Background: The herpes simplex virus replisome is composed of seven proteins encoded by the virus.
Results: Analyses of hybrid replisomes revealed species-specific interactions, and mutagenesis of UL8 identified amino acids required for DNA synthesis and UL52 primase interaction.
Conclusion: The herpesvirus replisome is a conserved molecular machine with species-specific interactions.
Significance: This work provides functional and mechanistic insights into herpesvirus replication.
Keywords: DNA Helicase, DNA Primase, DNA Replication, Herpesvirus, Herpes Simplex Virus, Viral Replication, UL29, UL5, UL52, UL8, UL9
Abstract
We have used oriS-dependent transient replication assays to search for species-specific interactions within the herpes simplex virus replisome. Hybrid replisomes derived from herpes simplex virus type 1 (HSV-1) and equine herpesvirus type 1 (EHV-1) failed to support DNA replication in cells. Moreover, the replisomes showed a preference for their cognate origin of replication. The results demonstrate that the herpesvirus replisome behaves as a molecular machine relying on functionally important interactions. We then searched for functional interactions in the replisome context by subjecting HSV-1 UL8 protein to extensive mutagenesis. 52 mutants were made by replacing single or clustered charged amino acids with alanines. Four mutants showed severe replication defects. Mutant A23 exhibited a lethal phenotype, and mutants A49, A52 and A53 had temperature-sensitive phenotypes. Mutants A49 and A53 did not interact with UL52 primase as determined by co-immunoprecipitation experiments. Using GFP-tagged UL8, we demonstrate that all mutants were unable to support formation of ICP8-containing nuclear replication foci. Extended mutagenesis suggested that a highly conserved motif corresponding to mutant A49 serves an important role for establishing a physical contact between UL8 and UL52. The replication-defective mutations affected conserved amino acids, and similar phenotypes were observed when the corresponding mutations were introduced into EHV-1 UL8.
Introduction
All herpesviruses rely on a common apparatus, a replisome, encoded by the virus for synthesis of new genomes (1). The structure and function of the herpesvirus replisome have been extensively characterized for herpes simplex virus type 1 (HSV-1). HSV-1 consists of a processive UL30-UL42 DNA polymerase and a UL5-UL8-UL52 helicase primase. The single-stranded DNA-binding protein ICP8 (infected cell protein 8) is encoded by the UL29 gene, and it acts during initiation of DNA replication as well as during propagation of the replication fork. The replisome is assembled on origins of replication activated by the origin-binding protein (OBP)3 encoded by the UL9 gene (2, 3). OBP is a superfamily II DNA helicase capable of site-specific unwinding of origins of DNA replication when assisted by ICP8 (4). Studies of other herpesviruses, such as EBV and CMV, have revealed orthologues to all replisome proteins except OBP. The mechanism for initiation of DNA replication thus differs between α-, β-, and γ-herpesvirus, but the replisome itself seems to be evolutionarily conserved.
Several functionally important protein-protein interactions between replisome components have been identified. The C terminus of the UL30 DNA polymerase interacts with the processivity subunit UL42 (5). The UL5 and UL52 subunits make up a functional helicase-primase complex (6, 7). UL8 binds to the UL5-UL52 dimer and facilitates unwinding of long stretches of DNA in the presence of ICP8 (8). A conserved C-terminal motif of OBP binds specifically to ICP8 (9).
Two of the components of the replisome, UL8 and UL42, display a more rapid evolution than the other components when the amino acid sequences are analyzed. UL42 appears to act as a monomer during HSV-1 replication, and it has the structure of a proliferating cell nuclear antigen monomer (10). UL8, on the other hand, has no sequence or structural homologues outside of the herpesvirus family. In addition to the well established interaction between UL8 and UL5-UL52, interactions with OBP, ICP8, and the UL30 DNA polymerase have also been proposed (8, 11, 12). These interactions have not been mapped in detail, and the functional consequences of the putative interactions remain unknown. However, the observations might indicate that UL8, a protein in itself devoid of enzymatic activities, might serve an integrative role in the herpesvirus replisome.
The replisome has been reconstituted in vitro, and it is possible to follow its assembly in vivo by observing the formation of replication foci and their growth into replication compartments (13–16). It has become increasingly clear that the herpesvirus replisome also has to cope with replication of damaged DNA templates and coordinate its function with the cellular system for DNA repair and recombination (17–19). Given the differential impact of viral DNA replication on expression of early-late (γ1) and true late (γ2) genes, it is possible that the replisome itself plays an important role for proper regulation of viral gene expression (20, 21, 22).
We have initiated a systematic search for additional and functionally significant interactions within the herpesvirus replisome. Here we report on such studies performed with hybrid replisomes reconstituted in vivo using expression plasmids encoding replication proteins from HSV-1 and EHV-1. An in depth search for interaction surfaces was carried out by systematic replacements of charged amino acids with alanines in UL8 and initially characterized in transient DNA replication assays. The replication-defective mutants were then further analyzed in co-immunoprecipitation experiments with UL5-UL52 and other replication proteins as well as the ability to co-localize with ICP8 in replication foci. The results show that a highly conserved sequence motif in UL8 is required for a physical interaction with the UL52 primase. In general, the replication-defective mutations affected conserved amino acids, and similar phenotypes were observed when the corresponding mutations were introduced into EHV-1 UL8. Our results suggest that the herpesvirus replisome operates as a functional unit requiring multiple species-specific interactions between the components of the replisome. Our results also indicate that these interactions are likely to occur in evolutionarily conserved regions of the proteins.
EXPERIMENTAL PROCEDURES
Cells and Viruses
BHK-21 cells were grown in Glasgow minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS). Equine herpesvirus 1 DNA was provided by Andrew Davison (Medical Research Council Virology Unit, Institute of Virology, Glasgow, UK).
Oligonucleotides, Plasmid Constructs, and Mutagenesis
DNA oligonucleotides were from Eurofins MWG GmbH. The plasmid pORI(WT), containing the HSV-1 origin of replication, and the plasmids expressing the seven essential HSV-1 DNA replication enzymes are described elsewhere (23, 24). pORI(EHV-1) contains EHV-1 oriS in a pUC19 vector. The expression plasmids for EHV-1 were obtained by PCR followed by TOPO cloning. The coding sequences were then transferred to the pE plasmid described previously (23). GFP-tagged HSV-1 UL8, pEmGFPUL8, was made by insertion of the coding sequence for Emerald GFP (Invitrogen) into the N terminus of HSV-1 UL8. Similarly, FLAG-tagged UL8, pFLAGUL8, was created by an oligonucleotide adding the sequence MDYKDDDDK to the N terminus of HSV-1 UL8. All UL8 mutants are described in supplemental Table 1, and a second series of derivatives is described in Table 1. HSV-1 UL8 mutants A1–A24, A47–A58, and all EHV-1 UL8 mutants were made using the QuikChange II site-directed mutagenesis kit (Stratagene). The remaining mutants were generated by two-step PCR mutagenesis. All regions amplified by PCR were verified by DNA sequencing.
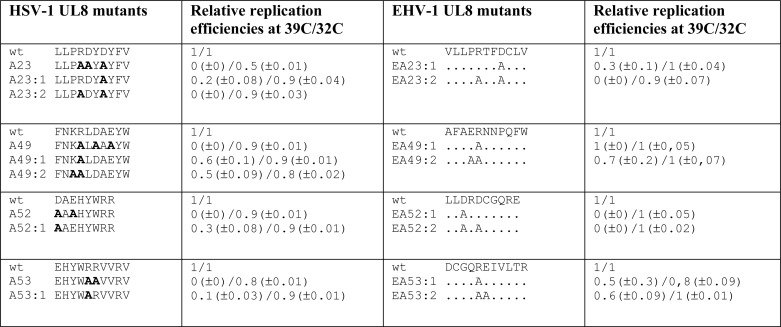
TABLE 1.
Mutations in HSV-1 and EHV-1 UL8 exhibiting replication-defective phenotypes
The amino acid substitutions made in the various mutants are shown in boldface type (see also supplemental Table 1). Replication experiments are documented in Figs. 8 and 9. Replication efficiencies in the presence of the UL8 mutants were normalized to those obtained in the presence of either wt HSV-1 UL8 or WT EHV-1 UL8. The values are derived from duplicate experiments such as those described in the legends to Figs. 8 and 9.
