Background: O2 pathways in animal hemoglobins and myoglobins are controversial.
Results: Ligands enter and exit sperm whale Mb and Cerebratulus lacteus Hb by completely different pathways.
Conclusion: Rational mutagenesis mapping can identify ligand migration pathways and provides experimental benchmarks for testing molecular dynamics simulations.
Significance: Globins can use either a polar gate or an apolar tunnel for ligand entry.
Keywords: Carbon Monoxide, Hemoglobin Myoglobin, Ligand-binding Protein, Molecular Dynamics, Mutagenesis Site-specific, Oxygen Binding, Cerebratulus lacteus, Sperm Whale
Abstract
Although molecular dynamics simulations suggest multiple interior pathways for O2 entry into and exit from globins, most experiments indicate well defined single pathways. In 2001, we highlighted the effects of large-to-small amino acid replacements on rates for ligand entry and exit onto the three-dimensional structure of sperm whale myoglobin. The resultant map argued strongly for ligand movement through a short channel from the heme iron to solvent that is gated by the distal histidine (His-64(E7)) near the solvent edge of the porphyrin ring. In this work, we have applied the same mutagenesis mapping strategy to the neuronal mini-hemoglobin from Cerebratulus lacteus (CerHb), which has a large internal tunnel from the heme iron to the C-terminal ends of the E and H helices, a direction that is 180° opposite to the E7 channel. Detailed comparisons of the new CerHb map with expanded results for Mb show unambiguously that the dominant (>90%) ligand pathway in CerHb is through the internal tunnel, and the major (>75%) ligand pathway in Mb is through the E7 gate. These results demonstrate that: 1) mutagenesis mapping can identify internal pathways when they exist; 2) molecular dynamics simulations need to be refined to address discrepancies with experimental observations; and 3) alternative pathways have evolved in globins to meet specific physiological demands.
Introduction
Most animal myoglobins (Mbs)4 and red cell hemoglobins (Hbs) bind O2 reversibly for storage or transport. The ligand covalently coordinates to the heme iron atom and is fully enclosed by amino acid side chains to prevent exposure to solvent and oxidation. In crystal structures, there is often no obvious pathway for ligand migration into the distal portion of the heme pocket shown in Fig. 1A. In 1966 after publication of the first high resolution globin structure, Perutz and Matthews (1) suggested that O2 enters Hb and Mb by a short channel gated by the distal histidine at the E7 helical position, and this idea was later supported by outward movement of the distal histidine when large ligands were bound to Mb (2, 3). However, in 1969, internal cavities were discovered in sperm whale myoglobin (SwMb) capable of binding xenon at moderate pressures (≤10 atm (4–9)). This observation led Petsko and co-workers (5) and others in the 1980s to suggest that instead of passing through the His(E7) gate, diatomic gases may enter and exit globins through apolar pathways between helices and the loops connecting them.
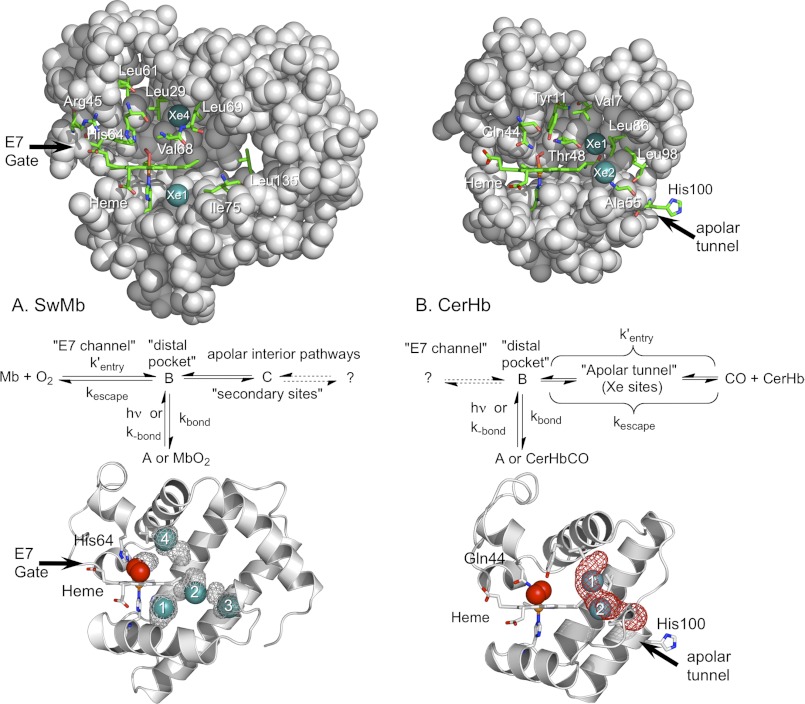
FIGURE 1.
Side views of WT CerHbO2 and SwMbO2. In both globins the molecules are oriented so that the planar heme group is viewed edge-on, its propionates are facing left and the “distal pocket” is above the heme. Portions of the protein matrix are removed so the interior view is clear. Xe atoms are depicted as teal spheres; key amino acids and the heme prosthetic groups are indicated by sticks with carbon in green, oxygen in red, and nitrogen in blue; the remainder of the protein structure is represented as gray spheres. A, upper panel, SwMbO2 with most of the E-helix removed (PDB code 2MGM) and Xe1 and Xe4 inserted into positions obtained from the metMb structure determined at high Xe pressures (PDB code 2W6W). Key distal pocket residues include Leu-29(B10), His-64(E7), Val-68(E11), and Phe-43(CD1). Additional mutants constructed in this study include mutations made at residues Arg-45(CD3), Leu-61(E4), Leu-69(E12), Ile-75(E18), and Leu-135(H12). Middle panel, mechanism for ligand binding to Mb based on Scott et al. (13, 38). Lower panel, cavity surfaces (gray mesh) in SwMbO2 (PDB code 2MGM), obtained using the surface function in PyMOL, are compared with the Xe atom positions found in metMb under high Xe pressures (PDB code 2W6W). As described in Pesce et al. (39), no apolar tunnels were found in SwMb using Surfnet (64). We also found no tunnel in SwMb using MOLE online 2.0 (65) using the default settings. The cavities found using PyMOL in SwMbO2 are not connected. Xe1–Xe4 atoms were inserted into the structure based on their positions in 2W6W. B, upper panel, WT CerHbO2 (PDB code 1KR7) with most of the E helix removed and bound Xe atoms inserted at their locations in L86A CerHbO2 equilibrated with Xe gas (PDB code 2XKH). Key distal pocket and apolar tunnel residues include Val-7(B6), Tyr-11(B10), Gln-44(E7), Thr-48(E11), Ala-55(E18), Ala-82(G8), Leu-86(G12), Leu-98(H9), and His-100(H11) in WT CerHbO2. Middle panel, mechanism for ligand binding to CerHb taken from Salter et al. (42) and Pesce et al. (43). Lower panel, ribbons drawing for WT CerHbO2 (1KR7) with an apolar tunnel (red mesh) computed using MOLE online 2.0 (65) with a probe radius of 3 and 1.25 Å interior threshold (default settings). Xe atoms were inserted based on their positions in PDB code 2XKH.
The current evidence for movement of photodissociated ligands out of the distal pocket and into the Xe1 and Xe4 sites in SwMb is clear and comes from time-resolved spectroscopic methods (10–13) and from direct observations by time-resolved x-ray crystallography (14–20). However, these measurements cannot detect whether ligands enter and exit the protein from these transient positions. Ligand movements out of the protein involve overcoming larger activation barriers, and the resultant unstable transition states cannot be seen directly. Consequently, entry and exit pathways have to be inferred either theoretically from molecular dynamics (MD) simulations or experimentally from the effects of mutagenesis on kinetic parameters for ligand entry and escape.
In 1990, Elber and Karplus (21) carried out locally enhanced simulations of ligand movement in SwMb and observed multiple trajectories for ligand migration out of the protein. In 2010, Elber (22) reviewed the most recent computational literature, which still indicates that there are multiple ligand pathways in animal myoglobins and hemoglobins and that the channel gated by the distal histidine is only a minor route for entry and exit (23–31).
In 1994, Huang and Boxer (32) used an elegant random mutagenesis protocol to create and screen a library of ∼1,500 single point mutants. Mutations were identified from colonies that produced lysates containing Mbs with kinetic parameters significantly different (>50%) from those of wild-type (WT) controls. Roughly 10% of the mutants examined showed differences, and a structural model of Mb was constructed to highlight the positions of the mutants showing altered kinetic parameters. In addition to positions near the bound ligand, other regions were highlighted, including interior positions near the Xe cavities and the C-terminal portion of the H helix. These results were interpreted to mean that multiple pathways for CO and O2 entry and exit exist, supporting the conclusions from the initial MD simulations (32).
