Abstract
Origins of replication present a paradox to evolutionary biologists. As a collection, they are absolutely essential genomic features, but individually are highly redundant and nonessential. It is therefore difficult to predict to what extent and in what regard origins are conserved over evolutionary time. Here, through a comparative genomic analysis of replication origins and chromosomal replication patterns in the budding yeasts Saccharomyces cerevisiae and Lachancea waltii, we assess to what extent replication origins survived genomic change produced from 150 million years of evolution. We find that L. waltii origins exhibit a core consensus sequence and nucleosome occupancy pattern highly similar to those of S. cerevisiae origins. We further observe that the overall progression of chromosomal replication is similar between L. waltii and S. cerevisiae. Nevertheless, few origins show evidence of being conserved in location between the two species. Among the conserved origins are those surrounding centromeres and adjacent to histone genes, suggesting that proximity to an origin may be important for their regulation. We conclude that, over evolutionary time, origins maintain sequence, structure, and regulation, but are continually being created and destroyed, with the result that their locations are generally not conserved.
Successful cell division requires accurate and complete DNA replication. The essentiality of DNA replication is reflected in the extraordinary conservation of the replication machinery across eukaryotes (Sclafani and Holzen 2007), the multilayered regulation of DNA replication ensuring that the entire genome is replicated only once during S phase (Blow and Dutta 2005; Truong and Wu 2011), and the numerous DNA repair and checkpoint pathways that engage at any flag of error (Sclafani and Holzen 2007). However, origins of replication—the sites where replication is initiated—vary across eukaryotes (Sclafani and Holzen 2007; Masai et al. 2010), are not all used in every cell cycle, and can be removed individually, as well as in large groups, without loss of cell viability (Dershowitz et al. 2007). This contrasting duality—collectively being essential for life, while individually free to diverge—raises the question of how, or if, origins of replication are conserved over evolutionary time.
Eukaryotic replication origins have been studied most thoroughly in the yeast Saccharomyces cerevisiae. The S. cerevisiae genome has 300–400 origins, each 100–500 bp in length, containing a functionally essential but not sufficient AT-rich consensus sequence, and residing in an intergenic space (Marahrens and Stillman 1992; Feng et al. 2006; Nieduszynski et al. 2007). Limited data from other yeast species have painted a partial and somewhat confusing view of how origins evolve. Origins in the sensu stricto Saccharomyces species (2–20 million years divergence from S. cerevisiae) are predicted to be nearly identical in sequence and location with S. cerevisiae origins (Yang et al. 1999; Nieduszynski et al. 2006). At 100–200 million years' divergence from S. cerevisiae, Kluyveromyces lactis also contains intergenic origins 100–500 bp in length, but its origins have a markedly different consensus sequence (Liachko et al. 2010). Furthermore, K. lactis origins are not conserved in location with origins in S. cerevisiae. Finally, origins in Schizosaccharomyces pombe (500 million to 1 billion years diverged from S. cerevisiae) are 500-bp to 2-kb AT-rich stretches of DNA lacking any consensus sequence (Segurado et al. 2003). It can roughly be inferred, then, that origins are only conserved in sequence and location over very short evolutionary distances but are conserved in AT content over great evolutionary distances.
The evolutionary history of S. cerevisiae is, however, marked by a whole genome duplication event (WGD) (Wolfe and Shields 1997; Kellis et al. 2004). Following the WGD, the gene content and genome size were reduced to nearly the original non-WGD genome size by deletion of duplicate genes and their intergenic regions (Kellis et al. 2004). As well, genome rearrangements occurred, generating novel chromosomes, mosaics of their non-WGD ancestors.
Given the turnover of intergenic regions and the nonessentiality of individual origins, it is not surprising that origins are not conserved in location between S. cerevisiae and K. lactis. It is, however, surprising that S. cerevisiae and K. lactis origins would contain different consensus sequences and could suggest that the WGD event caused a massive alteration in origin identity.
Here we aimed to better understand the evolution of origins by analyzing the replication origins in a yeast species more closely related to S. cerevisiae yet naïve to the WGD. To this end, we characterized origins and replication progression in Lachancea waltii (∼150 million years diverged from S. cerevisiae; Berbee and Taylor 2001) and performed a detailed comparative analysis with S. cerevisiae. Our analyses described here reveal that L. waltii replication origins are very similar to those found in S. cerevisiae at the levels of sequence, structure, and regulation. In position, however, few L. waltii origins show evidence of being conserved with S. cerevisiae, though the number of conserved origin locations is greater than would be expected by chance. From these results, we argue, first, that origins have been strongly conserved in sequence, structure, and regulation by the replication machinery. Second, we argue that origins may have been maintained in location only when they affected the expression or genomic stability of surrounding genes. Ultimately, our work implies that origin activity readily relocates throughout the genome to accommodate genomic change, necessitating the hypothesis that the genome is littered with sequences capable of mutating into functional origins.
Results
Identification of L. waltii sequences promoting plasmid replication
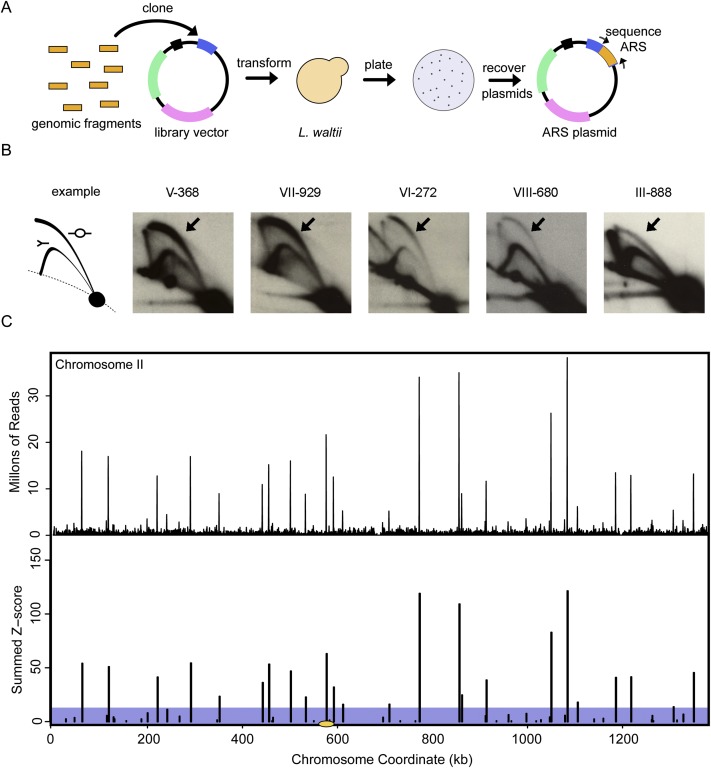
To find sequences that promote replication, we employed the classic autonomously replicating sequence (ARS) assay (Stinchcomb et al. 1979). We constructed a 25× genomic library for L. waltii, comprised of genomic inserts ranging in size from 350 to 1000 bp. The coverage of this library was confirmed by Illumina sequencing (see Supplemental Text). We transformed this library into L. waltii and scraped ∼46,770 colonies from plates that were selective for the genomic library plasmid (Fig. 1A). Plasmids were batch-purified from the pooled colonies, and the sequences of the inserts on the plasmids were determined by Sanger and Illumina sequencing. Two-dimensional (2D) gel electrophoresis analysis of genomic replication intermediates revealed bubble arcs at the chromosomal locations corresponding to candidate ARSs (Fig. 1B), confirming that our ARS assay was successful in identifying L. waltii origins.
Figure 1.
The ARS assay. (A) Sheared genomic DNA was cloned into a plasmid that contained a centromere but lacked a yeast origin of replication. The markers on the plasmid, indicated by boxes, in clockwise order and starting at the 1:00 position are LacZ (blue) multiple cloning site (contains SmaI, KpnI, and SacI sites), AmpR (pink), KanMXR (green), L. waltii CEN7 (black). Plasmids with genomic inserts were transformed into L. waltii and plated on G418. Colonies growing on G418 were presumed to have ARS elements in their inserts. These colonies were scraped and plasmids were extracted. Primers flanking the LacZ cloning site were used to identify the genomic insert (the ARS). (B) ARSs sequenced by Sanger sequencing were confirmed by genomic 2D gel analysis. The presence of a bubble arc (arrow) indicates that the sequence acts as a chromosomal origin. (C) All candidate ARSs were identified using Illumina sequencing. The top panel shows the raw sequencing data binned in 500-bp bins, shifting every 100 bp. The bottom panel shows the data after normalization against the genomic input library, removing all bins in the lower 97.5% of the data, summing adjacent remaining bins, and converting sequence read counts to Z-scores. Those remaining peaks with a summed Z-score of 12 or greater (above the shaded box) were scored as ARSs. The data for chromosome II are plotted with the centromere illustrated by a yellow ellipse. Plots for all chromosomes are shown in Supplemental Figure S1.
