Background: Mesothelin is a tumor differentiation antigen; its binding to tumor antigen CA-125 can lead to tumor metastasis.
Results: Structures of the Fab from a therapeutic antibody MORAb-009 and its complex with an epitope-containing fragment of mesothelin are obtained.
Conclusion: Overlapping binding sites for both CA-125 and MORAb-009 provides a basis for the antibody therapeutic effect.
Significance: This work represents the first experimental structure for mesothelin.
Keywords: Antibodies, Antigen, Cancer, Crystal Structure, X-ray Crystallography, Antibody, Mesothelin, Molecular Recognition
Abstract
Mesothelin is a tumor differentiation antigen that is highly expressed in many epithelial cancers, with limited expression in normal human tissues. Binding of mesothelin on normal mesothelial cells lining the pleura or peritoneum to the tumor-associated cancer antigen 125 (CA-125) can lead to heterotypic cell adhesion and tumor metastasis within the pleural and peritoneal cavities. This binding can be prevented by MORAb-009, a humanized monoclonal antibody against mesothelin currently under clinical trials. We show here that MORAb-009 recognizes a non-linear epitope that is contained in the first 64-residue fragment of the mesothelin. We further demonstrate that the recognition is independent of glycosylation state of the protein but sensitive to the loss of a disulfide bond linking residues Cys-7 and Cys-31. The crystal structure of the complex between the mesothelin N-terminal fragment and Fab of MORAb-009 at 2.6 Å resolution reveals an epitope encompassing multiple secondary structural elements of the mesothelin, including residues from helix α1, the loops linking helices α1 and α2, and between helices α4 and α5. The mesothelin fragment has a compact, right-handed superhelix structure consisting of five short helices and connecting loops. A residue essential for complex formation has been identified as Phe-22, which projects its side chain into a hydrophobic niche formed on the antibody recognition surface upon antigen-antibody contact. The overlapping binding footprints of both the monoclonal antibody and the cancer antigen CA-125 explains the therapeutic effect and provides a basis for further antibody improvement.
Introduction
Mesothelin is a cell surface protein that is normally found in mesothelial cells lining the pleura, pericardium, and peritoneum (1, 2) but is aberrantly expressed at a high level in a variety of cancers including mesothelioma, ovarian, pancreatic, and lung cancers (3–8). The human mesothelin gene (MSLN gene) encodes a 69-kDa precursor protein that is subsequently processed by the endoprotease furin to yield a 40-kDa glycosylphosphatidylinositol-anchored mesothelin (Msln)3 (2) and a 31-kDa megakaryocyte-potentiating factor (9) (Fig. 1A). Although the physiological function of mesothelin is unclear, studies have shown that it is capable of binding to the tumor antigen CA-125 (also known as MUC16) and mediates cell adhesion (10–12). CA-125 is a well documented biomarker for ovarian cancers (13), and the majority (88%) of mesothelioma cases are also CA-125-positive on the cell membrane (14), suggesting the possibility that binding of tumor-associated CA-125 to mesothelin on normal mesothelial cells lining the pleura or peritoneum can lead to heterotypic cell adhesion and tumor metastasis within the pleural and peritoneal cavities. By truncation and alanine replacement mutagenesis, the CA-125 binding site was mapped to a 64-residue fragment at the N terminus of mesothelin (12).
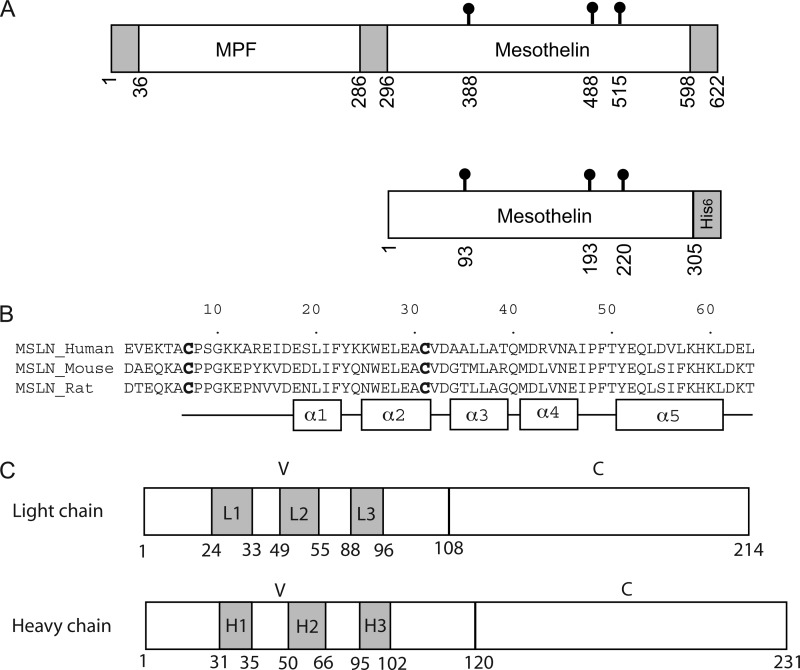
FIGURE 1.
Schematic diagrams of mesothelin and MORAb-009 Fab. A, the upper panel shows the precursor protein encoded by the human MSLN gene. The 622-residue precursor is subsequently processed by the endoprotease furin into the mature form of mesothelin containing residues from 296 to 598. For convenience, the mature mesothelin is renumbered from residue 1 to 303. The mature form of mesothelin together with two additional residues and a hexahistidine tag (the lower panel) was expressed for analysis. The three N-glycosylation sites in mesothelin (Asn-93, Asn-193, and Asn-220) are indicated. B, shown is sequence alignment of the N-terminal 64 residues of mesothelins from human, mouse, and rat. The conserved Cys-7 and Cys-31 that form a disulfide bridge are highlighted in boldface. Residues that form α-helices in the structure are shown as rectangles below the sequence alignment. C, schematics of Fab light and heavy chains are shown. Residue numbering is consistent with prior literature, and CDRs in shaded boxes are assigned according to the improved Chothia method. CDRs are labeled as L1, L2, and L3 for the light chain and H1, H2, and H3 for the heavy chain. The variable and constant domains are indicated as V and C, respectively. MPF, megakaryocyte potentiating factor.
The basis for anti-mesothelin cancer therapy is the observation that levels of antibodies specific for mesothelin are elevated in the sera of patients with mesothelioma and epithelial ovarian cancer and that this elevation is associated with high expression of mesothelin in tumors (15). Antibody response to mesothelin-expressing ovarian carcinoma cells may be responsible for reduction of tumor load and contribute to prolonged survival (16). Because mesothelin is specifically expressed at a significantly higher level in malignant tumors, development of an antibody against mesothelin is, therefore, of major importance in the field of cancer therapy (17). MORAb-009 is a promising antibody with potential clinical applications currently undergoing Phase II clinical trials. It is a chimeric IgG1/κ antibody that was generated by fusing the genes encoding the anti-mesothelin Fv (SS1 scFv) in-frame with human IgG1 and κ constant regions (17). Animal experiments have shown that application of MORAb-009 or its conjugate with pseudomonas exotoxin A in combination with chemotherapy leads to a marked reduction in tumor growth of mesothelin-expressing tumors (18, 19). Clinical studies demonstrated that it blocks the binding of mesothelin to CA-125 and thus could be used as a strategy to prevent tumor metastasis (20).