Building on Huang and Boxer's strategy, our group explored the E7 gate and Xe cavity pathways in SwMb by a rational mutagenesis mapping strategy in which small (Gly or Ala) and large (Phe or Trp) mutations were constructed in ligand-accessible cavities, along the major trajectories specified by the MD simulations and at some of the positions found in the random mutagenesis screen (13). Each of 90 different mutants was produced in milligram quantities; geminate and bimolecular rate parameters for ligand binding were measured with pure proteins; and in many cases, crystal structures were determined (33–38).
Our mutagenesis mapping results strongly indicate the following: 1) although O2 can move from the binding pocket into the Xe cavities, it must return to the distal pocket to exit the protein, and 2) the major pathway (≥75%) for O2 entry and exit is the short channel that opens by outward movement of the distal histidine side chain. The discrepancy between our mutagenesis map and that of Huang and Boxer (32) is not as great as it first appears. The largest changes in their random mutagenesis map were observed at or near the E7 gate, and most of the effects observed at remote positions can be rationalized as false positives due to mutations that destabilize the Mb structure, leading to lysates containing partially unfolded proteins with exposed or weakly bound heme groups (i.e. many of the Pro substitutions (32)). The multiple trajectories obtained from MD simulations are harder to reconcile and imply that either we cannot “find” internal pathways with a rational mutagenesis mapping strategy or the models and interaction potentials used in the simulations cannot accurately define ligand movements into solvent.
In 2002, Pesce et al. (39) reported the structure of the mini-hemoglobin from the sea worm Cerebratulus lacteus (CerHb). CerHb is a neuronal O2 storage protein, which surrounds the axons and brain of the Nemertean worm, and it rapidly releases O2 when the worm burrows into sand and becomes anaerobic while searching for prey (40). The analysis of the CerHb crystal structure revealed the presence of an apolar tunnel leading from the distal pocket to solvent, with an exit point between the E and H helices created by the loss of the N-terminal A helix (Fig. 1B). This tunnel was suggested to be the major route of entry and exit for O2 binding, and later work indicated that this “open” channel accounts for the very large association rate constants for O2 and NO binding to CerHb (200–300 μm−1 s−1), which are close to the values expected for diffusion-controlled processes (41).
Our initial mutagenesis and Xe binding studies of CerHb supported the original idea of Pesce et al. (39) that the internal tunnel is the major ligand pathway (39, 42, 43). Thus, we have used CerHb as a counter example to SwMb where the histidine gate mechanism for O2 binding appears to apply. However, Orlowski and Nowak (44) carried out simulations with CerHb and, as with SwMb, obtained multiple trajectories with the apolar tunnel being only one of them.
To resolve this discrepancy and determine whether the experimental mutagenesis mapping strategy can identify internal pathways, we constructed and examined the O2, CO, and NO ligand binding properties of 120 different mutants of CerHb. Rates of ligand entry and exit for this library of CerHb variants were compared with results for an expanded library of SwMb mutants, which extends the results of Scott et al. (13) to regions near the ends of the E and H helices.
Our goals were: 1) to verify the mutagenesis mapping strategy; 2) to define experimentally the ligand pathways in the two proteins; 3) to compare the results to MD simulations; and 4) to provide benchmarks for future theoretical work.
EXPERIMENTAL PROCEDURES
Hb and Mb Sample Preparations
WT and mutant recombinant CerHbs were expressed and purified as described previously using a synthetic gene with codon usage optimized for expression in Escherichia coli (39, 41). The WT SwMb gene and expression plasmid (pMb413) were constructed by Springer and Sligar (45) to optimize codon usage in E. coli. All new mutants were constructed using this gene in a pUC19-based expression vector and the Stratagene PCR-based QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Purification was achieved as described previously by Springer and Sligar (45, 46) and modified by Carver et al. (47). The WT control and the mutant Mbs have an extra N-terminal Met residue and a D122N substitution for crystallization in the P6 space group, neither of which has an effect on ligand binding (48). All experiments to determine rate constants were carried out in 0.1 m phosphate buffer, pH 7.0, 1.0 mm EDTA, 20 °C.
Measurement of Rate Constants for Ligand Association (kO2′, kCO′, and kNO′) and Dissociation (kO2 and kCO)
CO association time courses were measured after complete laser photolysis of SwMbCO and CerHbCO samples containing various [CO] under pseudo first order conditions as described previously (42). The association rate constants for NO binding to deoxy-SwMb and deoxy-CerHb were measured using a flow-flash multimixing apparatus and a 500-ns dye laser (42, 43, 49). In these experiments, SwMbCO or CerHbCO samples in deoxygenated buffer were mixed with various concentrations of NO and then photolyzed ∼10–20 ms after mixing, allowing measurement of time courses for NO binding to the newly formed deoxy-SwMb or deoxy-CerHb without the limitations of the low quantum yields of CerHbNO and SwMbNO complexes. O2 association and dissociation time courses were measured after complete laser photolysis of SwMbCO and CerHbCO samples in O2/CO mixtures using a 500-ns dye laser as described previously (42, 49, 50). Experimental procedures and fitting routines to obtain values for kO2′, and kO2 (kO2′ indicates oxygen association rate constant; and kO2 indicates oxygen dissociation rate constant) from analyses of the fast bimolecular binding and slow O2 displacement phases have been described in previous work (42, 49, 50).
Measurement of Geminate Recombination
Time courses for internal geminate CO and O2 recombination within CerHb and SwMb were measured at 436 nm after excitation with a 7-ns excitation pulse from a Lumonics YAG-laser system, using a Tektronix TDS3052 digitizing oscilloscope with nanosecond time resolution. Experimental procedures and fitting routines were followed as described previously (13, 42). Sample time courses are shown in Figs. 5–7. The results were analyzed and interpreted in terms of the previously published mechanisms for ligand binding to SwMb (13) and CerHb (42, 43), as described below.
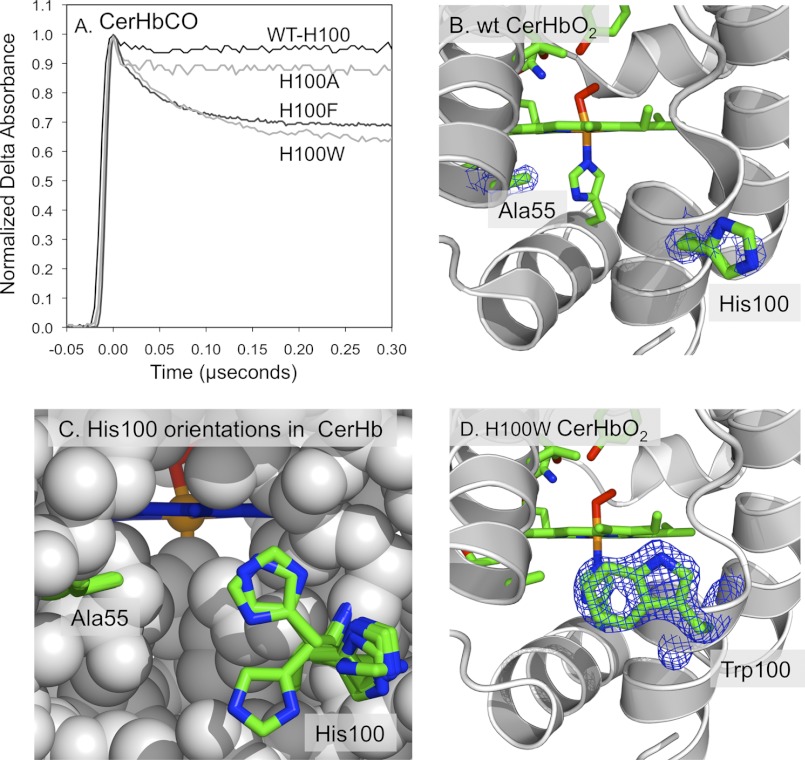
FIGURE 5.
His-100 is at the entrance to the apolar tunnel in CerHb. A, time courses for geminate recombination in His-100(H11) mutants of CerHbCO at 20 °C, pH 7.0. The fraction of geminate recombination increases 6-fold upon mutating the polar His to large aromatic amino acids. B, 2Fo − Fc electron density maps for Ala-55(E18) cutoff at 1.5 σ, and His-100(H11) cutoff at 0.7 σ in WT CerHbO2 (PDB code 1KR7). The polar imidazole side chain of His-100 at the H11 helical position is poorly defined, cannot be seen at the higher contour levels, and appears to extend out and interact with solvent molecules, keeping the channel open. C, positions of the His-100(H11) side chain in the structures for the O2 and aquomet complexes of WT CerHb (PDB codes 1KR7 and 2XKI), L86A CerHbO2 ± Xe (PDB codes 2XKG and 2XKH), T48V CerHbO2 (PDB code 1V07), A55W CerHbO2 (PDB code 2VYY), and A55F CerHbO2 (PDB code 2VYZ). D, 2Fo − Fc electron density map for the Trp-100(H11) cutoff at 1.5 σ in H100W CerHbO2 (PDB code 4F6I). The indole side chain is well defined.
