Abstract
Lymphatic filariasis is caused by the Brugia malayi parasite. Three new congeners of the depsipeptide WS9326A (1), WS9326C (2), WS9326D (3) and WS9326E (4), were isolated from Streptomyces sp. 9078 by using a B. malayi asparaginyl-tRNA synthetase (BmAsnRS) inhibition assay. WS9326D specifically inhibits the BmAsnRS, kills the adult B. malayi parasite, and does not exhibit significant general cytotoxicity to human hepatic cells, representing a new lead scaffold for antifilarial drug discovery.
Lymphatic filariasis (LF), one of the World Health Organization's (WHO) top ten neglected tropical diseases, is caused by the filarial nematode parasite Brugia malayi. LF affects more than 200 million people worldwide.1 Thus a top priority of the WHO is to search for new antihelminthic drugs that kill adult worms but exhibit fewer side effects than currently available medications such as albendazole and ivermectin. Because so few effective antifilarial drugs exist, the same drugs have been commonly used to treat both human and animal helminth diseases for more than three decades.2 Consequently, drug resistance has emerged clinically around the world, underscoring the clear need to identify antiparasite agents with new, alternative modes of action.3
Aminoacyl-tRNA synthetases (AARS) are a family of enzymes that play a key role in protein synthesis and thus AARS are one of the new molecular targets embraced by WHO for antiparasite drug discovery.4 In particular, the asparaginyl-tRNA synthetase (AsnRS) in B. malayi (i) is highly expressed in all stages of the parasite life cycle, (ii) is biochemically and structurally well characterized, and (iii) shows significant structural differences in comparison to human and other eukaryotic AARS.5
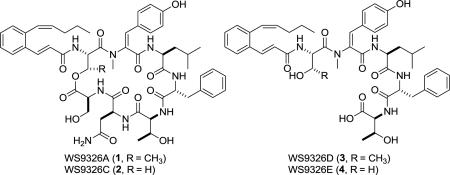
In an effort to discover new antifilarial drug leads, we recently completed a high throughout screening campaign, targeting the BmAsnRS. Of the ~73,000 extracts from a collection of 36,720 microbial strains screened, we identified 177 active strains. Previously, we reported the discovery of the tirandamycins (TAMs) from Streptomyces sp. 17944 and showed that TAM B was a potent and specific BmAsnRS inhibitor that efficiently killed the adult B. malayi parasite.6 We now report bioassay-guided fractionation of another active strain,Streptomyces sp. 9078, leading to the discovery of three new congeners of the known depsipeptide WS9326A (1),7 WS9326C (2), WS9326D (3), and WS9326E (4) (Figure 1). Importantly, 3, a moderate BmAsnRS inhibitor, efficiently kills the adult B. malayi parasite, representing another lead scaffold for antifilarial drug discovery.
Figure 1.
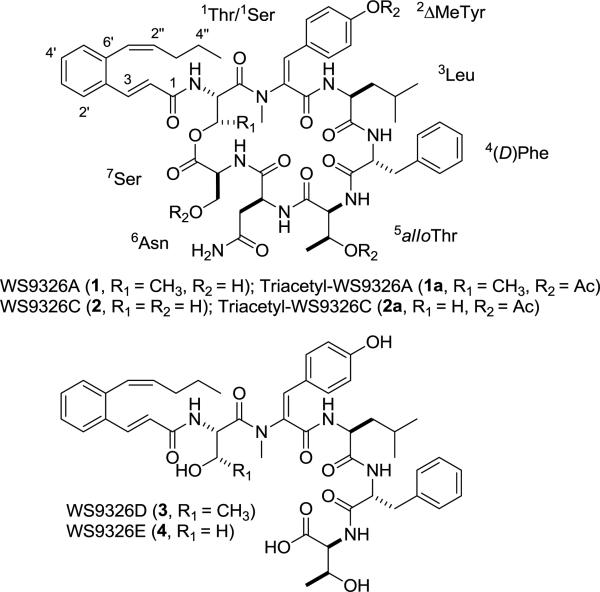
Structures of depsipeptide WS9326A (1)7 and the three new congeners, WS9326C (2), WS9326D (3), WS9326E (4), from Streptomyces sp. 9078.
