kb window (equivalent to P < 0.03, according to the random model) and excluding strong EZH2 sites (P < 10−5, corresponding to 6-9.7 depending on sample), we observed a large number of previously undetected sites, almost all of which were neither bivalent domains nor CpG islands (Fig. 3A). On ChrX, such “moderate” or “noncanonical” sites numbered at ∼1500 on d0 and grew to ∼4000 during XCI (d7). Moderate sites greatly exceeded strong sites in number and were mostly intergenic (∼2/3) (data not shown). Allelic profiles showed that the greatest gain occurred on X129 (Xi) (Fig. 3B). Although noncanonical sites also occurred on Chr13, their number was smaller and remained constant during cell differentiation. Furthermore, Chr13 sites showed little evidence of productive H3K27 trimethylation, whereas moderate ChrX sites were almost exclusively H3K27me3-marked (Fig. 3A,C). In fact, moderate sites on ChrX represented the lion's share of EZH2 activity during XCI. EZH2 and H3K27me3 levels summed over moderate sites far exceeded those over strong sites (Fig. 2D, left panel). The greatest change in H3K27me3 coverage between d0 and d7 was observed on these noncanonical sites (Figs. 2D, 3C, right panel). Taken together, these data indicate that massive acquisition of noncanonical EZH2 sites—both strong and moderate—defines XCI, and that noncanonical EZH2 sites (rather than bivalent sites) distinguish Xi from Xa and from autosomes. Though both bivalent and noncanonical sites are recruited during XCI, the latter best reflect the XCI-specific spread of PRC2 during cell differentiation.
Figure 3.
Moderate EZH2 sites are noncanonical PRC2 targets. (A) Characteristics of moderate EZH2 sites on d0 and d7. The portion of sites overlapping CpG islands is indicated by stripes. (B) Allelic skewing of moderate EZH2 sites. Numbers of sites skewing to Cast (blue) or 129 (red) are shown. Those with significant skewing (P < 0.05, norm. approx. of binomial) are shown in darker blue and red. (Gray bars) nonpolymorphic sites; (n/d) not determined. (C) Changes in EZH2 and H3K27me3 densities between d0 and d7. Medians (turquoise, Chr13; orange, ChrX), 25–75 percentile (box) and 10–90 percentile (error bars) are shown. (D) Metasite analysis for strong and moderate sites at indicated time points. Coverages were averaged over all strong (ChrX: 56, 147, 50 sites; Chr13: 79, 81, 83 sites for d0, d7, and MEFs) and moderate sites (ChrX: 1618, 4077, 1211 sites; chr13: 1241, 1041, 758 sites for d0, d7, and MEFs) and scaled from start to end. Profiles extend ±20 kb into flanks. Densities were normalized to the average site coverage over the chromosome.
Next, we experimentally validated the bioinformatically defined strong and moderate sites by performing allele-specific ChIP-qPCR on select loci from Chr13 and ChrX in d0/d7 ES cells and in MEFs (Supplemental Figs. S5, S6). Analysis of strong (constant and acquired) sites and moderate sites, revealed clear concordance between the ChIP-seq and ChIP-qPCR results for all tested epitopes, including EZH2, H3K27me3, and H3K4me3. First, strong sites (c-13, c-X1, c-X2) showed significantly greater enrichment by ChIP-qPCR than nearby moderate sites (m-13, m-X1, m-X2) (P < 0.05, one-tailed Mann-Whitney U-test). Second, allelic differences observed by ChIP-seq were reproduced by allele-specific ChIP-qPCR. Finally, results from three biological replicates were consistent and showed significantly higher EZH2 and H3K27me3 binding at both strong and moderate sites than observed for an IgG pull-down control and the negative control escapee locus, Kdm5c (P < 0.05, one-tailed Mann-Whitney U-test) (Supplemental Fig. S6). Thus, the ChIP-qPCR results validated our general approach to allele-specific ChIP-seq analysis.