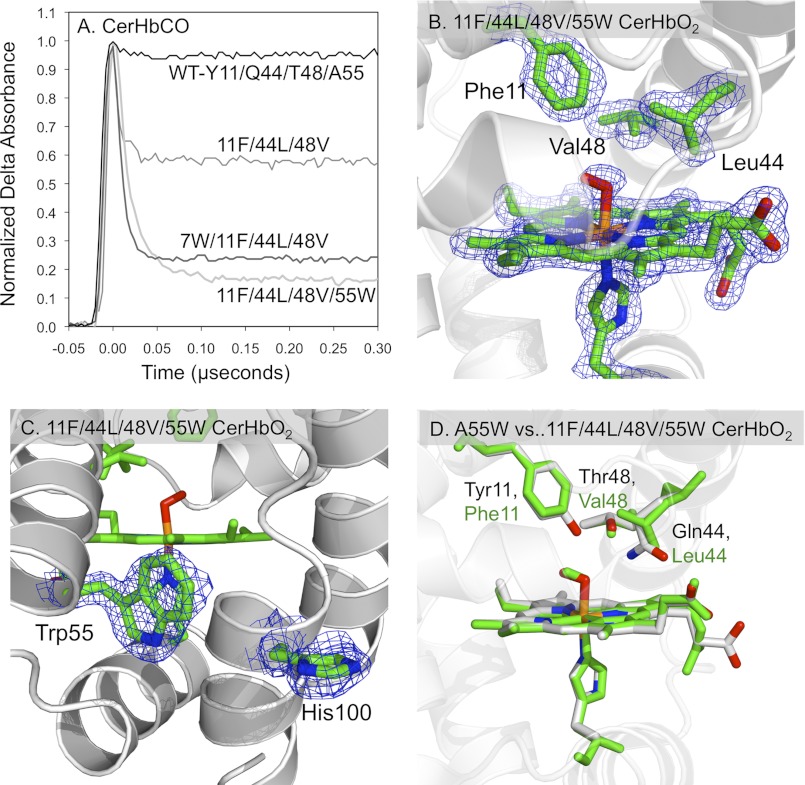
FIGURE 6.
Effects of an apolar active site on the structural and geminate recombination properties of CerHbCO. A, geminate rebinding in multiple mutants of CerHbCO with an apolar distal pocket containing Y11F/Q44L/T48V mutations at the B10, E7, and E11 helical positions, respectively, and Trp replacements at the Val-7(B6) and Ala-55(E18) tunnel positions. B, 2Fo − Fc electron density maps cutoff at 1.5 σ for the heme group, axial ligands, Phe-11(B10), Leu-44(E7), and Val-48(E11) in Y11F/Q44L/T48V/A55W CerHbO2 (PDB code 4F68). As shown, all three side chains are very well defined. C, 2Fo − Fc electron density maps for Trp-55(E18) cutoff at 1.5 σ and His-100(H11) cutoff at 1.0 σ in Y11F/Q44L/T48V/A55W CerHbO2 (PDB code 4F68). The side chain for His-100 is less well defined compared with the indole ring of Trp-55. D, comparison of the active sites of A55W and Y11F/Q44L/T48V/A55W CerHbO2. The Cα atoms of the structures for A55W (PDB code 2VYY) and quadruple CerHbO2 mutant (PDB code 4F68) were superimposed in PyMOL. The heme and active site side chains of the Y11F/Q44L/T48V/A55W CerHbO2 mutant are drawn as green sticks, whereas the heme and active site polar side chains of A55W CerHbO2 are drawn in CPK colors, with carbon, oxygen, nitrogen, and iron atoms being gray, red, blue, and orange, respectively. Positions 11, 44, 48, and 55 correspond to the B10, E7, E11, and E18 helical locations in the globin fold.
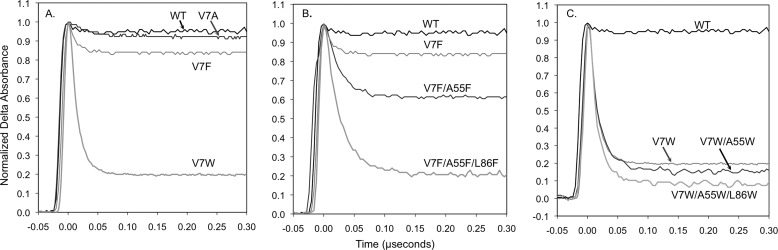
FIGURE 7.
Time courses for geminate recombination in multiple tunnel mutants of CerHb at 20 °C, pH 7.0. A, CO geminate rebinding in position Val-7(B6) mutants of CerHb. B, CO geminate rebinding in single, double, and triple Phe mutants in positions Val-7(B6), Ala-55(E18), and Leu-86(G12) mutants of CerHbCO. C, CO geminate rebinding in single, double, and triple Trp mutants in positions Val-7(B6), Ala-55(E18), and Leu-86(G12) mutants of CerHbCO.
Analysis of Bimolecular and Geminate Rate Parameters
In Scott et al. (13), bimolecular and geminate O2 bindings to SwMb were analyzed in terms of the three-step, side-path model shown in Fig. 1A. The rate constants for intramolecular rebinding, escape, and movement through the protein between sites B and C were obtained by fitting sets of time courses to this model using numerical integration algorithms described by Scott and Gibson (12). The geminate rebinding time courses for WT SwMb and many of the mutants can also be fitted well by the sum of two exponential processes and an offset reflecting the amount of escape to solvent. In this analysis, the rate of the fast phase equals kbond + kBC + kescape, the total fraction of geminate recombination, Fgem,total, equals kbond/(kbond + kescape), and the fraction of the slow phase, Fgem,slow, approximately equals kBC/(kbond + kBC + kescape), allowing an estimation of all three rate parameters (13). It should be noted that a simple two-step reaction analysis, using the correct Fgem,total, provides the same value of kentry′ and an only slightly larger kescape value.
The side-path mechanism was chosen for analysis of SwMb data because both Xe binding and mutations that fill the Xe1 and Xe4 cavities collapse the time courses to a single rapid phase with little or no change in the total fraction of geminate rebinding (12, 13). Similar results were obtained for geminate ligand rebinding to WT Scapharca inaequivalvis Hbl, where again, Xe binding and Phe and Trp replacements in internal cavities caused little change in the total fraction of geminate recombination (51). In the case of HbA subunits, single geminate phases for CO rebinding are observed at room temperature on nanosecond time scales and indicate that there are no side path C states, and again, the E7 channel appears to be the path for ligand entry and escape (49, 52). Thus, the scheme shown in Fig. 1A appears to apply to most animal hemoglobins with a tertiary and distal pocket structure similar to that of SwMb.
The fraction of photodissociated ligands that geminately rebind after photolysis is defined solely by kbond/(kbond+kescape) in the SwMb scheme in Fig. 1A. There is no dependence on the rates of interconversion of the C and B sites, which only affect the rates and amplitudes of the slower geminate phases. Thus, regardless of the complexity of the internal movements, the overall bimolecular rate constant for ligand binding to SwMb under physiological conditions ([C], [B], d[C]/dt, and d[B]/dt ≈ 0) is given by (13) Equation 1,
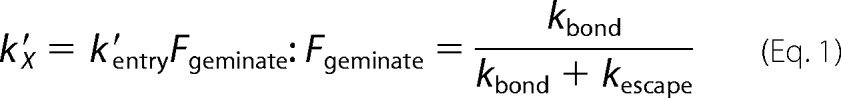 |
where X stands for O2, CO, or NO. In the scheme shown in Fig. 1A, kentry′ represents the second order rate constant for formation of state B, starting with ligand in the solvent phase. Values of kentry′ for O2 entry in SwMb were calculated from kO2′/Fgeminate and are listed in Table 3 for new mutants and in Scott et al. (13) for the remaining 90 variants.
TABLE 3.