The crude extract from a 9.6-L fermentation culture of Streptomyces sp. 9078 was subjected to sequential chromatography over SiO2 and Sephadex LH-20 columns, followed by semi-preparative HPLC over a C-18 column. Natural product isolation was guided by bioassay for inhibitory activity against the recombinant BmAsnRS, affording pure 1 (13.1 mg), 2 (5.2 mg), 3 (3.9 mg), and 4 (3.2 mg) as white powders, respectively [see Supporting Information (SI)]. Analysis of high resolution ESI-MS (HRESIMS) data and 1H and 13C NMR spectra of 1 and its triacetyl derivative 1a suggested 1 to be WS9326A, a depsipeptide that has been previously isolated from Streptomyces violaceoniger no. 9326.7 The identity of 1 was unambigously confirmed by extensive 1D and 2D NMR analysis of 1 and 1a (Tables S1 and S2), as well as comparison to the spectropic data reported previously.7
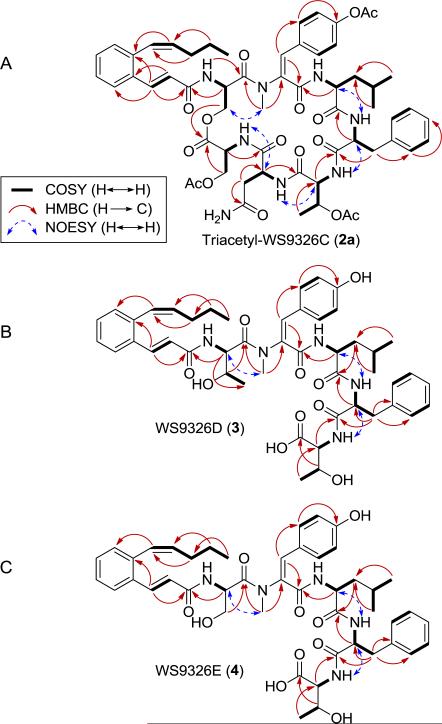
The molecular formula of 2 was determined to be C53H66N8O13 by HRESIMS, affording an [M + H]+ ion at m/z 1023.4842 (calculated [M + H]+ ion at m/z 1023.4827) and indicating that 2 differs from 1 (C54H68N8O13) by the absence of a CH2 unit. The 1H and 13C NMR spectra of 2 indicated that 2 exists as a mixture of two conformers in solution. A similar conformational mixture was known for 1, the triacetyl derivative of which (1a) however afforded a single conformer.7 Thus, 2 was simiarly converted into its triacetyl derivative (2a), the 1H and 13C NMR spectra of which in CDCl3 confirmed it as a single conformer. The structure of 2 was then established by careful comparison of the 1H and 13C NMR data between 2a and 1a (Tables 1, S1, and S2). The absence of a doublet methyl signal [δC 17.1 and δH 1.40 (3H, d, 6.30 Hz)] and a methine signal [δC 71.0 and δH 5.47 (1H, m)] with the concomitant presence of one new methylene signal [δC 63.8 and δH 4.43 (1H, m), 4.68 (1H, m)] led to the conclusion that the 1Thr residue in 1a was substituted by a 1Ser residue in 2a (Figure 1). This conclusion was further supported by key correlations observed in gHMBC, COSY, and NOSEY experiments of 2a (Figure 2A). Since the absolute stereochemistry of 1 was known,72, named as WS9326C, was assigned the same stereochemistry as that of 1 on the basis of its biosynthetic origin (Figure 1).
Table 1.