The potential application of MORAb-009 goes beyond its direct binding to mesothelin. Its Fv fragment is being tested as a carrier to deliver various anticancer agents to target cells. An anti-mesothelin recombinant immunotoxin, SS1-PE38 or SS1P, composed of the Fv portion of MORAb-009 (SS1) and a truncated form of Pseudomonas exotoxin (PE38) (21), was developed and evaluated in clinical studies (7). Despite this significant progress, an understanding at the atomic level of the mesothelin molecule and its interaction with MORAb-009 is still lacking. Here, we report the crystal structures of both the antigen-free Fab fragment of MORAb-009 and its complex with an N-terminal fragment of mesothelin.
EXPERIMENTAL PROCEDURES
Expression and Purification of Full-length Wild-type and Triple Mutant Mesothelin
Full-length cDNA of mesothelin was inserted into the baculovirus transfer vector pAcGP67B of BD BaculoGoldTM (BD Biosciences) in-frame with the hexahistidine tag at the C terminus. All mutations were made by PCR using the QuikChangeTM mutagenesis kit (Agilent Technologies, Inc., Wilmington, DE). The plasmid was co-transfected with linearized viral DNA into ∼2 million Sf9 cells, and the culture was gradually amplified to 10 liters of cultured insect cells for secretory expression of mesothelin. Culture media were collected and concentrated in a diafiltration device (Millipore, Billerica, MA) against a diafiltration solution containing 25 mm Tris, pH 7.5, 300 mm NaCl, and 10% glycerol. The sample was then mixed with nickel-nitrilotriacetic acid resin (Qiagen, Valencia, CA) pre-equilibrated with the same buffer supplemented with 10 mm imidazole. After washing with the diafiltration buffer supplemented with 50 mm imidazole, bound mesothelin was eluted in the presence of 100 mm imidazole. Fractions containing mesothelin were pooled and concentrated. Mesothelin was further purified by gel filtration using a Superdex 75 column equilibrated with 20 mm Tris, pH 8.0, 100 mm NaCl.
Blue Native PAGE (BN-PAGE) Analysis of Mesothelin-Fab Complex
The purified full-length wild-type or triple mutant mesothelin in 25 mm Tris, pH 7.5, 200 mm NaCl was mixed with MORAb-009 Fab and incubated at room temperature for 30 min to allow formation of the complex. The complex was then subjected to BN-PAGE analysis following the procedure as described in Ma and Xia (26).
Preparation of Fab Fragment from IgG
MORAb-009 IgG was obtained from Morphotek, Inc. (Exton, PA). Fab fragment was prepared using the Fab preparation kit from Thermo Scientific (Rockford, IL) and following the instructions from the manufacturer.
Expression of Thioredoxin-Mesothelin Fusion Protein and Various Truncates
The cDNA encoding the 64 N-terminal residues of mesothelin was inserted into the expression vector pET32a+ at the site after the coding region for thioredoxin (TrxA-Msln-(1–64)), which contains a hexahistidine tag N-terminal to mesothelin. Based on this plasmid, truncations from either the N or C terminus of mesothelin were made by PCR with the QuikChangeTM mutagenesis kit to construct a series of plasmids expressing thioredoxin (TrxA), TrxA-Msln-(1–10), TrxA-Msln-(1–20), TrxA-Msln-(1–30), TrxA-Msln-(1–40), TrxA-Msln-(1–50), TrxA-Msln-(1–59), TrxA-Msln-(1–61), TrxA-Msln-(7–64), TrxA-Msln-(14–64), and TrxA-Msln-(28–64). These constructs were transformed into Escherichia coli strain BL21(DE3), and expressions were induced by the addition of isopropyl-β-d-1-thiogalactopyranoside to a final concentration of 1 mm at 37 °C for 3 h. E. coli cells were then sampled and analyzed by SDS-PAGE followed by Western blot analysis probed using either MORAb-009 or anti-histidine antibodies. The C7S mutant was similarly made using QuikChangeTM site-directed mutagenesis kit.
Protein Expression and Purification of Msln-(7–64)
The plasmid for expressing an N-terminal fragment of mesothelin (Msln-(7–64)) was transformed into the E. coli strain Origami B. The culture was grown to an A600 of 0.8 before the expression of recombinant protein was induced at 37 °C for 4 h by adding isopropyl-β-d-1-thiogalactopyranoside to 1 mm. Cells were lysed by a French press in a lysis buffer containing 100 mm Tris, pH 7.5, 150 mm NaCl, and 1 mm PMSF at a pressure of 15,000 p.s.i. twice. The cell lysate was centrifuged at 15,000 × g for 30 min to remove unbroken cells, and the resulting supernatant was mixed with nickel-nitrilotriacetic acid resin (Qiagen) pre-equilibrated with washing buffer containing 25 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol, and 20 mm imidazole, pH 7.5, and incubated for 2 h at 4 °C. The resin was then transferred into a column. After washing the column with the washing buffer, the bound protein was eluted by the same buffer containing 300 mm imidazole. The eluant was concentrated and further purified by size-exclusion chromatography on a Superdex S-200 column equilibrated with running buffer containing 25 mm Tris, pH 7.5, 150 mm NaCl, and 2% glycerol. The fractions containing the mesothelin fragment were pooled and stored at 4 °C for later use.
Crystallization
Crystallization screenings were carried out robotically using a Mosquito liquid dispenser (TTP LabTech, Cambridge, MA) by the hanging-drop vapor diffusion method in a 96-well format and using commercial high-throughput screening kits (Hampton Research, Aliso Viejo, CA; Emerald Biosystems, Bainbridge Island, WA; Molecular Dimensions, Apopka, FL). Subsequent optimizations were done manually by mixing 1.5 μl of protein solution with 1.5 μl of precipitant solution, and the admixture was equilibrated against 500 μl of reservoir solution using the hanging-drop vapor diffusion method at 21 °C.
Crystallographic Analysis of Msln-(7–64)-Fab Complex and Isolated Fab
All x-ray diffraction data sets were collected at 100 K using beamline 22 (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory. Diffraction images were processed using the program HKL2000 (27). The Fab structure was solved by the molecular replacement (MR) method using the BALBES (28) program in the CCP4 program suite (29) and subsequently refined using Phenix (30) and REFMAC (31). The crystal structure of the complex of Fab and Msln-(7–64) was determined also by MR using MOLREP (32) and refined with REFMAC. All structure models were built using the program COOT (33). The atomic coordinates have been deposited in the Protein Data Bank as follows: PDB ID code 4F33 for the Fab fragment derived from MORAb-009 and 4F3F for the complex between Fab and mesothelin N-terminal fragment.
