Abstract
The use of tyrosine kinase inhibitors (TKI) has yielded great success in treatment of lung adenocarcinomas. However, patients who develop resistance to TKI treatment often acquire a somatic resistance mutation (T790M) located in the catalytic cleft of the epidermal growth factor receptor (EGFR) enzyme. Recently, a report describing EGFR-T790M as a germ-line mutation suggested that this mutation may be associated with inherited susceptibility to lung cancer. Contrary to previous reports, our analysis indicates that the T790M mutation confers increased Y992 and Y1068 phosphorylation levels. In a human bronchial epithelial cell line, overexpression of EGFR-T790M displayed a growth advantage over wild-type (WT) EGFR. We also screened 237 lung cancer family probands, in addition to 45 bronchoalveolar tumors, and found that none of them contained the EGFR-T790M mutation. Our observations show that EGFR-T790M provides a proliferative advantage with respect to WT EGFR and suggest that the enhanced kinase activity of this mutant is the basis for rare cases of inherited susceptibility to lung cancer.
Introduction
Recent work has identified a series of somatic mutations in exons 18 to 21 of epidermal growth factor (EGF) receptor (EGFR) that render lung tumors responsive to the gefitinib and erlotinib therapeutics (1–3). However, in patients that progress after drug treatment, it has been observed that a secondary “resistance” mutation is often acquired in exon 20 (4–6). This mutation, T790M, arises somatically in ~50% of these cases (4–6). In chronic myelogenous leukemia (CML) patients, an estimated 50% to 90% of tumors with acquired resistance have the analogous resistance mutation (T315I) in BCR-ABL (7). Interestingly, Bell et al. (8) report the first identification of the T790M mutation in the germ line of a European family that developed lung adenocarcinoma with bronchoalveolar (BAC) differentiation. This observation suggests that this mutation may be associated with genetic susceptibility to lung cancer and may underlie familial predisposition to the disease. The allele, although common in drug-treated tumors, seems extremely rare in the germ line of the general population, as the authors report no mutation observed in 782 alleles sequenced. Many groups have reported that the kinase activity of the EGFR-T790M–resistant mutant is indistinguishable from wild-type (WT) EGFR (4–6, 9, 10). What is somewhat perplexing is how T790M would confer susceptibility if its activity were identical to the WT molecule. Our analysis shows that T790M in fact does exhibit enhanced autophosphorylation at Y992 and Y1068, and this mutation is associated with a proliferative advantage in a human bronchial epithelial cell line. Interestingly, the mutation seems to be rare as it was not found in any of the familial or sporadic lung cancer populations we screened.
Materials and Methods
Cell lines
HEK293T, HEK293FT, and COS-7 cells were maintained in DMEM supplemented with 10% fetal bovine serum. Immortalized human bronchial epithelial cells (HBEC3-KT) were maintained in keratinocyte serum-free medium (with 50 μg/mL bovine pituitary extract and 5 ng/mL EGF). All cells were grown at 37°C in a humidified incubator with 5% CO2.
Plasmids, transfection, and viral infection
Mammalian expression plasmids encoding for human EGFR, EGFR-T790M, and EGFR-L858R were kind gifts from William Pao (Memorial Sloan-Kettering Cancer Center, New York, NY). HEK293T and COS-7 cells were transfected with the indicated EGFR plasmids using LipofectAMINE 2000 (Invitrogen). Gefitinib was obtained from Chemoprevention Branch, National Cancer Institute (Bethesda, MD) and added to cells 10 h before lysis.
To introduce WT and mutant EGFRs into HBEC3 cells, we used the pLenti6/V5 Directional TOPO Cloning kit (Invitrogen). Construction of pLenti-wt-EGFR was described previously (11). The T790M and L747_E749 deletion mutations were introduced into pLenti-wt-EGFR by using site-directed mutagenesis (Stratagene). pLenti-KRASV12 vector was constructed by cloning KRASV12 fragment from pBabe-KRASV12-hyg (11) into pLenti6/V5 vector. pLenti6/V5-GW/lacZ (Invitrogen) was used as a control. Viral transduction of HBEC3 cells was done following the manufacturer’s instructions. Briefly, the 293FT cells were transiently transfected with viral vector together with ViraPower (Invitrogen). Forty-eight hours after transfection, the supernatant of the 293FT cells was harvested and passed through a 0.45-μm filter and frozen at −80°C. The supernatant was used for infection after addition of 4 μg/mL polybrene (Sigma). Forty-eight hours after infection, drug selection of infected cells was started with 5 μg/mL blasticidin (Invitrogen) and continued for 7 days.
