Abstract
The bacterial community in the human gut has crucial health roles both in metabolic functions and in protection against pathogens. Phages, which are known to significantly affect microbial community composition in many ecological niches, have the potential to impact the gut microbiota, yet thorough characterization of this relationship remains elusive. We have reconstructed the content of the CRISPR bacterial immune system in the human gut microbiomes of 124 European individuals and used it to identify a catalog of 991 phages targeted by CRISPR across all individuals. Our results show that 78% of these phages are shared among two or more individuals. Moreover, a significant fraction of phages found in our study are shown to exist in fecal samples previously derived from American and Japanese individuals, identifying a common reservoir of phages frequently associated with the human gut microbiome. We further inferred the bacterial hosts for more than 130 such phages, enabling a detailed analysis of phage–bacteria interactions across the 124 individuals by correlating patterns of phage abundance with bacterial abundance and resistance. A subset of phages demonstrated preferred association with host genomes as lysogenized prophages, with highly increased abundance in specific individuals. Overall, our results imply that phage–bacterial attack–resistance interactions occur within the human gut microbiome, possibly affecting microbiota composition and human health. Our finding of global sharing of gut phages is surprising in light of the extreme genetic diversity of phages found in other ecological niches.
The human microbiome, representing the collection of all microbes that live on and within a human being, is composed of 10 times more cells than human cells (Eckburg et al. 2005; Walter and Ley 2011). These naturally occurring microbes, particularly those that reside in the human gut, are known to provide humans with crucial metabolic functions. They allow harvesting and storing energy from various dietary products, influence the development of the immune system, and protect from colonization by pathogens (Hooper et al. 2002; Dethlefsen et al. 2007). In a recent study by the MetaHIT Consortium (Metagenomics of Human Intestinal Tract) (Qin et al. 2010), DNA from fecal samples of 124 Europeans was sequenced to generate deep coverage of the human gut microbiome. Analysis of the assembled DNA fragments showed that the human gut microbial gene set is 150 times larger than the human gene complement (Qin et al. 2010).
In many studied ecosystems, phages outnumber bacterial cells by a factor of 10:1, posing significant predation pressure on their hosts (Chibani-Chennoufi et al. 2004). This phage pressure has been shown to play a crucial role in the evolution, diversity, and abundance of bacteria (Avrani et al. 2011; Stern and Sorek 2011). The richness and density of gut bacterial species and populations make the human gut an ideal ecological niche for phages. Indeed, the existence of phages in the human and animal gut has been demonstrated repeatedly using transmission electron microscopy (Letarov and Kulikov 2009), and virus-like particles (VLPs) were observed in high density on the surface of human gut mucosa. Furthermore, metagenomic sampling has shown that the majority of DNA viruses in the human gut are bacteriophages (Breitbart et al. 2003). However, research on the identity and prevalence of phages infecting gut-residing microbiota and of their effects on gut bacterial populations is still in its early days.
Recently, VLPs isolated from feces of four pairs of monozygotic twins and their mothers (Reyes et al. 2010), as well as six unrelated individuals (Minot et al. 2011), were sequenced over several time points. Both studies detected a largely unique and stable phage complement within each individual. These studies suggested that phages are rarely shared among individuals and that a predatory viral–microbial dynamic, as characterized in various other ecosystems, may be absent in the distal human gut.
Clustered regularly interspaced short palindromic repeats (CRISPR) loci, together with their associated cas genes, have been shown to constitute a defense system against propagation of phages and plasmids (Barrangou et al. 2007; Marraffini and Sontheimer 2008; Sorek et al. 2008). CRISPR loci are composed of short repeat sequences separated by hypervariable “spacer” sequences, usually sized 24–50 bp. In the first stage of bacterial defense, bacteria incorporate fragments of phage or plasmid genomes as novel spacers. These spacers are then transcribed into small RNAs, which together with the Cas protein complex guide the way to interfere with phage replication (van der Oost et al. 2009; Horvath and Barrangou 2010; Karginov and Hannon 2010; Marraffini and Sontheimer 2010). Thus, CRISPR spacers may be viewed as a database of fragments derived from phage and plasmid genomes. Indeed, CRISPR spacers have previously been used to identify phages in several metagenomic data sets, including microbiomes harvested from acid mine (Andersson and Banfield 2008), hot spring (Snyder et al. 2010), and human oral cavity (Pride et al. 2011) environments.
One of the major caveats of metagenomic analyses is that in most cases, microbes and viruses are sequenced together, thereafter making it difficult to distinguish between the two. Even when VLPs are isolated and sequenced directly, it is nearly impossible to identify the specific hosts of the isolated viruses, precluding the analysis of the relationship between virus and host. Here, we use the link between bacterial CRISPR spacers and the infecting mobile elements to study these interactions in the human gut.
To this end, we have analyzed the most extensive metagenomic sequencing data of the human gut microbiome produced to date (the MetaHIT set), composed of 576.7 billion bases of sequence (Qin et al. 2010). We extracted CRISPR spacers directly from the raw sequencing reads and then used these spacers as probes to search for phage genomic segments within the assembled sequences of the metagenomes (Fig. 1). This allowed us to (1) identify and characterize a large catalog of phages and mobile elements that infect bacteria in the human gut; (2) identify, for a subset of these phages, their bacterial hosts; and (3) perform a subject-wide analysis of phage–bacteria coexistence, by correlating patterns of phage abundance with patterns of bacterial abundance and resistance. These results were then compared across data sets obtained from additional microbiome sequencing projects, revealing the existence of a reservoir of both dormant and active phages frequently associated with the human gut microbiome.
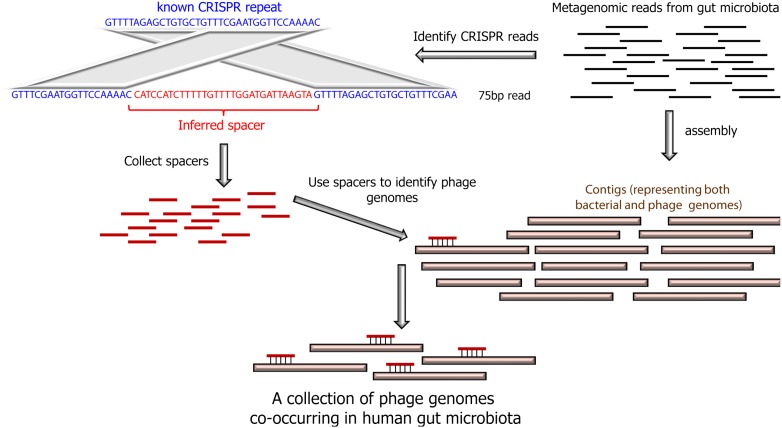
Figure 1.