To assess local spreading at strong and moderate sites, we plotted average EZH2 and H3K27me3 densities at binding sites ±20 kb of flanking sequence (Fig. 3D), with EZH2 sites being scaled from 0 to 1 between its start (S) and end (E) (metasite profile). On d0, no allelic differences were observed on ChrX and Chr13. On d7, strong allelic skew was observed on ChrX at both strong and moderate sites, and EZH2 and H3K27me3 densities were consistently greater on X129 (Xi) than on XCast (Xa), as expected. Interestingly, a flipped allelic skew was observed for strong sites in MEFs, occurring only in EZH2 levels and not in H3K27me3 levels. This is consistent with the notion that maintenance of H3K27me3 levels after the establishment of XCI requires only low levels of EZH2 at strong sites; on Xa, in contrast, higher levels of EZH2 may be continuously required for dynamic responses in X-linked gene expression. Most intriguingly, however, changes in EZH2 and H3K27me3 densities on d7 were greater at moderate than at strong X-linked sites and occurred not only within the sites but also across >20 kb of flanking sequence bidirectionally (Fig. 3D), supporting the idea that noncanonical moderate sites are central to the spreading of XCI.
Relationship of EZH2 sites to repetitive elements
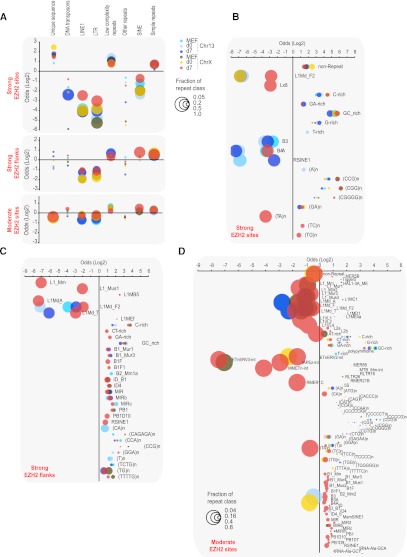
We investigated whether the PRC2 sites share underlying sequence motifs. Given the hypothesized roles for repetitive elements in spreading (Bailey et al. 2000; Lyon 2000; Wang et al. 2006; Chow et al. 2010), we examined correlations with repeat classes (Fig. 4). Our paired-end sequencing approach enabled unique alignments of a large number of repeat-containing reads by virtue of being paired with a nonrepetitive read. Strong sites were generally enriched in low-complexity repeats (d0, d7) (Z > 2.5, permutation-based random model) and simple repeats (d7), but most simple repeats were GC-rich motifs typically found at CpG islands and bivalent domains (Fig. 4A,B; Supplemental Tables S2, S3; Ku et al. 2008). Short interspersed nuclear elements (SINEs), long interspersed nuclear elements (LINEs), and long-terminal repeats (LTRs) were significantly underrepresented. (TC)n and (TG)n repeats were overrepresented specifically on ChrX, but <20% of strong EZH2 sites contained such repeats. Because flanking regions may play a role in spreading, we examined 3-kb flanks to either side of a strong site. Again, low-complexity and simple repeats were overrepresented, and LINE1s and LTRs were underrepresented on both ChrX and Chr13 (Fig. 4A,C; Supplemental Tables S2, S3). However, SINEs as a class were 1.7-fold enriched on ChrX in flanking regions, with enrichment occurring in >80% of flanking regions. This enrichment correlated with progression through XCI from d0 to d7. Moderate EZH2 sites similarly showed underrepresentation of LINE1s and LTRs, and modest overrepresentation of SINEs, low-complexity, and simple repeats (Fig. 4A,D; Supplemental Tables S2, S3).
Figure 4.