Bimolecular, geminate, and calculated rate parameters for new SwMb mutants located at positions near the E7 channel and near the ends of the E and H helices measured at pH 7.0, 20 °C
| Mutants | Bimolecular binding parametersa |
Observed geminate parametersa |
Calculated rate parametersa |
|||||
|---|---|---|---|---|---|---|---|---|
| kO2′ | kO2 | KO2 | kgem, O2 | Fgem, O2 | kbond | kescape | kentry′ | |
| μm−1s−1 | s−1 | μm−1 | μs−1 | μs−1 | μs−1 | μm−1s−1 | ||
| WT | 16 ± 3b | 14 ± 3 | 1.1 ± 0.2 | 12 ± 3 | 0.47 ± 0.09 | 5.7 ± 1.0 | 6.3 ± 1.0 | 34 ± 7 |
| Arg-45(CD3) | ||||||||
| Ala-45 | 7.7 | 30 | 0.26 | 5.8 | 0.31 | 1.8 | 4.0 | 25 |
| Trp-45 | 6.2 | 21 | 0.30 | 10 | 0.13 | 1.3 | 8.7 | 48 |
| Leu-61(E4) | ||||||||
| Ala-61 | 11 | 26 | 0.43 | 19 | 0.33 | 6.6 | 12 | 33 |
| Trp-61 | 8.8 | 11 | 0.80 | 15 | 0.24 | 3.6 | 11 | 37 |
| Leu-69(E12) | ||||||||
| Ala-69 | 28 | 13 | 2.2 | 22 | 0.46 | 10 | 12 | 61 |
| Trp-69 | 13 | 5.2 | 2.5 | 32 | 0.61 | 20 | 13 | 21 |
| Ile-75(E18) | ||||||||
| Ala-75 | 22 | 8.5 | 2.6 | 16 | 0.35 | 5.6 | 10 | 63 |
| Trp-75 | 16 | 8.8 | 1.8 | 16 | 0.43 | 6.9 | 9.1 | 37 |
| Leu-135(H12) | ||||||||
| Val-135 | 21 | 8.1 | 2.6 | 20 | 0.40 | 8.0 | 12 | 53 |
| Trp-135 | 23 | 6.5 | 3.5 | 18 | 0.33 | 5.9 | 12 | 70 |
a The values of KO2 were calculated from kO2′/kO2, and its standard deviation was calculated from the standard propagation of error formula. When fitting the O2 geminate data, single and double exponential fits were examined. In the case of kgem values, estimates from one exponential fits were used initially. In the case of Fgem values, two exponential fits were used to estimate the total Fgem,O2. Similar values for kentry′ and kescape were obtained from two exponential analyses as described under “Experimental Procedures.” Because no significant Renhance values were observed for these mutants, further more sophisticated analyses were not warranted.
b Rate constants in bold are taken from Ref. 13.
It is important to note that the experimental values of Fgem are model-independent and depend only on an accurate definition of the absorbance change for the total amount of geminate recombination. Thus, Equation 1 can also be used empirically to estimate the rate of entry into any globin, including CerHb, where in Fig. 1B binding involves a linear sequential set of ligand movements. As discussed by Scott et al. (13), the steady-state expression for kX′ using a linear scheme is still kX′ = kentry′ Fgem, but in this case kentry′ represents the bimolecular rate of entry into the first discrete site in the protein, which, in the case of CerHb, would be the apolar tunnel. However, if the barriers along the tunnel and from the tunnel into the distal pocket are small compared with bond formation, the mechanism reduces to a two-step process with a single geminate intermediate. Under these conditions, Fgem can again be approximated as kbond/(kbond + kescape).
In the case of CO binding to CerHb, there is evidence of a barrier between the tunnel and the distal pocket implying a linear three-step scheme. However, this extra complexity is only evident at low temperatures (53) or at room temperature when restrictions in the tunnel are completely removed as is observed in the L86A mutant (43).
When movement into and through the tunnel is restricted by mutagenesis, the geminate time courses become single exponentials and are readily analyzed as a simple two-step scheme (Equation 2),
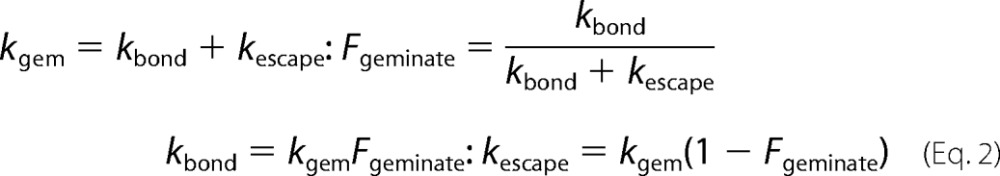 |
We used the expressions in Equations 1 and 2 to estimate values of kentry′, kescape, and kbond for CO binding to all of the mutants of CerHb examined (Tables 1 and 4 and supplemental Tables S1–S4). These estimations, although empirical, are the simplest and closest to the observed experimental data, making the maps shown in Fig. 4 experimentally defined and not model-dependent.
TABLE 1.
Selected rate constants displaying correlations between calculated rates of ligand entry, kNO′, and kO2′ in CerHb mutants measured at pH 7.0, 20 °C
| Mutantsa | kNO′ | kO2′ | kentry, CO′ |
|---|---|---|---|
| μm−1 | μm−1s−1 | μm−1s−1 | |
| WTa | 230 ± 30 | 230 ± 17 | 640 ± 400 |
| Val-7(B6) | |||
| V7A | 240 | 250 | 450 |
| V7F | 110 | 110 | 140 |
| V7W | 30 | 33 | 16 |
| Gln-44(E7) | |||
| Q44F | 110 | 20 | 26 |
| Thr-48(E11) | |||
| T48V | 80 | 30 | 27 |
| Ala-55(E18) | |||
| A55V | 140 | 180 | 260 |
| A55L | 47 | 72 | 37 |
| A55F | 48 | 61 | 39 |
| A55W | 45 | 63 | 34 |
| Leu-86(G12) | |||
| L86A | 200 | 230 | 250 |
| L86F | 110 | 120 | 150 |
| L86W | 68 | 97 | 30 |
| Phe-7/Phe-55/Phe-86 | 32 | 42 | 22 |
a The numbers for WT CerHb are based on the average of all determinations for the past 6 years (≥10 separate preparations). As described in the text, the error in kentry′ is very large for WT CerHb because Fgem is close to 0.0, poorly defined, and has an error of ±60%. When Fgem is ≥0.1, the error in kentry′ diminishes greatly to approximately ±20%. Complete sets of O2 and CO binding parameters for these and all other variants examined are given in supplemental Tables S1–S4. Some of the parameters in this table were taken from previous work (42, 43) or were re-measured.
TABLE 4.
Key rate and equilibrium parameters for O2, CO, and NO binding to wild-type and selected mutants of CerHb at pH 7, 20 °C
| CerHb varianta | kO2′ | kO2 | KO2 | kCO′ | Fgem, CO | kentry′(kNO′) | kescapeb |
|---|---|---|---|---|---|---|---|
| μm−1s−1 | s−1 | μm−1 | μm−1s−1 | μs−1 | |||
| WT | 230 ± 17 | 190 ± 25 | 1.2 ± 0.2 | 32 ± 5 | 0.05 ± 0.03 | 640 ± 400 (270) | 76 ± 40 |
| H100A | 210 | 230 | 0.93 | 30 | 0.05 | 570 | 72 |
| H100F | 99 | 110 | 0.90 | 17 | 0.29 | 59 | 14 |
| H100W | 77 | 92 | 0.84 | 17 | 0.34 | 50 | 7.0 |
| Y11F/Q44L/T48V | 340 | 270 | 1.6 | 165 | 0.40 | 410 | 44 |
| Y11F/Q44L/T48V/A55W | 70 | 50 | 1.4 | 27 | 0.85 | 30 | 7.5 |
| V7W/Y11F/Q44L/T48V | 82 | 54 | 1.5 | 40 | 0.77 | 52 | 22 |
| WT | 230 ± 17 | 190 ± 25 | 1.2 ± 0.2 | 32 ± 5 | 0.05 ± 0.03 | 640 ± 400 (270) | 76 ± 40 |
| V7A | 250 | 270 | 0.93 | 36 | 0.08 | 450 (240) | 30 |
| L86A | 230 | 150 | 1.5 | 25 | 0.10 | 250 (200) | 21 |
| V7A/L86A | 240 | 190 | 1.3 | 35 | 0.14 | 255 | 7.3 |
| V7F | 110 | 115 | 0.97 | 22 | 0.16 | 135 (110) | 54 |
| V7W | 33 | 14 | 2.4 | 13 | 0.82 | 16 (30) | 13 |
| L86F | 120 | 71 | 1.7 | 24 | 0.17 | 140 (110) | 77 |
| L86W | 97 | 60 | 1.6 | 16 | 0.54 | 30 (68) | 36 |
| A55F | 61 | 51 | 1.2 | 17 | 0.44 | 38 (48) | 5.0 |
| A55W | 63 | 66 | 0.95 | 15 | 0.44 | 34 (45) | 5.0 |
| V7F/L86F/A55F | 42 | 17 | 2.5 | 17 | 0.79 | 20 (32) | 8.2 |
| V7W/L86W/A55W | 23 | 3.0 | 7.6 | 7.2 | 0.93 | 8 | 4.1 |
a Complete sets of association and dissociation rate parameters and equilibrium constants for CO and O2 binding to these variants and others are given in supplemental Tables S1–S4.
b The values in parentheses in the second-to-last column are the values for kNO′, the bimolecular association rate constant for NO binding, which should approximate the computed rate of ligand entry, kentry′ (see supplemental Table S1). The errors in kentry′ and kescape for WT CerHb are very large due to the small value of Fgem, and this problem and the validity of the calculations are discussed under “Experimental Procedures.”