DNA Synthesis Analysis
BHK monolayers on 12-well plates were transfected with the indicated plasmids using Lipofectamine and Plus reagents as described by the manufacturer (Invitrogen). Cells were then cultured for 24 h at 39 °C, 48 h at 37 °C, or 72 h at 32 °C. Total DNA was isolated using the QIAmp DNA blood minikit (Qiagen) followed by overnight digestion with DpnI and HindIII. Cleaved DNA was separated by gel electrophoresis, blotted to a Hybond-H+ membrane (GE Healthcare) via alkaline transfer, hybridized to a HindIII-cleaved pUC19 radioactive probe, and visualized by autoradiography (25). All experiments were repeated at least twice.
To quantify replication efficiency, the ratio between replicated DNA and input DNA, as represented by the uppermost band on a Southern blot, was determined using a PhosphorImager or by densitometry of autoradiographs. Origin specificity was expressed as the ratio between replication efficiency for the cognate origin and the non-cognate origin.
Protein Analysis
BHK monolayers on 12-well plates were transfected at the indicated temperature, as described above, with 0.15 μg of PE5, PE52, and either the wild type or the mutant pFLAGUL8; when necessary, the total amount of DNA was adjusted to 0.45 μg with pUC19. At 12 h post-transfection, cells were lysed with M-PER mammalian protein extraction reagent (Thermo Scientific) supplemented with 0.15 m NaCl and Complete protease inhibitors mix (Roche Applied Science), followed by overnight immunoprecipitation at 4 °C with EZview red anti-FLAG M2 affinity gel (F7425, Sigma), as recommended by the manufacturer. Immunoprecipitates were eluted with Laemmli sample buffer, subjected to electrophoresis on a 4–15% SDS-polyacrylamide gradient gel, transferred to a PVDF membrane (Bio-Rad), and probed with primary rabbit polyclonal antibodies to HSV-1 UL5 and UL52 or primary mouse monoclonal antibodies to the FLAG sequence (F7425, Sigma-Aldrich), followed by goat anti-rabbit or mouse HRP-conjugated secondary antibodies (Pierce). Total expression levels of HSV-1 UL5 and UL52 and actin were analyzed by Western blot of the protein lysates in Laemmli buffer, using primary rabbit polyclonal antibodies to HSV-1 UL5 and UL52 or mouse monoclonal antibodies to actin (ab6276, Abcam). Membranes were stripped by using the Restore Western blot stripping buffer as recommended by the manufacturer (Thermo Scientific). All protein bands were detected by chemiluminescence using the Bio-Rad ChemiDoc XRS imager.
Immunofluorescence Analysis
BHK cells seeded on collagen I-coated 8-well culture slides (BD Biosciences) were transfected with the indicated plasmids and cultured at 39 °C as described above. At 6 h post-transfection, the cells were fixed with 3.7% paraformaldehyde, quenched with 0.2 m glycine, and permeabilized with 0.5% Triton X-100. The cells were then blocked with 3% FBS in phosphate-buffered saline (PBS) for 1 h at room temperature, followed by incubation with primary mouse monoclonal antibodies to ICP8 (ab20194, Abcam), diluted 1:200 in PBS with 3% FBS, for 1 h at room temperature. The subsequent incubation was with Alexa Fluor 568 goat anti-mouse IgG secondary antibodies (Molecular Probes), diluted 1:500 in PBS with 3% FBS, for 30 min at room temperature. 4′,6-Diamidino-2-phenylindole (DAPI) was used as the nuclear counterstain and as recommended by the manufacturer (Invitrogen). Images were from a Zeiss LSM700 Axio Observer.Z1 inverted confocal laser-scanning microscope located at the Center for Cellular Imaging at the Sahlgrenska Academy using a ×40/1.3 numerical aperture objective. Merging and RGB profile plots were done using the program ImageJ. Images were processed for publication using Adobe Photoshop CS3.
RESULTS
Hybrid Replisomes Reveal Species-specific Interactions
To look for functionally important interactions within the HSV-1 replisome, we settled for a comparison between HSV-1 strain 17 syn+ and an evolutionarily closely related virus EHV-1 strain Ab4. The viruses have similar origins of replication, and they both encode the conserved C-terminal of OBP responsible for an interaction with ICP8 (9). A BLAST search demonstrated 54% identities and 67% positives for the UL30 DNA polymerase and 49% identities and 65% positives for the single-stranded DNA-binding protein ICP8. In contrast, sequence similarities were lower for UL8 (30% identities and 45% positives) and for UL42 (27% identities and 47% positives). Transient transfection experiments with expression plasmids encoding these proteins and a plasmid containing HSV-1 oriS are commonly used to examine HSV-1 replication (23, 26). We have made a corresponding set of expression plasmids for EHV-1 using the same HCMV promoter and the SV40 polyadenylation signal as described previously (23). Transient replication assays in BHK cells were used to monitor the function of reconstituted HSV-1 and EHV-1 replisomes (Fig. 1).
FIGURE 1.
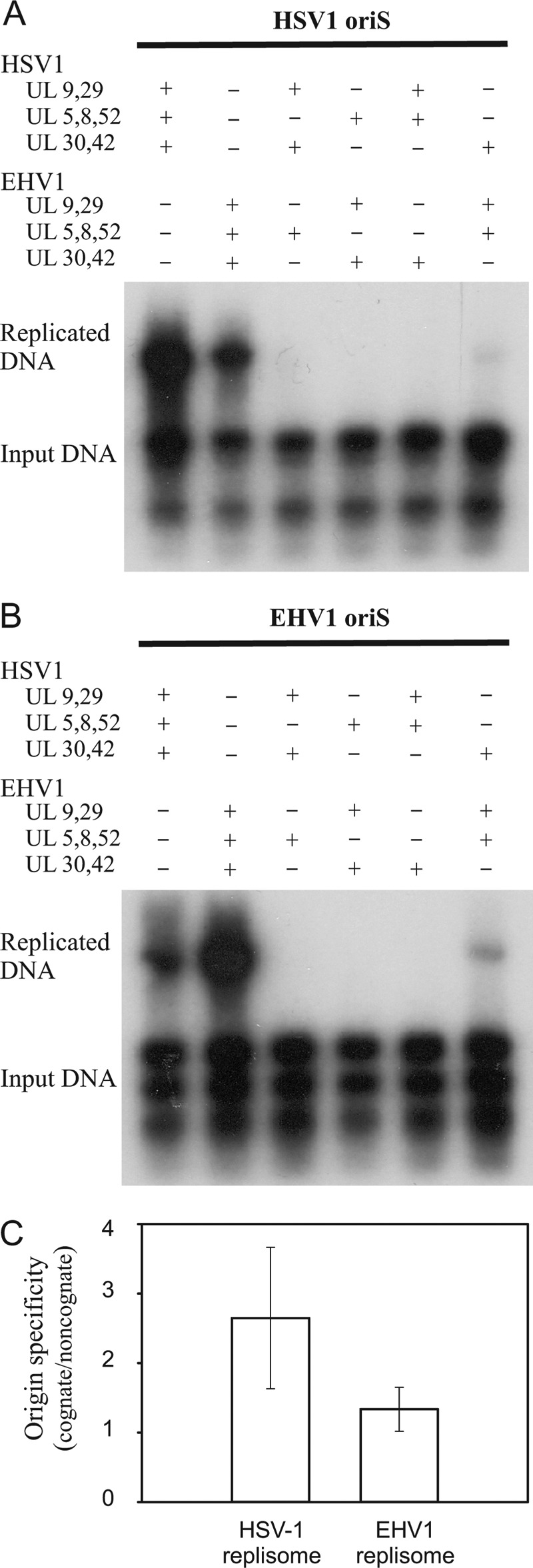
Replisomes from HSV-1 and EHV-1 rely on species-specific interactions between subcomplexes for function. A, autoradiograph from a transient replication experiment using pORI(WT) and expression plasmids encoding replisome components from HSV-1 and EHV-1 as indicated. B, autoradiograph from a transient replication experiment using pORI(EHV-1) and expression plasmids encoding replisome components from HSV-1 and EHV-1. C, comparison of origin specificity for the HSV-1 and EHV-1 replisomes. The values are the ratio of the replication efficiency for the cognate origin versus the non-cognate origin determined from three independent experiments. A ratio of 1 indicates no discrimination between the origins of replication. Error bars, S.D.
The herpesvirus replisome is composed of subcomplexes. Two interacting proteins, OBP and ICP8, activate the origins of replication; three proteins, UL5-UL8-UL52, make up the helicase-primase complex; and the third complex consists of catalytic UL30 DNA polymerase and its accessory factor UL42 (1). ICP8, a single-stranded DNA binding protein, is also continuously required at the replication fork. Here, we have tried to identify functionally significant interactions between the three subcomplexes (Fig. 1). Our results reveal that all three subcomplexes have to be derived from the same source, indicating biologically significant interactions between all subcomplexes. The only instance in which we note a residual activity in a hybrid replisome is when DNA polymerase UL30-UL42 is derived from HSV-1 and the remaining components come from EHV-1 and vice versa.