CRISPR spacers are used as probes to fish out phage genomes. In the MetaHIT metagenomics study (Qin et al. 2010), gut microbes were harvested from feces of 124 individuals, and DNA was sequenced to generate short reads (75 bp). These reads were then assembled into contigs, which mainly represent DNA of gut-residing bacteria, but potentially also contain DNA of phages associated with these bacteria. In the present study, CRISPR spacers were detected by searching for reads that match a known CRISPR repeat on both sides of the read. The spacers detected were then used to probe assembled contigs, and phage and mobile element contigs were identified as those showing high sequence identity with a spacer (but not with the CRISPR repeat).
Results
A catalog of phages in the human gut
In previous analyses of the assembled MetaHIT data (Qin et al. 2010), identification of CRISPR regions was not performed because the short Illumina reads and the repetitive nature of CRISPR arrays impeded CRISPR assembly. We used a specifically tailored CRISPR-extraction protocol on the raw sequence data of the 110 samples in which 75-bp reads were sequenced (Fig. 1; Methods). Overall, we identified a total of 52,267 spacers in the samples analyzed (Supplemental Data Set S1). On average, 475 unique spacers were found in each individual, with the number of identified spacers per sample ranging from 130 to 1017 (Supplemental Table S1). Based on simulations with known gut-residing bacterial genomes, our method detects ∼60%–100% of the actual number of spacers per sample in the most abundant gut bacteria, yet requires much higher sequencing depth to detect spacers from the less abundant bacterial species (Supplemental Fig. S1; Supplemental Text). Thus, the number of immunity spacers reported here is most likely an underestimate of the full repertoire of CRISPR spacers in the human gut. Taking this into consideration, these results suggest that human gut bacteria cumulatively hold a vast immune repository against phages and other mobile elements.
To identify assembled phage genomes in the MetaHIT data, we used the detected spacers as probes to search all assembled contigs from this sequencing project (Fig. 1; Methods). Accordingly, 18,188 spacers (34.8%) were found to match 47,561 contigs sized between 500 bp and 134 kb, which lacked the CRISPR repeat (Supplemental Data Set S2). We denote these contigs as mobile genetic element (MGE) contigs. Since the MGE contigs we identified may contain redundancy (e.g., the same phage genome was independently assembled in several different individuals), we performed clustering of the contig nucleotide sequences and used the longest contig of each cluster as its representative (Methods). This resulted in a collapse of the identified contigs into a nonredundant set of 11,639 MGE contigs, cumulatively totaling 77.3 Mb of DNA sequence (Supplemental Data Set S3).
To corroborate the origin of MGE contigs, we compiled a database of phage-only proteins (i.e., proteins exclusively associated with phage, nonbacterial functions; see Methods). We next performed bootstrap resampling of non-MGE MetaHIT contigs to produce 100 data sets of the same size as the MGE contigs data set, and found that on average 8.5% (±0.2%) of the contigs in these data sets match phage proteins. On the other hand, 23% of the MGE contigs matched such phage proteins, suggesting that the set of contigs we identified using the CRISPR spacers is highly enriched for phage-derived DNA (P < 0.01 based on bootstrap distribution).
To further establish that the contigs we identified are of phage origin, we used the data obtained in the study of Reyes et al. (2010) and the study of Minot et al. (2011), composed of the DNA sequence data of VLPs isolated from the feces of a total of 18 American individuals across several time points. We found that 20.1% of the Reyes VLP reads mapped to our nonredundant MGE contigs (with at least 85% identity over at least 85% of the read length), as did 28.6% of the Minot VLP reads. The results represent an average enrichment of fivefold and 8.5-fold, respectively, vis-à-vis 20 randomly selected similarly sized non-MGE contig sets (P < 0.05 based on bootstrap distribution). This strongly suggests that many of the contigs we identified represent partial or full phage genomes.
The MGE contigs we collected probably largely represent a mixture between DNA of phages, plasmids, and transposable elements, since all of these mobile elements were found to be targeted by CRISPR (Horvath and Barrangou 2010). To specifically study phage genomes, we first selected only MGE contigs sized 10 kb or more (Reyes et al. 2010) since these are expected to represent a significant portion of the phage genome and are also less likely to appear as disjoint fragments of the same phage. We then classified these large contigs as phage-originated if one or more predicted ORFs showed similarity to phage-only genes (Supplemental Data Set S4; Methods). We also included large MGE contigs that were significantly covered by the VLP-derived sequence data sets (Reyes et al. 2010; Minot et al. 2011), because these were directly extracted from virus-like particles (Methods). This classification resulted in 991 contigs, totaling 22.3 Mb, which most probably represent genomes of gut-residing phages. The analyses henceforth refer to these 991 phage contigs.
A reservoir of common phages associated with the human gut microbiome
We first attempted to infer the taxonomic classification of the CRISPR-targeted phage contigs. This was accomplished by matching predicted ORFs from the phage sequences to proteins of members of ICTV (International Committee on Taxonomy of Viruses)–defined families deposited on NCBI (Methods). Among the 304 (30.7%) of the phage contigs that were classified unambiguously, we found a vast abundance of Siphophages (78%), with a minority of Myophages and Podophages (11.5% and 6.5% of assigned contigs, respectively) (Supplemental Fig. S2; Supplemental Data Set S5). For 73% of classified contigs, the assignment was based on multiple concurring ORFs, suggesting reliable classification. These results show that all dominant gut phage types previously identified by fecal VLP sequencing (Breitbart et al. 2003; Reyes et al. 2010; Minot et al. 2011) are also targeted by the CRISPR systems of gut bacteria. We also checked for enrichment of gene functions in the phage sequences compared with bacterial genomes (Methods). Aside from phage structural genes, we found an abundance of DNA replication and phage metabolic functions (e.g., hydrolases, nucleotide synthesis), as well as restriction-modification systems and antibiotic resistance beta-lactamases (Supplemental Table S2). The latter is in line with studies suggesting that gut phages hold genes with a selective advantage to their host (Wang et al. 2010; Minot et al. 2011).