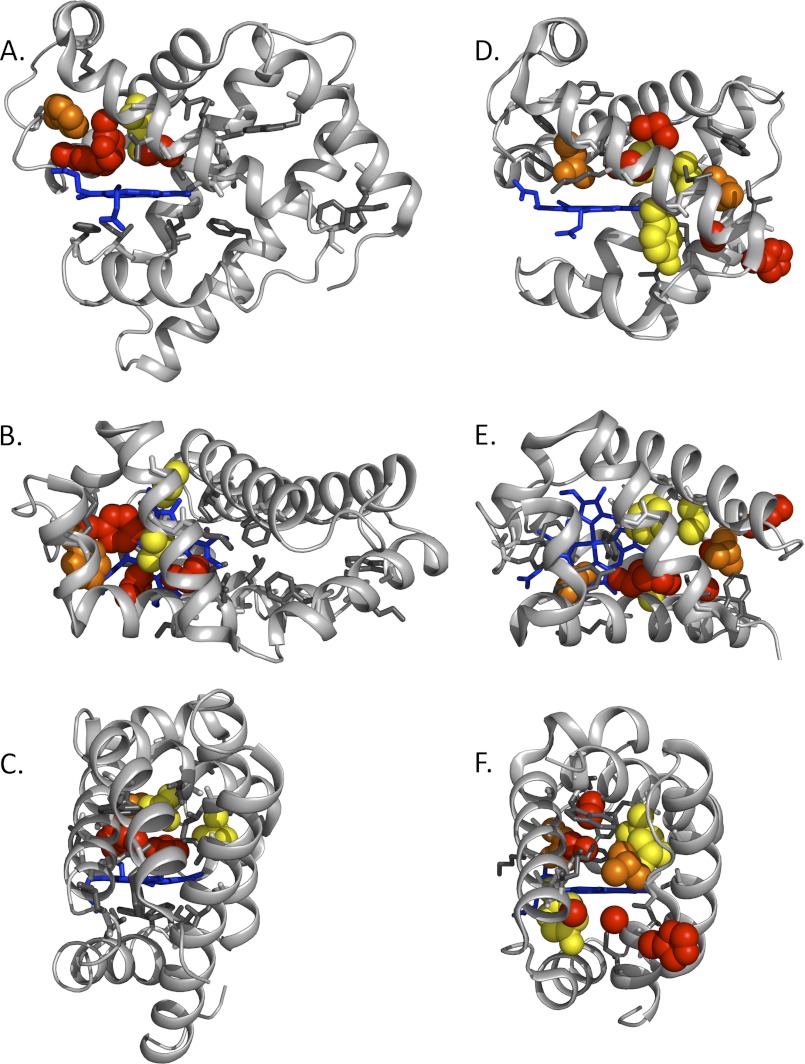
FIGURE 4.
Maps of the effects of large-to-small mutations on the rate of entry and escape in SwMb (PDB code 2MGK) and CerHb (PDB code 1KR7). The protein backbones are represented as silver ribbons, and the heme groups are shown as blue sticks. A–C, side, top, and back views of SwMb. D–F, side, top, and back views of CerHb and its ligand tunnel. Red spheres signify Renhance ≥ 1.5; orange spheres signify 1.5 > Renhance ≥ 1.0; yellow spheres signify 1.0 > Renhance ≥0.5; white or gray sticks signify 0.5 > Renhance (no significant effect). Numerical values are shown in Fig. 3 (13) and can be computed from the supplemental tables.
NO Association and the Validity of Using kentry′ and kescape
In principle, the calculated values of kentry′ and kescape should be used to map the ligand pathways quantitatively because these parameters factor out the differences in reactivity (kbond values) between the O2, CO, and NO. The values of these entry and exit parameters should be the same for all three diatomic gases because of their similar sizes and diffusion constants at room temperature. In the case of CO binding to WT CerHb, the relative errors in these calculated parameters are very large because the extent of CO geminate recombination is very small, ≤0.05, and it has an absolute error of approximately ±0.03, which is roughly the same for all values of Fgem measured. To reassure ourselves that the calculated kentry′ values are valid for the CerHb mutants, we examined the correlations between kentry′, kNO′, and kO2′ for some of the key variants (Table 1).
In contrast to CO, the fraction of internal NO recombination after photolysis is very large, ≥0.99 due to its high reactivity with iron. Once inside the protein, NO reacts immediately with the iron atom, with almost every internal collision between them being productive. As a result, the rate-limiting step for NO binding from solvent is the bimolecular rate of ligand entry into the protein, and the observed association rate constant, kNO′, should equal the calculated value of kentry′ (Equation 1, kX′ ≈ kentry′ when Fgem → 1.0).
As shown in Fig. 2A, there is a linear correlation between kentry′, calculated using kCO′ and Fgem,CO, and the experimentally measured value of kNO′ for a series of Val-7(B6), Gln-44(E7), Thr-48(E11), Ala-55(E18), and Leu-86(G12) CerHb mutants (Table 1). The error in kentry′ is very large for those mutants with large kCO′ and very small Fgem,CO values, making it hard to determine the slope of the correlation between kentry′ and kNO′. In addition, there appears to be a systematic overestimation of kentry′ for the CerHb variants with Fgem,CO < 0.1 (Fig. 2A). However, the correlation is strong for mutants where Fgem is larger due to partial blockage of the tunnel. Similar linear correlations between kentry′ and kNO′ were observed for all of the SwMb mutants reported in Scott et al. (13), verifying the kentry′ values for myoglobin.
FIGURE 2.
Correlation of calculated rates of ligand entry, kNO′, and kO2′ in CerHb. A, kentry′ versus kNO′; B, kO2′ versus kNO′ for Val-7(B6)Ala, -Phe, and -Trp; Thr-48(E11)Val; Ala-55(E18)Val, -Leu, -Phe, and -Trp; Leu-86(G12)Ala, -Phe, and -Trp; Gln-44(E7)Trp; and Val-7(B6)Phe/Ala-55(E18)Phe/Leu-84(G12)Phe CerHb mutants (supplemental Table S2). C, kentry′ versus kO2′, open circles, all single mutants are located in and along the apolar tunnel (supplemental Table S1); closed circles, multiple mutants are located in the apolar tunnel (supplemental Table S4).
As shown in Fig. 2B, there is a 1:1 correlation between kNO′ and kO2′ for the CerHb variants in Table 1 implying that O2 binding is also primarily limited by the bimolecular rate of entry into the protein. The only exceptions are the T48V mutant, where the loss of γ-OH at the E11 position releases the phenol side chain of Tyr(B10), allowing it to sterically inhibit iron-ligand bond formation, and the Q44F mutation, where the larger Phe(E7) side chain directly inhibits O2 binding at the iron atom. For both these mutants, overall O2 binding is slowed because bond formation is markedly inhibited. As a result, kbond becomes similar to kescape causing a decrease in kO2′ due to a decrease in Fgem and not the rate of entry (Equations 1 and 2). In the case of NO, its high reactivity with the iron atom is still much greater than that of ligand escape, keeping ligand entry as the rate-limiting step even for the T48V and Q44F CerHb mutants.
Fig. 2C shows the correspondence between the calculated ligand entry rate using overall and geminate CO binding data with the bimolecular rate constant for O2 binding. In this case, there is an ∼1:1 correspondence with all the mutants examined in this work, even though the scatter is fairly large. All of the results in Fig. 2 for CerHb and the correlations between kNO′ and kentry′ values for the SwMb mutants (13) support the use of calculated kentry′ and kescape values to map ligand pathways and demonstrate that similar conclusions would be obtained if kNO′ or kO2′ values were used directly in the mapping analysis instead of the calculated kentry′ values.
Crystallization, Data Collection, and Refinement of H100F, H100W, and Y11F/Q44L/T48V/A55W CerHb Variants
The O2, CO, and aquomet forms of H100F and H100W CerHb were crystallized using the sitting drop vapor diffusion method at a protein concentration of 25 mg/ml. The best crystals grew from solutions containing 2.3 m ammonium sulfate, 100 mm sodium citrate, pH 5.4, at 277 K. The aquomet form of the quadruple mutant with an apolar distal pocket was crystallized from solutions containing 200 mm sodium thiocyanate and 2.2 m ammonium sulfate. The O2 and CO forms of this multiple mutant were obtained by reduction of the ferric crystals in the presence of either air or 1 atm CO. These manipulations often caused high mosaicities and reversion to the ferric or completely deoxygenated reduced form. As a result, crystal manipulations and data collection were repeated several times.
In all cases crystals were transferred to appropriate storage solutions containing 2.8 m ammonium sulfate and 25% sucrose (w/v) immediately before data collection at 100 K. For CO complexes, the mounting solution was saturated with 1 atm of pure carbon monoxide.