The results also showed that the HSV-1 replisome preferred a template containing the cognate HSV-1 oriS as the initiator element (Fig. 1C). The EHV-1 replisome also displayed a preference, albeit less pronounced, for the cognate oriS from EHV-1 (Fig. 1C). The recognition sequence for the initiator protein OBP is identical for all α-herpesviruses (9). Thus, discrimination between the origins of replication must involve either the detailed assembly of initiator proteins on oriS or, perhaps, a downstream event, such as productive loading of helicase-primase. HSV-1 OBP is sensitive to the number of AT base pairs separating box I and II in oriS (24, 27). In EHV-1 oriS, the AT-rich spacer sequence contains four additional AT base pairs, which would make it an unfavorable substrate for HSV-1 OBP and explain the experimental result.
We also performed experiments where only single components have been replaced. The results show that it is not possible to replace any of the replisome components from HSV-1 with the corresponding component from EHV-1 and obtain a fully active replisome (Fig. 2). Some residual activity was observed when UL5 or UL8 were replaced. In addition, we have not found evidence for inhibition of DNA synthesis when a heterologous component is co-expressed with a full complement of replisome proteins, arguing that stable non-productive complexes do not form (results not shown).
FIGURE 2.
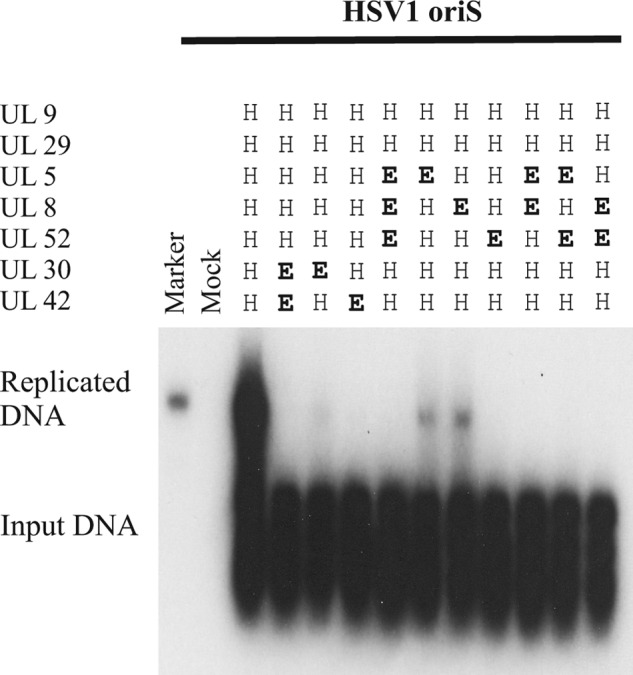
All components of the HSV-1 replisome form functionally significant species-specific contacts within the replisome. Shown is an autoradiograph from a transient replication experiment using pORI(WT) and expression plasmids encoding either HSV-1 or EHV-1 replication proteins as indicated. The marker is the HindIII-digested p2aori plasmid (36). The autoradiograph is intentionally overexposed to show low levels of replication.
Mutational Analysis of HSV-1 UL8
The UL8 protein appears to have no structural homologues outside the herpesviruses, and little information exists about functional domains in the protein (28). Results presented above and previously published observations demonstrate direct interactions between UL8 and UL5-UL52 as well as ICP8 (1, 8). It has also been suggested that UL8 interacts directly with OBP and the UL30 catalytic subunit of DNA polymerase (11, 12). To derive direct genetic information of such interactions, we settled for systematic mutagenesis of UL8 by replacing single and clustered charged amino acids with alanines. 52 mutants were made in which 1–4 closely situated charged amino acids were changed and initially examined in transient DNA replication experiments performed at 37 °C (Fig. 3 and supplemental Table 1). This strategy is likely to target primarily surface residues of the protein and therefore less likely to prevent proper folding of the protein. In fact, the vast majority of the mutants showed a WT phenotype. Mutants exhibiting reduced replication at 37 °C were also examined for a temperature-sensitive phenotype at 32 and 39 °C (Fig. 4). Mutant A23 was unable to support replication both at 32 and 39 °C and was considered to be a lethal mutant (Fig. 4 and Table 1). Mutants A49, A52 and A53 all had a strong ts phenotype (Fig. 4 and Table 1). The ts phenotype may reflect local temperature-dependent structural alterations or result from replication defects that might be compensated at a lower temperature. It was technically difficult to retrieve mutants from the middle part of UL8, and some unexpected mutants with interesting phenotypes are worth mentioning. Two variants of the mutant A28 containing the R378A and E379A substitutions were obtained: (i) mutant A28(G/E) with an additional G314E mutation and (ii) FLAG-tagged mutant version A28(Δ8) with an in-frame 8-amino acid deletion near the N terminus (amino acids 77–84). Mutant A28 has a WT phenotype, and A28(G/E) and A28(Δ8) have ts phenotypes for DNA replication (results not shown).
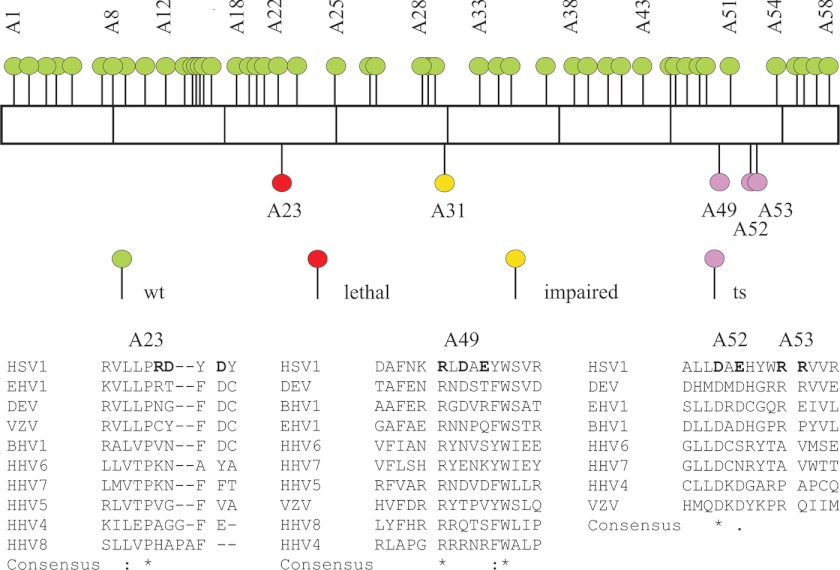
FIGURE 3.
Mutations in HSV-1 UL8 where alanines have replaced single or clustered charged amino acids. Top, each segment represents 100 amino acids of the 750-amino acid HSV-1 UL8 protein. Each mutant, as defined in supplemental Table 1, is represented by a colored symbol. Some of the mutants are indicated. The color code indicates the phenotype based on experiments in this article as follows: green, wild type; red, lethal; yellow, impaired; pink, temperature-sensitive. Bottom, the amino acid replacements made in HSV-1 UL8 for mutants A23, A49, A52, and A53 are shown in boldface type. A ClustalW alignment of UL8 sequences from 41 different herpesviruses was made. A selection from this alignment, including evolutionarily diverged α-herpesviruses and all human herpesviruses, is shown. The following Refsequences and/or accession numbers have been used: BHV1, bovine herpesvirus 1, NC_001847; DEV, anatid herpesvirus 1, duck enteritis virus, NC_013036.1; EHV-1, equine herpesvirus 1, NC_001491; HSV-1, human herpesvirus 1, herpes simplex virus type 1, NC_001806; VZV, human herpesvirus 3, varicella zoster virus, NC_ 001348.
FIGURE 4.