We proceeded to inspect the distribution of presence and abundance of the phage contigs we identified in the 124 sampled individuals. A phage contig was deemed present in a sample if it was covered by metagenomic reads from that sample throughout a significant portion (at least 70%) of the region bounded by proto-spacers and phage-only genes, while abundance was determined as coverage per kilobase per millions of reads (Methods; Supplemental Text). In each individual an average of 101 (10.2%) of the phages we identified were present, with the maximum reaching 181 (18.3%) in sample MH0006 (Supplemental Data Set S6). In general, individual viromes were characterized by a small number of highly abundant phages alongside a larger number of phages present in low abundance (Fig. 2B), in congruence with results from fecal VLP sequencing studies (Reyes et al. 2010; Minot et al. 2011).
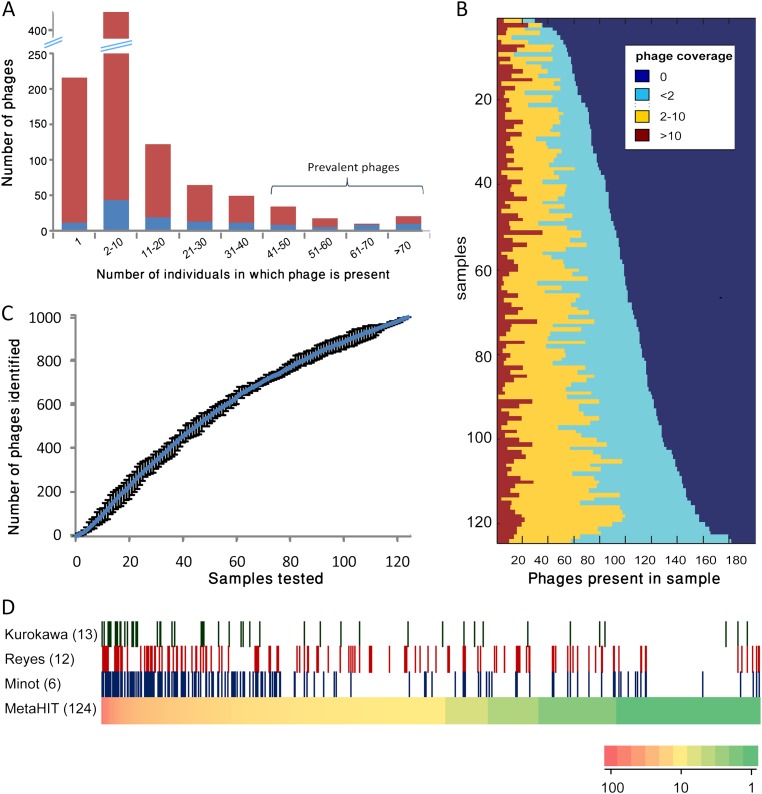
Figure 2.
Phage distribution across individuals and populations. (A) Phage abundance profile. (X-axis) Range of the number of samples in which a phage was present; (y-axis) number of phages whose prevalence is in the range specified. A phage was deemed present if it was significantly covered by metagenomic reads in the sample (Methods). (Blue) Phages shown to integrate into bacterial genome in at least one of the 124 samples; (red) phages with no evidence for integration. (B) Virome profiles of individual samples. Phages are sorted separately in each sample (row) according to their abundance in that sample. (Brown) Highly abundant phages; (yellow) phages of intermediate abundance; (light blue) phages of low abundance. Coverage is measured in reads per kilobase per millions of reads (RPKM). (C) Discovery rate of new phages as a function of number of samples investigated. Samples were added one at a time and in each step spacers were identified. Phage contigs matching to the cumulative set of identified spacers were counted as detected. This analysis was repeated 10 times with random sample order; the charts depict the mean values obtained over the 10 iterations, and bars demarcate maximum and minimum values. (D) Evidence for presence of phages identified in this study (based on MetaHIT data) (Qin et al. 2010) in VLP data derived from American individuals (Reyes et al. 2010; Minot et al. 2011), and in gut metagenomic data derived from Japanese individuals (Kurokawa et al. 2007). (Bottom row) 991 phage contigs as bars sorted by prevalence in the MetaHIT population (heatmap). In each of the top three rows, a bar is colored if the phage is significantly covered by reads from the data set listed. The number of individuals in each data set is in parentheses.
Interestingly, 78% of the phages we identified were present in two or more individuals (Fig. 2A), and the median number of individuals in which a phage contig appeared was 5, indicating that sharing of phages within the sampled population was not uncommon. Moreover, 29% of phage contigs were shared among at least 10% of the sampled population, with a small minority (2.9%) found in half of the samples or more. Many of these most abundant phages appeared as prophages integrated into their host genomes (Fig. 2A, and see below). Reads mapping to phage regions of shared contigs showed an average range of 3.6% between the maximum and minimum sequence identity across samples where they appeared (Supplemental Text).
Nevertheless, no coherent clustering of individuals based on their profile of phage contig presence was observed (Supplemental Fig. S3; Supplemental Data Set S7; Methods). This was mostly due to a generally sporadic pattern of presence of rarer phages across individuals, with most similarity between any two profiles explained by those phages relatively widely shared in the population. Moreover, abundance of a phage could vary more than 100-fold between individuals where it was present (Supplemental Data Set S6).
To test whether gut microbiota phages might be shared between more geographically distant individuals, we attempted to map the raw VLP metagenomic sequencing reads generated from American individuals in the Reyes et al. (2010) and Minot et al. (2011) studies onto the genomic DNA of the 991 phages we identified in the European MetaHIT samples (Methods). We found 123 (12.4%) MetaHIT phage contigs that had significant coverage of VLP sequence data from the Reyes et al. study, and 162 (16.3%) MetaHIT contigs that had significant coverage from the Minot et al. study. Since these contigs were found to be covered by data derived from virus-like particles, they largely represent phages shown to become active, at least in human feces. To further query whether gut viromes can be shared between other geographically distant populations, we examined the metagenomic study of gut microbiota from 13 Japanese individuals (Kurokawa et al. 2007). Fifty (5%) of our phage contigs were significantly covered by metagenomic sequences collected from Japanese individuals. We note that these samples were sequenced at much lower depth, likely leading to significant underestimation of viral sequences in their gut metagenome (Supplemental Fig. S4; Supplemental Text).