RESULTS
Identification of the Minimal Fragment Containing the Epitope Recognized by MORAb-009
Fully processed, full-length mesothelin has 303 amino acid residues, starting from the precursor protein Glu-296 to Asp-598 (Fig. 1A). For convenience, we renumber the residues in mature mesothelin starting with Glu-1 and ending with Asp-303. Full-length mesothelin with two additional C-terminal residues followed by a hexahistidine tag is expressed as a secretory protein in baculoviral-infected insect cells and purified to homogeneity (Fig. 2A). This protein is highly glycosylated and capable of binding to MORAb-009 (Fig. 2B, left panel) with a dissociation constant (Kd) estimated at 5 nm by isothermal titration calorimetry (data not shown). We made a triple mutant by removing three predicted N-glycosylation sites, N93Q, N193Q, and N220Q. The purified triple mutant had a significantly reduced molecular mass (Fig. 2A) but maintained its binding to the Fab fragment shown by BN-PAGE (Fig. 2B, right panel) analysis.
FIGURE 2.
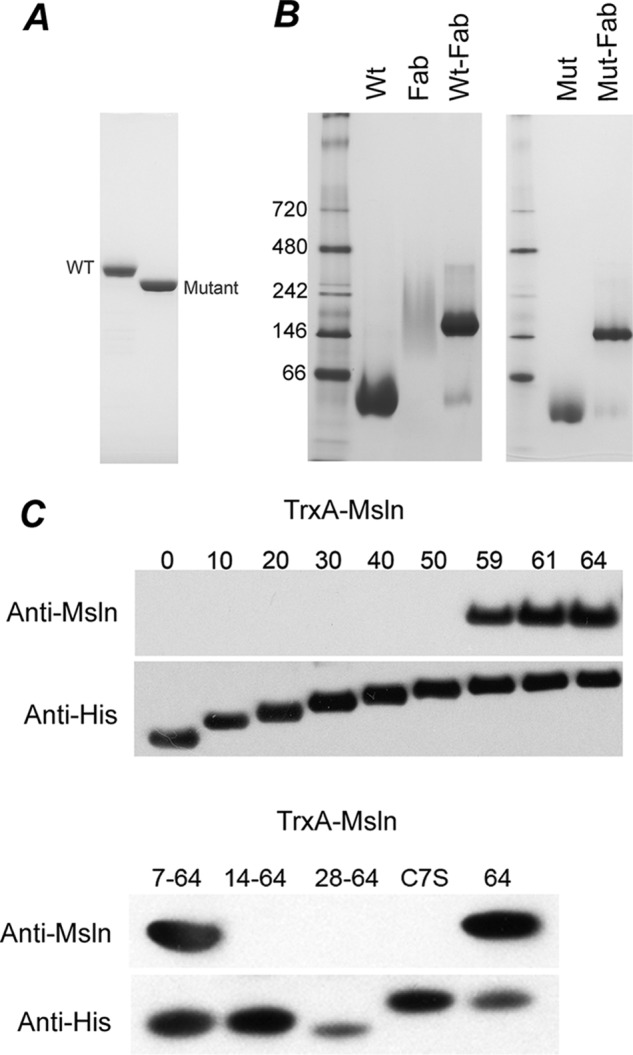
Binding of MORAb-009 to full-length mesothelin and identification of the minimal epitope-containing fragment recognized by MORAb-009. A, SDS-PAGE analysis of the purified wild-type mesothelin and triple mutant expressed in insect cells is shown. The wild-type mesothelin is glycosylated, whereas the N93Q/N193Q/N220Q triple mutant shows a significant downward shift in molecular weight. B, BN-PAGE analysis of MORAb-009 and its complexes with wild-type and triple mutant mesothelin is shown. When mixed with MORAb-009 Fab, both wild type and triple mutant can form a complex as shown by the single band on the BN-PAGE shifted to a much higher molecular weight compared with the mesothelin alone. The Fab fragment shows a broad smeared band on the BN-PAGE gel. C, identification of the minimal fragment of mesothelin for MORAb-009 binding is shown. Cell extracts from E. coli expressing TrxA-fused mesothelin fragments were analyzed by Western blot using MORAb-009 as the primary antibody. The expression levels of these fragments were confirmed using antibody against the hexahistidine tag. The number of residues included in each fragment is indicated. The C7S in the lower panel is the C7S mutant of TrxA-Msln-(1–64), whereas the 64 indicates the fragment for TrxA-Msln-(1–64).
Previously, it was reported that a 64-residue N-terminal fragment of mesothelin is the minimal size for CA-125 interaction and that this interaction can be blocked by the monoclonal antibody SS1 Fv (12) from which MORAb-009 is derived. We constructed a vector to express in E. coli the 64-residue fragment of mesothelin fused C-terminally to TrxA. We found by Western blot that the purified fusion protein was highly reactive to MORAb-009. Using this construct, we made a series of C-terminal truncates in approximately 10-residue decrements (Fig. 2C, top panel). The results showed that mesothelin N-terminal fragments up to 50 residues in length did not react with MORAb-009. When the size was increased to 59 residues or longer, the antibody binding was restored. To further narrow down the boundaries that define the minimal fragment for binding, the protein was truncated from the N terminus, and the results showed that the minimal mesothelin fragment to react with MORAb-009 is between residues 7 and 59. Further truncation from the N terminus led to production of fragments non-reactive to the antibody (Fig. 2C, bottom panel). However, the Kd, as measured by isothermal titration calorimetry, of this N-terminal fragment of mesothelin indicated a 5-fold reduction in binding affinity to the antibody (data not shown).
An alignment of available mesothelin sequences from mammals shows that the two cysteine residues (Cys-7 and Cys-31) in the N-terminal fragment are conserved, indicating a possible disulfide bond (Fig. 1B). To test the presence of this disulfide bridge, we mutated residue Cys-7 to serine. Indeed, the mutation abolished its ability to bind MORAb-009 (Fig. 2C, lower panel), underlining the importance of this disulfide linkage in maintaining the structural integrity of mesothelin.
Structure Determination for the Fab-Mesothelin Complex and Isolated Fab
To investigate the interaction between mesothelin and MORAb-009 at atomic resolution, we made a new construct expressing the minimal fragment from residue Cys-7 to Leu-64 and fused a hexahistidine tag at its C terminus (Msln-(7–64)). This fragment was expressed and purified and then mixed with the Fab fragment from MORAb-009. The resulting complex was purified by size-exclusion chromatography and crystallized. Crystals of the Msln-(7–64)-Fab complex diffracted x-rays to better than 2.5 Å resolution and a data set processed to 2. 61 Å resolution was obtained (Table 1). To facilitate the structure determination of the complex by the MR method using the structure of the antibody as a phasing template, we needed to crystallize the Fab fragment alone and determine its structure first.
TABLE 1.