Summary of 1H (700 MHz) and 13C (175 MHz) NMR Data for 2a in CDCl3 and 3 and 4 in d6-DMSOa
| 2a |
3 |
4 |
|||||
|---|---|---|---|---|---|---|---|
| positionb | δH (J in Hz) | δ C | δH (J in Hz) | δ C | δH (J in Hz) | δ C | |
| Acyl | 1 | 166.1, s | 165.3, s | 165.5, s | |||
| 2 | 6.98, 1H, d (16.0) | 122.2, d | 6.88, 1H, d (15.7) | 123.3, d | 6.71, 1H, d (15.7) | 123.3, d | |
| 3 | 7.91. 1H, d (16.0) | 140.0, d | 7.54, 1H, d (15.7) | 137.1, d | 7.55, 1H, d, 15.7) | 137.0, d | |
| 1′ | 133.6, s | 138.0, s | 138.0, s | ||||
| 2′ | 7.56, 1H, d (7.80) | 126.3, d | 7.62, 1H, d (6.86) | 125.9, d | 7.58, 1H, d (7.63) | 125.9, d | |
| 3′ | 7.18, 1H, t, (7.60) | 127.0, d | 7.36, 1H, t (7.00) | 129.0, d | 7.37, 1H, t (7.21) | 129.4, d | |
| 4′ | 7.29, 1Hc | 128.9, d | 7.33, 1H, t (6.72) | 127.4, d | 7.33, 1H, t (7.35) | 127.8, d | |
| 5′ | 7.20, 1H, d (7.80) | 129.9, d | 7,19, 1He | 129.7, d | 7.20, 1Hf | 130.5, d | |
| 6′ | 138.4, s | 137.0, s | 137.0, s | ||||
| 1″ | 6.55, 1H, d (11.4) | 126.9, d | 6.53, 1H, d (11.5) | 126.9, d | 6.51, 1H, d (10.8) | 127.4, d | |
| 2″ | 5.80, 1H, dt (11.5, 7.40) | 134.9, d | 5.79, 1H, m | 134.1, d | 5.78, 1H, m | 134.1, d | |
| 3″ | 2.00, 2H, m | 30.5, t | 1.97, 2H, m | 30.0, t | 1.98, 2H, m | 29.0, t | |
| 4″ | 1.35, 2H, m | 22.7, t | 1.36, 2H, m | 22.1, t | 1.36, 2H, m | 22.9, t | |
| 5″ | 0.80, 3H, t (7.50) | 13.8, q | 0.90, 3H, t (7.33) | 13.6, q | 0.81, 3H, t (7.28) | 13.7, q | |
| 1Ser/1Thr | NH | 7.65, 1H, d (7.80) | 8.58, 1H, brs | 8.64, 1H, brs | |||
| α | 5.45, 1H, m | 50.9, d | 5.01, 1H, m | 52.9, d | 5.05, 1H, m | 51.3, d | |
| β | 4.43, 1H, m | 63.8, t | 4.11, 1H, m | 66.6, d | 3.56, 1H, m | 61.3, t | |
| 4.68, 1H, m | 3.80, 1H, m | ||||||
| γ | 1.05, 3H, brs | 20.0, q | |||||
| CO | 169.5, s | 170.9, s | 170.9, s | ||||
| 2ΔMeTyr | NMe | 3.57, 3H, s | 40.4, q | 2.88, 3H, s | 34.0, q | 2.86. 3H, s | 33.4, q |
| α | 138.8, s | 130.7, s | 129.4, s | ||||
| β | 6.84, 1H, s | 128.2, d | 6.60, 1H, s | 131.6, d | 6.70, 1H, s | 130.7, d | |
| 1 | 131.0, s | 123.5, s | 123.3, s | ||||
| 2,6 | 7.29, 2Hc | 129.9, d | 7.28, 2H, d (7.63) | 129.3, d | 7.19, 2Hf | 130.7, d | |
| 3,5 | 7.05, 2Hd | 121.9, d | 6.67, 2H, d (7.07) | 115.1, d | 6.67, 2H, d (7.98) | 115.2, d | |
| 4 | 150.9, s | 158.4, s | 158.6, s | ||||
| CO | 166.0, s | 164.9, s | 165.3, s | ||||
| CO (Ac) | 169.6, s | ||||||
| Me (Ac) | 2.30, 3H, s | 21.1, q | |||||
| 3Leu | NH | 8.08, 1H, d (8.20) | 7.28, 1H, brs | 7.35, 1H, d (9.35) | |||
| α | 4.45, 1H, m | 51.1, d | 4.35, 1H, m | 51.5, d | 4.39, 1H, m | 51.5, d | |
| β | 1.28, 1H, m | 38.0, t | 1.20, 2H, m | 40.0, t | 1.20, 2H, m | 39.2, t | |
| 1.64, 1H, m | |||||||
| γ | 1.00, 1H, m | 24.2, d | 1.15, 1H, m | 23.9, d | 0.85, 1H, m | 24.0, d | |
| δ | 0.71, 3H, d (6.60) | 23.0, q | 0.70, 3H, brs | 21.7, q | 0.73, 3H, d (6.20) | 22.1, q | |
| 0.79, 3H, d (7.10) | 21.5, q | 0.70, 3H, brs | 22.7, q | 0.73, 3H, d (6.20) | 22.5, q | ||
| CO | 170.9, s | 172.4, s | 173.2, s | ||||
| 4Phe | NH | 7.94, 1H, d (8.10) | 8.49, 1H, brs | 8.44, 1H, brs | |||