After filtering the ARS Illumina data and extracting the sequences corresponding to ARSs (see Methods and Supplemental Text), we identified 182 ARS candidates (Fig. 1C; Supplemental Fig. S1; Supplemental Data set S1). Additional ARS assays found three of these ARSs to be false positives and uncovered one false negative. Sanger sequencing identified 36 ARSs. All but three of these ARSs were represented in our Illumina data. Two of the three ARSs not recovered are located in nonunique regions of the genome (rDNA and mating locus; see the Supplemental Text for a discussion of these ARSs) and one is a weak ARS (LwARSVIII-680) that only produces transformant colonies on plates after an additional day of growth on selective medium. We finalized our ARS list with 183 ARS sequences.
Characterization of chromosome replication dynamics and origin usage in L. waltii
While the ARS assay identified sequences that promote plasmid replication in L. waltii, it does not distinguish which sequences function robustly in L. waltii chromosomes nor the time during S phase that replication initiates from these sequences. Therefore, we next performed two genome-wide assays aimed at characterizing replication dynamics and origin usage in L. waltii.
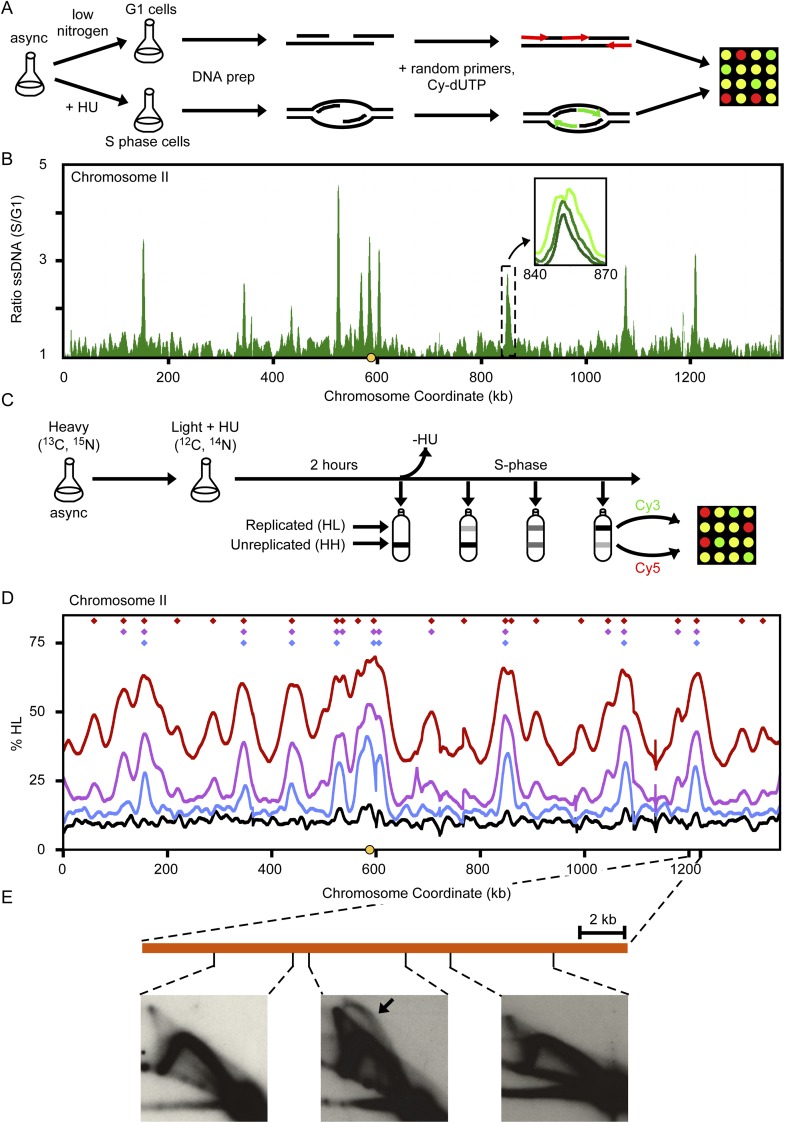
We first identified chromosomal origins that fire early in S phase. In wild-type S. cerevisiae and Schizosaccharomyces pombe cells, single-stranded DNA (ssDNA) accumulates at early-firing replication origins when cells enter S phase in the presence of hydroxyurea (HU) (Feng et al. 2006). Hence, by mapping the sites of ssDNA formation in L. waltii in the presence of HU, we can determine which ARSs are early-firing origins, as well as examine their organization across the genome.
We incubated a logarithmically growing culture of L. waltii cells in HU and harvested timed samples (Fig. 2A). Starting at 120 min after addition of HU, we see significant peaks of ssDNA at discrete locations scattered across the genome (Fig. 2B; Supplemental Fig. S2). Over time, these peaks gradually spread into neighboring regions, consistent with the slow movement of replication forks away from the origins of replication (Fig. 2B, inset). We identified 93 statistically significant peaks (see Methods) that we designate as early-firing origins of replication (Supplemental Data set S2). For discussion here, we term these origins HU-positive (HU-pos) ARSs.
Figure 2.
The ssDNA and density transfer assays. (A) Outline of ssDNA-based mapping of early-firing origins. L. waltii cells were treated with HU (200 mM) to enrich for ssDNA around origins of replication, or with low nitrogen medium to maintain cells in G1. ssDNA regions were labeled by random primed labeling without template denaturation and hybridized to a microarray. (B) The ratio of ssDNA in S/G1 is plotted for chromosome II. Early-firing origins are revealed as peaks in the plot. (Inset) Broadening in ssDNA peaks as S phase progresses. Plots for all chromosomes are shown in Supplemental Figure S2. (C) Outline of density transfer experiment to monitor replication dynamics. L. waltii cells were pregrown in a heavy isotope medium and then transferred to a light isotope medium containing HU (100 mM). After 2 h, HU was removed and cells were collected over the course of the S phase. DNA isolated from these samples were fragmented and subjected to ultracentrifugation to separate the heavy-heavy (HH), unreplicated DNA from the heavy-light (HL), replicated DNA. The HH and HL DNAs for each sample were labeled and competitively hybridized on a microarray. (D) Replication of chromosome II as revealed by the density transfer. The different colored lines correspond to samples taken at different times in the S phase: black (arrest), blue (15% HL), purple (25% HL), red (45% HL). The centromere is shown by a yellow circle on the x-axis. Color-coded diamonds above the plots indicate locations and samples in which origin activity (peaks of HL DNA) was detected. Plots for all chromosomes are shown in Supplemental Figure S3. (E) 2D gel analysis across a representative HL DNA peak confirms that the peak contains an origin.
To view replication dynamics of the entire chromosome, we used the density-transfer to array technique (Raghuraman et al. 2001; Alvino et al. 2007; McCune et al. 2008). HU was used to promote the accumulation of cells in early S phase. We collected timed samples throughout S phase for flow cytometry, slot blotting, and microarray (Fig. 2C). The plot of % HL DNA (replicated molecules) across chromosome II from cells held in HU and for three timed samples collected after the removal of HU are shown in Figure 2D. In HU, no significant peaks of replicated DNA are evident on chromosome II. Following removal of HU, DNA of hybrid density (HL) appeared initially at just a handful of sites—the earliest replicating regions of the genome that were identified by the ssDNA assay (cf. Fig. 2B and D, Supplemental Figs. S2 and S3). At later times, additional peaks of replicated DNA became prominent identifying later firing origins.
To confirm that origins of replication reside within these hybrid density peaks, we performed 2D gel analysis on 10 overlapping restriction fragments that tile across the early peak seen on the density transfer profile for chromosome II centered at position 1210 kb. We identified a single fragment containing an active chromosomal origin (representative 2D gels shown in Fig. 2E). This fragment also corresponds to the local maximum identified by the ssDNA assay and was recovered in the ARS assay. In total, the density transfer replication profiles suggest there are ∼200 chromosomally active origins in L. waltii with 174 being confidently identified following the methods of Alvino et al. (2007).