Statistics on x-ray diffraction data sets and refined structural models
| Data set | Fab | Msln-(7–64)-Fab |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.0 | 1.0 |
| Space group | P41212 | P64 |
| Cell dimension (Å,°) | a = b = 140.6, c = 282.0, α = β = γ = 90 | a = b = 146.2, c = 80.9, α = β = 90, γ = 120 |
| Resolution (Å) | 39.1-1.75 (1.81-1.75)a | 50.0-2.61 (2.7-2.61) |
| No. of observations | 1,436,883 | 186,593 |
| No. of unique reflections | 262,387 | 29,264 |
| Redundancy | 5.5 (2.1) | 6.4 (2.4) |
| Completeness (%) | 92.0 (70.7) | 97.8 (80.0) |
| Rmergeb (%) | 9.2 (40.7) | 10.1 (44.2) |
| Mean I/σ(I) | 19.5 (1.4) | 22.4 (1.8) |
| Refinement | ||
| No. of reflections | ||
| Working set | 229,079 | 27,763 |
| Test set | 2,630 | 1,485 |
| Rcrystal (%) | 20.2 | 18.9 |
| Rfree (%) | 22.7 | 23.5 |
| Residues | ||
| Heavy chain | 880 | 216 |
| Light chain | 848 | 211 |
| Mesothelin | 0 | 59 |
| Water | 1,328 | 47 |
| PEG | 6 | - |
| Average B-factor (Å2) | 32.3 | 56.8 |
| Root mean square deviations | ||
| Bond length (Å) | 0.007 | 0.019 |
| Bond angle (°) | 1.1 | 2.2 |
| Ramachandran plot statistics | ||
| Most favored region | 1346 (90.9%) | 370 (87.3%) |
| Allowed regions | 130 (8.8%) | 53 (12.5%) |
| Disallowed region | 4 (0.3%) | 1 (0.2%) |
a Numbers in the parentheses are statistics for outermost shells.
b Rmerge is defined as Σ|Ihi − 〈Ih〉|/ΣIh,i, where Ihi is the intensity for ith observation of a reflection with Miller index h, and 〈Ih〉 is the mean intensity for all measured Ihs and Friedel pair.
The structure of isolated MORAb-009 Fab was solved by MR using a template Fab structure (PDB code 1A6T) from a neutralizing monoclonal antibody against human rhinovirus 14 and was refined to 1.75 Å resolution (Table 1). There are four Fab fragments in a crystallographic asymmetric unit; each fragment includes 212 residues (2–213) for the light chain and 220 residues (1–220) for the heavy chain. The four non-crystallographic symmetry-related Fab fragments showed nearly identical conformation, with root mean square deviations from pair-wise superposition ranging from 0.167 to 0.434 Å using all 432 Cα atoms. Therefore, we used in all subsequent analysis the fragment containing chain E (light chain) and chain F (heavy chain), which had the best overall electron density.
The coordinates of the MORAb-009 Fab fragment were used as a search model to solve the structure of the complex of Fab and Msln-(7–64). The resulting MR phases allowed calculation of difference Fourier maps that revealed additional electron densities attributable to residues of the N-terminal fragment of mesothelin. The atomic model for the structure of the complex was refined to 2.6 Å resolution (Table 1, Fig. 3A) and consists of 211 residues from the light chain (2–212), 220 residues from the heavy chain (1–220), and 59 residues from mesothelin (6–64), including an extra N-terminal residue, Met-6. The electron density for residues in the range 135–138 of the heavy chain is not well defined probably due to a high degree of flexibility. Residue Thr-51 of the light chain of MORAb-009 is an outlier in Ramachandran plot, which persisted in all five independently determined Fab structures reported here regardless of whether it is in complex with the antigen or not. This residue is part of the cdr2 that has no contact with the antigen; it is located in a tight turn and appears to be conformationally rigid.
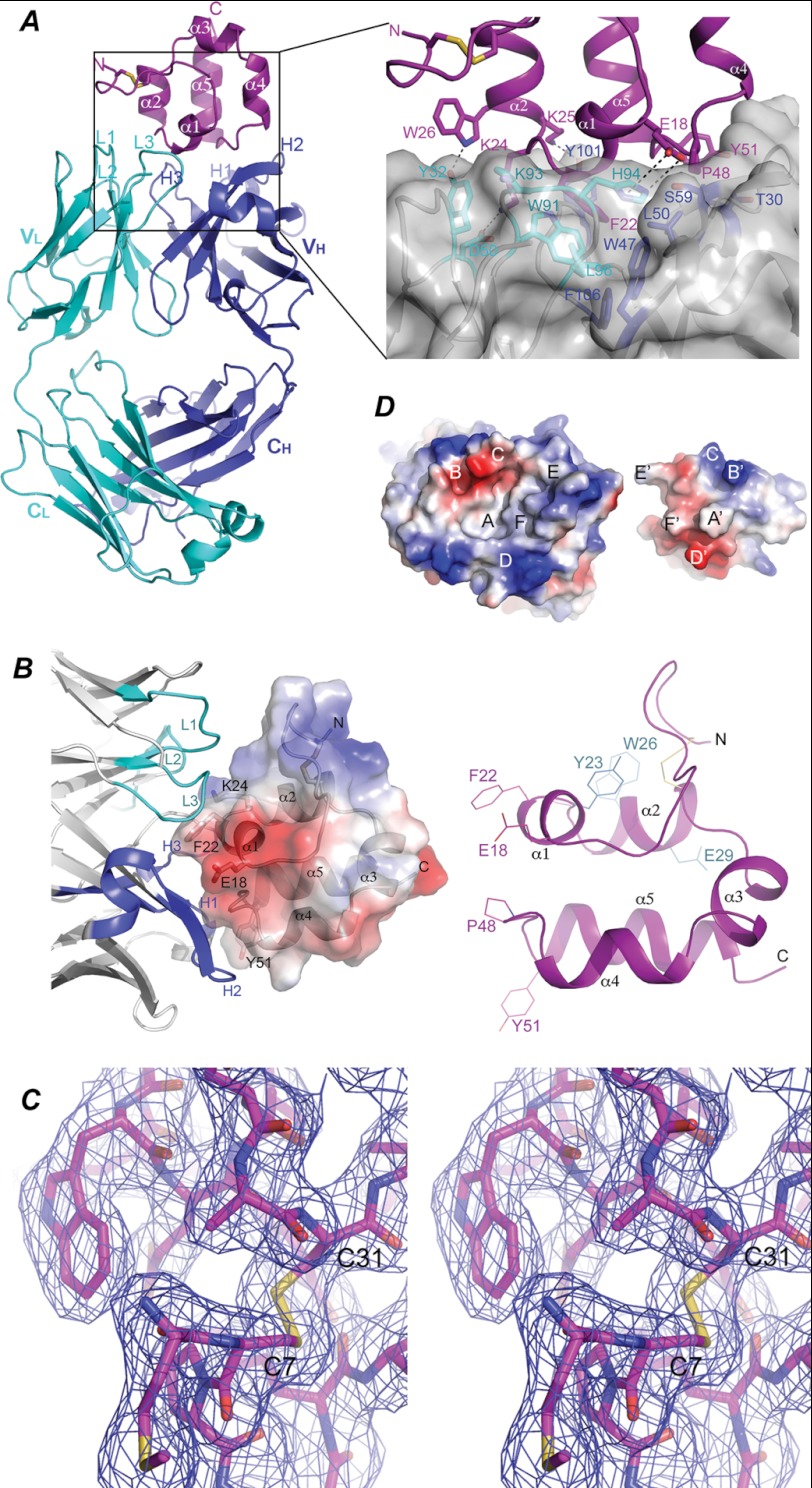
FIGURE 3.