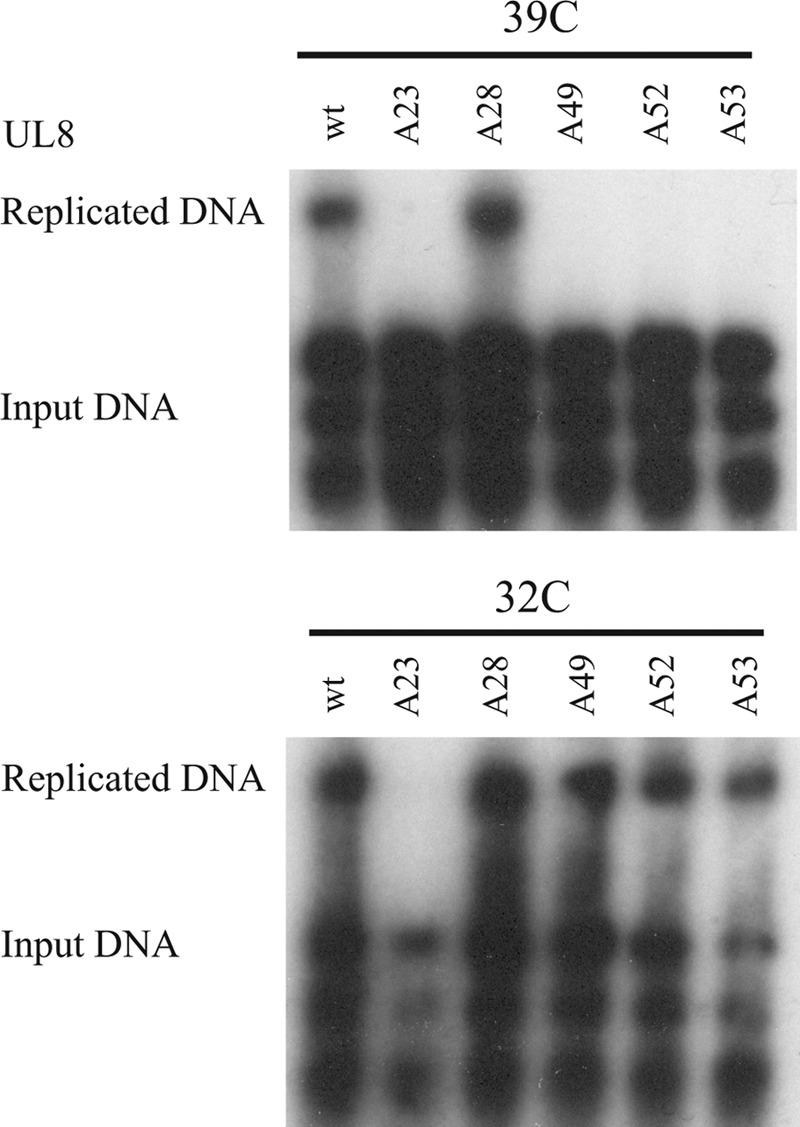
Replication defective mutants in HSV-1 UL8. Shown is an autoradiograph of a transient replication experiment in which the expression plasmid encoding WT UL8 has been replaced by expression plasmids encoding mutants A23, A28, A49, A52, and A53. The experiment was performed at 39 (top) and 32 °C (bottom).
To facilitate further analyses of the replication defects, we made GFP- and FLAG-tagged versions of the wild type UL8 and the mutants mentioned in Table 1. We first looked at the ability of N-terminally tagged WT UL8 to support transient DNA replication in comparison with untagged UL8. The results indicate that both tagged versions supported oriS-dependent DNA synthesis as efficiently as WT UL8 (Fig. 5). They were thus considered suitable for further analysis of DNA replicating defects caused by mutations in UL8.
FIGURE 5.
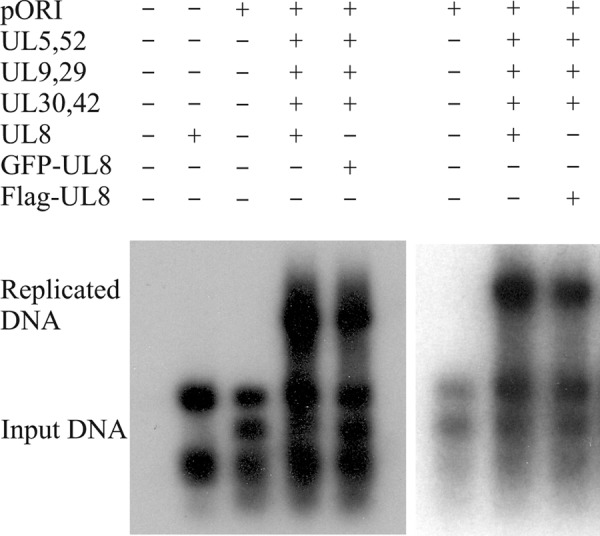
GFP- and FLAG-tagged UL8 support DNA replication. Shown is an autoradiograph from a transient replication experiment using pORI(WT) and expression plasmids encoding HSV-1 replication proteins as indicated. The replication efficiencies for WT UL8, GFP-tagged, and FLAG-tagged UL8 when expressed as the ratio between replicated DNA and input DNA (see “Experimental Procedures”) were 1.0, 1.1, and 1.0.
Effects of Mutations on Interactions with UL5 and UL52
UL8 is a part of the trimeric helicase-primase complex UL5-UL8-UL52 (29). It has been noted that a subcomplex can be formed containing the UL5 helicase and UL52 primase (6, 7). The subcomplex retains all enzymatic activities, and UL8 can bind reversibly to the UL5-UL52 subcomplex. One functional role for UL8 is to promote a direct interaction between helicase-primase and the single-stranded DNA-binding protein ICP8 and thereby facilitate unwinding of long stretches of duplex DNA in the presence of ICP8 (8). The protein-protein interactions within the UL5-UL8-UL52 complex have not been mapped, and we, therefore, tested to what extent the replication-defective mutants of UL8 were able to interact with UL5-UL52. Cells were transfected with expression vectors encoding UL5, UL52, and either FLAG-tagged WT UL8 or the FLAG-tagged UL8 mutants. Cell extracts were prepared and used for co-immunoprecipitation experiments using an anti-FLAG antibody. The immunoprecipitate was analyzed by Western blotting using an anti-UL5-UL52 antiserum (Fig. 6). We found that WT UL8 and the mutant derivatives were pulled down with similar efficiencies, indicating that the mutations were not affecting the solubility of the proteins (Fig. 6, A and B, top panels). Small amounts of UL5 and UL52 could be detected in pull-down experiments performed in cells transfected only with pE5 and pE52, indicating either poor solubility or nonspecific interactions between UL5 and UL52 with the beads (Fig. 6A, third lane). However, using pFLAGUL8 together with pE5 and pE52, we observed a selective enrichment UL52 of in the immunoprecipitate using the anti-FLAG antibody (Fig. 6A, fourth lane). A much less intense band was observed for UL5, indicating less UL5 in the precipitate as compared with the levels seen in the extract (Fig. 6A, bottom). These observations agree with a previous report (11). When the FLAG-tagged UL8 mutants were analyzed at 39 °C, we observed that mutants A23 and A28(Δ8) were able to pull down UL52. On the other hand, FLAG-tagged mutants A49 and A53 did not efficiently pull down UL52 at 39 °C, suggesting that the mutations directly affect interactions between UL8 and UL52. Mutant A52 had a slightly impaired interaction with UL52 (Fig. 6A). The absence of clear effects of the mutations on a direct interaction with UL5 helicase might indicate that the interaction between UL5 and UL8 is not very stable under the experimental conditions used (11). To test this possibility, we performed similar experiments in extracts from cells transfected with expression vectors for FLAG-tagged UL8 and either UL52 or UL5 at 37 °C. We found that that FLAG-tagged WT UL8 readily pulled down UL52 (Fig. 6B, top). Mutants A49 and A53 still failed to bring down UL52 in an immunoprecipitate (Fig. 6B, top). Under identical conditions, we could not detect specific pull-down of UL5 by FLAG-tagged WT UL8 or mutant UL8 (results not shown). It might be of interest to note that the expression levels of UL52 were reduced in extracts from cells transfected with replication-defective mutants of UL8 (Fig. 6B, bottom). This observation might indicate that improperly formed complexes between UL52 and UL8 are unstable.
FIGURE 6.