A cryo-cooling N2 system was used to maintain low temperature (100 K) in the environment of the mounted crystals to reduce radiation damage. Complete x-ray diffraction data sets for each of the CerHb variants were obtained from single crystals using CuKα radiation (λ = 1.5418 Å) from a Rigaku RUH3R rotating anode x-ray generator operated at 50 kV and 90 mA and a Rigaku R-AXIS IV++ image plate detector (Rigaku Americas Co.). Data were collected, scaled, and reduced using d*TREK software (54).
The program PHENIX (55) was used for both structure solution and refinement. The structures of the H100F and H100W CerHb derivatives (PDB codes 4F6B, 4F6D 4F6F, 4F6G, 4F6I, and 4F6J) were solved by difference Fourier syntheses using the structure of wild-type CerHb (PDB code 1KR7) with the ligand and solvent molecules omitted. The structures of the Y11F/Q44L/T48V/A55W CerHb mutants (PDB codes 4F68 and 4F69) were also solved by difference Fourier syntheses using the structure of the A55W CerHb mutant (PDB code 2VYY). An initial round of simulated annealing was used to calculate unbiased electron density maps to confirm the correct placement of the mutated residues. This process was followed by several refinement macrocycles with maximum likelihood as the target and included bulk solvent correction and anisotropic scaling of the data, individual coordinate refinement with minimization, and individual isotropic ADP refinement interspersed with manual fitting. The ligands were modeled into the electron density maps at this point, and refinement was continued with restraints imposed on the expected Fe2+-CO, Fe2+-O2, or Fe3+-H2O geometries. Solvent molecules were then included, and in a few cases, combined TLS and individual ADP refinements were carried out in the final stages of refinement.
Map fitting and other manipulations with molecular models were performed using the graphic software COOT (56). The accession codes for the models, crystal parameters, and statistics of x-ray data collection and refinement are provided in Table 2. Figs. 1, 4, 5, and 6 were prepared using the PyMOL Molecular Graphics System, version 1.2r3pre (Schrödinger, LLC).
TABLE 2.
Data collection and refinement statistics for CerHb mutants
| Y11F/Q44L/ T48V/A55W CerHbO2 | Y11F/Q44L/ T48V/A55W CerHbCO | H100F met-CerHb | H100F CerHbO2 | H100F CerHbCO | H100W met-CerHb | H100W CerHbO2 | H100W CerHbCO | |
|---|---|---|---|---|---|---|---|---|
| PDB entry code | 4F68 | 4F69 | 4F6B | 4F6D | 4F6F | 4F6G | 4F6I | 4F6J |
| Crystal data | ||||||||
| Resolution range | 24.66–1.80 Å | 27.0–1.60 Å | 24.55–1.65 Å | 27.0–1.80 Å | 27.04–1.56 Å | 24.55–1.64 Å | 27.0–1.56 Å | 30.24–1.45 Å |
| Space group | P 212121 | P 212121 | P 212121 | P 212121 | P 212121 | P 212121 | P 212121 | P 212121 |
| Unit cell parameters | a = 42.62, b = 43.55, c = 59.75 Å | a = 42.64, b = 43.36, c = 59.5 Å | a = 42.68, b = 43.47, c = 59.50 Å | a = 42.53, b = 43.49, c = 59.50 Å | a = 42.37, b = 43.55, c = 59.41 Å | a = 42.68, b = 43.48, c = 59.48 Å | a = 42.15, b = 43.40, c = 59.70 Å | a = 42.10, b = 43.51, c = 59.49 |
| Reflections (measured/unique) | 25,517/10,643 | 38,297/14,641 | 45,856/13,828 | 23,325/10,039 | 47,165/14,607 | 46,250/13,789 | 37,156/12,733 | 39,047/16,823 |
| Completeness | 98.8% (100%)a | 97.0% (89.7%) | 98.0% (98.1%) | 93.7% (94.0%) | 90.0% (62.3%) | 98.0% (98.8%) | 95.0% (68.6%) | 83.8% (46.8%) |
| 〈I〉/σ〈I〉 | 14.3 (4.1) | 11.7 (2.6) | 19.7 (7.0) | 40.0 (11.2) | 15.9 (6.6) | 20.0 (9.1) | 11.1 (2.0) | 12.1 (1.8) |
| Redundancy | 2.4 (2.3) | 2.6 (2.1) | 3.3 (3.4) | 2.5 (2.6) | 3.2 (2.8) | 3.3 (3.5) | 2.4 (1.9) | 2.3 (1.2) |
| Rmerge | 3.8% (17.7%) | 4.8% (24.0%) | 4.9% (11.4%) | 4.6% (10.1%) | 4.6% (16.0%) | 4.9% (26.0%) | 4.4% (33.0%) | 4.0% (27.9%) |
| Refinement | ||||||||
| Resolution range | 24.66–1.80 Å | 27.0–1.60 Å | 24.55–1.65 Å | 27.0–1.80 Å | 27.04–1.56 Å | 24.55–1.64 Å | 27.0–1.56 Å | 30.24–1.45 Å |
| R-factor | 20.3% | 20.6% | 18.0% | 16.0% | 19.3% | 18.4% | 19.8% | 19.8% |
| Rfree | 22.1% | 23.3% | 20.2% | 19.7% | 21.7% | 20.6% | 21.4% | 21.9% |
| Root mean square deviations from ideal values | ||||||||
| Bond length | 0.009 Å | 0.008 Å | 0.007 Å | 0.025 Å | 0.014 Å | 0.007 Å | 0.010 Å | 0.010 Å |
| Bond angles | 0.956° | 0.973° | 0.943° | 0.922° | 1.004° | 0.966° | 0.902° | 0.900° |
| Residues in most favorable region | 95.7% | 96.8% | 94.7% | 94.7% | 96.8% | 94.7% | 95.7% | 94.7% |
| Residues in additional allowed region | 4.3% | 3.2% | 5.3% | 5.3% | 3.2% | 5.3% | 4.3% | 5.3% |
a Parameters in parentheses are for the outer resolution shell.
RESULTS
Mutagenesis Mapping Strategy
The distal pockets, internal Xe-binding cavities, and tunnels within SwMb and CerHb are shown in Fig. 1. Key amino acid side chains near the E7 gate (Gln-44 in CerHb and His-64 in SwMb), at the Xe-binding sites, and along the apolar tunnel in CerHb between the E and H helices are shown as sticks and labeled in the upper panels of Fig. 1. From these side views, it is clear that the continuous apolar tunnel in CerHb is not present in SwMb due both to the presence of the A helix and to the occurrence of larger amino acid side chains located along the interiors of the E, G, and H helices in SwMb, which separate the various internal Xe cavities.
To define experimentally the ligand entry/exit pathway in CerHb, we constructed a series of replacements with small (Ala and Val) to large (Trp and Phe) amino acids at positions in or along the E7 channel, the apolar tunnel, and other regions reported to be important, based on previous simulations and experimental work with recombinant Mbs and Hbs. We examined 101 single point mutations at 22 different positions in CerHb, and the overall rate constants for O2 and CO binding were measured using rapid mixing, laser photolysis methods with O2/CO mixtures, and flow-flash techniques as described in Salter et al. (42) and Pesce et al. (43). Internal CO recombination within each mutant was also measured using a 7-ns YAG laser. The rate, kgem, and the fraction, Fgem, of geminate recombination and the overall bimolecular rate constant for CO binding, kCO′, were used to estimate the first order rate constant for ligand escape (kescape = kgem,CO(1 − Fgem,CO)) following its photodissociation from the heme iron and the bimolecular rate of ligand re-entry from solvent (kentry′ = kCO′/Fgem,CO) (42, 43). In key cases, the calculated value of kentry′ was verified by measuring the bimolecular rate constant for NO association as described under “Experimental Procedures” (Table 1 and Fig. 2). Complete sets of O2 and CO kinetic parameters for all the CerHb variants examined are listed in the supplemental Tables S1–S4.
Underlying Assumptions
The first premise of our mutagenesis approach is that larger and smaller apolar amino acid side chains should impose larger and smaller kinetic barriers, respectively, for both ligand entry and escape if they are located along the primary pathway for ligand binding (13, 42). Thus, the overall bimolecular rate constant of ligand entry for a mutant with a small amino acid, kentry,small′, should be significantly larger than the entry rate constant for a mutant with a large amino acid, kentry,large′, at positions located along the migration pathway that leads to bond formation. Similar relationships should occur for the ligand escape rate constants for the same set of mutants. To quantify these effects, we computed the overall rate enhancement effect, Renhance, of a large-to-small mutation as shown in Equation 3 (13),
 |
Large-to-small replacements at positions on the major ligand pathway will increase both kentry′ and kescape, which will lead to positive values for both logarithmic terms in Equation 3 and a large value of Renhance when added together. In contrast, mutations at positions distant from the ligand trajectory should have little or no effect on entry and escape.