Molecular interactions between mesothelin and Fab. A, shown is a ribbon diagram of the structure of the Msln-(7–64)-Fab complex. The mesothelin fragment is colored in magenta. The N-terminal disulfide bond is illustrated as sticks in yellow. The Fab light chain and heavy chain are colored in cyan and blue, respectively. Variable and constant domains of the light chain (VL and CL) and the heavy chain (VH and CH) are indicated. CDRs of the light chain are labeled as L1, L2, and L3, respectively, and those of heavy chain are labeled H1, H2, and H3, respectively. Inset, shown is an enlarged view of the hydrophobic and aromatic-aromatic interactions between Phe-22 of mesothelin and Fab. Residues involving in interacting with Phe-22 of mesothelin are labeled. B, a close up view of the structure of Msln-(7–64) is shown. Right panel, the five α-helices are labeled as α1, α2, α3, α4, and α5, sequentially, from the N to C terminus. Residues important for interacting with Fab are labeled in magenta, and those for interacting with CA-125 are shown in cyan. Left panel, a semitransparent electrostatic potential surface is plotted covering the structure of the fragment with positive and negative potentials in blue and red, respectively. CDRs for the light chain are colored in cyan, and those for the heavy chain are in blue. C, a stereoscopic pair shows a region of mesothelin in stick model around the disulfide bond between Cys-7 and Cys-31. Overlapping this model is a 2Fo − Fc map contoured at 1.0 σ level showing the quality of electron density. D, the electrostatic potential surfaces show the chemical and shape complementarity between the interacting mesothelin and Fab. The mesothelin molecule is intentionally flipped by 180° to expose the interacting surfaces, revealing a concave surface on the side of Fab and a convex one on the mesothelin side. The two surfaces are in perfect shape and chemical complementation. Complementing pairs on both surfaces are labeled as A-A′, B-B-, etc.
The Structure of Mesothelin N-terminal Fragment
The structure of mesothelin N-terminal fragment represents the first experimental model for mesothelin and its mammalian homologs. The structural model of Msln-(7–64) consists of an N-terminal loop (residues 7–17) followed by five consecutive α-helices named sequentially as α1 (18–22), α2 (25–31), α3 (34–39), α4 (41–46), and α5 (51–63), which are linked by short loops (Figs. 1B and 3, A and B). These secondary structural elements spiral into a right-handed, two-turn superhelix; helices α2 and α5 are nearly parallel to each other. The structure is stabilized by a disulfide bridge between the N-terminal Cys-7 and Cys-31 at the end of helix α2, which has well defined electron density (Fig. 3C) and is consistent with both the mutagenesis data (Fig. 2C) and the observed sequence conservation for these two residues (Fig. 1B). The N-terminal fragment of mesothelin possesses a dipole moment roughly running along the axis of the superhelix. Searching an existing protein structure data base did not identify any structure with significant structural similarity.
Interactions between Fab and Mesothelin
As shown in Fig. 3A, the complex is formed with binding contacts from both the light and heavy chains of Fab, although the majority of interactions are contributed from the heavy chain. The epitope recognized by MORAb-009, as revealed by the structure, consists of two non-consecutive antigenic determinants; the first is centered on helix α1 and extends on both sides from Glu-18 of the N-terminal loop to the beginning of helix α2 (Trp-26). The second contains the loop between helices α4 and α5 (Fig. 3, A and B). On the opposite side of the mesothelin epitope are residues from complementarity-determining regions (CDRs) of the Fab, forming a slightly concave surface that surrounds the epitope. The numbering of the Fab sequence in this work follows the convention used in a previous publication (17). However, in determining the CDRs for both heavy and light chains of MORAb-009, the improved Chothia method of antibody numbering was used (22), and the result was applied to the current numbering (Fig. 1C). Among the six CDRs, CDR3 of the light chain and CDR2 and CDR3 of the heavy chain make major contributions to the binding of mesothelin, whereas the rest makes considerably fewer contacts with the antigen.
The antigen and antibody interaction buries a total of 1,746 Å2 of solvent-accessible surface area from both proteins (945 Å2 for mesothelin, 303 Å2 for the light chain, and 497 Å2 for the heavy chain), which corresponds to an interacting surface of ∼870 Å2, a number that is roughly in the middle of the average of 700–1100 Å2 observed for antibody-antigen interactions (23). There are clear chemical and shape complementarities between the two interacting surfaces, as visualized in Fig. 3D. Specifically, several large residues that project toward the antibody form a prominent part of the epitope; residues Glu-18, Phe-22, and Lys-24 are from the α1 helix, whereas Pro-48 and Tyr-51 are from the loop between helices α4 and α5 (Fig. 3, A, inset, B, and D). Each residue fits snuggly into a niche on the antibody surface with matched hydrophobic or ionic interactions. For example, the side chain of residue Phe-22 of mesothelin extends into a hydrophobic niche formed by residues Trp-91, His-94, and Leu-96 of the light chain and by residues Trp-47, Leu-50, and Phe-106 of the heavy chain (Fig. 3A, inset). Clearly, residue Phe-22 plays an important role in specific recognition by the antibody and contributes significantly to the binding affinity. There are extensive electrostatic attractions and hydrogen bonding interactions between mesothelin and Fab. Although the hydrophobic interactions are near the center of the interacting surface, charged or polar interactions are peripheral (Fig. 3D). Prominent polar interactions are found between pairs of residues as in Lys-24 of mesothelin and Asp-49 of L2, Glu-18 of mesothelin and His-94 of L3, as well as in Tyr-51 of mesothelin and Thr-30 of H1. Other interactions occur between mesothelin and Fab via main chain atoms or ordered water molecules (data not shown).
Conformational Adaptation in Fab upon Binding to Mesothelin
A structure superposition of the isolated Fab with that in complex with mesothelin gave a root mean square deviation of 0.346 Å over 359 Cα atoms demonstrated a globally stable conformation for the Fab in two different crystal environments (Fig. 4A). Indeed, Fab molecules in both structures share the same elbow angle of 138.8° (24). However, significant structural deviations do occur, particularly for CDRs and associated residues making direct contacts with the antigen. The largest movement observed on the backbone of Fab occurs in CDR3 of the heavy chain (H3) with 5 Å movement for the main chain Cα atom of Tyr-101 (Fig. 4, A and B), opening up a hydrophobic niche at the light and heavy chain interface for mesothelin binding. Accompanying this large movement of main chain are the observed side-chain conformational changes for a number of residues. For example, in the free form the side chains of residues Tyr-54 of H1 and Tyr-101 of H3 point toward each other but are rotated away from each other in the complex with mesothelin, shifting in positions by as much as 9.0 and 11.6 Å, respectively (Fig. 4B). Both main- and side-chain movements open up a pocket to accommodate the incoming mesothelin molecule and simultaneously allow formation of new contacts between mesothelin and the antibody such as the one between Tyr101 of H3 and Lys-25 of mesothelin. Residues of the light chain also undergo significant rearrangements in response to antigen binding, albeit less than those observed in the heavy chain. The side chain of residue Trp-91 of L3 moves into the hydrophobic niche to be part of its floor and interacts with Phe-22 of the mesothelin (Fig. 4B). A smaller shift was observed for Tyr-32 of L1 to interact with Trp-26 of mesothelin. Based on our crystallographic data, these observed conformational changes in the binding of mesothelin by antibody appear to fit the mechanism of induced fit model. However, it does not exclude the possibility of selecting a preexisting antibody conformation by mesothelin for binding in solution. Interestingly, residues undergoing large conformational changes upon binding of the ligand have higher relative B factors in the structure of the free form, suggesting these residues are intrinsically flexible or potentiated for interacting with antigen.