Concordance among the three L. waltii replication assays
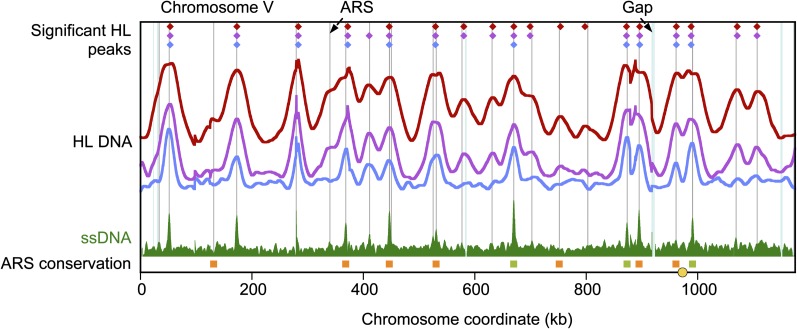
We see excellent agreement among the ARS assay, ssDNA maps, and replication profiles (Fig. 3; Supplemental Figs. S4, S5). All of the ssDNA origins are represented by peaks in the density transfer experiment, 84 of which are peaks computationally called following the methods of Alvino et al. (2007). As well, 81 of the 93 ssDNA origins are found by the ARS assay; 137 of 183 ARSs coincide with computationally identified hybrid density peaks, and thus are clearly chromosomally used. The % HL values in the 25% HL density transfer sample at ssDNA origins are significantly greater than both the genome average and the % HL at ARS locations that are not ssDNA origins (P-value < 10−15, Welch two sample t-test) (Supplemental Fig. S6). We therefore have confirmation that the locations identified in the ssDNA assay are early-firing origins. With the total sum of these three assays, we created a tabulation of 195 L. waltii replicative sequences (Supplemental Data set S2). Based on the density transfer profile, we estimate that our list is missing a maximum of 20 origin locations. For further discussion on the overlap of the assays, see the Supplemental Text. In most of the analyses that follow, we excluded the two ARSs that lie outside of the canonical, assembled L. waltii genome: the rDNA ARS and the mating locus ARS (see Supplemental Text).
Figure 3.
All L. waltii replication data for chromosome V. Profiles of % HL and HL DNA peak locations (color-coded as in Fig. 2) are shown above the ssDNA profile. (Gray vertical lines) ARS locations. (Blue vertical lines) Sites that are redundant in the genome and cannot be mapped by Illumina sequencing data. (Filled squares) L. waltii ARSs that show syntenic conservation with ARSs in one or more other species (S. cerevisiae, L. kluyveri, and K. lactis). Orange, green, or brown squares indicate conservation with one, two, or all three of these species, respectively. (No instances of conservation with all three species were seen on this chromosome.) The yellow circle at the bottom represents the centromere. Plots for all chromosomes are shown in Supplemental Figure S4.
L. waltii ARSs are small sequences in large, divergently transcribed intergenic regions
Using our list of 195 L. waltii origin sequences, we were able to define the composition of L. waltii origins/ARSs and compare them with their functional counterparts in S. cerevisiae. The L. waltii ARSs have sizes between 250 and 1000 bp, mirroring the size range of ARSs recovered from similar assays in S. cerevisiae (Nieduszynski et al. 2007). As in S. cerevisiae (Wyrick et al. 2001; Nieduszynski et al. 2006), L. waltii ARSs are located in large intergenic regions (∼1.4 kb compared with an average of 0.6 kb). One L. waltii ARS (LwARSI-1118) is located at the 24-bp overlap of two divergently transcribed genes, raising the possibility that one or both of these ORFs have a misidentified start codon. Specifically, we find that L. waltii ARSs are depleted from convergently transcribed intergenic regions (P-value < 10−5, hypergeometric test), but are enriched in divergently transcribed intergenic regions (P-value = 0.0083, hypergeometric test) (Supplemental Table S1). This intergenic region preference is slightly reversed for S. cerevisiae, where ARSs have no preference for divergent intergenic regions, are depleted from co-directional intergenic regions, and are enriched in convergent intergenic regions (MacAlpine and Bell 2005; Nieduszynski et al. 2006; Yin et al. 2009). As a further point of comparison, K. lactis ARSs have no preference for intergenic region type (Liachko et al. 2010).
L. waltii ARSs contain an essential consensus sequence that is highly similar to that of S. cerevisiae ARSs
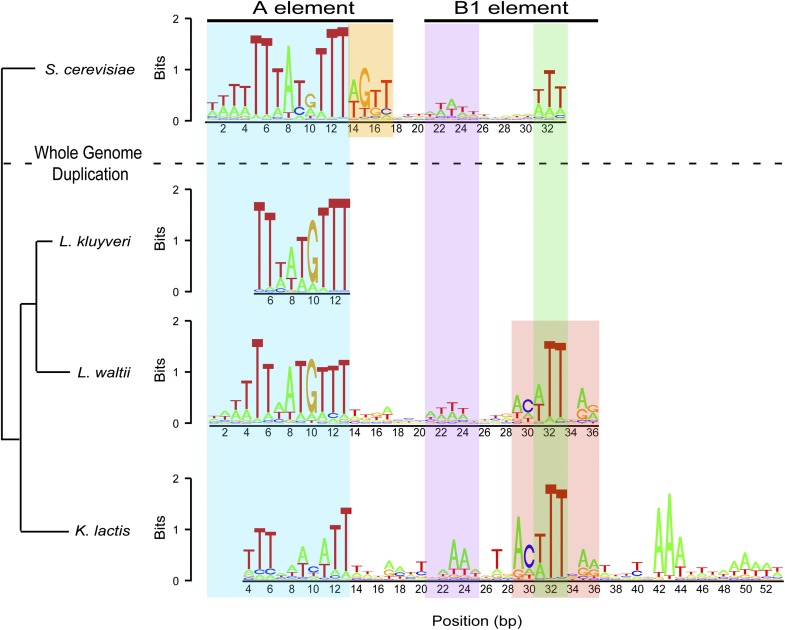
S. cerevisiae origins are characterized by an AT-rich 11- to 17-bp ARS consensus sequence (ACS) termed the A element (Theis and Newlon 1997; Nieduszynski et al. 2006), which is recognized by the origin recognition complex (ORC) making it essential for origin function. We analyzed the L. waltii ARSs for a consensus sequence and found a 13-bp consensus with remarkable similarity to the S. cerevisiae A element (Fig. 4, blue box)—an AT-rich ACS with a central ATG sequence on the T-rich strand. Unlike in S. cerevisiae, though, the 17-bp extended A element (Fig. 4, orange box) is not information rich.
Figure 4.
ACS alignment for S. cerevisiae, L. kluyveri, L. waltii, and K. lactis. The consensus sequence for L. waltii ARSs as compared with those in S. cerevisiae, L. kluyveri, and K. lactis are plotted showing the T-rich strand. (Blue box) 13-bp A element; (orange box) extended 17-bp A element. The purple, green, and red boxes show the B1 element. The S. cerevisiae ACS was taken from Eaton et al. (2010), the L. kluyveri ACS was taken from Liachko et al. (2011), and the K. lactis ACS was taken from Liachko et al. (2010). The tree phylogeny is based on Jeffroy et al. (2006).
Recently, Eaton et al. (2010) performed a more in-depth analysis of motifs within S. cerevisiae ARSs and recovered an additional motif downstream from the ACS. This motif corresponds to the B1 element, which is also bound by ORC (Rao and Stillman 1995; Rowley et al. 1995; Lee and Bell 1997; Chang et al. 2008). By extending our motif search in L. waltii ARSs to 40 bp, we discovered an element parallel to the S. cerevisiae B1 element (Fig. 4). Like the S. cerevisiae B1, this putative L. waltii B1 element is composed of two smaller motifs, the first of which has a weak AT-rich signal (Fig. 4, purple box), and the second of which is marked by three highly conserved T/A's (Fig. 4, green box). We observe in L. waltii an additional few base pairs on either side of the putative L. waltii B1 element (Fig. 4, red box). Interestingly, this expanded B1 element is also observed in K. lactis; 189 of the 195 ARSs have at least one match to the A and B1 element motif (see Supplemental Data sets S2 and S3 for the best and all matches to the ACS for each ARS).
To test if this motif is required for L. waltii DNA replication, we mutated the 13-bp putative A element in nine ARSs and repeated the ARS assay (Table 1; Supplemental Data set S4). We found that disruption of these bases completely abolished ARS function. Some ARSs we tested had multiple matches to the ACS, but in each case it required deletion of one specific match—not necessarily the best one—to abolish ARS function. Given that there are over 10,000 matches to the 13-bp consensus sequence in the L. waltii genome, we can conclude that the consensus sequence is not sufficient for initiating DNA replication. We have not tested the L. waltii B1 element for essentiality. We expect it to be important for ARS function but nonessential as is the case for S. cerevisiae (Marahrens and Stillman 1992; Bell 1995).
Table 1.