| α | 4.70, 1H, m | 54.9, d | 4.72, 1H, m | 53.9, d | 4.74, 1H, m | 53.8, d | |
| β | 2.79, 1H, m | 36.7, t | 2.71, 1H, m | 38.0, t | 2.72, 1H, m | 37.4, t | |
| 3.13, 1H, dd (13.8, 8.20) | 3.05, 1H, m | 3.05, 1H, m | |||||
| 1 | 137.2, s | 137.1, c | 137.0, s | ||||
| 2,6 | 7.12, 2H, d (7.50) | 129.2, d | 7.20, 2He | 130.7, d | 7.30, 2H, d (7.14) | 129.7, d | |
| 3,5 | 6.99, 2H, t (7.70) | 128.4, d | 7.22, 2He | 127.8, d | 7.22, 2Hf | 129.0, d | |
| 4 | 7.05, 1Hd | 126.9, d | 7.15, 1H, t (6.51) | 126.0, d | 7.14, 1H, t (7.28) | 126.9, d | |
| CO | 173.4, s | 170.9, s | 170.9, s | ||||
| 5allo Thr | NH | 7.28, 1H, d (8.40) | 8.10, 1H, brs | 8.07, 1H, brd (6.23) | |||
| α | 4.93, 1H, dd (8.10, 3.60) | 56.0, d | 4.04, 1H, m | 58.5, d | 4.02, 1H, m | 58.8, d | |
| β | 5.28, 1H, m | 69.1, d | 3.65, 1H, m | 67.0, d | 3.68, 1H, m | 67.6, d | |
| γ | 1.18, 3H, d (6.60) | 15.1, q | 0.98, 3H, brs | 20.0, q | 0.97, 3H, brs | 20.1, q | |
| CO | 168.1, s | 170.4, s | 170.5, s | ||||
| CO (Ac) | 170.5, s | ||||||
| Me (Ac) | 1.79, 3H, s | 20.7, q | |||||
| 6Asn | NH | 7.65, 1H, d (7.80) | |||||
| α | 4.86, 1H, td (9.00, 4.40) | 49.3, d | |||||
| β | 2.63, 1H, dd (15.2, 4.30) | 36.0, t | |||||
| 2.81, 1H, m | |||||||
| γCO | 173.1, s | ||||||
| CO | 173.4, s | ||||||
| 7Ser | NH | 7.19, 1H, d (7.21) | |||||
| α | 4.26, 1H, dd (11.3, 5.30) | 52.7, d | |||||
| β | 4.42, 1H, m | 62.4, t | |||||
| 4.58, 1H, dd (13.2, 7.70) | |||||||
| CO | 170.2, s | ||||||
| CO (Ac) | 170.9, s | ||||||
| Me (Ac) | 2.06, 3H, s | 20.8, q | |||||
Assignments were based on COSY, HSQC, HMBC, and NOSEY experiments
Numbering followed literature precedence7
Overlapping signals
Overlapping signals
Overlapping signals
Overlapping signals
Figure 2.
Key COSY, HMBC, and NOESY correlations supporting the structures of (A) WS9326C (2) and triacetyl-WS9326C (2a), (B) WS9326D (3), and (C) WS9326E (4).
The molecular formula of 3 was determined to be C47H59N5O10 by HRESIMS, affording an [M + H]+ ion at m/z 854.4357 (calculated [M + H]+ ion at m/z 854.4340). Analysis of the 1H and 13C NMR spectra indicated that 3 is a linear lipopeptide consisted of five amino acids. The structures of the five amino acids were determined by 1D and 2D NMR, the sequence of which was assigned in the order of –NH-1Thr-2ΔMeTyr-3Leu-4(D)Phe-5alloThr-CO2H with the assistance of gHMBC and NOESY (Table 1 and Figures 1 and 2B). The identity of the 3-[2-(1(Z)-pentenyl)phenyl]-2(E)-propenoyl moiety was readily evident upon analysis of its 1H and 13C NMR spectra (Table 1) and in comparison with the same moiety in 1 and 2 (Tables S1 and S2). The regiochemistry of the acyl moiety was assigned on the basis of key gHMBC correlations between Hα-1Thr [δH 5.01 (1H, m)] and C-1 [δC 165.3 (s)] of the acyl unit of 3, establishing that the acyl moiety is attached to α-C of 1Thr via an amide linkage (Figure 2B). Thus 3, named as WS9326D, could be envisaged as a biosynthetic intermediate of 1, thereby sharing the same absolute stereochemistry (Figure 1).