The second premise of our mapping approach is that ligand binding and single point mutations have only minor effects on overall globin structure. Ligand binding to Mb does produce structural changes involving replacement of distal pocket water, small outward movements of the distal histidine, and alterations of the proximal pocket and minor tilting of the heme to accommodate in-plane movement of the iron atom. However, none of these changes have a significant effect on the overall tertiary structure and the internal Xe cavities shown in Fig. 1A (14, 19, 36, 37). Recently, Germani et al. (57) have shown that CO and O2 binding to deoxy-CerHb causes even smaller changes in the heme cavity and has no effect on the size or shape of the apolar tunnel.
Our previous work with distal pocket mutants of CerHb also showed very little change in overall structure (41), and crystallographic studies of the mutations at the 55(E18) (42) and 100(H11) positions (this report) near the solvent entrance to CerHb have little effect on overall globin structure or the tunnel. In contrast, Xe addition to WT CerHbO2 crystals causes loss of diffraction, and we could only determine the structure of Xe bound to L86A CerHb where the tunnel cavity near the internal G12 position is enlarged. Similarly, we have been unable to crystallize V7F, V7W, L86F, and L86W variants presumably because the larger aromatic side chains expand the tunnel and unfavorably alter inter-protein contacts in the crystal lattice. However, all standard UV-visible, FTIR, and CD spectroscopic properties of these tunnel mutants are very similar to those of the WT protein.
In addition, mutations near the bound ligand can have large effects on overall ligand binding parameters due to either direct steric hindrance of bond formation or electrostatic stabilization of the bound ligand. In most cases, these effects are manifested as large changes for the internal rate of iron-ligand bond formation or dissociation (kbond and k−bond in Fig. 1) and do not significantly alter the rates for entry and escape (13, 38). In some cases, the results can be very complex, particularly for mutants containing Q44H and Q44W replacements (supplemental Tables S2 and S3) (42). However, in general, the assumption that single point mutations are conservative with respect to overall globin tertiary structure appears to hold for both CerHb and SwMb (33, 36–38, 41–43, 48).
We also constructed and characterized eight new SwMb mutants to augment the data from Scott et al. (13) and to better complement the set of CerHb mutants located near the exit point of its apolar tunnel and at position CD3 near the His(E7) gate (Table 3). These SwMb mutations included R45W, L61W, L69A, L69W, I75A, I75W, L135A, and L135W at the CD3, E4, E12, E18, and H12 helical positions. Estimates of the rates of entry and escape in SwMb were measured using O2 instead of CO, as described in Scott et al. (13). The iron atom in Mb is less reactive due to proximal constraints, and as a result, very little internal CO recombination is observed at room temperature in ordinary buffers. In contrast, WT Mb shows ∼50% geminate O2 recombination, and the rate is slow enough (kgem ≈10–20 μs−1, t½ ≥30–60 ns) to be measured readily using a 7-ns YAG laser excitation pulse. In the case of CerHb, O2 geminate recombination is too fast to measure accurately with WT and most mutant proteins (kgem ≥130 μs−1, t½ ≤5 ns) (42).
Rate Enhancement Maps
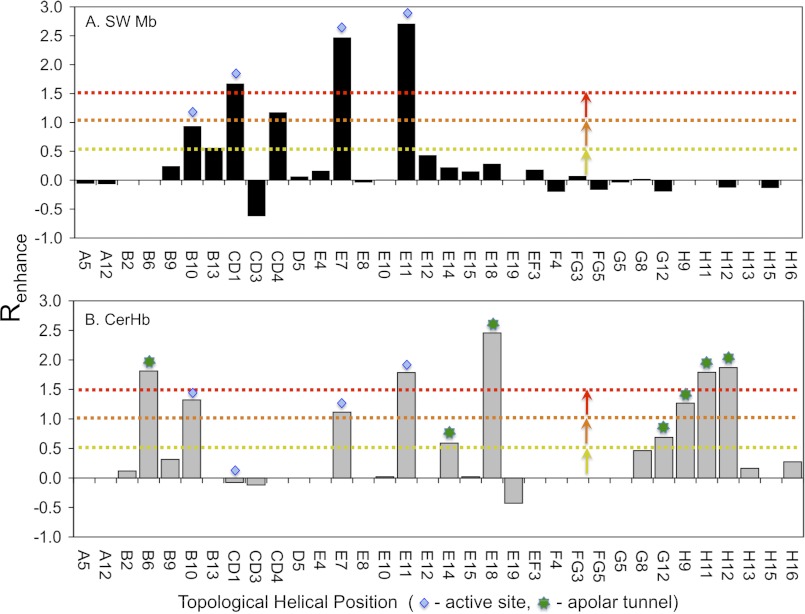
Bar graphs of the rate enhancements caused by large-to-small mutations, Renhance, versus sequence position are shown in Fig. 3 for both SwMb and CerHb. In both cases, significant effects are observed at the active site positions where the amino acid side chains interact directly with bound ligands (see B10, CD1, E7 and E11 bars labeled with blue diamonds in Fig. 3). However, beyond those regions the differences between Mb and CerHb are dramatic.
FIGURE 3.
Effects of Ala or Val to Phe or Trp mutations on entry and escape in CerHb and SwMb. Blue diamonds denote helical positions typically associated with the E7 gate pathway. Green asterisks are associated with positions along the apolar tunnel pathway. Data for SwMb were taken from this study and Ref. 13. The enhancement factor, Renhance, on the y axis is defined in Equation 3, and the topological helical position is given along the x axis.
In the case of Mb, large rate enhancements are only observed at or near the E7 channel or in the back of the distal pocket where ligands are initially captured before binding to the heme iron atom, i.e. positions 29(B10), 32(B13), 43(CD1), 46(CD4), 64(E7), and 68(E11). No significant enhancements of the rates of entry and escape are observed at positions around the Xe1 cavity (i.e. positions 89(F4), 99(FG5), 104(G5), and 138(H15)), in regions equivalent to the apolar tunnel in CerHb (i.e. between the E, G, and H helices), or at other remote locations suggested by previous MD simulations (Fig. 3A). In contrast, large rate enhancements for entry into and escape from CerHb are observed for replacements lining the apolar tunnel shown in Fig. 1B, including positions near the C-terminal ends of the E and H helices and at position Val-7(B6) where the distal pocket connects to the apolar tunnel (bars labeled with green asterisks, Fig. 3B).
The magnitudes of the Renhance parameters are mapped onto the three-dimensional crystal structures of WT SwMb and CerHb in Fig. 4 (13). The order of Renhance effects is red > orange > yellow spheres at native amino acid positions (see legend to Fig. 4). The white or gray sticks in Fig. 4 indicate positions where little or no effect is observed on kentry′ and kescape for large-to-small amino acid substitutions.
The results are quite clear for both proteins. Only positions near the E7 gate or in the distal heme pocket are highlighted in SwMb, and no rate enhancements are observed along alternative pathways, including the Xe pockets or other interior apolar regions (Fig. 4, A–C). In contrast, mutagenesis mapping clearly highlights the internal tunnel in CerHb (Fig. 4, D–F), proving experimentally that ligands enter and exit through this pathway. This conclusion is particularly evident in Fig. 4F where the tunnel between the E and H helices is clearly seen and circumscribed by red, orange, and yellow side chains. The same view for SwMb (Fig. 4C) shows that this pathway is completely blocked by the A helix.
Identifying the Solvent Exit of the Apolar Tunnel
Three further tests of the apolar pathway in CerHb were performed. First, we explored the solvent-exposed exit aperture of the apolar tunnel at the ends of the E and H helices; second, we examined the influence of polar interactions at the E7 gate; and third, we filled the apolar channel with aromatic amino acid side chains to completely block this pathway.
In WT CerHbO2, the polar imidazole side chain of His-100 at the H11 helical position is extended out into solvent and does not appear to block entry into the protein (Figs. 1B and 5B) (42). As expected, removing the imidazole at this position by a H100A mutation has little effect on ligand binding; however, both Phe and Trp mutations at this position cause ∼3-fold decreases in both kO2′ and kO2, with little change in affinity (Table 4). These same aromatic amino acid mutations also cause large increases in the fraction of CO geminate recombination, from ∼0.05 for WT CerHbCO to ∼0.34 for H100W (Fig. 5A), and a marked decrease in kgem,CO due to an ∼10-fold decrease in kescape (Table 4). All of these effects suggest that large Phe and Trp side chains at the H11 helical position in CerHb rotate back to the protein surface to partially cover the solvent exit of the ligand tunnel and restrict ligand escape.