Mutations in the ACS remove ARS function in L. waltii and S. cerevisiae

L. waltii ACSs show A-T asymmetry and are depleted for nucleosomes
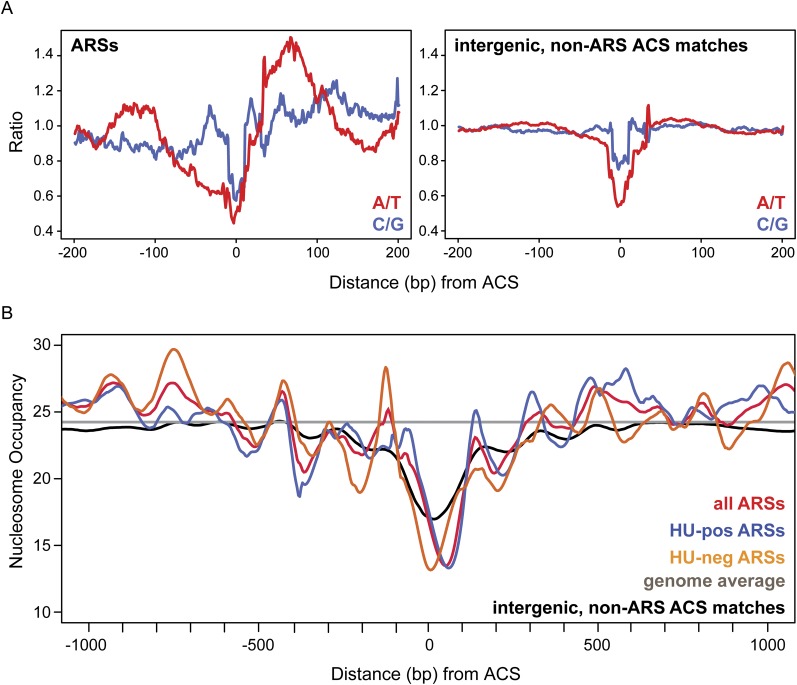
In addition to the A and B1 elements, S. cerevisiae origins are also comprised of additional A-rich B elements lacking a defined consensus sequence (Eaton et al. 2010; Chang et al. 2011). These sequences are located between 50 and 100 bp downstream from the T-rich ACS and as a result create an apparent A-T asymmetry at the ACS (Breier et al. 2004; Eaton et al. 2010). To determine whether this A-T asymmetry is a feature of L. waltii origins, we plotted the A/T and C/G ratios in 20-bp windows surrounding the ACS present in ARSs. We observe a gradual reduction in the A/T ratio approaching the T-rich ACS from 100 bp upstream (Fig. 5A, left). Immediately following the ACS itself, the A/T ratio increases sharply and an A-rich region is achieved between 50 and 100 bp downstream from the ACS. This result is specific to origins as this A-T asymmetry was not observed in non-ARS, intergenic ACS matches (Fig. 5A, right).
Figure 5.
A-T asymmetry and the nucleosome profile surrounding the L. waltii ACS. (A) The ratios of A/T and C/G bases around the L. waltii ACS are shown. All sequences were plotted such that the ACS begins at position 0 and are oriented such that the T-rich ACS strand is plotted. (Left plot) ACSs present in ARSs; (right plot) ACS matches found in intergenic, non-ARS locations. (B) The nucleosome profile surrounding the L. waltii ACS is shown. All nucleosome data are orientated as in A. The colored lines show the nucleosome profile: red, all ARSs; blue, HU-pos ARSs; orange, HU-neg ARSs; black, intergenic non-ARS ACS matches; gray, genome-wide average.
AT-rich sequences are observed to affect nucleosome positioning (Yuan et al. 2005; Field et al. 2008; Kaplan et al. 2009). The S. cerevisiae ACS in particular depletes nucleosomes at the ACS, irrespective of the direction of transcription, and positions nucleosomes on either side (Berbenetz et al. 2010; Eaton et al. 2010). Using the published nucleosome data for L. waltii (Tsankov et al. 2010), we investigated the nucleosome profile around L. waltii origins/ARSs. The nucleosome profile at L. waltii ARSs matches that seen for S. cerevisiae (Fig. 5B). Moreover, we observed that the nucleosome depletion at the ACS is skewed 50-100 bp to the right of the ACS within the A-rich region exactly as is seen in S. cerevisiae (Berbenetz et al. 2010; Eaton et al. 2010). However, unlike in S. cerevisiae (Eaton et al. 2010), HU-pos and HU-neg ARSs showed slightly different nucleosome profiles (Fig. 5B, blue vs. orange lines, respectively): While the HU-pos ARS nucleosome profile showed the skew in nucleosome depletion, the HU-neg ARSs did not. This profile was more similar to that seen for intergenic, non-ARS ACS matches throughout the genome (Fig. 5B, black line). This difference was not due to a difference in the A/T composition at the ACS in HU-neg ARSs: HU-neg ARSs still displayed the A-rich region 50-100 bp downstream from the ACS (data not shown). HU-neg ARSs include late origins and ARSs that do not fire on the chromosome. While we caution against overinterpretation, it is possible that these results indicate that nucleosome positioning may influence either the ability of an ARS to fire in the chromosomal context or its time of firing.
S. cerevisiae is able to replicate a plasmid with an L. waltii ARS
Given the great similarity between ARSs in S. cerevisiae and L. waltii, we wondered whether an L. waltii ARS would be able to function in S. cerevisiae and vice versa. To test this idea, we transformed S. cerevisiae with a set of L. waltii ARSs. Two-thirds of these ARSs supported plasmid maintenance in S. cerevisiae either very well or partially (Table 1; Supplemental Data set S4). The top scoring L. waltii ACSs within these ARSs are the best match to the S. cerevisiae ACS. Among the four L. waltii ARSs that did not function in S. cerevisiae, one has the L. waltii and S. cerevisiae ACSs at different locations and one has no match at all to the S. cerevisiae ACS (Table 1; Supplemental Data set S4). The possibility remained, however, that for those L. waltii ARSs that were able to function in S. cerevisiae, different consensus sequences were being recognized by the two species. To determine if S. cerevisiae and L. waltii utilize the same sequence for replication, we tested whether ARSs with a mutated L. waltii ACS were functional in S. cerevisiae. For two of the three ARSs tested in S. cerevisiae, plasmid maintenance, as measured by colony forming ability, was abolished upon mutation of the L. waltii ACS (Table 1; Supplemental Data set S4). With the third ARS (LwARSVI-772), the ACS mutation only reduced ARS function in S. cerevisiae. We presume that, for this origin, the ACS used by S. cerevisiae differs from the essential ACS used by L. waltii. Therefore, S. cerevisiae is generally able to replicate a plasmid using the same fragment and essential sequence as L. waltii. Additionally, we tested a set of S. cerevisiae ARSs in L. waltii and found that L. waltii is able to use about two-thirds of S. cerevisiae ARSs (Supplemental Data set S4). Overall, we have sequence, structure, and now functional evidence demonstrating that L. waltii and S. cerevisiae ARSs are highly similar.
L. waltii centromeres are early replicating and telomeres are late replicating
We next examined how L. waltii ARSs are organized in the genome. The average spacing between adjacent ARSs along L. waltii chromosomes (∼52 kb) is intermediate in value between the ARS spacing in S. cerevisiae (∼30 kb) and K. lactis (∼71 kb) (Supplemental Fig. S7). The early (HU-pos) ARSs in L. waltii occur in clusters (P-value = 0.0106, two-sample Kolmogorov-Smirnov test on distribution density values; P-value = 0.0105, test on mean value) similar to the clustering of early origins in S. cerevisiae (McCune et al. 2008). HU-neg ARSs in L. waltii do not display clustering (P-value = 0.1424, two-sample Kolmogorov-Smirnov test on distribution density values; P-value = 0.1855, test on mean value). However, as previously mentioned, we cannot be confident that all HU-neg ARSs are able to fire on the chromosome.
We observed that landmark locations in the L. waltii genome replicate at characteristic times in S phase. Centromeres are among the first regions of the L. waltii genome to be replicated while telomeres are among the last. At the 25% HL sample in the density transfer experiment, the 25-kb region around centromeres had an average of 44% HL while 25 kb from the ends of the assembled chromosomes, which cover the subtelomeric regions and at least some of the complete telomeres, had an average of 23.4% HL. Both of these values are significantly different from the average % HL of the entire genome at this time in S phase, 26.1% (P-value < 10−15, Welch two sample t-test) (Supplemental Fig. S8). A visual inspection of the chromosomes makes it apparent that not only are centromeres early replicating and telomeres late replicating, but also that the ARSs closest to centromeres are early firing and those closest to telomeres are late firing (Fig. 3; Supplemental Fig. S4). The only telomere displaying a high % HL (42.5%) at this time in S phase is the left telomere on chromosome VII. The centromere on chromosome VII is 78 kb from the left end of the chromosome, and its presence may explain why this telomere replicates early (see Discussion). These observations of early replicating centromeres and late replicating telomeres are identical to what has been observed for S. cerevisiae (McCarroll and Fangman 1988; Raghuraman et al. 2001; McCune et al. 2008). Together, these results demonstrate that, while origin spacing differs between the two species, the temporal organization of replication is similar.