Relationship of repetitive elements to EZH2 sites. (A) Repeat classes enriched or depleted in strong sites, ±3 kb flanking strong sites, and moderate sites are plotted by their level of enrichment (positive log2 odds ratio) or depletion (negative). Different time points and chromosomes, as marked. Bubble sizes indicate the fraction of sites (scale: 0.05, 0.2, 0.5, 1.0) containing a given repeat class (if enriched) or lacking it (if depleted). For example, LINE1 sequences are depleted across the board in most sites, hence their bubbles are large. Only statistically significant (Z > 2.5) enrichment or depletion is shown (bubble sizes smaller than 0.05 indicate insignificance). The full data are listed in Supplemental Table S2 for further reference. (B–D) Log2 odds ratios for significant (Z > 2.5) enrichment and depletion of specific repeat types at strong sites (B), flanking (±3 kb) strong sites (C), and moderate sites (D). The full data are listed in Supplemental Table S3 for further reference.
We performed similar analyses with known transcription factor consensus motifs. Several motifs were overrepresented in EZH2 sites, but none were significantly enriched over a random model that took into account CpG content and proximity to gene promoters (data not shown). De novo motif discovery using MEME (Bailey et al. 2006) retrieved only simple and low-complexity motifs, consistent with our repeat analysis. Taken together, while these data do not exclude a role for LINEs in other aspects of XCI (Chow et al. 2010; Namekawa et al. 2010), they do not support a direct role of LINEs in spreading EZH2. Positive association is instead seen for SINEs, low-complexity, and simple repeats.
A hierarchy of EZH2 sites
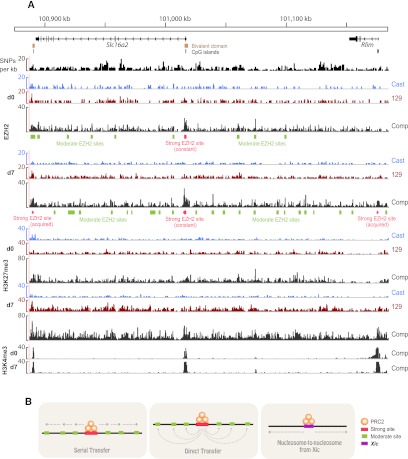
What is the relationship between strong and moderate sites and how does spreading occur between them? Exemplified by the Slc16a2-Rlim (formerly Rnf12) region (Fig. 5A) and the Mamld1 locus (Supplemental Fig. S7), bivalent domains show high EZH2 coverage on d0. During XCI, new strong sites were acquired (nonbivalent for Mamld1 in Supplemental Fig. S7, two bivalent ones in Fig. 5), and multiple closely spaced moderate sites appeared near preexisting strong sites, concurrently with increased density of H3K27me3. With one strong site for every 20–40 moderate sites, we hypothesize that strong sites might serve as recruiting “hubs” from which EZH2 is passed onto adjacent moderate sites. Three models of spreading are possible (Fig. 5B). EZH2 may be relayed from a strong site to multiple adjacent moderate sites through “serial transfer.” Alternatively, a strong site could relay EZH2 to moderate sites via “direct transfer” through long-range 3D interactions. Finally, moderate and strong sites may not be functionally linked, and spreading could instead occur nucleosome-by-nucleosome from the Xic.
Figure 5.
Developmental profile of the Slc16a2-Rlim region. (A) Composite (comp) and allelic (Cast, 129) ES d0 and d7 profiles, showing strong and moderate EZH2 sites across the Slc16a2-Rlim region. SNP and Indel density per 1-kb window, CpG islands and bivalent domains are shown above the epitope tracks. (B) Three potential models for spreading of EZH2.