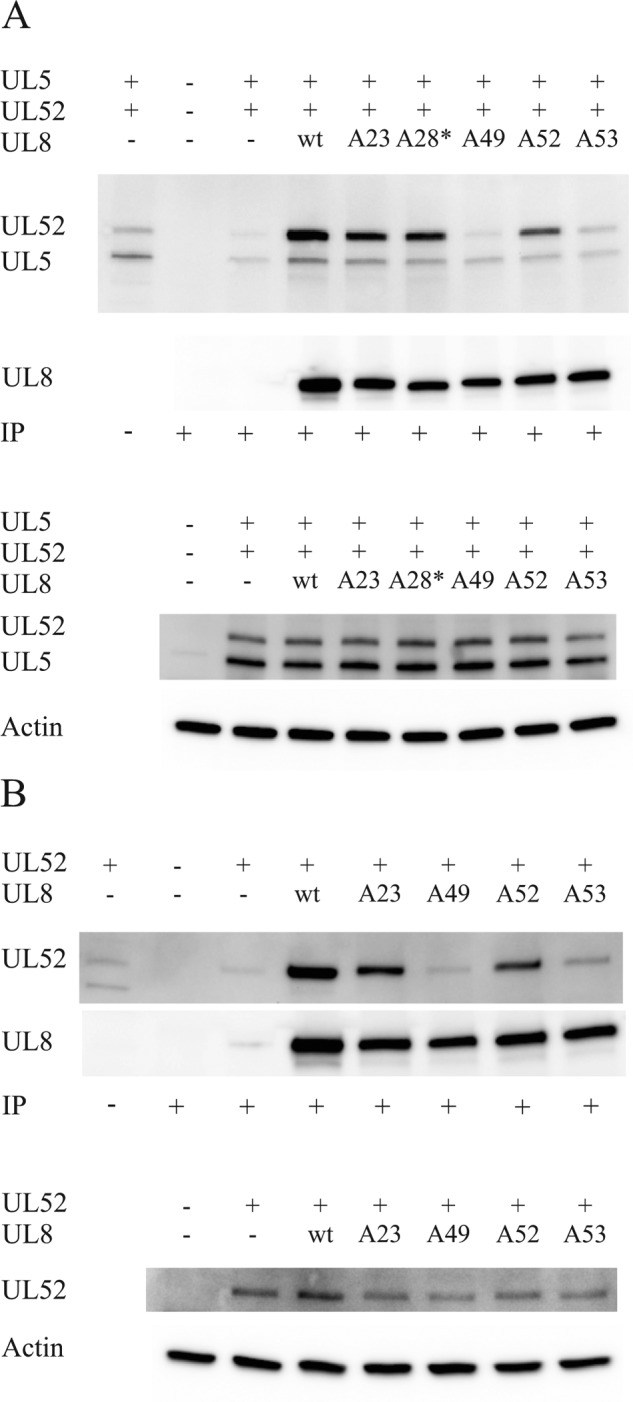
Effects of replication-defective mutations on interactions between FLAG-tagged UL8 and UL52. A, co-immunoprecipitation (IP) experiments using extracts from cells transfected at 39 °C with expression plasmids encoding FLAG-UL8 and mutant derivatives thereof together with expression plasmids encoding HSV-1 UL5 and UL52 as indicated. The blots were probed with different antibodies as indicated on the left. Top, Western blot from a co-immunoprecipitation experiment probed with an antiserum against UL5 and UL52 and, after stripping, with an anti-FLAG antibody. Lane 1, one-twentieth of the cleared extract used in the co-immunoprecipitation experiment represented by lane 3; lane 2, extract from non-transfected cells; lane 3, extract from cells transfected only with UL5 and UL52; lanes 4–9, extracts from cells transfected with FLAG-tagged WT or mutant versions of UL8 together with UL5 and UL52 as indicated. The lane labeled A28* here denotes the mutant A28(Δ8), which also has an 8-amino acid deletion (amino acids 77–84). Bottom, Western blots showing the expression of UL5 and UL52 as well as actin in the extracts used for co-immunoprecipitation. B, co-immunoprecipitation experiments using extracts from cells transfected at 37 °C with expression plasmids encoding FLAG-UL8 and mutant derivatives thereof together with an expression plasmid encoding UL52 alone. The blots were probed with different antibodies as indicated on the left. Top, Western blot from a co-immunoprecipitation experiment probed with an antiserum against UL5 and UL52 and, after stripping, with an anti-FLAG antibody. Lane 1, one-twentieth of the cleared extract used in the co-immunoprecipitation experiment represented by lane 3; lane 2, extract from non-transfected cells; lane 3, extract from cells transfected only with UL52; lanes 4–8, extracts from cells transfected with FLAG-tagged WT or mutant versions of UL8 together with UL52 as indicated. Bottom, Western blots showing the expression of UL52 as well as actin in the extracts used for co-immunoprecipitation.
We have also performed identical experiments using extracts from cells transfected at 32 °C. These experiments gave the same results as those performed with extracts from cells transfected at 39 °C. It should be noted that immunoprecipitations were performed at 4 °C in all instances. Because efficient DNA replication is supported by mutants A49, A52, and A53 at 32 °C, the observations, together with studies of the intracellular distribution of mutant UL8 examined by confocal microscopy (see below), seem to exclude the possibility that gross structural perturbations were induced in mutants A49, A52, and A53 and favor an interpretation where the mutations affect a local structure required for protein-protein interactions.
We have also tried to examine putative interactions between UL8 and other replication proteins, such as OBP, ICP8, and the UL30 subunit of DNA polymerase, using the identical experimental protocol. However, we failed to detect specific enrichment of these proteins in immunoprecipitates with FLAG-tagged WT UL8 as well as with mutant versions of UL8 (results not shown).
Intracellular Distribution of UL8
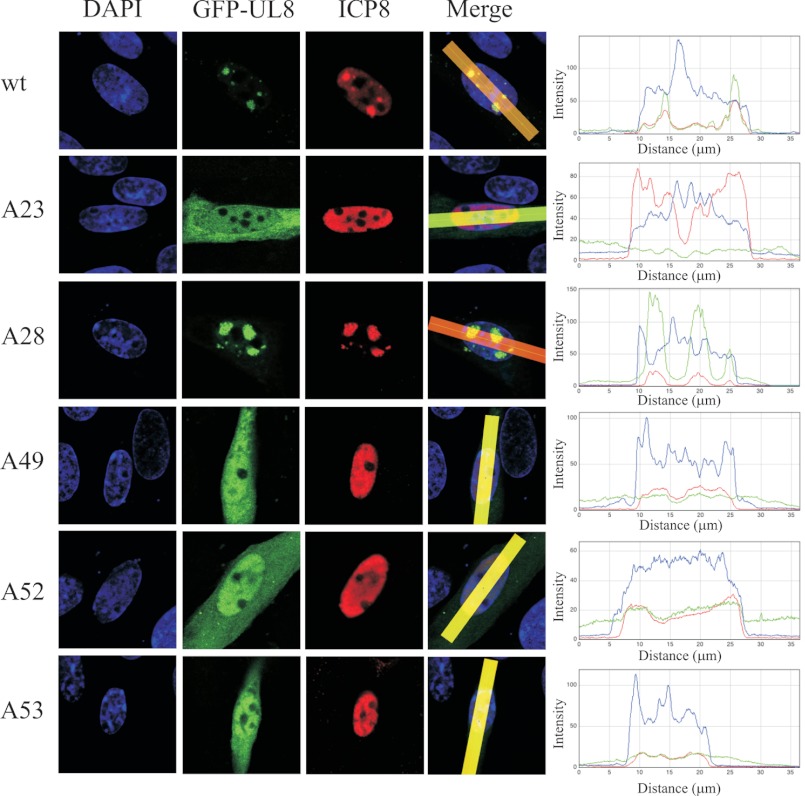
We have looked at the cellular distribution of UL8 in the context of DNA replication. Cells were transfected with a plasmid containing HSV-1 oriS, pORI(WT), and six expression plasmids: pE9, pE29, pE5, pE52, pE30, and pE42. In addition, expression plasmids encoding either wild type GFP-UL8 or GFP-tagged mutant versions of UL8 were included in the transfection mixture. After incubation at 39 °C for 6 h, the cells were fixed and stained with DAPI and an anti-ICP8 antibody and subsequently analyzed by confocal microscopy (Fig. 7). The results reveal that wild type GFP-tagged UL8 as well as mutant A28, which has a WT phenotype, supported formation of intranuclear replication foci in which both UL8 and ICP8 co-localized. Mutant A23 did not support formation of replication foci, and nuclear localization was also impaired. Mutants A49, A52, and A53 also failed to produce replication foci, but GFP-UL8 was still able to translocate to the nucleus, although a substantial fraction still remained in the cytoplasm. The ability of the mutant proteins to enter the nucleus suggests that they are not grossly misfolded.
FIGURE 7.
Effects of UL8 mutations on cellular distribution. Cells were transfected with pORI(WT) together with plasmids expressing GFP-UL8, WT, or mutants, as indicated, and the remaining six HSV-1 replication proteins. At 6 h post-transfection at 39 °C, cells were fixed and stained with DAPI and an anti-ICP8 antibody detected with an Alexa Fluor 568 secondary antibody. The cells were then examined by confocal microscopy using a ×40 objective. Merging and profile scans were made using ImageJ (right-hand panels). To facilitate visual inspection, the images were subsequently adjusted in Adobe Photoshop using the auto levels command.