Few origins are conserved across yeast species
Given that the sequences of origins and replication of chromosomal domains are similar between S. cerevisiae and L. waltii, we wondered whether these two yeasts' origins might be conserved in physical location. Unlike genes, however, origins have no individual identity—we cannot tell if two origins share a common ancestor based on homology. We therefore asked whether origins might be conserved with respect to neighboring genes and hence syntenic between S. cerevisiae and L. waltii. To do so, we mapped S. cerevisiae ARSs onto the L. waltii genome (see Methods and Supplemental Fig. S9). We then counted the number of L. waltii ARSs existing where the S. cerevisiae ARSs mapped. As all mappings were completed using intergenic regions, we excluded LwARSI-1118, in addition to the L. waltii rDNA ARS and the mating type locus ARS, from this analysis. The locations of the L. waltii origins were also randomized to determine what degree of overlap of ARSs we should expect by chance alone.
We find that 55 of 193 L. waltii ARSs overlap with S. cerevisiae ARSs and this degree of overlap is highly significant (P-value < 0.0001, permutation test, see Methods) (Supplemental Data set S5). These L. waltii ARSs were split between HU-pos (35) and HU-neg ARSs (20) and both groups significantly overlapped with S. cerevisiae ARSs (HU-pos ARSs, P-value < 0.0001; HU-neg ARSs, P-value = 0.0305; permutation test). There was some degree of correlation in the replication timing of these L. waltii–S. cerevisiae conserved origins (ρ = 0.3229, Spearman's rank correlation, P-value < 0.02), and 63% of the origins agree in HU status.
Our initial scheme for mapping S. cerevisiae ARSs was fairly liberal and allowed a single S. cerevisiae ARS to map twice in L. waltii and on two completely different chromosomes when the S. cerevisiae ARS occurs next to a break in synteny. To see if different results would be obtained by enforcing a more stringent mapping of S. cerevisiae ARSs in L. waltii, we repeated the above analysis but only used those S. cerevisiae ARSs that mapped to a single location in L. waltii. Even using this strict synteny mapping of S. cerevisiae ARSs, we still find ARSs to significantly overlap between the two species (P-value < 0.0001, permutation test).
Ultimately, we wish to know if an origin in S. cerevisiae and L. waltii are derived from a common ancestor. While we have attempted to glean such information by seeing whether ARSs from the two species map to the same location in L. waltii, such overlaps do not necessarily imply evolutionary descent. To estimate the frequency that two ARSs mapping to the same location might actually be evolutionarily related, we performed the identical procedure using tRNAs. For tRNAs we were able to determine not only if two tRNAs are syntenic but also whether they encode the same tRNA anticodon. We find 94 of 214 L. waltii tRNAs are predicted to be conserved (Supplemental Data set S5). While there are many differences between tRNAs and ARSs, if the two share a similar false positive rate in homology assignment, we predict that 41 L. waltii ARSs are truly homologous with an S. cerevisiae ARS. We fully recognize that this false positive rate assumption is flawed but consider it a useful benchmark to assess how many ARSs may truly be related by descent.
We were also curious to see how conserved ARSs are between L. waltii and the non-WGD yeasts K. lactis and L. kluyveri. We find that 28 of 193 L. waltii ARSs overlap with K. lactis ARSs, and this overlap is highly significant (P-value = 0.0001, permutation test) (Supplemental Data set S5). Using a K. lactis tRNA correction of 16.5%, we expect 23 of these matches to be homologous. Of the known 84 L. kluyveri ARSs (Liachko et al. 2011), 73 of which map into L. waltii, 14 are conserved with L. waltii (P-value < 0.0001, permutation test) (Supplemental Data set S5). Applying a tRNA correction of 8.1%, we estimate 13 known L. kluyveri ARSs to be homologous with L. waltii ARSs.
To summarize our findings, we estimate that ∼21% of L. waltii ARSs are conserved with S. cerevisiae ARSs, 12% of L. waltii ARSs are conserved with K. lactis, and 18% of L. waltii ARSs are conserved with L. kluyveri (Table 2). These values are very similar to the percent of ARSs in S. cerevisiae that appear to have been maintained in duplicate copy following the WGD (Table 2).
Table 2.
Conservation of genes, tRNAs, and ARSs in multiple genome comparisons

Origins may be conserved through their effects on surrounding genes
Unlike genes, any individual origin is believed to be redundant with neighboring origins and therefore nonessential (Dershowitz et al. 2007). For these reasons, it is unclear what the selective forces might be that act to preserve any single origin over evolutionary time. While very few ARSs appear to be conserved between L. waltii and S. cerevisiae, more are conserved than can be explained by random chance. We infer there may be some characteristic in common among these origins that promoted their evolutionary conservation. We did not find anything significant with regard to replication time or surrounding gene expression of conserved ARSs (see Supplemental Text). By GO term analysis, we did observe enrichment of conserved ARSs, around the genes encoding histones (Supplemental Table S2). Conserved ARS LwARSII-573 is adjacent to the genes for histone subunits H31 and H41, LwARSII-588 is by genes for histone subunits H2B2 and H2A2, and LwARSII-851 is by the genes for histone subunits H2A1 and H2B1. Interestingly, LwARSII-573 is conserved with both S. cerevisiae and K. lactis, and LwARSII-851 is conserved with S. cerevisiae and L. kluyveri (the adjacent intergenic region in K. lactis contains an ARS).
The strong conservation of ARSs around histone genes suggests a relationship between replication and the transcriptional regulation of these genes. In S. cerevisiae the expression of HTA1 (H2A1), HTA2 (H2A2), HTB1 (H2B1), and HTB2 (H2B2) are dependent on DNA replication (Lycan et al. 1987; Omberg et al. 2009). Expression of these genes decreases in the absence of replication. In addition, the entire family of histone genes in S. cerevisiae replicates earlier than the other genes (Raghuraman et al. 2001). We note that all three of these L. waltii ARSs are early firing. Together, these data argue that there exists a selective advantage for cells that are able to replicate their histone genes early in S phase. It is likely that the early doubling of histone mRNA templates ensures efficient production of histones prior to the duplication of the rest of the genome. The relationship between the histone genes and origins may be exceptional for we did not observe genome-wide correlations in replication and gene expression (see Supplemental Text).
We also observed enrichment of conserved ARSs within 100, 50, or 25 kb of centromeres (P-values ≤ 0.01, resampling test) (Supplemental Fig. S4). As previously discussed, L. waltii, like S. cerevisiae, centromeres reside within clusters of early replicating origins. Indeed, 13 of the 14 conserved ARSs closest to the centromeres in L. waltii are early replicating. Centromeres have been discovered to regulate the firing time of nearby origins (see Discussion). In doing so, they may also create a genomic environment that promotes origin conservation.
Origins may be conserved by promoting gene amplifications or genome rearrangements
As we identified 74 L. waltii origins that appear to be conserved in location with S. cerevisiae, K. lactis, and/or L. kluyveri we wondered if origins may be conserved via some other effect on surrounding genes. For example, we note that origins are conserved around all three L. waltii hexose transporter genes. This family of genes underwent a massive gene amplification in the lineage leading to S. cerevisiae and is suspected to support a fermentative, glucose-dependent lifestyle (Conant and Wolfe 2007). Furthermore, it is observed that, under experimental conditions of limiting glucose, these genes readily amplify (Brown et al. 1998; Dunham et al. 2002; Gresham et al. 2008; Kao and Sherlock 2008). Interestingly, of the 17 S. cerevisiae hexose transporter genes, all but four are within a gene or two of an origin. Therefore, the expansion of the hexose transporter genes in S. cerevisiae likely maintained the proximity of these genes to origins.
We and others have reported that gene amplifications and other genome rearrangements are frequently bounded by origins (Di Rienzi et al. 2009; Gordon et al. 2009; Liachko et al. 2010). Moreover, we have suggested that one mechanism by which gene amplification occurs depends directly on proximity to an origin of replication (Brewer et al. 2011). These data raise the possibility that the expansion of the hexose gene family was dependent on proximity to an origin, which in turn promoted the maintenance of these origins.
If origins can be conserved by facilitating gene amplifications, then conserved ARSs should be associated with genomic breakpoints in synteny. Remarkably, we find that almost half of the ARSs conserved between S. cerevisiae and L. waltii are within 1 kb of an S. cerevisiae–L. waltii breakpoint and that this association is highly significant (P-value = 0.0017, MEDM permutation test) (Table 3; Supplemental Table S3). This association is not recapitulated in analyzing breakpoints and conserved ARSs between L. waltii and K. lactis, but as the total number of conserved ARSs between these species is very low (8), sample size may explain this result.
Table 3.