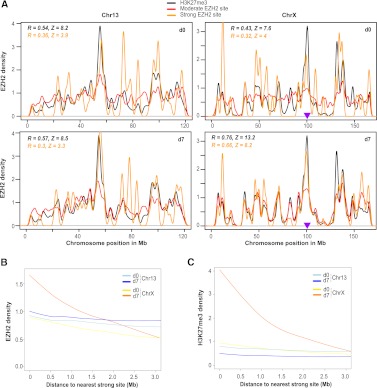
To gain insight, we calculated densities of strong and moderate EZH2 sites and H3K27me3 over the lengths of Chr13 and ChrX (Fig. 6A) and analyzed positional correlations and significance (Pearson R and Z) between moderate EZH2 sites and strong sites, as well as moderate sites and H3K27me3. On d0, these correlations had only borderline significance on both Chr13 and ChrX. On d7, correlations were unchanged for Chr13 but increased greatly on ChrX (note overlapping red, orange, and black lines from d0 to d7). The correlation of H3K27me3 with moderate sites was especially strong (black, R = 0.76, Z = 13.2) and was highly significant even at small bin sizes (0.5 Mb). These data contain two separable observations regarding possible modes of XCI spreading: First, the lack of height order for distinct peaks (i.e., largest to smallest from Xic to telomere) in EZH2 and H3K27me3 densities over ChrX on d7 argues against nucleosome-by-nucleosome spreading of XCI, which might be expected to manifest as a continuous chromosomal gradient descending from the Xic. Second, the XCI-specific increase in positional correlation illustrates that moderate sites cluster near strong sites on ChrX on d7, an event highly unlikely to be due to chance (Z = 8.2). Thus, we suggest a functional link between strong and moderate EZH2 sites, perhaps with strong sites seeding moderate sites in their vicinity to spread H3K27me3.
Figure 6.
Spread of H3K27me3 from EZH2 sites occurs in a distance-dependent manner. (A) EZH2 and H3K27me3 densities from d0 and d7 were binned over Chr13 and ChrX positions to obtain correlation coefficients and Z scores from a permutation-based random model. Pearson R coefficients are indicated for the correlation of strong/moderate EZH2 sites (orange values) and H3K27me3/moderate EZH2 sites (black values). A purple triangle marks the location of the Xic. (B,C) Smoothed trend lines of EZH2 (B) or H3K27me3 (C) densities at a given distance from the nearest strong site, on indicated days and chromosomes. Densities are plotted over a sliding 1-kb window with steps of 200 bp.
Both serial and direct transfer models (Fig. 5B) might predict a sensitivity of spreading to physical distance. To ask how EZH2 and H3K27me3 densities of a sliding window change as a function of distance from the nearest strong EZH2 site, we plotted trend lines of EZH2 and H3K27me3 densities over megabase-scale distances (Fig. 6B,C). On Chr13, EZH2 and H3K27me3 densities did not correlate with distance from the nearest strong site on either d0 or d7. The d0 ChrX profile looked nearly identical to those of Chr13. On d7, however, EZH2 and H3K27me3 densities increased dramatically around strong sites (near 0) (Fig. 6B,C), consistent with metagene and metasite profiles (Figs. 1, 3). Intriguingly, these densities decayed with distance from the nearest strong site. EZH2 densities dropped threefold down to baseline by 3 Mb, and H3K27me3 densities dropped fourfold over the same distance. These observations revealed a gradient of EZH2 binding and catalytic activity around strong sites, arguing for distance-dependent effects at the megabase-scale specific to ChrX during XCI.
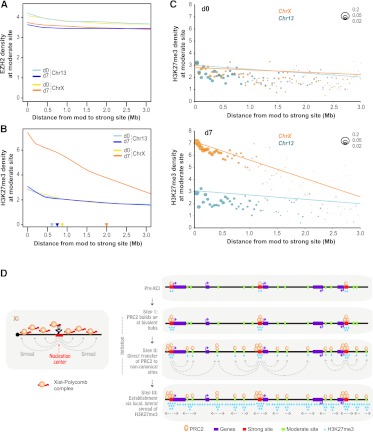
Are EZH2 and H3K27me3 densities of a given moderate site also sensitive to physical distance between strong and moderate sites? On Chr13, similar to trend lines for overall coverage densities (Fig. 6B,C), EZH2 and H3K27me3 densities at moderate sites did not correlate with distance to the nearest strong site on either d0 or d7 (Fig. 7A,B). This was also the case for ChrX on d0. Analysis of ChrX on d7 revealed that the H3K27me3 trend line for moderate sites (Fig. 7B) displayed a similar decay (Fig. 6C) in H3K27me3 density with increasing distance to the nearest strong site. Importantly, the EZH2 trend line showed no significant distance-dependent effect at all over 0–3 Mb (Fig. 7A,B).