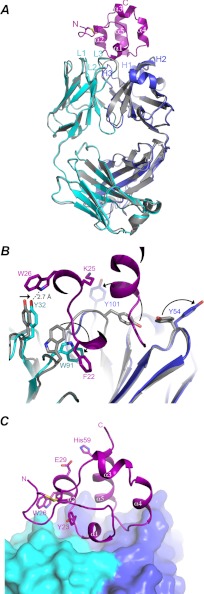
FIGURE 4.
Conformational adaptation in Fab upon binding to mesothelin. A, shown is superposition of the antigen-free Fab to the one bound with mesothelin. The light and heavy chains of the antigen bound Fab are shown as ribbon diagrams in cyan and blue, respectively; the free Fab is shown in gray. The bound mesothelin is shown as a ribbon model in magenta. CDRs of both the light chain and the heavy chain are labeled. B, shown are conformational changes in side chains of Fab upon binding to mesothelin. Mesothelin is displayed in magenta. The residues involving in significant conformational transitions are shown in stick model, and direction of the transition from the antigen free to antigen bound form for each residue is indicated by a black arrow. C, shown is the putative CA-125 binding site in mesothelin. Sitting on top of the Fab, which is shown as molecular surface in cyan and blue, respectively, for the light and heavy chains, is the mesothelin fragment depicted as a ribbon diagram in magenta. Residues Tyr-23, Trp-26, and Glu-29, shown in stick models and labeled, are the proposed CA-125-interacting residues. Residue His-59 is not involved in CA-125 binding.
DISCUSSION
The Recombinant Msln-(7–64) Represents the Native Conformation of Full-length Mesothelin and MORAb-009 Recognizes a Non-linear Epitope
The crystal structure of the complex reveals that Msln-(7–64) is a tightly folded, compact molecule that seems to be the smallest fragment maintaining a native-like structure essential for antibody recognition. In fact, this N-terminal fragment is so stable that it survives non-reducing electrophoresis in the presence of SDS and is still recognized by the antibody in subsequent Western blot analysis. Indeed, the N-terminal loop and the C-terminal helix α5 are crucial for the stability of the Msln-(7–64) fragment and cannot be further truncated from either end, although they have little contact with the Fab. This argument is further strengthened and extended by the observation that the Fab-mesothelin interaction is abolished when the disulfide bond between residues Cys-7 and Cys-31 is eliminated in the C7S mutant.
It was previously reported that the cancer antigen CA-125 recognizes the N-terminal region of mesothelin and the recognition site is conformationally sensitive (12). The monoclonal antibody SS1, the predecessor of MORAb-009, is capable of interrupting CA-125 binding to mesothelin. In this work we demonstrate unambiguously by truncation, cysteine mutagenesis, and crystallographic experiments that the antigenic epitope for the monoclonal antibody MORAb-009 is also non-linear and conformationally sensitive (Figs. 2 and 3). The epitope is bipartite, consisting of residues from helix α1 and a loop between helices α4 and α5 and has a footprint of 870 Å2.
Mechanism of Disruption of CA-125 Binding to Mesothelin by MORAb-009
Although the physiological function of mesothelin remains obscure, its N-terminal portion appears particularly immunogenic. In particular, the N-terminal part has been shown to be responsible for CA-125 binding, suggesting a likely route for cancer metastasis. Site-directed mutagenesis has demonstrated that residue Tyr-23 of mesothelin is critical for CA-125 binding; residues Trp-26 and Glu-29 may also play a role, whereas His-59 is not involved in binding (12). Mapping these residues onto the structure of the mesothelin and Fab complex, we are able to show that the CA-125 binding region partially overlaps with the binding epitope of MORAb-009 and centers perhaps on the loop between helices α1 and α2 (Fig. 4C). In the complex structure, residue Tyr-23 of mesothelin is mostly buried by the surrounding residues of mesothelin and by CDR L3 of Fab, with only the -OH group exposed to the surface. In the free form, however, the Tyr-23 would most likely be exposed to the surface and is accessible for interacting with CA-125 or with the antibody. Thus, MORAb-009 exerts its action to block attachment of CA-125 by competing for binding to the same region, making Tyr-23 inaccessible for CA-125 interaction. Both residues Trp-26 and Glu-29 of mesothelin are constituents of helix α2 and have exposed side chains in the complex. Although the side chain of Trp-26 does form an H-bond with the hydroxyl group of Tyr-32 of the Fab light chain, Glu-29 does not have any contact with the antibody. Thus the partially destroyed CA-125 binding activity by mutating either residue indicates proximity of these residues to the binding site. Residue His-59 is on the other side of the putative CA-125 binding site, consistent with the mutagenesis data that its mutation does not interfere with CA-125 binding.
Correlation of Mesothelin Binding Energy with Specific Mutations on MORAb-009
The efficacy of therapeutic antibodies or immunotoxins has been shown to correlate with their binding affinities and, for solid tumors, also with their sizes. Using the phage display affinity maturation technique, mutations were introduced to residues on the L3 CDR loop of the parental antibody of MORAb-009, namely the mouse antibody SS (17). A series of mutations were identified that showed improved affinity for mesothelin and cytotoxicity to cultured mesothelin-expressing cancer cells (Table 2). Different from its parental line, MORAb-009 or SS1 bears two mutations (G93K and Y94H) that give rise to a significantly higher affinity for mesothelin with a Kd of 0.72 nm over the parental Kd of 11 nm (Table 2), as measured by the surface plasmon resonance method. The structure shows that both residues are part of the hydrophobic niche that embraces the side chain of Phe-22 of mesothelin. Although Lys-93 provides its main chain atoms to the pocket, His-94 uses both main- and side-chain atoms. His-94 additionally forms an H-bond with Ser-59 of the heavy chain; both His-94 and Ser-59 join forces to interact with mesothelin Glu-18 with distances of 3.5 and 2.6 Å, respectively, for His-94 and Ser-59 of the heavy chain.
TABLE 2.
Correlation between structure and activity of mesothelin-interacting antibodies
| Antibody | Mutations | Kda | Cytotoxicitya | Estimated relative free energyb |
|---|---|---|---|---|
| nm | kcal/mol | |||
| SSc | Ser-92, Gly-93, Tyr-94, Leu-96 | 11 | 1 | −110 |
| SS1d | G93K, Y94H | 0.72 | 13 | −131 |
| D8 | S92G, G93F, Y94N | 0.3 | 11 | −100 |
| C10 | S92G, G93S, Y94H | 0.2 | 11 | −75 |
| E4 | L96T | 3 | 2 | −93 |
a Data were obtained from the Ref. 17.
b Free energies for the mesothelin-Fab complex and for mesothelin and Fab alone were estimated in the program Phenix without refinement. The energy difference between individual components and the complex was taken as the estimated relative free energy upon antigen-antibody association.
c SS, parental mouse monoclonal antibody.
d SS1, MORAb-009.