Association of breakpoints in synteny between S. cerevisiae and L. waltii with S. cerevisiae ARSs

Discussion
Here we have provided the first comprehensive, high resolution, genome-wide look at origins and replication dynamics in a budding yeast other than S. cerevisiae. We have performed this study in a yeast that is 150 million years diverged from S. cerevisiae and did not experience the WGD. This work affords us the capacity to understand the essential characteristics of replication in budding yeast and to determine to what degree origins are permitted to change during massive genome restructuring. Our results demonstrate that the sequence and structure of origins as well as the dynamics of chromosome replication are well conserved, but that the genomic location of origins is not conserved, with the exception of instances where the origin may have had an impact on the expression or stability of surrounding genes.
The implications of the similarity in ARS sequence and nucleosome structure are straightforward: The element that constitutes an origin has remained largely the same over the divergence of L. waltii and S. cerevisiae. Conservation of origin sequence and structure was almost certainly driven by conservation in the protein machinery that recognizes the ACS within the origin, namely the ORC.
Notable differences do exist between the ACSs in these two yeasts. The extended 17-bp A element in L. waltii is not as rich in information content as that in S. cerevisiae, and the L. waltii B1 element contains additional bases not reported in the S. cerevisiae B1 element. It is tempting to speculate that these differences may reflect differences in ORC binding between the two species. In particular, we predict that the L. waltii ACS makes fewer contacts with Orc2p and more contacts with Orc5p when compared with S. cerevisiae (Lee and Bell 1997). These L. waltii ORC subunits are not more diverged from their respective S. cerevisiae ORC subunits compared with the other L. waltii ORC subunits so we cannot at this time evaluate whether there is any substance to this hypothesis. We do note that the few L. waltii ARSs that fail to function in S. cerevisiae are less well matched with the S. cerevisiae ACS at the B1 element than at the A element. This observation is in line with what has been previously reported for S. cerevisiae: Deviations in sequences outside of the ACS, especially the B1 element, can affect the function of the ARS (Chang et al. 2011).
Intriguingly, in considering the ACSs of S. cerevisiae, L. waltii, K. lactis (Liachko et al. 2010), and L. kluyveri (Fig. 4; Liachko et al. 2011), it becomes apparent that these noted differences in the L. waltii ACS are not unique to L. waltii. L. kluyveri and K. lactis also show only the smaller A element, and the expanded B1 element of L. waltii is found in K. lactis. In considering the phylogeny of these yeasts, it thus appears that the ACS has changed gradually and steadily over evolutionary time.
Despite the sequence and structural similarity of origins, we did not find origins to be well conserved in location. Combined with knowledge that, when origins are lost, cryptic, “backup” origins may be able to fire (Dershowitz et al. 2007; Blow et al. 2011), our work suggests that the genome is littered with weak, potential origin “seeds.” In support of this idea, both the S. cerevisiae and L. waltii genomes have over 10,000 matches to the ACS sequence. We envision that, if strong origin sequences were removed during the genome reduction following the WGD, these seeds may have evolved to serve as origins. Based on our analysis of ARS conservation between the closely related yeasts L. waltii and L. kluyveri, it would appear that origin sites mutate in and out of function very rapidly.
It is difficult to address what changes occurred to convert a sequence from a potential to a functional origin. Since origins do not occur in all types of intergenic regions with equal frequency, the transcriptional direction of neighboring genes may affect where an origin can appear. However, intergenic region preference differs across yeast species: S. cerevisiae ARSs preferentially reside in convergently transcribed intergenic regions (MacAlpine and Bell 2005; Nieduszynski et al. 2006; Yin et al. 2009), K. lactis ARSs have no preference (Liachko et al. 2010), and L. waltii and S. pombe origins prefer divergent intergenic regions (Segurado et al. 2003). Thus transcription from neighboring genes is not likely to be a universal determinant of origin location.
The need for plasticity in origins over evolutionary time may explain why origins in many studied eukaryotes lack strong sequence determinants (for review, see Mechali 2010). By lacking sequence determinants, potential origin sites are able to be redundant in the genome, thus giving origins the ability to appear and disappear in step with genome evolution. Hence, it is ultimately the paucity of origin sequence determinants that solves the paradox of how origins persist in the genome and ensure faithful DNA replication.
The strong similarity seen in the general temporal patterns of chromosome replication in S. cerevisiae and L. waltii speaks not to what constitutes an origin but rather to how origin firing is regulated. Given our finding that origin location is not conserved, the only way for telomeres to remain late firing and centromeres to be early firing is for origin firing to be controlled by the genomic environment rather than being encoded within the origin itself. This hypothesis is in fact already supported in the literature. Telomeres have been shown to delay origin activation in S. cerevisiae: ARSs moved to a telomere show a delay in their firing time and ARSs moved away from a telomere show an advance in firing time (Ferguson and Fangman 1992). The telomere effect has been demonstrated to extend ∼30 kb from telomeres (Ferguson and Fangman 1992). Internal late-firing origins appear to be regulated by histone deacetylation (Vogelauer et al. 2002; Aparicio et al. 2004; Knott et al. 2009). More recently, centromeres have been shown to have the opposite effect in both S. cerevisiae (Pohl et al. 2012) and Candida albicans (Koren et al. 2010). In this regard, L. waltii chromosome VII presents an interesting case. The centromere on chromosome VII is ∼80 kb from the presumed telomere, which replicates early. This observation suggests that the centromere early replication effect extends into the telomere and overrides its replication delay effect. The question of what determines origin-firing time in S. cerevisiae has yet to be pinpointed, though chromatin state likely explains much of it (Diller and Raghuraman 1994; Stevenson and Gottschling 1999; Bell and Dutta 2002; Vogelauer et al. 2002; Zappulla et al. 2002; Aparicio et al. 2004; Hayashi et al. 2009; Knott et al. 2009). The existence of early replicating clusters in L. waltii supports the hypothesis that chromatin also controls replication to at least some degree in L. waltii.
As there is no observable defect in the loss of a single origin, it is very hard to envision a selective pressure acting directly on an individual origin. However, if the origin had an effect on the transcription of the surrounding genes or the genomic stability of the region, a clear selective pressure arises. Our results suggest that origins may in fact be conserved when they impact neighboring genes. First, we found that the origins adjacent to histone genes are conserved, suggesting that these origins were conserved because they enhance the expression of these genes. Second, we find that half of S. cerevisiae conserved origins are at sites of genome rearrangements and observe a peculiar enrichment of origins around the amplified hexose transporter genes.
An additional clear case of an origin promoting a duplication and translocation event was found for the L. waltii rDNA ARS: We discovered that the rDNA ARS and the surrounding intergenic region have duplicated and migrated to another chromosome (see Supplemental Text). It is remarkable that the sequence of this intergenic region is almost identical to that within the rDNA locus. This observation implies that the duplication and translocation event were recent.
The work described here indicates that the composition of replication origins is resistant to change, but that genome evolution forces origin activity to be repositioned throughout the genome. We observed limited cases where origins may have been maintained in location and believe these represent unique situations in which the origin itself has an effect on the surrounding genes. We believe that it is the simplicity and redundancy of the essential origin sequence that guarantees distribution of origins throughout the genome to ensure the genome remains faithfully replicated regardless of how the genome reshapes itself.
Methods
Yeast strains, growth conditions, and media
L. waltii type strain ATCC56500, the ura3 L. waltii strain (Di Rienzi et al. 2011), and the S. cerevisiae type strain S288C were used. Standard rich (YEP with dextrose or glycerol), synthetic (YC), nitrogen starvation medium (−N), dense isotope, and G418 selective media have been previously described (McCune et al. 2008; Di Rienzi et al. 2011). L. waltii liquid cultures were incubated at 23°C and plates at 30°C.
L. waltii genomic library construction
L. waltii genomic DNA was obtained from logarithmically growing cultures using the method described at http://fangman-brewer.genetics.washington.edu/DNA_prep.html. This DNA was randomly sheared by sonication, end-repaired, and size-selected for fragments between 350 and 1000 bp using standard procedures. The DNA was cloned into the previously described L. waltii centromeric plasmid, which has a KanMX marker but lacks an origin of replication for L. waltii (Fig. 1A; Di Rienzi et al. 2011). Ligations were transformed into Escherichia coli cells and plasmid DNA was harvested. See the Supplemental Text for specific details.
ARS assay
The L. waltii genomic library was transformed into L. waltii as previously described (Di Rienzi et al. 2011) with selection on YEPD + G418 plates. After three days, yeast colonies were scraped from plates and resuspended in –N medium. To recover plasmids, 25 μL cell pellets were used in the high efficiency yeast plasmid rescue protocol described at http://labs.fhcrc.org/gottschling/Yeast%20Protocols/yplas.html. Multiple reactions were combined and concentrated by a standard ethanol precipitation creating the L. waltii ARS library. ARS assays in S. cerevisiae were performed using the same protocol as for L. waltii. To test S. cerevisiae ARSs on the S. cerevisiae URA3:CEN plasmid, YIP5-5 (Ferguson et al. 1991), ura3 S. cerevisiae, and L. waltii cells were transformed and plated on YC-ura plates.