Colony formation assays
Liquid colony formation assays were done as described previously (11). Briefly, 200 viable cells were plated in triplicate on 100-mm plates and cultured in keratinocyte serum-free medium supplemented with 50 μg/mL of bovine pituitary extract without EGF. Surviving colonies were counted 10 days later after staining with methylene blue, and colonies >3 mm in diameter were counted.
Western blot and antibodies
Cell lysates were prepared in LDS sample buffer (Invitrogen) and electrophoresed on NuPAGE gels (Invitrogen). Protein was transferred to polyvinylidene difluoride membranes and blotted using the antibodies as indicated: anti-EGFR (Cell Signaling Technology), anti–EGFR-Y1068 (Cell Signaling Technology), anti–EGFR-Y992 (Cell Signaling Technology), anti–mitogen-activated protein kinase (MAPK; Cell Signaling Technology), anti–phosphorylated MAPK (Thr202/Tyr204; Cell Signaling Technology), anti–phosphorylated AKT (Ser473; Cell Signaling Technology), anti-AKT (Cell Signaling Technology), anti–α-tubulin (Santa Cruz Biotechnology), and anti–cyclin D1 (Santa Cruz Biotechnology).
EGFR exon 20 genotyping
Amplification of human EGFR exon 20 was done via standard PCR methods using forward (5′-GACACTGACG-TGCCTCTCC-3′) and reverse (5′-TTATCTCCCCTCCCCGTATC-3′) primers. PCR products were electrophoresed on agarose gels, purified, and subjected to standard DNA sequencing. The EGFR-T790M mutation is deduced by the genotype (C/T) at position 86 in exon 20.
Lung cancer DNAs
The Genetic Epidemiology of Lung Cancer Consortium (GELCC) has accrued over 700 families with three or more first-degree relatives with lung cancer (12), of which this study genotyped 237 individual probands. Paraffin blocks of lung tumors (45) with BAC differentiation (and 32 corresponding normals), in addition to the fresh frozen lung tumor DNAs, were obtained from the Washington University Tissue Procurement Center as paraffin blocks. For the blocks, genomic DNA was prepared by slicing several paraffin curls and adding xylenes to dissolve the paraffin. The remaining tissue was washed with ethanol, dried, and resuspended in PCR buffer (0.5% Tween 20, 0.5% NP40) and digested in proteinase K (1 mg/mL) at 55°C overnight. Chloroform/isoamyl alcohol (24:1) was added to the digest and soluble DNA was isolated from the aqueous phase.
Results
To address previous observations that EGFR-T790M activity is the same as the WT molecule, we did transient transfections of EGFR and EGFR-T790M expression constructs in COS-7 and 293T cells. Overexpression of EGFR and the T790M mutant in these cell lines clearly indicated that the phosphorylation status at Y1068 is enhanced with respect to WT (Fig. 1A–E). Tyr1068 is an EGFR autophosphorylation site that couples receptor activation to Ras signaling (13). Interestingly, our observations are in disagreement with previously published reports (4–6, 9, 10). Our data also show that phosphorylation levels between WT and T790M are indistinguishable at higher expression levels (Fig. 1C), yet differences are more apparent at lower expression levels. As a control, we also showed the drug-resistant nature of EGFR-T790M in comparison with WT EGFR (Fig. 1D). We further addressed the activation status of MAPK, a downstream molecule phosphorylated and activated by EGFR. We observed that phosphorylated MAPK (Thr202/Tyr204) levels were enhanced in the T790M mutant compared with WT EGFR (Fig. 1E).
Figure 1.
Phosphorylation of Y1068 is enhanced in EGFR-T790M. HEK293 cells (A) and COS-7 cells (B) were transfected with the indicated DNAs and lysed 48 h after transfection. Lysates were immunoblotted with anti-EGFR and anti–EGFR-Y1068 as indicated. Phosphorylation of Y1068 is enhanced in the EGFR-T790M mutant. C, phosphorylation and activation of EGFR-T790M are evident at lower levels of protein expression compared with higher levels in COS-7 cells (lane 3 versus lane 5). D, gefitinib does not affect the phosphorylation status of T790M-EGFR. E, phosphorylation of MAPK is enhanced in EGFR-T790M–expressing (versus EGFR-WT expressing) cells. Cells were stimulated with EGF (10 ng/mL) for 5 min.