Overall, 246 (24.8%) of the phages we identified in the MetaHIT European data were shared by at least one other data set. As may be expected, these phages also tended to be significantly more abundant within the MetaHIT population itself (Fig. 2D), with median prevalence of 16.5 individuals compared with a median prevalence of four individuals for phages not shown to exist in another data set (P < 10−15, Mann-Whitney U-test). Nevertheless, 30% of phages present in another data set appeared in five or less MetaHIT individuals (Supplemental Data Set S6), so that even phages that seldom appeared in the MetaHIT population were found in geographically distant individuals.
We next checked if multiple samples harbored CRISPR spacers targeting each of the 991 phage contigs we detected, regardless of whether the phage itself was detected in the sequencing data of the samples. Indeed, a majority of phages (72%) were targeted by spacers found in bacteria from multiple individuals, with the maximum number of individuals targeting a single phage reaching 43 (Supplemental Data Set S6). The observation that gut microbiota across individuals possess spacers targeting some of the same phages may reflect a combination of shared bacterial strain ancestry and ongoing pressure maintaining or shaping gut bacterial CRISPR arrays to respond to these shared phages.
Overall, our results point to the existence of a reservoir of phages frequently associated with the gut microbiome. We note, however, that the variable nature of the data sets examined—in terms of number of individuals sampled, sequencing depth, and phage detection methods—precludes a reliable quantitative estimate of the exact degree of sharing in the overall gut phage cohort.
We next attempted to assess the comprehensiveness of the phage set we identified in the MetaHIT data (Fig. 2C; Methods). Discovery rates of new phages using CRISPR spacer targeting did not reach saturation, indicating that additional phages are expected to be found with more individuals sampled. However, the curve shows that rate of discovery of new phages diminishes with addition of new samples. For example, the average number of new phages detected in each individual among the first 20 individuals sampled is 11.1, but the rate drops to 4.4 new phages/sample among the last 20 individuals.
Phage–bacteria infection–resistance interactions
To gain insight into the contribution of phages to the composition of gut microbial communities, it is first essential to understand which phage infects which bacteria. However, the nature of metagenomic studies, where complex communities of microorganisms are sequenced together, makes such phage–host assignment highly challenging (Tadmor et al. 2011). We set out to use CRISPR arrays as a connecting link enabling the identification of specific bacteria targeted by specific phages (Fig. 3A–C). It is well established that CRISPR arrays acquire spacers in a nonsymmetrical, unidirectional manner, so that new spacers are always inserted next to the CRISPR “leader” sequence. Thus, different isolates of the same bacterial strain are more likely to share some of the leader-distal spacers but to differ in their leader-proximal spacer content, depending on their most recent encounters with phages (Tyson and Banfield 2008). We were able to demonstrate that this principle holds true for gut bacteria based on the MetaHIT data set (Supplemental Table S3), suggesting that attack–resistance events between bacteria and phages can lead to adaptation of CRISPR arrays in the gut.
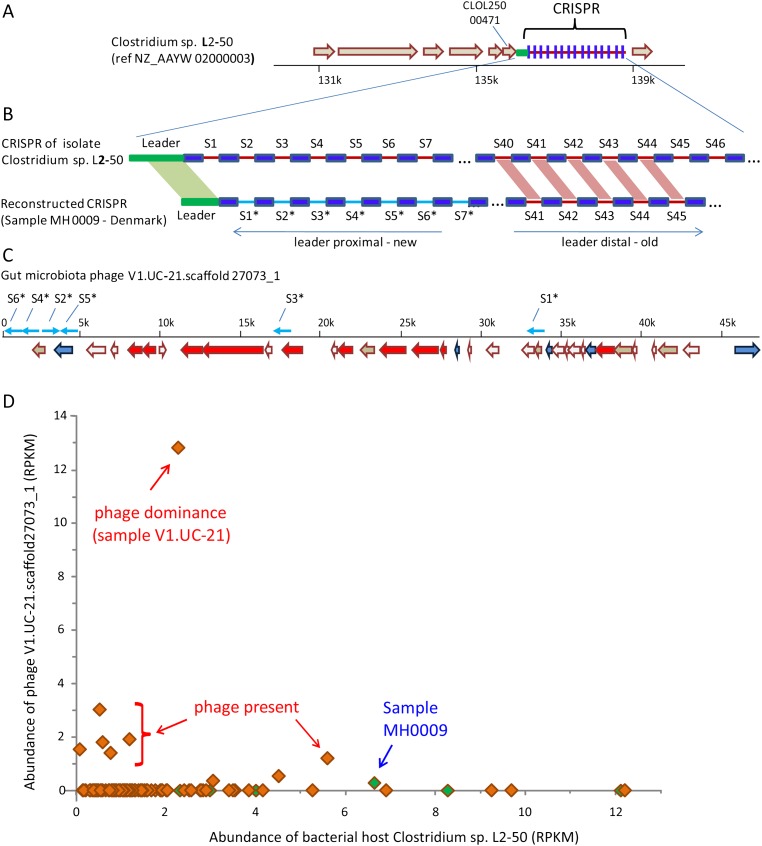
Figure 3.
Phage/host infection and resistance interactions. (A) The CRISPR region in the isolate gut bacterium Clostridium sp. L2-50, which was sequenced as part of the Human Microbiome Project (HMP) (Turnbaugh et al. 2007). Shown is a 10-kb region from the draft assembled genome. (Block arrows) Annotated genes. (B) A CRISPR array reconstructed from metagenomic sequence reads of sample MH0009 partially matches the Clostridium sp. L2-50 array. (Dark blue boxes) CRISPR repeats; (red and cyan lines) spacers. Spacers are numbered according to their position in the array relative to the leader sequence. Spacers show identity in sequence and in order at the leader-distal region (“old” spacers), while leader-proximal spacers (newly acquired) differ between the arrays. (C) Contig V1.UC-21.scaffold27073_1 was identified as a phage in this study, because it is hit by multiple spacers. (Block arrows) Genes with colors denoting function; (red) phage-specific genes; (blue) DNA replication genes; (white) genes of unknown function; (brown) genes of other functions. (Cyan arrows) Positions where spacers from the reconstructed array in panel B show identity with the phage sequence (spacer hits not drawn to scale). All drawn spacers fully match or have one mismatch with the phage sequence. (D) Abundance of bacterial host versus phage in MetaHIT samples. X- and y-axes represent abundance of the bacterial host and phage, respectively, measured in RPKM. Each data point represents a European individual sampled as part of the MetaHIT gut microbiota project (Qin et al. 2010). Green-filled samples are those in which our analysis found a spacer that matched the phage sequence. Sample MH0009, in which the CRISPR array in panel B was reconstructed, is identified.