Sanger sequencing
Plasmid DNA and Primer35 and Primer36, which directly flank the SmaI cloning site, were used to sequence the library insert by Sanger sequencing. All primer sequences are available in Supplemental Table S4.
Illumina sequencing
Genomic DNA inserts were amplified from the library vector using primers IlluminaLibF and IlluminaLibR (Supplemental Table S4), which contain the Illumina adapters followed by the library vector sequences that directly flank the cloning site. See the Supplemental Text for details of the PCR reactions. These DNA fragments were then subjected to 76-bp paired-end sequencing using an Illumina GAIIx. The genomic library was sequenced twice in two separate Illumina runs (SRA accession SRP008333) and the ARS library in one (SRA accession SRP008333).
Illumina sequence data analysis
Illumina reads were mapped onto the L. waltii genome using the software Bowtie (Langmead et al. 2009). Only reads mapping uniquely in the L. waltii genome were analyzed. The L. waltii genome used here includes the assembled L. waltii genome described previously (Di Rienzi et al. 2011), the L. waltii mating locus, the 2 μm plasmid (Chen et al. 1992), and the rDNA locus (described here; see Supplemental Text). For aligning paired-end reads, an insert size between 250 and 1100 was allowed in Bowtie. When single-read sequencing data were used, sequences were extended 500 bp to estimate the complete insert.
Fragments that contain ARSs were identified in the sequencing data through a filtering scheme. The ARS library Illumina sequence data were converted into sequence abundance in overlapping windows across the genome, normalized against the genomic library, and finally evaluated using a cutoff based on sequence abundance to determine which recovered fragments correspond to ARSs. This filtering method incorporates the observations that ARS sequences are clustered and in the upper tail of the distribution for sequence abundance. For complete details on the filtering process see Supplemental Text and Supplemental Figure S10.
L. waltii microarray design
A custom Agilent microarray (Agilent Technologies order number G4497A, archived in NCBI GEO under accession no. GPL15109) with ∼33,000 unique L. waltii probes was designed. See Supplemental Text for further details.
ssDNA mapping
L. waltii cells were grown in YEPD to mid-log phase (OD660 0.4). HU was added to a final concentration of 200 mM (see Supplemental Text). Timed 300 mL samples were harvested at 120, 180, and 240 min after HU addition and arrested with 6 mL of 10% sodium azide and 48 mL of 0.5 M EDTA. The G1 control sample was produced by transferring logarithmically growing L. waltii into medium low in ammonium sulfate and incubating for 20 h before harvesting cells. G1 and S phase cells were processed for ssDNA labeling as per Feng et al. (2007), and competitively hybridized to the custom L. waltii microarray. Hybridizations and scanning were performed by the University of Washington Center for Array Technology according to the manufacturers' instructions. Data were normalized and the S/G1 ratios calculated as previously described (Feng et al. 2006). The data were Lowess-smoothed using a 6-kb window, unless the window contained a gap predicted to be >750 bp according to Kellis et al. (2004). Statistically significant peaks were calculated as previously described (Feng et al. 2006; McCune et al. 2008), and peaks that achieved statistical significance in at least five of six microarray data sets were included in the final list of HU-pos ARSs. The ssDNA microarray data are available at NCBI GEO under accession no. GSE35253 and the processed data are provided in Supplemental Data set S6.
Density transfer
Dense isotope transfer experiments were used to generate genome-wide replication timing data as previously described (Raghuraman et al. 2001) with modifications described in the Supplemental Text. Array hybridizations and scanning were performed by the University of Washington Center for Array Technology according to the manufacturers' instructions. Data were normalized to the percent replication calculated from the slot blots as previously described (Alvino et al. 2007). Data were then Lowess-smoothed using a 20-kb window, except where windows contained gaps predicted to be >750 bp according to Kellis et al. (2004). Smoothed % HL values from two microarray hybridizations (dye swaps) were averaged with the exception of the 25% HL sample for which dye swaps were not available. Peaks in % HL profiles and the timed samples in which those peaks became significant were identified as described by Alvino et al. (2007). The HU arrest sample was used as a control for the baseline variation in % HL. The density transfer microarray data are available at NCBI GEO under accession no. GSE35155 and the processed data are provided in Supplemental Data set S7.
Motif searches
The MEME (Bailey and Elkan 1994) Suite v4.1.1 was downloaded from http://meme.nbcr.net/. A second order Markov model of the background was built from L. waltii intergenic sequences. Motifs of size 20–40 bp were searched assuming the motif is present zero or once per sequence (the ZOOPS model) and enriched over the background. Motif diagrams were generated using WebLogo (Crooks et al. 2004). MAST (Bailey and Gribskov 1998) with a P-value cutoff of 0.001 and an E-value cutoff of 100 was used to find matches to the motif in ARSs and all intergenic regions. The S. cerevisiae origin motif position weight matrix and background model were obtained from Eaton et al. (2010).
ARS mutagenesis
The putative motif was replaced with an EcoRV site using a standard PCR based method detailed in the Supplemental Text. Mutations were verified by sequencing.
Nucleosome profiles
Inferred nucleosome occupancy for asynchronously growing mid-log phase L. waltii cells were taken from Tsankov et al. (2010). Only nucleosome positions for the assembled L. waltii genome (Di Rienzi et al. 2011) were used. Four regions in the genome showed double the nucleosome occupancy of the surrounding regions. These discrepancies, likely due to copy number artifacts, were corrected (see Supplemental Text). Nucleosomes mapping within 1 kb of the start of the T-rich strand of the ARS motif were plotted using the loess.smooth function in the R statistical environment using 1000 points (evaluation parameter) and a smoothness parameter (span) of 0.035.
Clustering analysis of origins
The numbers of ARSs in a row that were all HU-pos (recovered from ssDNA assay) or all HU-neg were counted to define the cluster size. The cluster size null distributions were determined by shuffling the ARS type labels 10,000 times. Distributions were converted into density values. Separately, the mean cluster size from the real distribution was compared against the null distribution of means generated from the simulations.
Gene expression data
Gene expression values for asynchronously growing mid-log phase L. waltii cells were taken from Tsankov et al. (2010). Expression values were averaged over the length of each gene so that each gene was assigned a single expression value.
S. cerevisiae replication timing data
Replication timing at the 10-min intervals for the S. cerevisiae genome was taken from McCune et al. (2008). Values were averaged between array replicates, and the average % HL for each gene was calculated.
Enrichment analysis
Each L. waltii chromosome sequence was broken into 5-kb segments generating a total of 2054 bins across the entire genome. L. waltii ARSs (described here) and tRNAs (Di Rienzi et al. 2011) were assigned to bins using their midpoint. Each bin was scored for the presence or absence of a feature. An enrichment test was performed on the overlap of two features using the hypergeometric distribution as the null distribution. P-values < 0.05 were considered as evidence of a correlation.
GO term enrichment analysis
GO term enrichment of genes surrounding ARSs was determined using the GO∷TermFinder software (Boyle et al. 2004) available on the AmiGO website (Carbon et al. 2009). To assign GO terms to L. waltii ORFs, all L. waltii ORFs were converted to their S. cerevisiae homologs following the annotations of Byrne and Wolfe (2005). The four ORFs directly surrounding an ARS were analyzed for enrichment. GO terms with P-value < 0.05 and with at least two genes present in the data set were considered significant.
Syntenic comparison of ARSs and tRNAs
Synteny of ARSs and tRNAs between L. waltii and S. cerevisiae, K. lactis, or L. kluyveri was accomplished by (1) anchoring S. cerevisiae, K. lactis, L. kluyveri ARSs/tRNAs onto their neighboring genes in their native genome, (2) defining the space in which the ARS/tRNA could exist in the L. waltii genome, and (3) checking for an overlap of these ARSs/tRNAs with L. waltii ARSs/tRNAs (see Supplemental Fig. S9). Significance of overlap between species was determined by permuting the location of L. waltii ARSs/tRNAs using a previously described permutation algorithm (Di Rienzi et al. 2009). P-values < 0.05 were considered significant. Full details of this method are provided in the Supplemental Text and all syntenic blocks and ARS/tRNA mappings are described in Supplemental Data sets S8, S9.
Breakpoint analysis
Breakpoints between L. waltii and the putative ancestral yeast, the Ancestor (Gordon et al. 2009), were taken from Di Rienzi et al. (2009). L. waltii–K. lactis breakpoints were mapped using the methods described previously for the L. waltii–Ancestor breakpoints. Associations of breakpoints and genomic features (ARSs and tRNAs) were performed using the Minimal Endpoint Distance Measures method accompanied by a simulation to randomize breakpoints as was previously performed for L. waltii–Ancestor breakpoints (Di Rienzi et al. 2009).