We also tested whether EGFR-T790M expression in an immortalized human bronchial epithelial cell (HBEC3) line affected growth properties of these cells. Cells were transduced by lentivirus expressing EGFR and EGFR mutants and selected for stable integration as done previously (11). Two hundred cells were seeded on plates without EGF supplementation and allowed to grow for 10 days. We observed that the EGFR-T790M–expressing cells had a greater number of large (≥3 mm in diameter) colonies compared with WT EGFR but less than the activated L747-E749 deletion (Fig. 2A and B). These results indicate that EGFR-T790M does in fact provide a growth advantage over WT EGFR. When we analyzed the status of EGFR phosphorylation at Y992 and Y1068 in these cells, we saw an increase in the EGFR-T790M mutant (Fig. 2C), consistent with what was observed in HEK293T and COS-7 cells (Fig. 1A–D).
Figure 2.
Colony formation growth assay for EGFR-T790M. Viral transduction and selection of HBEC cells stably expressing EGFR were done as described previously (11). A, cells (200) were plated in triplicate and grown in keratinocyte serum-free medium (with 50 μg/mL bovine pituitary extract and without EGF). After 10 d, cells were stained with methylene blue. B, stained cell colonies >3 mm in diameter were counted. *, P < 0.0001, two-tailed t test. C, Western blots for EGFR, EGFR-Y1068, EGFR-Y992, MAPK, MAPK (T202/Y204), AKT, AKT (S473), cyclin D1, and α-tubulin in the HBEC stable cell lines were done as indicated.
To address possible downstream signaling mechanisms that are affected by mutant EGFR, we looked at the phosphorylation/activation status of MAPK and AKT (Fig. 2C). In contrast to our observations in 293T cells, we did not see MAPK activation induced by T790M. However, this is consistent with previously published results where no obvious changes are seen with the deletion mutant, yet colony formation is enhanced (11). Transcriptional changes, such as up-regulation of cyclin D1 levels, were recently reported to be a result of EGFR mutant signaling (14); however, we observed no changes induced by the mutants (Fig. 2C).
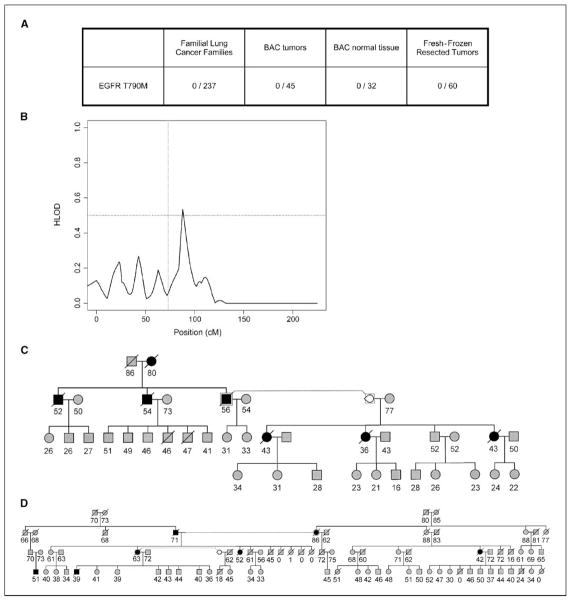
If EGFR-T790M is a human susceptibility allele, it is necessary to determine its prevalence in families with high susceptibility to lung cancer. With these families, a set of 52 extended pedigrees was used to show linkage on chromosome 6q23-25 (12). A concerted effort by the GELCC has attempted to identify the gene(s) associated with this susceptibility. We proposed that if the T790M mutation is a common variant for predisposition to lung cancer, it could potentially be enriched in probands from lung cancer families. Genomic DNA from 237 probands representing lung cancer families with more than three affected individuals was analyzed. PCR amplification and DNA sequencing of the resistance mutation in exon 20 were done. Interestingly, we did not observe the T790M mutation in any of the family probands analyzed or in any of 60 random fresh-frozen resected lung tumors (Fig. 3A). This would suggest that the T790M mutation is not enriched in our population and is likely a minor contributor to genetic susceptibility in familial lung cancers. Furthermore, in the aforementioned GELCC linkage study, the analysis of 52 families (and family subsets within) did not reveal a significant logarithm of odds score on or near 7p11, where the EGFR gene is located, which also suggests that the mutation is not a major contributor to familial predisposition to lung cancer (Fig. 3B; ref. 12). Representative families used in this study are indicated in Fig. 3C and D. We further tested to see if T790M might in fact be solely responsible for predisposition to adenocarcinomas with BAC differentiation. We sequenced 45 BAC tumors and 32 of the corresponding normal tissues and did not observe the T790M mutation in any of these samples (Fig. 3A). This further suggests the rarity of this potential predisposing mutation.