Figure 7.
A hierarchy of EZH2 sites, with limited EZH2 translocation and long-range H3K27me3 gradient. (A) Smoothed trend lines of EZH2 densities at a moderate (mod) site at given distances (x-axis) from the nearest strong site, on indicated days and chromosomes. (B) Smoothed trend lines of H3K27me3 densities at a moderate (mod) site at given distances (x-axis) from the nearest strong site, on indicated days and chromosomes. Color-coded triangles denote distance-at-half-maximum for each chromosomal time point. (C) Depiction of the number of moderate (mod) sites at given distances from the nearest strong site on d0 and d7 for Chr13 and ChrX. Scale: Bubble sizes represent fractions of 0.2, 0.05, and 0.02 of all mod sites. Each graphed bubble represents the median of sites in 40-kb bins. Pearson correlation coefficients (R) and Z scores (Fisher transformed) are shown. (D) Model and summary.
Moderate sites tend to be located close (<1 Mb) to strong sites (Figs. 6A, 7C). This trend was especially pronounced on ChrX on d7 (Fig. 7C), with the largest fraction of all sites clustered within a distance of 0.5 Mb from the closest strong site, suggesting acquisition of moderate sites during XCI in the vicinity of strong sites. Importantly, H3K27me3 densities of moderate sites decay with increasing distance to the closest strong site only on ChrX on d7, as seen for both trend lines fitted to raw data (Fig. 7A,B) as well as medians of densities binned over set distances (40-kb bins) (Fig. 7C). For a numerical characteristic of this long-range effect, we conservatively estimated the radius of the “sphere of influence” of strong EZH2 sites on neighboring moderate sites by calculating the distance-at-half-maximum for the density trend lines. For Chr13 on both days and ChrX on d0, the range was small (0.6–0.8 Mb) (Fig. 7B, triangles). In contrast, the range for ChrX on d7 was threefold greater (2.0 Mb). These data show that, during XCI, there is significant growth of moderate sites around strong sites and that, furthermore, there is a substantial increase in the sphere of influence of a given strong site.
The general EZH2 and H3K27me3 gradient (Fig. 6B,C) likely resulted from clustering of moderate sites around strong sites (Fig. 7C). Importantly, the H3K27me3 density of each moderate site decreased with increasing distance from the closest strong site, specifically on ChrX on d7 (Fig. 7B,C). The lack of a similar gradient for EZH2 (Fig. 7A) suggests that EZH2 is not relayed in serial fashion from strong site to proximal moderate site and on to distal moderate site, because EZH2 densities of moderate sites would decay with increasing distance. Instead, the data are most consistent with the notion that strong sites are directly seeding moderate sites nearby, transferring discrete quantities of EZH2 directly from strong to moderate sites, rather than spreading EZH2 linearly in a nucleosome-to-nucleosome fashion. Therefore, of the three potential models in Figure 5B, we favor a direct transfer model in which XCI spreading occurs in a three-step process (Fig. 7D).