Data access
Illumina sequencing data from this study are available at the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP008333. The microarray from this study is available at the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GPL15109, and the data are available under accession numbers GSE35155 and GSE35253.
Acknowledgments
This work was accomplished only through the aid and consultation of numerous people. We thank David Collingwood for his aid in processing the L. waltii array data, and Joseph Sanchez for sequencing the region on chromosome I bearing similarity to the rDNA locus. We are grateful to Charles Grant for teaching us how to use MEME, Ruolan Qiu for aiding in the construction of the L. waltii genomic library, Jay Shendure and Choli Lee for conducting the Illumina sequencing, the Center for Array Technologies in Seattle for microarray hybridization and scanning, and Joe Hiatt and Carlos Araya for advice in preparing DNA for Illumina sequencing. We also thank the members of the Brewer/Raghuraman and Dunham labs for their advice and encouragement. Finally, we would like to thank the anonymous reviewers for their many helpful and constructive comments. This work was made possible by the University of Washington Royalty Research Foundation. B.J.B and M.K.R. were supported by a National Institute of General Medical Sciences grant (GM18926), W.S.N. was supported by a National Center for Research Resources award (P41 RR0011823), S.C.D. was supported by the Genetics Predoctoral Training Program, and K.C.L. was supported by a postdoctoral fellowship from the American Cancer Society.
Author contributions: S.C.D. performed the ARS assay. K.C.L. performed the ssDNA and density transfer experiments. K.C.L. performed the ARS mutagenesis. K.C.L. and S.C.D. generated the ACS logos. S.C.D. performed all computational analyses, generated the nucleosome profiles, assembled the rDNA locus, performed the cross species tests of ARSs between S. cerevisiae and L. waltii, and performed all evolutionary conservation analyses. T.M. designed the L. waltii microarray. W.S.N. provided assistance with MEME. S.C.D., K.C.L., T.M., M.K.R., and B.J.B. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138248.112.
References
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK 2007. Replication in hydroxyurea: It's a matter of time. Mol Cell Biol 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Gribskov M 1998. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Bell SP 1995. Eukaryotic replicators and associated protein complexes. Curr Opin Genet Dev 5: 162–167 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Berbee ML, Taylor JW 2001. Fungal molecular evolution: Gene trees and geologic time. In The Mycota: Systematics and evolution (ed. D McLaughlin, E McLaughlin), pp. 229–245. Springer-Verlag, Berlin [Google Scholar]
- Berbenetz NM, Nislow C, Brown GW 2010. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet 6: e1001092 doi: 10.1371/journal.pgen.1001092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A 2005. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA 2011. How dormant origins promote complete genome replication. Trends Biochem Sci 36: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G 2004. GO:TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier AM, Chatterji S, Cozzarelli NR 2004. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol 5: R22 http://genomebiology.com/2004/5/4/R22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Payen C, Raghuraman MK, Dunham MJ 2011. Origin-dependent inverted-repeat amplification: A replication-based model for generating palindromic amplicons. PLoS Genet 7: e1002016 doi: 10.1371/journal.pgen.1002016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF 1998. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol 15: 931–942 [DOI] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH 2005. The Yeast Gene Order Browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15: 1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S 2009. AmiGO: Online access to ontology and annotation data. Bioinformatics 25: 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Theis JF, Miller J, Nieduszynski CA, Newlon CS, Weinreich M 2008. Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW sequence within the origin recognition complex binding site. Mol Cell Biol 28: 5071–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, May CD, Hoggard T, Miller J, Fox CA, Weinreich M 2011. High-resolution analysis of four efficient yeast replication origins reveals new insights into the ORC and putative MCM binding elements. Nucleic Acids Res 39: 6523–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Cong YS, Wesolowski-Louvel M, Li YY, Fukuhara H 1992. Characterization of a circular plasmid from the yeast Kluyveromyces waltii. J Gen Microbiol 138: 337–345 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH 2007. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol 3: 129 doi: 10.1038/msb4100170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz A, Snyder M, Sbia M, Skurnick JH, Ong LY, Newlon CS 2007. Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements. Mol Cell Biol 27: 4652–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Collingwood D, Raghuraman MK, Brewer BJ 2009. Fragile genomic sites are associated with origins of replication. Genome Biol Evol 2009: 350–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Lindstrom KC, Lancaster R, Rolczynski L, Raghuraman MK, Brewer BJ 2011. Genetic, genomic, and molecular tools for studying the protoploid yeast, L. waltii. Yeast 28: 137–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diller JD, Raghuraman MK 1994. Eukaryotic replication origins: Control in space and time. Trends Biochem Sci 19: 320–325 [DOI] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci 99: 16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ 2006. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol 8: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Raghuraman MK, Brewer BJ 2007. Mapping yeast origins of replication via single-stranded DNA detection. Methods 41: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BM, Fangman WL 1992. A position effect on the time of replication origin activation in yeast. Cell 68: 333–339 [DOI] [PubMed] [Google Scholar]
- Ferguson BM, Brewer BJ, Reynolds AE, Fangman WL 1991. A yeast origin of replication is activated late in S phase. Cell 65: 507–515 [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216 doi: 10.1371/journal.pcbi.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet 5: e1000485 doi: 10.1371/journal.pgen.1000485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet 4: e1000303 doi: 10.1371/journal.pgen.1000303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H 2009. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol 11: 357–362 [DOI] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H 2006. Phylogenomics: The beginning of incongruence? Trends Genet 22: 225–231 [DOI] [PubMed] [Google Scholar]
- Kao KC, Sherlock G 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet 40: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavare S, Aparicio OM 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Tsai HJ, Tirosh I, Burrack LS, Barkai N, Berman J 2010. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet 6: e1001068 doi: 10.1371/journal.pgen.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Bell SP 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol 17: 7159–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Bhaskar A, Lee C, Chung SC, Tye BK, Keich U 2010. A comprehensive genome-wide map of autonomously replicating sequences in a naive genome. PLoS Genet 6: e1000946 doi: 10.1371/journal.pgen.1000946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Tanaka E, Cox K, Chung SC, Yang L, Seher A, Hallas L, Cha E, Kang G, Pace H, et al. 2011. Novel features of ARS selection in budding yeast Lachancea kluyveri. BMC Genomics 12: 633 doi: 10.1186/1471-2164-12-633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycan DE, Osley MA, Hereford LM 1987. Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol Cell Biol 7: 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Bell SP 2005. A genomic view of eukaryotic DNA replication. Chromosome Res 13: 309–326 [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817–823 [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M 2010. Eukaryotic chromosome DNA replication: Where, when, and how? Annu Rev Biochem 79: 89–130 [DOI] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL 1988. Time of replication of yeast centromeres and telomeres. Cell 54: 505–513 [DOI] [PubMed] [Google Scholar]
- McCune HJ, Danielson LS, Alvino GM, Collingwood D, Delrow JJ, Fangman WL, Brewer BJ, Raghuraman MK 2008. The temporal program of chromosome replication: Genomewide replication in clb5Δ Saccharomyces cerevisiae. Genetics 180: 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M 2010. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Nieduszynski CA, Knox Y, Donaldson AD 2006. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev 20: 1874–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Hiraga S, Ak P, Benham CJ, Donaldson AD 2007. OriDB: A DNA replication origin database. Nucleic Acids Res 35: D40–D46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omberg L, Meyerson JR, Kobayashi K, Drury LS, Diffley JF, Alter O 2009. Global effects of DNA replication and DNA replication origin activity on eukaryotic gene expression. Mol Syst Biol 5: 312 doi: 10.1038/msb.2009.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl TJ, Brewer BJ, Raghuraman MK 2012. Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae. PLoS Genet 8: e1002677 doi: 10.1371/journal.pgen.1002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL 2001. Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Rao H, Stillman B 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci 92: 2224–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF 1995. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J 14: 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F 2003. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE 1999. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev 13: 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Struhl K, Davis RW 1979. Isolation and characterisation of a yeast chromosomal replicator. Nature 282: 39–43 [DOI] [PubMed] [Google Scholar]
- Theis JF, Newlon CS 1997. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc Natl Acad Sci 94: 10786–10791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong LN, Wu X 2011. Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol 3: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ 2010. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol 8: e1000414 doi: 10.1371/journal.pbio.1000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M 2002. Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Yang C, Theis JF, Newlon CS 1999. Conservation of ARS elements and chromosomal DNA replication origins on chromosomes III of Saccharomyces cerevisiae and S. carlsbergensis. Genetics 152: 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Deng W, Hu L, Kong X 2009. The impact of nucleosome positioning on the organization of replication origins in eukaryotes. Biochem Biophys Res Commun 385: 363–368 [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Sternglanz R, Leatherwood J 2002. Control of replication timing by a transcriptional silencer. Curr Biol 12: 869–875 [DOI] [PubMed] [Google Scholar]