We therefore used the spacer-containing metagenomic reads identified above to assemble 1160 sample-specific, partial CRISPR arrays of between six and 50 spacers long (Supplemental Data Set S8; Methods). We then searched for arrays that showed consecutive matching “old” spacers in a CRISPR array in one of 350 human microbiome isolate genomes that were fully sequenced (Fig. 3A,B; Turnbaugh et al. 2007). Such a match can assign CRISPR arrays to specific bacteria because the CRISPR region is highly variable among bacterial strains (Tyson and Banfield 2008). Thus, the presence of multiple shared “old” spacers ensures a recent common ancestor from which the fully sequenced isolate and the strain present in the metagenomic sample were derived. Although horizontal transfer of CRISPR arrays (Minot et al. 2011) could potentially result in similar spacer sharing, such an event is likely much rarer than simple spacer acquisition and is further discounted when the genomic coordinates of the array in the sequenced isolate and the strain present in the sample are identical (as in Fig. 3).
Spacers present in the CRISPR array are derived from DNA of phages that previously infected the host (Sorek et al. 2008). Spacers found in our bacteria-assigned arrays therefore match phages that target these same bacteria (Fig. 3B,C). Using this linkage between bacteria–CRISPR–phage, we were able to assign 31 phage contigs to 11 different bacterial hosts (Supplemental Data Set S6). Fourteen of these phages (45%) target species of Bacteroidetes and Parabacteroidetes, consistent with the documented abundance of these groups in the human gut (Eckburg et al. 2005). Additional species for which specific phages were identified, many of which belong to the gut-abundant Clostridia class, included Clostridium sp. L2-50, Dialister invisus, Ruminococcus sp. 5_1, Roseburia intestinalis, Bifidobacterium adolescentis, and Acidaminococcus sp. D21 (Supplemental Data Set S6).
To gain further understanding of the relationship between phage and host distribution in gut microbial communities, we next examined the abundance profiles of phages and their inferred bacterial hosts across the MetaHIT fecal samples from 124 individuals (Methods) (Fig. 3D). For most phages (66%), the distribution showed lower levels of phage as compared with its bacterial host across the majority of samples where the phage existed, with occasions of relative phage dominance in specific individuals, defined here as when phage abundance significantly exceeds that of its bacterial host (Fig. 3D). Such phage dominance over its host suggests that in those individuals the phage might be bursting at or near the time of sampling. However, the lack of time-point data in our study does not allow verification of this hypothesis.
We next examined the correlation between evidence for CRISPR resistance and phage prevalence among the 110 samples in which CRISPR spacers could be extracted. For most (84.8%) sample–phage pairs where the phage was targeted by a spacer, the sample did not harbor the same phage. This suggests the potential effectiveness of CRISPR in excluding targeted phages. Nevertheless, the non-negligible number of sample–phage pairs where the phage and a targeting spacer coexist could suggest an attack–resistance interaction at the time of sampling, or association of the phage with a sensitive host subpopulation while another subpopulation carries the immunity spacer.
We also found positive correlation between the existence of a spacer 100% identical to a phage sequence and the existence of that phage in the same gut sample. In 21.2% of cases where a sample had one or more spacers fully matching to a phage contig, the targeted phage was present in the sample (although usually at low levels), as compared with only 13% of cases where spacers with a lower match were found (P < 10−10, Chi-square for independence). Spacers matching the phage precisely suggest a more recent acquisition since not enough time has elapsed for accumulation of mismatches, particularly taking into account the typically fast pace of viral evolution. A possible interpretation of this result is therefore that these spacers were recently acquired in response to phage predation, and active phages still coexist with resistant bacteria in the same sample.
Lysogenic life style of microbiota phages
It was previously suggested that human gut viromes are dominated by temperate phages, capable of lysogenic life cycles (Breitbart et al. 2003; Reyes et al. 2010; Minot et al. 2011). Our data indeed show a significant subset of phages capable of a lysogenic lifestyle in the human gut: In 244 (24.6%) of the phage contigs we identified, an ORF with homology with an integrase or a recombinase was detected, indicative of ability for integration into a bacterial genome (Methods; Supplemental Data Set S4). To further identify evidence for a lysogenic life cycle, we looked for cases in which the assembled phage contigs also contained flanking sequences showing identity with one of 350 sequenced human microbiome genomes. For 135 phage contigs (13.6%), such evidence of prophage integration into a bacterial genome was found in at least one of the 124 individuals sampled (Fig. 1A; Supplemental Data Set S6). Taxonomically, the hosts of these prophages belonged primarily to the gut dominant phyla Bacteroidetes and Firmicutes, in congruence with previous observations (Supplemental Data Set S6; Reyes et al. 2010). In all cases in which the bacterial host of the phage was inferred both through a CRISPR array and through prophage integration, inferences were in close agreement (Supplemental Data Set S6).