The molecular formula of 4 was determined to be C46H57N5O10 by HRESIMS, yielding an [M + H]+ ion at m/z 840.4194 (calculated [M + H]+ ion at m/z 840.4183) and differing from 3 by the absence of a CH2 unit. The structure of 4 was established by direct comparison of the 1H and 13C NMR data between 4 and 3 (Table 1). The absence of a methyl signal [δC 20.0 and δH 1.05 (3H)] and a methine signal [δC 66.6 and δH 4.11 (1H, m)] with the concomitant presence of one new methylene signal [δC 61.3 and δH 3.56 (1H, m), 3.80 (1H, m)] revealed that the 1Thr residue in 3 was substituted by a 1Ser residue in 4 (Figure 1). This conclusion was further supported by key correlations observed in gHMBC, COSY, and NOSEY experiments of 4 (Figure 2C). Hence, 4, named as WS9326E, could be viewed as a biosynthetic intermediate of 2 with the same absolute stereochemistry (Figure 1).
Each of the purified compound was reevaluated for their inhibitory activity against BmAsnRS using the recently reported nonradioactive assay.8 This pretransfer editing assay exploits L-aspartate β-hydroxamate, a novel asparagine substrate mimic, to drive the enzymatic activity of AsnRS and maximize production of inorganic phosphate that can be measured by its reaction with malachite green (SI). While 1 and 2 showed little activity at 100 μM, moderate inhibition against BmAsnRS was observed for 3 and 4 with apparent IC50s estimated to be 50 μM (for 3) and 75 μM (for 4), respectively (Figure S1). We next tested 3 for its ability to kill adult B. malayi worms in vitro following the published procedure.6, Live adult B. malayi worms were maintained in 6-well plates, and 3, varying from 10 nM to 50 μM, was added to selected wells with both albendazole (100 μM), a known LF drug,3 and TAM B, the new LF drug lead we have discovered previously from S. sp. 17944,6 as positive controls (SI). Remarkably, 3 kills the adult B. malayi worms rapidly within 24 hours and can efficiently kill the adult worms within 10 days at concentration as low as 10 nM (Figure S2); worm death was unambiguously confirmed from simple paralysis by the modified MTT assay6,9 (Figure S3) (SI). Finally, 3 was evaluated using human HepG2 cells for general cytotoxicity, which was defined as >50% cell death at 24 hours as measured by the MTT assay for mitochondrial activity,9 and 3 was found to be nontoxic at concentration as high as 100 μM (Figure S4) (SI).
Natural products remain the best sources of drugs and drug leads, however, natural products are underrepresented unfortunately in all small molecule libraries currently available.10 Depsipeptide 1 was first isolated as a tachykinin antagonist from S. violaceoniger no. 9326 two decades ago.7 Guided by the innovative high throughput screening targeting BmAsnRS, we now report that 3, a new congener of 1, is a novel BmAsnRS inhibitor, efficiently kills the adult B. malayi parasite, and does not exhibit significant general cytotoxicity to human hepatic cells. Therefore, 3 represents another new lead scaffold for antifilarial drug discovery. These findings, together with our early report of TAM B as a promising antifilarial drug lead,6 underscore the great promise of our strategy in screening microbial natural products as BmAsnRS inhibitors for antifilarial drug discovery. The fact that these leads can be produced in sufficient quantities by scale up microbial fermentation and that their biosynthetic machinery could be subjected to combinatorial biosynthetic strategies for titer improvement and structural diversity should greatly facilitate follow up mechanistic and preclinical studies, thereby realistically developing these promising leads into potential clinical drugs.
Supplementary Material
Acknowledgment
We thank the Filariasis Research Reagent Resource (FR3), Division of Microbiology and Infectious Diseases, NIAID, NIH for providing adult B. malayi. This work was supported in part by NIH grants A1053877 (M.K.) and GM086184 (B.S.) as well as the Natural Products Library Initiative at TSRI.