Figure 3.
EGFR genotyping and linkage analysis for chromosome 7 with SimWalk2. A, the EGFR-T790M mutation was not observed in familial lung cancer blood DNA, BAC tumors and normals, or fresh-frozen resected tumors. B, the genetic position for EGFR is at ~73 cM (based on interpolation), between markers D7S1818 (70 cM) and D7S3046 (79 cM). Simwalk2 analysis was based on the parametric method using the genetic model reported by Moscatello et al. (18). Fifty-five pedigrees with five or more affected members were used. C and D, two representative pedigrees of GELCC collected families with predisposition to lung cancer. In (C) and (D), ●. (females) or ■ (males) represent lung, throat, or laryngeal cancer. Numbers below each individuals, sample numbers; numbers in brackets, individual ages at the time the pedigree was constructed; slashes, dead individuals.
Discussion
Several lines of evidence exist that lend support to T790M as a putative susceptibility allele. T790M has been detected somatically in tumors that have not been treated with gefitinib or erlotinib, which may suggest that this mutant may have arisen during drug-free cancer progression (15). Furthermore, the NCI-H1975 BAC cell line, which has never undergone tyrosine kinase inhibitor treatment, has both activating L858R and resistant T790M mutations, suggestive that T790M may be growth promoting (5). Bell et al. (8) also have observed that the T790M mutation seems to occur in cis with the activating mutations. Perhaps more persuasive is that the analogous resistance mutation in CML patients, BCR-ABL-T315I, displays increased in vitro kinase activity (16). Similarly, the analogous mutations in Src (T341M) and FGFR1 (V561M) also result in increased phosphorylation and activation (10). Why T790M in EGFR does not reportedly function in a similar manner is unclear.
Our results suggest that the T790M mutation may in fact provide a proliferative advantage in normal cells by increasing kinase activity and downstream signaling. Our data suggest that EGFR-T790M does in fact exhibit higher kinase activity than WT molecule in HEK293T and COS-7 cell lines. Overexpression in our HBEC3 cell line showed increased tyrosine phosphorylation at Y992 and Y1068 and increased colony formation. Increased proliferation is most evident in the absence of EGF in the medium, suggesting that the activating T790M EGFR mutation is involved in the proliferative effects observed. EGFR kinase activity is essential for oncogenic transformation. EGFR extracellular domain deletion mutants and overexpression of EGFR are commonly found in human cancers (17, 18). Our data also show that phosphorylation levels between WT and T790M are indistinguishable at higher expression levels (Fig. 1C), yet differences are more apparent at lower expression levels. We believe this may be an explanation why others have seen no difference between EGFR and T790M.
EGFR signaling activates many pathways that lead to proliferative advantages. The Ras/MAPK pathway is activated via ligation of Grb2 to an activated EGFR molecule. In our 293T overexpression system, we observed that MAPK activity is increased by T790M versus WT EGFR. However, a significant effect of T790M on these downstream kinases was not evident in our HBEC3 system, which might suggest that other signaling/transcriptional events are responsible for the proliferative changes. Recent work has revealed transcriptional changes in cyclin D1, may be a key response to gefitinib resistance by T790M and susceptibility to the irreversible inhibitor CL-387,785 (14). We did not observe changes in cyclin D1 levels in the HBEC3 system and believe other, yet unidentified, mechanisms exist to account for the proliferative advantages caused by mutant EGFR.
A recent study, using a mutant-enriched PCR system, revealed the presence of the T790M mutation as a minor clone in non–small cell lung cancer tumors (19). It is suggested that these clones are selected for during gefitinib treatment and are enriched in the resistant tumor but do not provide the main proliferative function for oncogenesis. Our observations suggest that T790M as a germ-line mutation (i.e., in all cells) may provide the mild proliferative push for lung cancer development.
Many lines of evidence suggest the existence of a limited number of genetic factors that control susceptibility to lung cancer (20–22). However, due to a high-case fatality rate (5-year survival rate of 15%), obtaining biospecimen samples for DNA analysis is particularly difficult (23). A collaborative effort of GELCC has accrued DNA from families with lung, throat, and laryngeal cancers since the early 1990s. At present, 771 families with three or more first-degree relatives affected with lung cancer have been collected, of which 11% have sufficient family-wide biospecimen availability for any future studies. Subsequent sequencing of 237 families with predisposition to lung cancer, 45 BAC tumors and 60 fresh-frozen resected tumors, did not reveal any mutations, suggesting that T790M is likely a rare mutation. Nevertheless, our data provide a basis for EGFR-T790M as a rare susceptibility allele in human lung cancer.