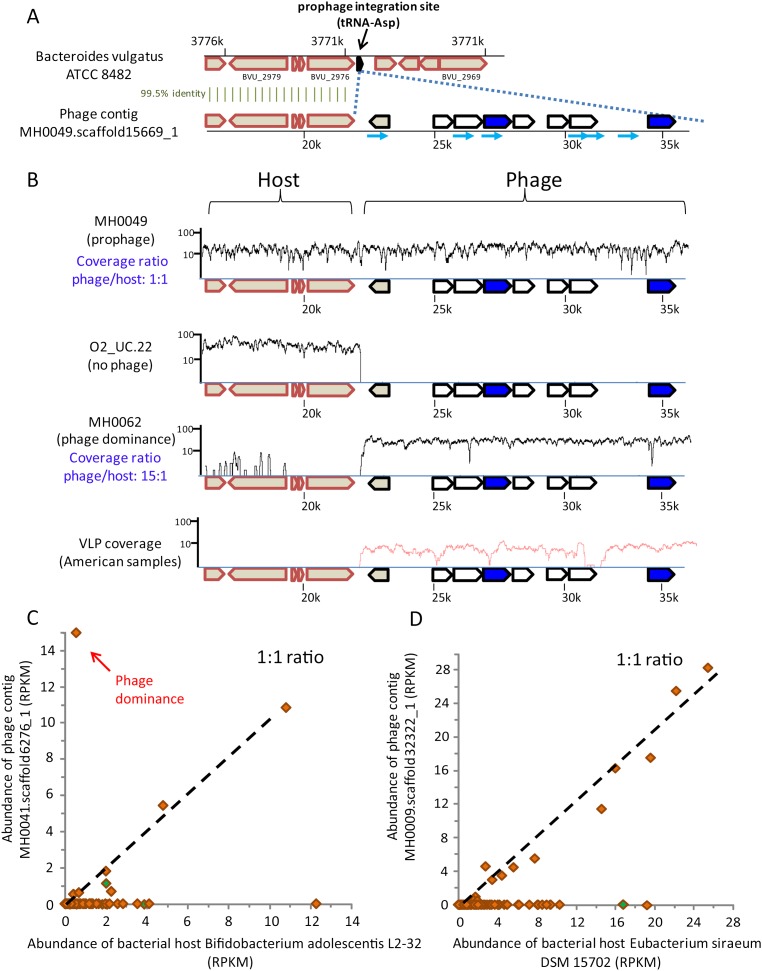
Figure 4, A and B, presents an example for such prophage integration. This example most probably represents a prophage that was assembled in the context of the genome in which it had integrated. The first 22 kb of phage contig MH0049.scaffold15669_1, which was assembled in the Danish sample MH0049, is 99.5% identical to a region in the sequenced genome of Bacteroides vulgatus ATCC 8482. The prophage integration site resides within a tRNA gene, a common site for prophage insertions (Fouts 2006). The coverage of this phage contig in sample MH0049 (Fig. 4B) indicates that in this individual, the phage was carried within the bacterial genome, and this is also supported by 12 metagenomic reads spanning the integration junction. In other individuals, the phage does not appear to be integrated: For example, in the Spanish sample O2_UC.22, the phage is missing, while coverage patterns in sample MH0062 suggest that the phage is dominant relative to its host and therefore might be bursting (Fig. 4B). While we cannot rule out the possibility that the latter pattern is the result of lysogeny in a different host, coverage of reads derived from VLPs sampled from American individuals (Reyes et al. 2010; Minot et al. 2011) supports that this phage is commonly active (Fig. 4B). Such VLP support for potential lytic activity of phages observed as integrated in host genomes was found for 35% of these 135 phages.
Figure 4.
Lysogenic lifestyles of gut microbiota phages. (A) The 5′ end of phage contig MH0049.scaffold15669_1, which was assembled in sample MH0049 (Danish individual), has a 99.5% identity in the sequenced genome of Bacteroides vulgatus ATCC 8482. (Block arrows) Genes; (cyan-colored arrows) spacers matching the phage contig. (B) Coverage of phage contig MH0049.scaffold15669_1 by MetaHIT metagenomic reads from three samples. (X-axis) Position on phage contig; (y-axis) read coverage (log scale). (Red curve) Coverage of VLP reads from Reyes et al. (2010). (C,D) Abundance of phages MH0041.scaffold6276_1 and MH0009.scaffold32322_1 and their respective bacterial hosts in MetaHIT samples, indicative of lysogeny as a preferred lifestyle. The x- and y-axes represent abundance of host and phage, respectively. Each data point represents a European individual sampled as part of the MetaHIT gut microbiota project (Qin et al. 2010). Green-colored samples are the ones in which a spacer matched the phage sequence. “Phage dominance” indicates samples where the phage is suspected to have become active. The correlation coefficient of phage and host abundances for samples where both existed is 0.4 and 0.98, respectively.
Some of the lysogenic phages in our data show significant preference to be associated with the genomes of their bacterial targets (Fig. 4 C,D). We detected 57 lysogenic phages whose abundance was in high correlation with the abundance of their host (correlation coefficient > 0.2; P < 0.01). Indeed, 28 of these phages (49%) also appeared as prophages in the genome of the fully sequenced bacterial isolate. Therefore, the abundance measured for such phages in many cases probably reflects the abundance of the host genome in which they are integrated. Nevertheless, rare instances of individuals with deviation from the correlation between phage and host abundance, such that phage abundance exceeds host abundance, suggest that these prophages can occasionally become predominantly lytic (Fig. 4C). Our results further strengthen previous observations (Reyes et al. 2010) that integrated, lysogenized prophages form a significant reservoir for phages active in the very distal human gut.
Discussion
We have used the CRISPR anti-phage immune system of bacteria to analyze a large metagenomic data set of 124 individuals and provide a wide perspective on the bacteriophages resident in the human gut. The large number of individuals sequenced to produce this data set and the ability of CRISPR spacers to reliably identify phage genomes across the analyzed population, including those not necessarily highly active in feces at the time of sampling, have allowed us to accumulate a significant, although not exhaustive, set of relevant phages. We then used this set as a benchmark to examine the question of phage sharing across individuals using various additional data sets, which have only been looked at separately before.
Our results suggest the existence of a non-negligible common reservoir of phages that is spread among unrelated individuals residing in distant geographical locations. While this appears to contrast with findings of previous studies (Reyes et al. 2010; Minot et al. 2011) that individual gut virome profiles are overall dissimilar, there is in fact no contradiction between the two observations. First, the larger number of samples and deeper sequencing depth of the MetaHIT data partially account for the differing results. Indeed, several VLP-derived contigs from those studies that were independently identified by us were not shared across individuals in the original data set but appeared in a substantial share of the MetaHIT samples (Supplemental Table S4). More interestingly, while VLPs represent active phages (at least in feces), our study could identify sharing of phages not necessarily active at the time of sampling, possibly due to lysogenization. These apparently dormant phages can become activated in specific individuals under particular circumstances, which may explain the low sharing reported on phage contigs assembled in the fecal VLP sequencing studies (Supplemental Table S4).
Based on the diminishing slope found in our rate-of-discovery analysis (Fig. 2C), we propose that there may be a relatively limited pool of common phages present in many individuals along with a much larger set of rarer phages that will require significantly more sequencing to reach saturation. However, we note that it is difficult to compare these results with predictions on global phage diversity (Rohwer 2003) since our method can only detect phages targeted by the bacterial CRISPR immune system and is tilted toward discovery of phages associated with the more abundant gut bacterial species. Furthermore, our method excludes both smaller-sized phage genomes, such as Microviridae whose presence in the human gut has been repeatedly demonstrated (Breitbart et al. 2003; Krupovic and Forterre 2011; Minot et al. 2011), and RNA phages, which are absent from the DNA-based MetaHIT data. The latter concern is mitigated by the finding that the majority of RNA viruses in the human gut are not bacteriophages (Zhang et al. 2006).