Footnotes
Supporting Information Available. Detailed experimental procedures, biological evaluations of 3 (Figures S1 to S4), and assorted 1D and 2D NMR spectra for 1, 1a, 2, 2a, 3, and 4 (Tables S1, S2 and Figures S5 to S28). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.a WHO . In: Lymphatic Filariasis: Reasons for Hope. Dzenowagis J, editor. World Health Organization; Geneva: 1997. [Google Scholar]; b Ridley RG, Kita K. Expert Opin. Drug Discovery. 2007;2(Supl. 1):S1. doi: 10.1517/17460441.2.S1.S1. [DOI] [PubMed] [Google Scholar]
- 2.Woods DJ, Lauret C, Geary TG. Expert Opin. Drug Discovery. 2007;2:S25–S33. doi: 10.1517/17460441.2.S1.S25. [DOI] [PubMed] [Google Scholar]
- 3.a Geary TG. Trends Parasitol. 2005;21:530–532. doi: 10.1016/j.pt.2005.08.014. [DOI] [PubMed] [Google Scholar]; b Molyneux DH, Bradley M, Hoerauf A, Kyelem D, Taylor MJ. Trends Parasitol. 2003;19:516–522. doi: 10.1016/j.pt.2003.09.004. [DOI] [PubMed] [Google Scholar]; c Pritchard RK. Expert Opin Drug Discovery. 2007;2:S41–S52. doi: 10.1517/17460441.2.S1.S41. [DOI] [PubMed] [Google Scholar]; d Fox LM, Furness BW, Haser JK, Desire D, Brissau J-M, Milord M-D, Lafontant J, Lammie PJ, Beach M. J. Am. J. Trop. Med. Hyg. 2005;73:115–121. [PubMed] [Google Scholar]; e Kron MA, Marquard K, Hartlein M, Price S, Lederman R. FEBS Lett. 1995;374:122–124. doi: 10.1016/0014-5793(95)01092-s. [DOI] [PubMed] [Google Scholar]
- 4.Kron MA, Ramirez BL, Ramirez Y. Expert Opin. Drug Discovery. 2007;2:S1–S8. doi: 10.1517/17460441.2.S1.S75. [DOI] [PubMed] [Google Scholar]
- 5.a Merritt EA, Arakaki TL, Gillespie JR, Larson ET, Kelley A, Mueller N, Napuli AJ, Kim J, Zhang L, Verlinde CL, Fan E, Zucker F, Buckner F, Van Voorhis WC, Hol WG. J. Mol. Biol. 2010;397:481–94. doi: 10.1016/j.jmb.2010.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Crepin T, Peterson F, Hartlein M, Kron MA, Page MG. Curr. Drug Discovery Technol. 2012;8:66–75. doi: 10.2174/157016311794519947. [DOI] [PubMed] [Google Scholar]; c Kron MA, Petridis M, Milev Y, Leykam J, Härtlein M. Mol. Biochem. Parasitol. 2003;129:33–39. doi: 10.1016/s0166-6851(03)00080-x. [DOI] [PubMed] [Google Scholar]
- 6.Yu Z, Vodanovic-Jankovic S, Ledeboer N, Huang S, Rajski SR, Kron M, Shen B. Org. Lett. 2011;13:2034–2037. doi: 10.1021/ol200420u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Hayashi K, Hashimoto M, Shigematsu N, Nishicawa M, Ezaki M, Yamashita M, Kiyoto S, Okuhara M, Kohsaks M, Imanaka H. J. Antibiot. 1992;45:1055–1063. doi: 10.7164/antibiotics.45.1055. [DOI] [PubMed] [Google Scholar]; b Hashimoto M, Hayashi K, Murai M, Fujii T, Nishikawa M, Kiyoto S, Okuhara M, Kohsaka M, Imanaka H. J. Antibiot. 1992;45:1064–1070. doi: 10.7164/antibiotics.45.1064. [DOI] [PubMed] [Google Scholar]; c Shigematsu N, Hayashi K, Kayakiri N, Takase S, Hashimoto M, Tanaka H. J. Org. Chem. 1993;58:170–175. [Google Scholar]
- 8.Danel F, Caspers P, Nuoffer C, Hartlein M, Kron MA, Page MG. Curr. Drug Discovery Technol. 2010;8:66–75. doi: 10.2174/157016311794519947. [DOI] [PubMed] [Google Scholar]
- 9.Comley JCW, Res MJ, Turner CH, Jenkins DC. Int. J. Parastol. 1989;19:77–83. doi: 10.1016/0020-7519(89)90024-6. [DOI] [PubMed] [Google Scholar]
- 10.Newman DJ, Cragg GM. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.