Acknowledgments
Grant support: NIH grants U01CA76293 (Genetic Epidemiology of Lung Cancer Consortium), R01CA058554, R01CA093643, R01CA099147, R01CA099187, R01ES012063, R01ES013340, R03CA77118, R01CA80127, P30ES06096, P50CA70907 (Specialized Program of Research Excellence), and N01HG65404 and Department of Defense VITAL grant. This study was supported in part by NIH, the Intramural Research Programs of National Cancer Institute, and National Human Genome Research Institute.
We thank W. Pao (Memorial Sloan-Kettering Cancer Center, New York, NY) for providing mammalian expression plasmids encoding for human EGFR, EGFR-T790M, and EGFR-L858R; R. Lubet (Chemoprevention Branch, National Cancer Institute, Bethesda, MD) for providing gefinitib; and J. Clark, Q. Chen, and M. Watson for their assistance in various aspects of this work.
References
- 1.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–70. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–53. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 8.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–6. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 9.Blencke S, Ullrich A, Daub H. Mutation of threonine 766 in the epidermal growth factor receptor reveals a hotspot for resistance formation against selective tyrosine kinase inhibitors. J Biol Chem. 2003;278:15435–40. doi: 10.1074/jbc.M211158200. [DOI] [PubMed] [Google Scholar]
- 10.Blencke S, Zech B, Engkvist O, et al. Characterization of a conserved structural determinant controlling protein kinase sensitivity to selective inhibitors. Chem Biol. 2004;11:691–701. doi: 10.1016/j.chembiol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 11.Sato M, Vaughan MB, Girard L, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–28. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 12.Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am J Hum Genet. 2004;75:460–74. doi: 10.1086/423857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojas M, Yao S, Lin YZ. Controlling epidermal growth factor (EGF)-stimulated Ras activation in intact cells by a cell-permeable peptide mimicking phosphorylated EGF receptor. J Biol Chem. 1996;271:27456–61. doi: 10.1074/jbc.271.44.27456. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi S, Shimamura T, Monti S, et al. Transcriptional profiling identifies cyclin D1 as a critical downstream effector of mutant epidermal growth factor receptor signaling. Cancer Res. 2006;66:11389–98. doi: 10.1158/0008-5472.CAN-06-2318. [DOI] [PubMed] [Google Scholar]
- 15.Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–23. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto M, Kurosu T, Kakihana K, Mizuchi D, Miura O. The two major imatinib resistance mutations E255K and T315I enhance the activity of BCR/ABL fusion kinase. Biochem Biophys Res Commun. 2004;319:1272–5. doi: 10.1016/j.bbrc.2004.05.113. [DOI] [PubMed] [Google Scholar]
- 17.Garcia de Palazzo IE, Adams GP, Sundareshan P, et al. Expression of mutated epidermal growth factor receptor by non-small cell lung carcinomas. Cancer Res. 1993;53:3217–20. [PubMed] [Google Scholar]
- 18.Moscatello DK, Holgado-Madruga M, Godwin AK, et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55:5536–9. [PubMed] [Google Scholar]
- 19.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–8. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 20.Sellers TA, Bailey-Wilson JE, Elston RC, et al. Evidence for mendelian inheritance in the pathogenesis of lung cancer. J Natl Cancer Inst. 1990;82:1272–9. doi: 10.1093/jnci/82.15.1272. [DOI] [PubMed] [Google Scholar]
- 21.Yang P, Schwartz AG, McAllister AE, Swanson GM, Aston CE. Lung cancer risk in families of nonsmoking probands: heterogeneity by age at diagnosis. Genet Epidemiol. 1999;17:253–73. doi: 10.1002/(SICI)1098-2272(199911)17:4<253::AID-GEPI2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Chen PL, Sellers TA, Bailey-Wilson JE, Rothschild H, Elston RC. Segregation analysis of smoking-associated malignancies: evidence for mendelian inheritance. Am J Hum Genet. 1991;49 doi: 10.1002/ajmg.1320520311. [DOI] [PubMed] [Google Scholar]
- 23.Bunn PAJ, Soriano A. New therapeutic strategies for lung cancer: biology and molecular biology come of age. Chest. 2000;117:163–8S. doi: 10.1378/chest.117.4_suppl_1.163s. [DOI] [PubMed] [Google Scholar]