While previous studies on the gut microbiome have identified common clusters of bacterial species termed enterotypes (Arumugam et al. 2011), our study did not show a similar trend for human gut phages (Supplemental Fig. S2; Methods). This incongruence may be explained by the greater diversity of phages, the much more limited resolution of taxonomic classification of phages, and the partial nature of the phage data set studied here.
Our findings also imply that CRISPR spacers are actively acquired in response to phages in the human gut. First, the large number of unique phage-matching spacers we detected is suggestive of the numerous events in which such interactions have occurred. Second, we have identified cases in which the most recently acquired spacers in the rapidly evolving CRISPR array perfectly match a phage co-occurring in the same gut, at apparently depressed levels (e.g., Fig. 3), as well as absolutely no conservation of the most recently acquired spacer in a particular array across individuals (Supplemental Table S3). Finally, our data suggest that CRISPR spacers usually successfully exclude targeted phages, while showing positive correlation of highly matching spacers (thus also likely to be recently acquired) with targeted phages about to be completely excluded. Combined, this evidence suggests that there is an ongoing “immune” interaction between phages and their bacterial hosts within the gut microbial community, whose potential effect remains to be fully characterized. It is possible that the rapid nature of this dynamic is what has made it so difficult to capture in previous studies. Notably, although the typical ecological manifestations of predatory interactions were observed in numerous niches, past studies did not report on such dynamics in the gut, and this study was not suited to examine it directly due to the lack of time-series data.
It is also not entirely clear how to reconcile phage–bacteria infection–resistance interactions through the CRISPR system with the apparent abundance of lysogenic phages in the gut, which this study also supports. This suggests there may be some form of protection of the bacterial genome (harboring the prophage) from the CRISPR system (Stern et al. 2010). A recent study (Edgar and Qimron 2010) has shown that acquisition of spacers against a lysogenized phage can lead to bacterial cell death but may also prevent prophage induction and subsequent cell lysis to the benefit of bacteria under certain circumstances. Spacers against a phage not yet integrated were shown to prevent lysogenization (Edgar and Qimron 2010). This study and our findings suggest the CRISPR system may have important roles in regulating phage–bacteria interactions even when they are not primarily lytic.
It is clear, therefore, that further study is warranted to unravel the seemingly unique ecology of the human gut microbiome in this respect. Our identification of the gut bacterial targets for more than 130 phages, based on evidence for integration into the bacterial host genome (Fig. 4) or on CRISPR-derived phage–host assignment (Fig. 3), now opens the window for future, detailed time-course studies on the dynamic effect of phages on individual bacterial species and on the gut microbial consortium as a whole.
Methods
Inference of CRISPR spacers from short sequencing reads
The PILER-CR CRISPR inference program (Edgar 2007) was run on 1144 sequenced bacterial genomes and 392 sequenced reference genomes from the human gut, and the consensus repeat was extracted from all arrays inferred. All repeat sequences were queried using BLASTN against MetaHIT (Qin et al. 2010) sequencing reads of length 75 bp. Spacers were inferred when there were two BLAST hits with an e-value threshold of 0.01 at both ends of a read, or there was a BLAST hit on one end of the read and an at least 6 bp identical to the repeat (which we dubbed an “anchor”) on the opposite end. A spacer was inferred between such repeat matches if its size was 20 bp or more. Spacers in each individual with at least 90% identity along at least 90% of their length were clustered together.
From spacers to mobile genetic element (MGE) contigs
Inferred spacers were used to query all assembled MetaHIT contigs using BLASTN with an e-value threshold of 10−4. Contig hits underwent a filtering procedure to remove those that resembled CRISPR arrays. To obtain a nonredundant set, contigs <50 kb were clustered using uclust (Edgar 2010) requiring 80% identity over at least 80% of the contig length. Contigs >50 kb were clustered using BLASTN with the same parameters. The longest member of each cluster was selected as a representative for downstream analyses.
Identification of phage contigs
Gene prediction using Glimmer3.02 (Salzberg et al. 1998) was performed on the nonredundant MGE contigs. Predicted genes were then searched against the Conserved Domains Database (CDD) (Marchler-Bauer et al. 2011) using rpsblast with e-value threshold 0.05. An MGE contig was deemed a phage contig if its size was 10 kb or more and the CDD description of at least one of its annotated genes contained the word “phage,” “holin,” or “tail.” Forty-six contigs with an abundance of nonphage genes were discarded. Additional phage contigs were identified if >40% of their sequence was covered by VLP reads from Reyes et al. (2010) and Minot et al. (2011) with the parameters detailed below.
Taxonomic classification of phage contigs
All viral proteins were downloaded from NCBI (July 2011). BLASTP with an e-value threshold of 10−5 was performed between all proteins annotated in the phage contigs and the viral phage proteins, with only the best match taken per phage contig protein. Hits to viral proteins from the genome of a member of an ICTV-defined family were used to classify a contig as belonging to that viral family. Ambiguous classification was called if two or more genes in a phage contig were homologous to proteins of different viral families.
Enrichment of gene functions in phage contigs
COG functions for all COG-annotated genes in HMP GI-tract-related bacteria were downloaded from the IMG/HMP site (http://www.hmpdacc-resources.org/cgi-bin/imgm_hmp/main.cgi) in December 2011. Frequency of each COG function in the bacterial set was compared with the frequency of the same function among all COG-annotated genes found in the pooled set of 991 phages. Only COGs appearing more than 10 times in the phage set were considered.
Abundance of phages across samples
Coverage calculations were limited to the window encompassing the region defined by the spacer hits (proto-spacers) and phage-associated genes. This was in order to make sure only phage abundance was measured if it was integrated in a bacterial genome. Metagenomic reads from each sample were mapped to phage contigs using BLASTN, requiring at least 80% of the read to align against the contig with at least 85% identity. To declare a phage contig exists in a particular sample, the phage region of the contig had to be covered by at least one read per base pair and at least 70% of the phage-region bases had to be covered (if the phage region was <2 kb, it was extended by 2 kb upstream and downstream). See the Supplemental Text for discussion on the parameters chosen for the sharing analysis. Coverage is reported in reads per kilobase per millions of reads (RPKM). Visual data on phage abundance across samples are found at http://www.weizmann.ac.il/molgen/Sorek/microbiome_phages/.