The atomic details of the Fab-Msln-(7–64) complex provide the structural basis for changes in affinities for other variants. Mutants S92G/G93F/Y94N (D8), S92G/G93S/Y94H (C10), and L96T (E4) showed higher affinity with Kd values of 0.3, 0.2, and 3 nm, respectively (Table 2). From the structure (Fig. 3A, inset), all these mutations altered the binding pocket to various extents for Phe-22 of mesothelin. Residue Phe-93 in D8 may interact with Tyr-23 of mesothelin. Residue Gly-92 in both D8 and C10 probably makes the L3 loop more flexible to promote a conformational change for better interactions with Phe-22 of mesothelin. The residue Leu-96 forms the floor of the Phe-22 binding niche, and mutation L96T clearly alters the interaction by providing an additional dipolar interaction between the phenyl ring of Phe-22 and the hydroxyl group of Thr-96.
It should be pointed out that based on the structure of the Msln-(7–64)-Fab complex, we estimated relative free energies for binding of mesothelin by various SS variants, and the antibody SS1, which is equivalent to MORAb-009, clearly stands out as having the lowest free energy for binding to mesothelin (Table 2). It did not escape our notice that all previous efforts in improving the antibody concentrated on the L3 loop. Our structural data show, on the contrary, that the majority of interactions between mesothelin and antibody are in fact provided by CDRs of the heavy chain, optimization of which could provide further improvement for the affinity of antibody toward mesothelin.
Homologous Proteins and Molecular Modeling Studies of Mesothelin
The mesothelin superfamily of proteins, which includes mesothelin, mesothelin precursor, megakaryocyte potentiating factor, stereocilin, and otoancorin were predicted to have superhelical structures with Armadillo or Armadillo-type repeats (25). Functionally, these repeats have been suggested to interact with carbohydrate moieties of extracellular glycoproteins (25). The structure of the N-terminal 64 residues reported here is indeed superhelical even though a detailed comparison showed large deviations.
A hybrid model was constructed by replacing the N-terminal fragment of the full-length mesothelin model with our experimental structure, and the result (Fig. 5) shows that with its C terminus anchored to the cell membrane, the N-terminal portion of mesothelin offers the largest portion of accessible surface of the mesothelin molecule for interaction. This hybrid model reveals additional interactions between residues from the heavy chain of Fab and those from the third layer of helices in the mesothelin model, thus offering a possible explanation for the observed 5-fold difference in Kd values between MORAb-009 binding to the full-length and the minimal fragment of mesothelin.
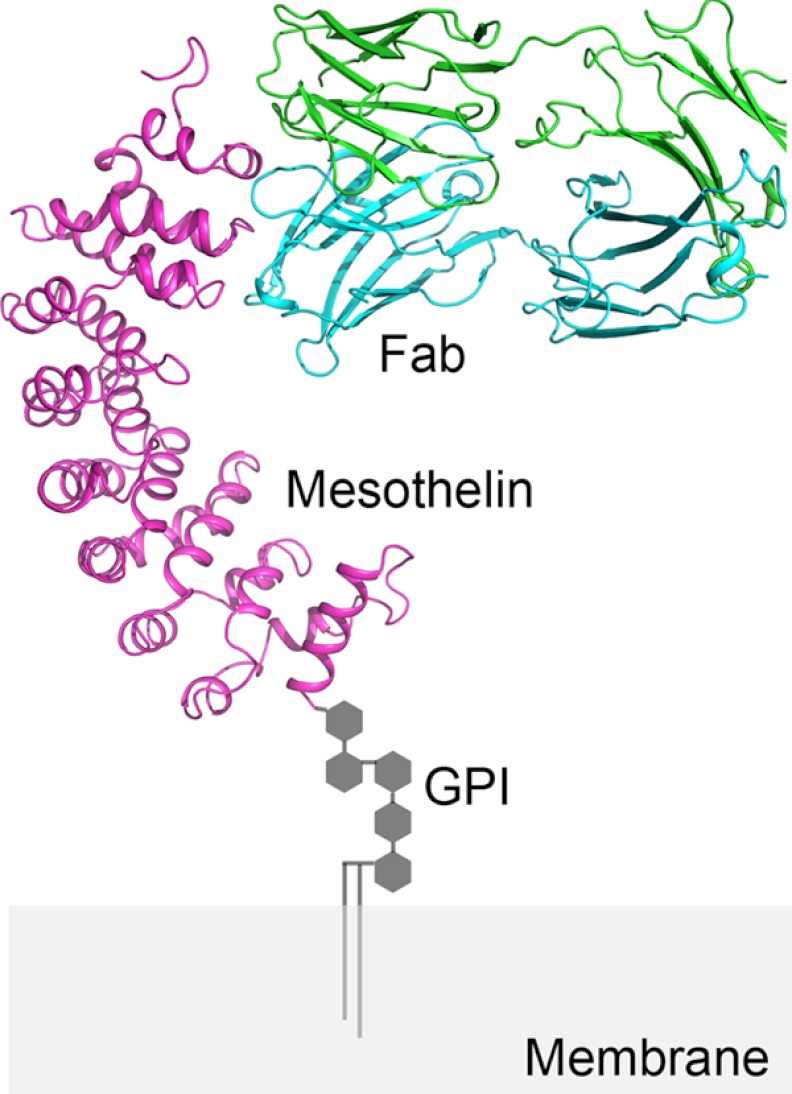
FIGURE 5.
A model of MORAb-009 binding to mesothelin displayed on the cell surface. Based on the previously published full-length mesothelin model, a hybrid model was constructed by replacing the N-terminal fragment of mesothelin with the experimental structure Msln-(7–64). The C terminus is anchored to the cell membrane through a glycosylphosphatidylinositol (GPI) linker.
Acknowledgments
We thank the beam line staff of the SER-CAT at Advanced Photon Source, Argonne National; & Laboratory for assistance in data collection. The initial construct for insect cell expression was kindly provided by Dr. Dimiter S. Dimitrov of NCI, National Institutes of Health. We thank Drs. Byungkook Lee and Mitchell Ho of NCI, National Institutes of Health for comments on the manuscript. We also thank George Leiman for editorial assistance during the preparation of this manuscript.
This work was supported, in whole or in part, by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. This work was also supported by a Cooperative Research and Development Agreement CRADA with Morphotek.
The atomic coordinates and structure factors (codes 4F33 and 4F3F) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Msln
- mesothelin
- Msln-(7–64)
- the N-terminal fragment of mesothelin from residues 7–64
- BN-PAGE
- blue native PAGE
- CDR
- complementarity determining region
- MR
- molecular replacement
- TrxA
- thioredoxin.