Discussion
Here, we have used genetically marked hybrid cell lines with high SNP density to produce chromatin profiles of Xa and Xi at high resolution. The allele-specific ChIP-seq approach has enabled us to define Polycomb binding sites on Xi and follow the spread of PRC2 and H3K27me3 during XCI. From these data, several major conclusions can be drawn regarding the pattern of Polycomb-mediated silencing on Xi. First, XCI spreading is governed by a hierarchy of two types of PRC2 sites. “Canonical” sites typically contain CpG islands, associate with bivalent genes, and have high EZH2 density (∼100/150 of strong EZH2 sites match this description). “Noncanonical” sites are typically intergenic and lack H3K4me3 or CpG islands. Noncanonical sites greatly outnumber canonical sites by 40:1. Our analysis did not uncover consensus motifs for EZH2 binding, although the modest enrichment of SINEs and simple repeats merits future study. It appears unlikely that LINEs play a direct role in EZH2 recruitment, but they may play roles in other steps of XCI (Chow et al. 2010; Namekawa et al. 2010).
We propose a three-step model for XCI spreading (Fig. 7D). PRC2 is initially recruited by Xist RNA to the previously identified “nucleation center” within Xist exon 1 (Fig. 7D, left panel; Jeon and Lee 2011). From the nucleation center, PRC2 spreads to ∼150 strong sites (Fig. 7D, right panel). Notably, even in the pre-XCI state (d0), many bivalent domains (∼50) already demonstrate strong PRC2 binding (P < 10−5), but H3K27me3 does not spread beyond the domain. Thus, PRC2 is poised at canonical hubs, but its catalytic activity spreads only after the initiation of XCI when Xist RNA spreads over ChrX. Because Xist is not expressed on d0, initial recruitment to bivalent sites is likely an Xist-independent phenomenon, akin to PRC2 recruitment to any autosomal locus. Cell differentiation and formation of an Xist cloud over the future Xi expands PRC2's sphere of influence, by both adding coverage to existing bivalent sites and by de novo recruitment of strong sites along the X in cis.
From these strong sites, PRC2 spreads locally via thousands of noncanonical sites recruited en masse specifically to the Xi (Fig. 7D, right panel). Significantly, between d0 and d7, noncanonical sites attract the bulk of total EZH2 activity on ChrX. The rise of noncanonical sites around a strong site (Figs. 6A, 7C) seems unlikely to be due to chance and suggests a functional link between their recruitment and spreading of XCI on a local scale. We considered two relay modes. One mode involves serial transfer from strong site to adjacent moderate sites (Fig. 5B, left panel); proximal moderate sites would then spread PRC2 onward to increasingly distal moderate sites. An alternative mode involves direct transfer from strong site to several moderate sites in the general vicinity (within ∼1 Mb) (Fig. 5B, middle panel). The former seems less likely, because EZH2 densities do not differ on average between a moderate site and its preceding neighbor; indeed, similar EZH2 densities were observed along a potential serial “chain” (Fig. 7A). Although EZH2 densities did not change significantly, H3K27me3 showed a distance-dependent decrease (Fig. 7B), suggesting that EZH2 activity decreased away from the closest moderate site, perhaps reflecting the passage of time since transfer (longer for proximal sites that were seeded earlier than distal sites). These observations lead us to favor a direct transfer model, though we stress that our current data cannot rule out a serial transfer model or other models. Interestingly, the rise of moderate sites coincides with accumulation of Xist RNA along the future Xi, consistent with the idea that recruitment of moderate sites is Xist RNA-dependent. Future work will be directed at integrating the Polycomb sites with recent results suggesting that Xist may prevent Xi from snapping back into the ordered three-dimensional conformation of Xa (Splinter et al. 2011). We consider this report to be relevant to our results, because disruption of an ordered chromosome conformation by Xist may expose recipient loci to stochastic and transient collisions with strong PRC2 hubs, resulting in direct transfer of EZH2.
In summary, we have identified a hierarchy of PRC2 spreading elements on ChrX and provided a first hint of how Polycomb silencing may be propagated from the Xic. An open question is how Xi spreading elements relate to Polycomb response elements (PRE), which remain poorly defined in mammals (Simon and Kingston 2009). Also unknown is how bivalent and noncanonical sites are selected as spreading elements. The new understanding provided herein and in recent works will serve as a foundation for future studies.