Mutations Affecting DNA Replication Occur in Conserved Regions of UL8
When we looked at the mutations affecting DNA replication, we observed that they either involved evolutionarily conserved amino acids in UL8 or were in close association with conserved motifs (Fig. 3). A second series of mutations was thus made in order to look at the role of the conserved amino acids, and they were examined for their effects on DNA replication at 32 and 39 °C (Table 1 and Fig. 8A). The derivative A23:1, in which alanine replaced a conserved aspartic acid, showed a severe reduction in replication at 39 °C. A double mutant, A23:2, behaved as a strong ts mutant, unable to support replication at the non-permissive temperature. The derivatives of the triple mutant A49 showed moderately reduced replication at 39 °C (Fig. 8A and Table 1). Alanine substitutions of a conserved aspartic acid in A52 and of a moderately conserved arginine in A53 both severely reduced replication at 39 °C. We conclude that the conserved amino acids in mutants A23, A52, and A53 all are important for efficient DNA replication.
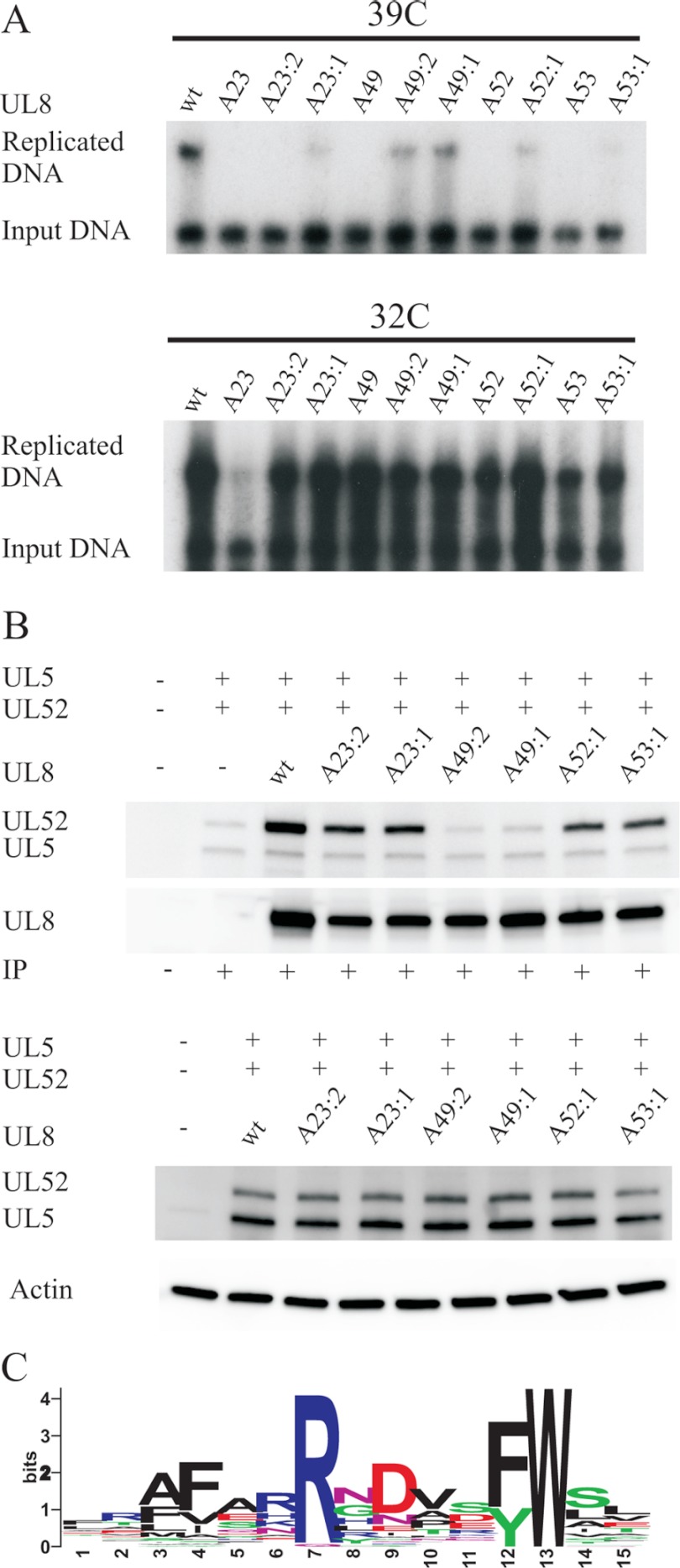
FIGURE 8.
Conserved amino acids in HSV-1 UL8 are functionally important. A, autoradiographs from transient replication experiments performed at 39 °C for 24 h and 32 °C for 72 h. Cells were transfected with plasmids encoding WT or single and double mutants derived from previously characterized mutants in UL8 as indicated, pORI(WT), and the remaining six HSV-1 expression plasmids. The mutants are defined in Table 1. B, co-immunoprecipitation experiments using extracts from cells transfected at 39 °C with expression plasmids encoding FLAG-UL8 or mutant derivatives thereof together with expression plasmids encoding HSV-1 UL5 and UL52 as indicated. The blots were probed with different antibodies as indicated on the left. Top, Western blot from a co-immunoprecipitation experiment probed with an antiserum against UL5 and UL52 and, after stripping, with an anti-FLAG antibody. Lane 1, extract from non-transfected cells; lane 2, extract from cells transfected only with UL5 and UL52; lanes 3–9, extracts from cells transfected with FLAG-tagged WT or mutant versions of UL8 together with UL5 and UL52 as indicated. Bottom, Western blots showing the expression of UL5 and UL52 as well as actin in the extracts used for co-immunoprecipitation. C, a ClustalW alignment of UL8 sequences from 41 different herpesviruses was used to create a sequence logo for a conserved motif corresponding to mutant 49 (37, 38).
To examine the effects of the single and double mutants on the interaction with UL52 primase, we performed additional co-immunoprecipitation experiments (Fig. 8B). Again, we found that WT UL8 and the mutant derivatives were pulled down with similar efficiencies, indicating that the mutations were not affecting the solubility of the proteins (Fig. 8B, top). We found that only mutants A49:1 and A49:2 showed a pronounced reduction in the ability to specifically pull down UL52. The result may imply that the region targeted by the A49 mutations is directly required for binding of the UL8 protein to UL52 primase.
We also investigated, by bioinformatics, the extent of sequence conservation in the region affected by the A49 mutations and found a highly conserved sequence motif present not only in α-herpesviruses but also in β- and γ-herpesviruses (Figs. 3 and 8C). The motif consists of a stretch of seven charged or polar amino acids surrounded by phenylalanine or tyrosine on both sides. In addition, a ubiquitously conserved tryptophan is found at the downstream end. Within the polar stretch of amino acids, an arginine residue conserved in 40 of 41 examined protein sequences was identified.
We next examined if mutations of the conserved amino acids in EHV-1 also gave rise to replication-defective phenotypes. A new set of mutations replacing the conserved amino acids with alanines in EHV-1 UL8 was made (Table 1). Expression plasmids encoding the UL8 mutants were then used in transient replication experiments (Fig. 9). Our results show that the mutants EA23:1, EA23:2, EA52:1, and EA52:2 were all strongly defective for DNA replication at 39 °C. From our analysis of HSV-1 UL8, these mutations appear not to affect the interaction with primase but still affect residues essential for DNA synthesis. Furthermore, mutants EA23:1, EA52:1, and EA52:2 also had a strong ts phenotype (Table 1) (Fig. 9). Mutants EA23:2 and EA53:1 showed replication defects at both temperature. For EA53:1, replication at 32 °C seemed to be most affected.
FIGURE 9.
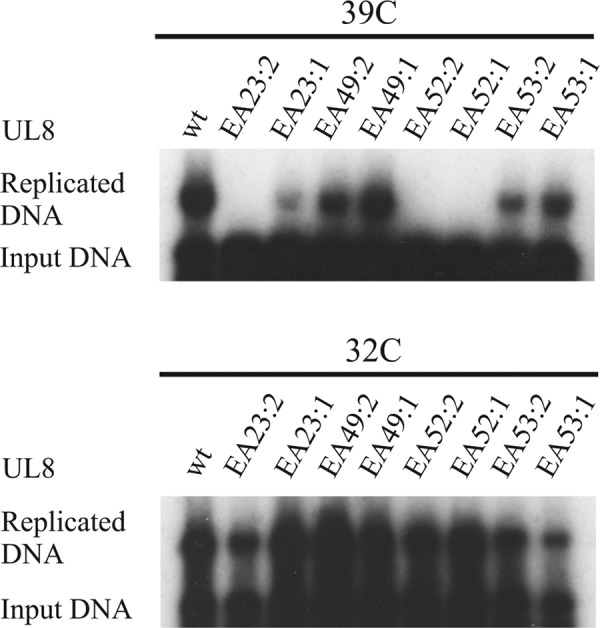
Functional correlation between conserved amino acids in HSV-1 UL8 and EHV-1 UL8. Autoradiographs of transient replication experiments performed at 39 °C for 24 h (top) and 32 °C for 72 h (bottom). Cells were transfected with plasmids encoding WT or mutant versions of EHV-1 UL8 as indicated, pORI(EHV-1), and the remaining six EHV-1 expression plasmids. The mutants are defined in Table 1.