REFERENCES
- 1. Chang K., Pastan I., Willingham M. C. (1992) Isolation and characterization of a monoclonal antibody, K1, reactive with ovarian cancers and normal mesothelium. Int. J. Cancer 50, 373–381 [DOI] [PubMed] [Google Scholar]
- 2. Chang K., Pastan I. (1996) Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc. Natl. Acad. Sci. U.S.A. 93, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hassan R., Kreitman R. J., Pastan I., Willingham M. C. (2005) Localization of mesothelin in epithelial ovarian cancer. Appl. Immunohistochem. Mol. Morphol. 13, 243–247 [DOI] [PubMed] [Google Scholar]
- 4. Hassan R., Laszik Z. G., Lerner M., Raffeld M., Postier R., Brackett D. (2005) Mesothelin is overexpressed in pancreaticobiliary adenocarcinomas but not in normal pancreas and chronic pancreatitis. Am. J. Clin. Pathol. 124, 838–845 [PubMed] [Google Scholar]
- 5. Argani P., Iacobuzio-Donahue C., Ryu B., Rosty C., Goggins M., Wilentz R. E., Murugesan S. R., Leach S. D., Jaffee E., Yeo C. J., Cameron J. L., Kern S. E., Hruban R. H. (2001) Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas. Identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE). Clin. Cancer Res. 7, 3862–3868 [PubMed] [Google Scholar]
- 6. Ordóñez N. G. (2003) Value of mesothelin immunostaining in the diagnosis of mesothelioma. Mod. Pathol. 16, 192–197 [DOI] [PubMed] [Google Scholar]
- 7. Hassan R., Bera T., Pastan I. (2004) Mesothelin. A new target for immunotherapy. Clin. Cancer Res. 10, 3937–3942 [DOI] [PubMed] [Google Scholar]
- 8. Ho M., Bera T. K., Willingham M. C., Onda M., Hassan R., FitzGerald D., Pastan I. (2007) Mesothelin expression in human lung cancer. Clin. Cancer Res. 13, 1571–1575 [DOI] [PubMed] [Google Scholar]
- 9. Kojima T., Oh-eda M., Hattori K., Taniguchi Y., Tamura M., Ochi N., Yamaguchi N. (1995) Molecular cloning and expression of megakaryocyte potentiating factor cDNA. (1995) Molecular cloning and expression of megakaryocyte potentiating factor cDNA. J. Biol. Chem. 270, 21984–21990 [DOI] [PubMed] [Google Scholar]
- 10. Rump A., Morikawa Y., Tanaka M., Minami S., Umesaki N., Takeuchi M., Miyajima A. (2004) Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J. Biol. Chem. 279, 9190–9198 [DOI] [PubMed] [Google Scholar]
- 11. Gubbels J. A., Belisle J., Onda M., Rancourt C., Migneault M., Ho M., Bera T. K., Connor J., Sathyanarayana B. K., Lee B., Pastan I., Patankar M. S. (2006) (2006) Mesothelin-MUC16 binding is a high affinity, N-glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Mol. Cancer 5, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaneko O., Gong L., Zhang J., Hansen J. K., Hassan R., Lee B., Ho M. (2009) A binding domain on mesothelin for CA125/MUC16. J. Biol. Chem. 284, 3739–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bast R. C., Jr., Xu F. J., Yu Y. H., Barnhill S., Zhang Z., Mills G. B. (1998) CA 125. The past and the future. Int. J. Biol. Markers 13, 179–187 [DOI] [PubMed] [Google Scholar]
- 14. Bateman A. C., al-Talib R. K., Newman T., Williams J. H., Herbert A. (1997) Immunohistochemical phenotype of malignant mesothelioma. Predictive value of CA125 and HBME-1 expression. Histopathology 30, 49–56 [DOI] [PubMed] [Google Scholar]
- 15. Ho M., Hassan R., Zhang J., Wang Q. C., Onda M., Bera T., Pastan I. (2005) Humoral immune response to mesothelin in mesothelioma and ovarian cancer patients. Clin. Cancer Res. 11, 3814–3820 [DOI] [PubMed] [Google Scholar]
- 16. Yen M. J., Hsu C. Y., Mao T. L., Wu T. C., Roden R., Wang T. L., Shih IeM. (2006) Diffuse mesothelin expression correlates with prolonged patient survival in ovarian serous carcinoma. Clin. Cancer Res. 12, 827–831 [DOI] [PubMed] [Google Scholar]
- 17. Chowdhury P. S., Pastan I. (1999) Improving antibody affinity by mimicking somatic hypermutation in vitro. Nat. Biotechnol. 17, 568–572 [DOI] [PubMed] [Google Scholar]
- 18. Hassan R., Ebel W., Routhier E. L., Patel R., Kline J. B., Zhang J., Chao Q., Jacob S., Turchin H., Gibbs L., Phillips M. D., Mudali S., Iacobuzio-Donahue C., Jaffee E. M., Moreno M., Pastan I., Sass P. M., Nicolaides N. C., Grasso L. (2007) Preclinical evaluation of MORAb-009, a chimeric antibody targeting tumor-associated mesothelin. Cancer Immun. 7, 20. [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., Xiang L., Hassan R., Paik C. H., Carrasquillo J. A., Jang B. S., Le N., Ho M., Pastan I. (2006) Synergistic antitumor activity of taxol and immunotoxin SS1P in tumor-bearing mice. Clin. Cancer Res. 12, 4695–4701 [DOI] [PubMed] [Google Scholar]
- 20. Hassan R., Schweizer C., Lu K. F., Schuler B., Remaley A. T., Weil S. C., Pastan I. (2010) Inhibition of mesothelin-CA-125 interaction in patients with mesothelioma by the anti-mesothelin monoclonal antibody MORAb-009. Implications for cancer therapy. Lung Cancer 68, 455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pastan I., Hassan R., Fitzgerald D. J., Kreitman R. J. (2006) Immunotoxin therapy of cancer. Nat. Rev. Cancer 6, 559–565 [DOI] [PubMed] [Google Scholar]
- 22. Abhinandan K. R., Martin A. C. (2008) Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol. Immunol. 45, 3832–3839 [DOI] [PubMed] [Google Scholar]
- 23. Sundberg E. J., Mariuzza R. A. (2004) Antibody structure and recognition of antigen. Molecular Biology of B Cells, (Alt F. W., Honjo T., Neuberger M. S., eds) Vol. VII, pp. 491–509, Elsevier Academic Press, London [Google Scholar]
- 24. Stanfield R. L., Zemla A., Wilson I. A., Rupp B. (2006) Antibody elbow angles are influenced by their light chain class. J. Mol. Biol. 357, 1566–1574 [DOI] [PubMed] [Google Scholar]
- 25. Sathyanarayana B. K., Hahn Y., Patankar M. S., Pastan I., Lee B. (2009) Mesothelin, stereocilin, and otoancorin are predicted to have superhelical structures with ARM-type repeats. BMC Struct. Biol. 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma J., Xia D. (2008) The use of blue native PAGE in the evaluation of membrane protein aggregation states for crystallization. J. Appl. Crystallogr. 41, 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Otwinowski Z., Minor W. (1997) Processing of X-ray Diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) BALBES. A molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 30. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]