Methods
Cell culture and ES cell differentiation
For MEFs, female F1 (129S1 × CAST/EiJ) embryos were harvested on E13.5, and fibroblasts were outgrown, immortalized with SV40 large T antigen, and cloned, and those carrying an inactive X129 were used. Female 16.7 ES cells (Lee et al. 1999) and TsixTST/+ (Ogawa et al. 2008) have been described. Differentiating ES cells were grown without LIF for 4 d, then plated for outgrowth until d7.
ChIP-seq
ChIP samples were prepared and immunoprecipitated as described (Lee et al. 2006), using antibodies against H3K4me3 (Abcam, ab8580), H3K36me3 (Abcam, ab9050), H3K27me3 (Abcam, ab6002), EZH2 (Active Motif, 39639), and RNA polymerase II phosphorylated on Serine 5 of the C-terminal domain (Abcam, ab5131). ChIP DNA concentration was measured using the Quant-iT Picogreen dsDNA Assay kit (Invitrogen). DNA for Illumina sequencing was prepared according to Illumina instructions with minor modifications (ultrapure T4 DNA ligase [Enzymatics] for ligation, room-temperature purification of gel slices using QIAquick [Qiagen] spin columns, and amplification using Phusion [NEB] GC buffer). Paired-end sequencing was carried out for 2 × 36 cycles on a Genome Analyzer II (Illumina).
Allele-specific analysis
Mouse genome sequencing data (129S1/SvImJ and CAST/EiJ) from the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/resources/mouse/genomes/) aligned to the C57BL/6J reference genome (NCBI mm9) was screened for high quality single nucleotide polymorphisms (SNPs) and insertions/deletions indels. Variant genomes were constructed from mm9 using a total of 17,879,569 SNPs, 726,387 indels for CAST/EiJ, and 4,551,690 SNPs, 224,271 indels for 129S1/SvImJ. In total, the resulting CAST/EiJ and 129S1/SvImJ genomes differ in 22,095,665 SNPs and 948,567 indels. Read pairs were aligned to both genomes allowing for up to ∼5 mismatches at high-quality bases or 2–4 small gaps using novoalign (www.novocraft.com). Each uniquely aligned pair to the CAST/EiJ genome was compared to the corresponding pair aligned to the 129S1/SvImJ genome. Pairs that differed significantly in alignment score due to mismatches/gaps were classified as allele-specific and the better alignment retained. Pairs with identical alignment scores or scores that differed only slightly due to fragment length penalties were classified as neutral. Each experiment yielded three tracks: Cast, 129, and composite (neutral, 129, Cast combined).
Generation of coverage maps and enrichment segments
Alignment coordinates were mapped to mm9 to permit comparisons to mm9 annotations. To calculate coverage, unique fragments defined by paired reads were included, discarding duplicate fragments. Coverage was normalized by input. Chromosomal segments that are likely to be enriched for a chromatin epitope were defined by analyzing significantly enriched overlapping 1-kb windows. The significance of the coverage enrichment in a window was determined based on the null model of paired-end fragments randomly shuffled across the chromosome (see Supplemental Material for details).
Metagene and metasite profiles
The profiles of average coverage density over genes (metagene profiles) and over segments (metasite profiles) were constructed using normalization of profile densities so that the area under the curve was proportional to the average gene/segment coverage of this chromosome (see Supplemental Material for details).
Estimates of density map correlations
Correlations between chromosomal density maps were calculated as Pearson correlation coefficients of coverages in nonoverlapping windows, with estimates of statistical significance based on a random permutation null model (see Supplemental Material for details).
Estimates of allelic skew
Allelic skew of coverage at a given segment was analyzed by comparing allele-specific coverages at this segment. The significance of skew was estimated based on the normal distribution of effective numbers of allele-specific fragments (see Supplemental Material for details).
Repeat enrichment analysis
The significance of repeat enrichment or depletion was estimated against the distribution of RepeatMasker repeat numbers in the segments shuffled along the chromosome (see Supplemental Material for details).