Surprisingly, mutants A49:1 and A49:2, both affecting the highly conserved arginine residue, had very limited effects on DNA synthesis. It is possible that overexpression of proteins in transient replication assays might compensate for a poor interaction with the UL52 primase, as observed for HSV-1 UL8. Together, these observations indicate that the molecular interactions and functions of UL8 are conserved in evolution despite considerable changes in the primary sequence.
DISCUSSION
We have addressed, in a systematic manner, functionally significant interactions in the herpesvirus replisome. We show, using hybrid replisomes derived from herpes simplex virus type 1 and equine herpesvirus 1, that replication of their genomes depends on species-specific recognition and activation of origins of DNA replication but even more so on species-specific protein-protein interactions within the replisome (Figs. 1 and 2). It therefore seems that, despite significant mechanistic similarities, the machineries performing this task have diverged to the extent that genetic exchange involving its components will not be possible. It is likely that the driving force behind the evolution of the replisome is adaptation to the host and not solely to the replication efficiency. We have limited information about how quickly herpesviruses develop to reach a state where genetic exchange becomes unlikely. An intertypic recombinant in which the HSV-1 genome has acquired the left-hand 18% of the HSV-2 genome, including the UL5 and UL8 genes, has significantly reduced primase activity and reduced neurovirulence (30). The effect can be attributed to the UL5 and not the UL8 component of hybrid helicase-primase (31). Thus, already for evolutionarily closely related viral species, such as HSV-1 and HSV-2, the malfunction of hybrid molecular machines poses a restriction on genetic exchange.
We observe that the HSV-1 replisome clearly prefers HSV-1 oriS, and, conversely, the EHV-1 replisome shows a slight preference for EHV-1 oriS (Fig. 1C). It therefore appears that loading of replisomes on herpesvirus chromosomes tends to be species-specific despite the use of identical recognition sequences for the UL9 origin-binding protein and a similar arrangement of its binding sites (9). It is, however, not unexpected, because mutations that alter the distance between boxes I and II in HSV-1 oriS by half a helical turn have been shown to form a stable but apparently inactive complex at the origin of replication, causing a significant reduction of DNA synthesis (32, 24). The structural features of the origin-binding protein that allow discrimination between origins of replication are not known.
Our studies of hybrid replisomes support the idea that a herpesvirus replisome behaves as an integrated molecular machine despite the fact that it cannot be isolated as a stable complex. The observation points at a number of yet identified molecular interactions of functional importance. For example, the HSV-1 UL9 origin-binding protein assisted by the single-stranded DNA-binding protein ICP8 is capable of activating the viral origin of replication, but it cannot support the formation of productive replication forks consisting of EHV-1 proteins (Fig. 1). A conceivable explanation could be that species-specific interactions exist between ICP8 and helicase-primase. However, neither the HSV-1 origin-binding protein nor the EHV-1 counterpart can be combined with the remaining replication proteins from a heterologous source to support replication, suggesting that, in addition to the conserved motif, WPXXXGAXXFXXL, species-specific interactions also affect binding of ICP8 to OBP or contribute to OBP-mediated loading of some other replisome component (results not shown).
A more puzzling observation concerns the interaction between DNA polymerase and other replication proteins. It is known that for HSV-1 and EHV-1, the UL30 catalytic subunit interacts via its extreme C terminus with the processivity subunit UL42 (10, 33). We observe here that neither UL30 nor UL42 can be replaced individually by a heterologous counterpart, suggesting that the interaction between UL30 and UL42 is species-specific (Fig. 2). Although it has been reported that the UL 30 DNA polymerase interacts with UL8, the functional importance of the interaction is unknown (12). DNA polymerase does not, for example, stimulate the helicase activity during leading strand synthesis using a preformed replication fork. T7 DNA polymerase appears to work just as well (34). Less is known about DNA polymerase on the lagging strand. It is possible that some mechanism will exist that facilitates coupling between synthesis of primers and the subsequent delivery of primers to the UL30-UL42 DNA polymerase (35). Such a mechanism might involve interactions between the DNA polymerase and one or more components of helicase-primase.
The considerations discussed above and previous experimental observations suggesting interactions between UL8 and ICP8, between UL8 and UL9, and between UL8 and UL30 make UL8 an interesting candidate for a closer examination. It has been shown that deletion of 23 amino acids from the N terminus or 33 from the C terminus abolishes the ability of UL8 to support DNA synthesis (28). However, these studies provide little insight into functional domains of the protein. We have performed a systematic study of UL8 using a protocol in which either single or clustered charged amino acids were subjected to site-directed mutagenesis and replaced by alanines. We have screened 52 mutants for their ability to support DNA synthesis. The vast majority of these mutants exhibited a wild type phenotype. However, because our initial screening was at 37 °C, it may still be that some mutants have a phenotype at a different temperature. Five mutants with distinct phenotypes were identified (Fig. 3 and Table 1). The properties of the mutants were characterized in transient replication assays, by confocal microscopy, and by co-immunoprecipitation. In summary, mutant A23, with R254A, D255A, and D257A substitutions, failed to support DNA replication at 32 and 39 °C, and its localization to the nucleus was impaired. However, it was capable of binding to the UL52 primase subunit of the helicase-primase. The single point mutation of the conserved Asp-257 residue reduced DNA replication, and the double mutant R254A/D257A, displayed a strong ts phenotype (Table 1). The mutations might affect the structure of UL8 or an interaction with cellular or viral factors. A molecular explanation is yet to be found. Mutant A31 showed reduced replication but was not further analyzed here (supplemental Table 1). Mutants A49, A52, and A53 were all ts mutants clustered near the C terminus. Interestingly, mutants A49 and A53 did not interact with UL52, whereas for A52, the interaction with UL52 was slightly impaired. These three mutants all were found in the nucleus as well as in the cytoplasm, but they were unable to support the formation of replication foci. A variant of the mutant A28, A28(Δ8), with an in-frame deletion of 8 amino acids (amino acids 77–84) near the N terminus, still bound UL52 (Fig. 6). Together, our results strongly suggest that UL52 interacts with the C-terminal part of UL8 in a way that is specifically affected in mutants A49 and A53. We were unable to detect interactions between WT UL8 or mutant derivatives thereof with UL5 as well as OBP, ICP8, and the UL30 subunit of DNA polymerase using our standard co-immunoprecipitation assay (results not shown). It is of interest to note that a previous report also had difficulties in identifying a robust complex between UL5 and UL8 (11).
Extended mutagenesis of UL8 indicates that a highly conserved sequence motif in UL8 affected by the mutations in A49 is specifically required for the interaction with UL52 primase, as detected by co-immunoprecipitation. In contrast, mutations in A52 had strong temperature-dependent effects on DNA synthesis but very little effect on the interaction with UL52 primase. Finally, for the mutant A53 and its single mutant derivative A53:1, we observed strong temperature-dependent effects on DNA synthesis. However, although the A53 mutant did not bind to UL52 in a co-immunoprecipitation experiment, the single mutant A53:1 was able to interact with UL52. Together, this genetic analysis hints at complex and dynamic interactions between UL8 and UL52 primase at the replication fork. This is expected if we consider that the UL52 primase binds intermittently to ICP8-coated single-stranded DNA during primer synthesis, whereas it is likely to remain attached to the UL5 helicase moving in the opposite direction on the lagging strand template. A putative role for UL8 might then be to regulate the conformation and the activity of UL52.
Interestingly, when the corresponding mutations are introduced into EHV-1 UL8, the replication phenotypes become very similar. This observation suggests that the molecular interactions and functions of UL8 are conserved in evolution despite considerable changes in the primary sequence.
To conclude, our results argue that the herpesvirus replisome acts as an integrated molecular machine relying on species-specific but evolutionarily conserved molecular interactions between its components. Once the individual components from EHV-1 have been purified, it should be possible to dissect the functional role of each of the interactions in biochemical assays employing hybrid replisomes and mutant versions of the replication proteins. It would also be of considerable interest to transfer the ts mutations to the virus genome and analyze the effects on virus replication. Such studies may also help to elucidate how DNA replication is coordinated with repair and recombination as well as gene regulation.