Similarity of phage existence profiles
Phage existence profiles for pairs of samples were compared using binary asymmetric distance: the proportion of phages that exist in only one sample out of all phages that exist in at least one of the samples in the pair.
Rate-of-discovery analysis
MetaHIT samples were added one at a time. After each sample was added, a tally was made of all unique phage contigs in the accumulated samples that were detectable using the spacers found in those accumulated samples. The analysis was repeated 10 times with random sample order (Fisher-Yates shuffling). Fourteen of the samples could not contribute spacers due to shorter sequencing reads.
Distribution of phage contigs in other data sets
Reads sequenced in all individuals and time points in each study (Kurokawa et al. 2007; Reyes et al. 2010; Minot et al. 2011) were used to form a study pool. Each pool was mapped to MetaHIT phage contigs with BLASTN using a cutoff of at least 85% identity over at least 85% of the read length. A phage contig was determined to exist in a data set if the phage region (as previously defined) was covered by at least one read per base pair, and at least 70% of bases in the region were covered. For the VLP data sets, contigs that failed the above test were subjected to a second test with the same set of parameters across the entire length of the contig since the location of read mapping in this case did not need to be strictly controlled.
Abundance of HMP bacteria
HMP genomes deposited in IMG in March 2011 were queried for COG functions corresponding to 31 universal single-copy genes (Ciccarelli et al. 2006). BLASTN with -F F flag and an e-value threshold of 0.00005 was performed using all reads from each sample against the universal single copy genes from organisms for which at least 25 of the 31 gene sequences were available. Only the best BLAST hit was taken for each read and only if at least 80% of the read aligned against a bacterial gene with at least 85% identity. Coverage was measured in RPKM.
Assembly of CRISPR arrays
All reads that matched the set of known CRISPR repeats using BLASTN with an e-value of 0.01 were gathered from each of the 110 individuals in the MetaHIT data that had 75-bp read sequences. The QSRA short-read assembler (Bryant et al. 2009) was then used to assemble these reads, in each individual separately. To facilitate assembly of repetitive CRISPR loci, reads were considered for extension of an assembled array only if they matched at least the last 60 bases of the growing contig (option -u), or, if not enough of these existed, at least the last 50 bases (option -l).
Evidence for prophage integration
BLASTN with default parameters was used to align phage contigs onto the set of HMP genomes. Prophage integration was determined if at least 1000 bases of the phage contig were aligned to the bacterial genome with at least 95% identity, and the alignment was localized to one or both ends of the phage contig.
Acknowledgments
We thank Alejandro Reyes and Samuel Minot for their gracious help in providing supplementary VLP sequencing data. We also thank Debbie Lindell, Eyal Weinstock, Dvir Dahary, Eytan Ruppin, Hila Sberro-Livnat, Asaf Levy, Omri Wurtzel, Gil Amitai, and Shany Doron for comments on the manuscript. R.S. was supported, in part, by the ERC-StG program (grant 260432), the Leona M. and Harry B. Helmsley Charitable Trust, and by a DIP grant from the Deutsche Forschungsgemeinschaft. A.S. was the beneficiary of a postdoctoral grant from the AXA research fund. E.M. was supported, in part, by a fellowship from the Edmond J. Safra Bioinformatics Program at Tel Aviv University. I.T. was supported by the Clore Center at the Weizmann Institute of Science.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138297.112.
References
- Andersson AF, Banfield JF 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050 [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. 2011. Enterotypes of the human gut microbiome. Nature 473: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D 2011. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature 474: 604–608 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F 2003. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185: 6220–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DW Jr, Wong WK, Mockler TC 2009. QSRA—a quality-value guided de novo short read assembler. BMC Bioinformatics 10: 69 doi: 10.1186/1471-2105-10-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H 2004. Phage–host interaction: An ecological perspective. J Bacteriol 186: 3677–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA 2007. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA 2005. Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2007. PILER-CR: Fast and accurate identification of CRISPR repeats. BMC Bioinformatics 8: 18 doi: 10.1186/1471-2105-8-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461 [DOI] [PubMed] [Google Scholar]
- Edgar R, Qimron U 2010. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J Bacteriol 192: 6291–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE 2006. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34: 5839–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI 2002. How host–microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22: 283–307 [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ 2010. The CRISPR system: Small RNA-guided defense in bacteria and archaea. Mol Cell 37: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Forterre P 2011. Microviridae goes temperate: Microvirus-related proviruses reside in the genomes of Bacteroidetes. PLoS ONE 6: e19893 doi: 10.1371/journal.pone.0019893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14: 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letarov A, Kulikov E 2009. The bacteriophages in human- and animal body-associated microbial communities. J Appl Microbiol 107: 1–13 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. 2011. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD 2011. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res 21: 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Sun CL, Salzman J, Rao N, Loomer P, Armitage GC, Banfield JF, Relman DA 2011. Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res 21: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F 2003. Global phage diversity. Cell 113: 141. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Delcher AL, Kasif S, White O 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res 26: 544–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JC, Bateson MM, Lavin M, Young MJ 2010. Use of cellular CRISPR (clusters of regularly interspaced short palindromic repeats) spacer-based microarrays for detection of viruses in environmental samples. Appl Environ Microbiol 76: 7251–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6: 181–186 [DOI] [PubMed] [Google Scholar]
- Stern A, Sorek R 2011. The phage–host arms race: Shaping the evolution of microbes. Bioessays 33: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Keren L, Wurtzel O, Amitai G, Sorek R 2010. Self-targeting by CRISPR: Gene regulation or autoimmunity? Trends Genet 26: 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R 2011. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science 333: 58–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI 2007. The human microbiome project. Nature 449: 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol 10: 200–207 [DOI] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34: 401–407 [DOI] [PubMed] [Google Scholar]
- Walter J, Ley RE 2011. The human gut microbiome: Ecology and recent evolutionary changes. Annu Rev Microbiol. 65: 411–429 [DOI] [PubMed] [Google Scholar]
- Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK 2010. Cryptic prophages help bacteria cope with adverse environments. Nat Commun 1: 147 doi: 10.1038/ncomms1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, Hibberd ML, Liu ET, Rohwer F, Ruan Y 2006. RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biol 4: e3 doi: 10.1371/journal.pbio.0040003 [DOI] [PMC free article] [PubMed] [Google Scholar]