Analysis of coverage in long-range vicinity of EZH2 sites
The analysis of coverage trends in the megabase-scale vicinity of strong EZH2 sites was based on the coverage of 1-kb windows with 200-bp shifts over the entire chromosome. For each window, the closest strong EZH2 segment was determined, and the window coverage was plotted against the distance to the closest segment. The trends of moderate site coverage around the strong sites were analyzed in a similar fashion.
Data access
ChIP-seq data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE36905. In addition, full lists of EZH2 sites on ChrX and Chr13 are presented in Supplemental Tables S4 and S5.
Acknowledgments
We thank members of our laboratory and the Bernstein laboratory for many valuable discussions. We also thank B.A. Chapman for bioinformatics suggestions. This work was supported by the German Research Foundation DFG (S.F.P.), the MGH ECOR Fund (E.Y., Y.J.), a Korean Research Foundation grant (Y.J.), and the NIH (RO1-GM090278, J.T.L.). J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.133751.111.
References
- Bailey JA, Carrel L, Chakravarti A, Eichler EE 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc Natl Acad Sci 97: 6634–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW 2006. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Disteche CM 2010. Escape from X inactivation in mice and humans. Genome Biol 11: 213 doi: 10.1186/gb-2010-11-6-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF 1992. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- Cantrell MA, Carstens BC, Wichman HA 2009. X chromosome inactivation and Xist evolution in a rodent lacking LINE-1 activity. PLoS ONE 4: e6252 doi: 10.1371/journal.pone.000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404 [DOI] [PubMed] [Google Scholar]
- Chow JC, Yen Z, Ziesche SM, Brown CJ 2005. Silencing of the mammalian X chromosome. Annu Rev Genomics Hum Genet 6: 69–92 [DOI] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141: 956–969 [DOI] [PubMed] [Google Scholar]
- Chureau C, Chantalat S, Romito A, Galvani A, Duret L, Avner P, Rougeulle C 2011. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet 20: 705–718 [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB 1996. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler SM, Riggs AD 1983. Mammalian X-chromosome inactivation. Annu Rev Genet 17: 155–190 [DOI] [PubMed] [Google Scholar]
- Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, et al. 1998. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 7: 737–742 [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Nakagawa S 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19: 469–476 [DOI] [PubMed] [Google Scholar]
- Helbig R, Fackelmayer FO 2003. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma 112: 173–182 [DOI] [PubMed] [Google Scholar]
- Jeon Y, Lee JT 2011. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146: 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CM, Newall AE, Brockdorff N, Nesterova TB 2002. Enox, a novel gene that maps 10 kb upstream of Xist and partially escapes X inactivation. Genomics 80: 236–244 [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol 12: 815–826 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF 1999. Imprinting and X-chromosome inactivation. Results Probl Cell Differ 25: 73–90 [DOI] [PubMed] [Google Scholar]
- Lyon MF 2000. LINE-1 elements and X chromosome inactivation: A function for “junk” DNA? Proc Natl Acad Sci 97: 6248–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG 2009. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res 19: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT 2010. Two-step imprinted X inactivation: Repeat versus genic silencing in the mouse. Mol Cell Biol 30: 3187–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT 2008. Intersection of the RNA interference and X-inactivation pathways. Science 320: 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Pullirsch D, Hartel R, Kishimoto H, Leeb M, Steiner G, Wutz A 2010. The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development 137: 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius B, Shi C, Hengshuo L, Sandhu KS, Radomska KJ, Rosen GD, Lu L, Kullander K, Williams RW, Jazin E 2010. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics 11: 614 doi: 10.1186/1471-2164-11-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, et al. 2011. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25: 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Willard HF, Mukherjee S, Furey TS 2006. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol 2: e113 doi: 10.1371/journal.pcbi.0020113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Gribnau J 2007. X inactivation Xplained. Curr Opin Genet Dev 17: 387–393 [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20: 614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]