Supplementary Material
Acknowledgment
Confocal microscopy was performed at the Center for Cellular Imaging at the Sahlgrenska Academy, University of Gothenburg.
This work was supported by grants from the Swedish Cancer Foundation and the Swedish Research Council (to P. E.).

This article contains supplemental Table 1.
- OBP
- origin-binding protein.
REFERENCES
- 1. Muylaert I., Tang K. W., Elias P. (2011) Replication and recombination of herpes simplex virus DNA. J. Biol. Chem. 286, 15619–15624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Elias P., O'Donnell M. E., Mocarski E. S., Lehman I. R. (1986) A DNA-binding protein specific for an origin of replication of herpes simplex virus type 1. Proc. Natl. Acad. Sci. U.S.A. 83, 6322–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Olivo P. D., Nelson N. J., Challberg M. D. (1988) Herpes simplex virus DNA replication. The UL9 gene encodes an origin-binding protein. Proc. Natl. Acad. Sci. U.S.A. 85, 5414–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aslani A., Olsson M., Elias P. (2002) ATP-dependent unwinding of a minimal origin of DNA replication by the origin-binding protein and the single-strand DNA-binding protein ICP8 from herpes simplex virus type I. J. Biol. Chem. 277, 41204–41212 [DOI] [PubMed] [Google Scholar]
- 5. Bridges K. G., Hua Q., Brigham-Burke M. R., Martin J. D., Hensley P., Dahl C. E., Digard P., Weiss M. A., Coen D. M. (2000) Secondary structure and structure-activity relationships of peptides corresponding to the subunit interface of herpes simplex virus DNA polymerase. J. Biol. Chem. 275, 472–478 [DOI] [PubMed] [Google Scholar]
- 6. Calder J. M., Stow N. D. (1990) Herpes simplex virus helicase-primase. The UL8 protein is not required for DNA-dependent ATPase and DNA helicase activities. Nucleic Acids Res. 18, 3573–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dodson M. S., Lehman I. R. (1991) Association of DNA helicase and primase activities with a subassembly of the herpes simplex virus 1 helicase-primase composed of the UL5 and UL52 gene products. Proc. Natl. Acad. Sci. U.S.A. 88, 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falkenberg M., Bushnell D. A., Elias P., Lehman I. R. (1997) The UL8 subunit of the heterotrimeric herpes simplex virus type 1 helicase-primase is required for the unwinding of single strand DNA-binding protein (ICP8)-coated DNA substrates. J. Biol. Chem. 272, 22766–22770 [DOI] [PubMed] [Google Scholar]
- 9. Olsson M., Tang K. W., Persson C., Wilhelmsson L. M., Billeter M., Elias P. (2009) Stepwise evolution of the herpes simplex virus origin-binding protein and origin of replication. J. Biol. Chem. 284, 16246–16255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zuccola H. J., Filman D. J., Coen D. M., Hogle J. M. (2000) The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell 5, 267–278 [DOI] [PubMed] [Google Scholar]
- 11. McLean G. W., Abbotts A. P., Parry M. E., Marsden H. S., Stow N. D. (1994) The herpes simplex virus type 1 origin-binding protein interacts specifically with the viral UL8 protein. J. Gen. Virol. 75, 2699–2706 [DOI] [PubMed] [Google Scholar]
- 12. Marsden H. S., McLean G. W., Barnard E. C., Francis G. J., MacEachran K., Murphy M., McVey G., Cross A., Abbotts A. P., Stow N. D. (1997) The catalytic subunit of the DNA polymerase of herpes simplex virus type 1 interacts specifically with the C terminus of the UL8 component of the viral helicase-primase complex. J. Virol. 71, 6390–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Falkenberg M., Lehman I. R., Elias P. (2000) Leading and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proc. Natl. Acad. Sci. U.S.A. 97, 3896–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stengel G., Kuchta R. D. (2011) Coordinated leading and lagging strand DNA synthesis by using the herpes simplex virus 1 replication complex and minicircle DNA templates. J. Virol. 85, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quinlan M. P., Chen L. B., Knipe D. M. (1984) The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36, 857–868 [DOI] [PubMed] [Google Scholar]
- 16. Livingston C. M., DeLuca N. A., Wilkinson D. E., Weller S. K. (2008) Oligomerization of ICP4 and rearrangement of heat shock proteins may be important for herpes simplex virus type 1 prereplicative site formation. J. Virol. 82, 6324–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohni K. N., Livingston C. M., Cortez D., Weller S. K. (2010) ATR and ATRIP are recruited to herpes simplex virus type 1 replication compartments even though ATR signaling is disabled. J. Virol. 84, 12152–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muylaert I., Elias P. (2010) Contributions of nucleotide excision repair, DNA polymerase η, and homologous recombination to replication of UV-irradiated herpes simplex virus type 1. J. Biol. Chem. 285, 13761–13768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weller S. K. (2010) Herpes simplex virus reorganizes the cellular DNA repair and protein quality control machinery. PLoS Pathog. 6, e1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Honess R. W., Roizman B. (1974) Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14, 8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Honess R. W., Roizman B. (1975) Regulation of herpesvirus macromolecular synthesis. Sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. U.S.A. 72, 1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mavromara-Nazos P., Roizman B. (1987) Activation of herpes simplex virus 1 γ 2 genes by viral DNA replication. Virology 161, 593–598 [DOI] [PubMed] [Google Scholar]
- 23. Stow N. D., Hammarsten O., Arbuckle M. I., Elias P. (1993) Inhibition of herpes simplex virus type 1 DNA replication by mutant forms of the origin-binding protein. Virology 196, 413–418 [DOI] [PubMed] [Google Scholar]
- 24. Gustafsson C. M., Hammarsten O., Falkenberg M., Elias P. (1994) Herpes simplex virus DNA replication. A spacer sequence directs the ATP-dependent formation of a nucleoprotein complex at oriS. Proc. Natl. Acad. Sci. U.S.A. 91, 4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yao X. D., Elias P. (2001) Recombination during early herpes simplex virus type 1 infection is mediated by cellular proteins. J. Biol. Chem. 276, 2905–2913 [DOI] [PubMed] [Google Scholar]
- 26. Wu C. A., Nelson N. J., McGeoch D. J., Challberg M. D. (1988) Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J. Virol. 62, 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aslani A., Simonsson S., Elias P. (2000) A novel conformation of the herpes simplex virus origin of DNA replication recognized by the origin-binding protein. J. Biol. Chem. 275, 5880–5887 [DOI] [PubMed] [Google Scholar]
- 28. Barnard E. C., Brown G., Stow N. D. (1997) Deletion mutants of the herpes simplex virus type 1 UL8 protein. Effect on DNA synthesis and ability to interact with and influence the intracellular localization of the UL5 and UL52 proteins. Virology 237, 97–106 [DOI] [PubMed] [Google Scholar]
- 29. Crute J. J., Tsurumi T., Zhu L. A., Weller S. K., Olivo P. D., Challberg M. D., Mocarski E. S., Lehman I. R. (1989) Herpes simplex virus 1 helicase-primase. A complex of three herpes-encoded gene products. Proc. Natl. Acad. Sci. U.S.A. 86, 2186–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barrera I., Bloom D., Challberg M. (1998) An intertypic herpes simplex virus helicase-primase complex associated with a defect in neurovirulence has reduced primase activity. J. Virol. 72, 1203–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bloom D. C., Stevens J. G. (1994) Neuron-specific restriction of a herpes simplex virus recombinant maps to the UL5 gene. J. Virol. 68, 3761–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lockshon D., Galloway D. A. (1988) Sequence and structural requirements of a herpes simplex viral DNA replication origin. Mol. Cell Biol. 8, 4018–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loregian A., Case A., Cancelotti E., Valente C., Marsden H. S., Palù G. (2006) Cloning, expression, and functional characterization of the equine herpesvirus 1 DNA polymerase and its accessory subunit. J. Virol. 80, 6247–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Falkenberg M., Elias P., Lehman I. R. (1998) The herpes simplex virus type 1 helicase-primase. Analysis of helicase activity. J. Biol. Chem. 273, 32154–32157 [DOI] [PubMed] [Google Scholar]
- 35. Sherman G., Gottlieb J., Challberg M. D. (1992) The UL8 subunit of the herpes simplex virus helicase-primase complex is required for efficient primer utilization. J. Virol. 66, 4884–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao X. D., Matecic M., Elias P. (1997) Direct repeats of the herpes simplex virus a sequence promote nonconservative homologous recombination that is not dependent on XPF/ERCC4. J. Virol. 71, 6842–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schneider T. D., Stephens R. M. (1990) Sequence logos. A new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo. A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.