The imidazole ring of the naturally occurring His-100 side chain is poorly defined in the WT CerHbO2 structure, with only a portion of the expected electron density of the ring apparent in the calculated map (Fig. 5B). In the structures containing the A55W replacement, the His-100 chain is better defined but in a different orientation due the large indole ring at the E18 position, which restricts the motion at the H11 helical position (i.e. see Fig. 6C). Fig. 5C shows the conformation of His-100 in seven different CerHb structures in both reduced and oxidized forms. The side chain appears to show free rotation about the Cα–Cβ bond with no specific energy minimum, presumably because of equally favorable interactions with solvent water molecules. As a result, the entrance into the apolar channel is kept open. When Phe or Trp is inserted into this position, the more apolar aromatic rings appear to be forced into the entrance of the channel by a hydrophobic effect and restrict ligand entry and escape (Table 4 and Fig. 5A).
To verify this interpretation, we crystallized and determined structures of the H100F and H100W CerHb mutants in reduced O2- and CO-bound forms and in the oxidized state (Table 2). In both cases, the aromatic side chains rotated back onto the protein surface, partially entering and blocking the tunnel from the H11 helical position (Fig. 5D). The Trp-100 and Phe-100 side chain conformations have ∼100% occupancy, are well defined by electron density (Fig. 5D), and account for the decreases in rates of ligand entry and exit and the increase in geminate CO recombination observed for H100F and H100W mutants (Table 4 and Fig. 5A).
In WT CerHb, the small residue Ala-55 at E18 is located on the opposite side of the entrance to the apolar channel from His-100 at H11. Substitution of a Trp for Ala at this position also blocks ligand entry, decreases kentry′ and kescape, and traps photodissociated ligands inside the protein, increasing Fgem (Table 4 and Figs. 3B and 6C) (42). Similar large decreases in kentry′ and kescape and increases in Fgem occur when Ala-101 at H12 is replaced with Trp (Fig. 3B and supplemental Table S1). Combined, these mutagenesis results unambiguously locate the entrance for ligand movement into CerHb between the ends of the E and H helices at positions E18, H11, and H12.
Constructing an Apolar Distal Pocket
Pesce et al. (39, 43) suggested that extensive polar interactions between Tyr-11(B10), Gln-44(E7), Lys-47(E10), and Thr-48(E11) limit opening of the E7 gate, causing ligands to use the apolar tunnel for entry and exit. To test this idea, we examined ligand binding to a triple CerHb mutant with a completely apolar active site containing Y11F/Q44L/T48V mutations at the B10, E7, and E11 helical positions, respectively. As shown in Table 4, rates of O2 binding to and dissociation from this triple mutant increase by only 50%, and there is little change in the computed values of kentry′ and kescape compared with WT CerHb. The extent of geminate recombination does increase to ∼0.40 (Fig. 6A) due to a marked ∼7-fold increase in the rate of covalent bond formation (kbond) compared with WT CerHb (from ∼4.0 to ∼30 μs−1 for the mutant, see supplemental Table S4). However, when the A55W mutation is added to this triple mutant to block exit from the apolar tunnel (Fig. 6C), there are marked ∼5-fold decreases in kO2′ and kO2; the fraction of geminate CO rebinding increases to 0.85, and the calculated value of kentry′ decreases from ∼400 to ∼30 μm−1 s−1 (Table 4 and Fig. 6A). Thus, even when the distal pocket is made apolar, ligands still appear to enter and exit the protein through the apolar tunnel, and blocking the entrance to the tunnel markedly inhibits bimolecular binding.
We determined the crystal structure of the Y11F/Q44L/T48V/A55W mutant to examine whether the apolar substitutions cause significant changes in the tertiary structure of CerHb. Electron density maps for the active site amino acids of the quadruple mutant and Trp-55 at the tunnel entrance are shown in Fig. 6, B and C, respectively. A comparison of the active site of the quadruple mutant to that of the single A55W mutant is shown in Fig. 6D. The structures overlay remarkably well with a root mean square deviation of 0.17 Å for the Cα atoms, and the side chains of the B10, E7, and E11 amino acids have very similar conformations despite the changes in polarity (note that the root mean square deviation was 0.15 Å for the Cα atoms of WT CerHb and the quadruple mutant). The major difference is that there is more “free” space surrounding the bound O2 atoms due to the loss of the Oζ atom of Tyr-11(B10) and the Nϵ or Oϵ atoms of Gln-44(E7). This loss of steric hindrance adjacent to the bound ligand accounts for the increase in the rate and fraction of geminate recombination after laser photolysis (Fig. 6A and Table 4). Instead of being pushed away by the larger Tyr(B10) and Gln(E7) side chains, photodissociated CO can remain near the center of the heme group and more rapidly rebinds than in WT CerHb. However, the large, ≥10-fold decrease in the rate of entry caused by the A55W mutation at E18, even when the active site is apolar, argues strongly that most ligands (≥90%) still enter CerHb through the apolar channel (kentry′ values in Table 4).
Completely Blocking the Tunnel
As a final test of the map for CerHb, we attempted to completely block ligand movement through the apolar tunnel. To prove that entry into the distal pocket from the tunnel is at the B6 position, we mutated Val-7(B6) to Ala, Phe, and Trp. As shown in Fig. 7, introducing an indole ring at this position traps CO inside the distal pocket increasing both Fgem and kgem dramatically. At the same time, the V7W replacement markedly decreases kentry′, kO2′, and kNO′ and the rate of O2 dissociation (Table 4), demonstrating that this position is on the pathway for ligand binding. This conclusion is verified by the large increase in Fgem and a decrease in kentry′ when the V7W mutation is added to the apolar pocket (Y11F/Q44L/T48V) CerHb mutant (Fig. 6A and Table 4).
The V7A/L86A CerHb double mutant was constructed as a control for a completely open channel with Ala at the three key positions along the apolar tunnel, 7(B6), 55(E18), and 86(G12), respectively (WT CerHb has Ala at position 55(E18)). This double mutant has kinetic parameters very similar to those of WT CerHb (Table 4). In contrast, the V7F/A55F/L86F and V7W/A55W/L86W triple mutants show dramatic ≥10-fold decreases in kO2′, kO2, and kentry′ and ≥10-fold increases in Fgem,CO, from ∼0.05 for WT CerHb to 0.80 and 0.93 for the triple Phe and Trp mutants, respectively (Table 4 and Fig. 7). The computed Renhance value comparing Ala-7/Ala-55/Ala-86 versus Trp-7/Trp-55/Trp-86 is 3.1, indicating a 3-fold increase in the free energy barrier to ligand escape and entry. These data show unambiguously that the apolar tunnel in CerHb can be closed by mutagenesis, reducing the measured bimolecular rate constants for O2 and NO binding and the calculated rate for ligand entry to less than 10% of the WT values.
DISCUSSION
Single versus Multiple Pathways
The experimental mutagenesis results in Figs. 2–7, Table 4, and supplemental Tables S1–S4 are very clear. Ligands enter and exit CerHb through the apolar tunnel as originally suggested by Pesce et al. (39). When three Phe or Trp replacements at positions 7(B6), 55(E18), and 86(G12) are used to fill the entire tunnel, the bimolecular rate of ligand entry decreases dramatically. An estimate of the rate of ligand entry when the apolar channel is closed was calculated from the average of the kentry′, kNO′, and kO2′ values for the V7F/A55F/L86F and V7W/A55W/L86W mutants (Table 4) and is 25 ± 12 μm−1 s−1. An estimate of the value for ligand entry into CerHb with an open tunnel was computed from the average of the kentry′, kNO′, and kO2′ values for WT CerHb and the V7A, L86A, and V7A/L86A mutants (Table 4) and is equal to 300 ± 130 μm−1 s−1.
If the triple aromatic mutations at the B6, E18, and G12 helical positions completely close the tunnel, the residual bimolecular rate of 25 μm−1 s−1 applies to ligand binding through alternative routes. This value suggests that in WT CerHb ≥92% of the ligand molecules enter through the apolar tunnel and only 8% by alternative routes. Interestingly, when the tunnel is blocked, the value of kentry′ is only slightly smaller than the estimated rates of ligand entry into WT (His-64(E7)) and Gln-64(E7) SwMb, which are 34 and ∼60 μm−1 s−1, respectively (13). Thus, the limiting value of 25 μm−1 s−1 for the triple aromatic CerHb mutants could represent ligand movement through the Gln(E7) gate, although this conclusion is speculative.
In the case of SwMb, all the positions that regulate entry and escape are located in the distal pocket or at the heme propionate-solvent interface (Figs. 3 and 4, A–C). As pointed out by Scott et al. (13) and Olson et al. (38), when the effects of Xe binding and selected mutations are examined individually, the arguments in favor of ligand movement through the E7 gate are even stronger. There are no changes in the fraction of geminate O2 recombination, the calculated bimolecular rate constants for ligand entry, or the observed association rate constants for O2 and CO binding to WT SwMb when the protein solution is pressurized with 5–10 atm of Xe. Similarly, when Trp substitutions are made in the Xe1 and Xe4 sites, there are small or no changes in kentry′, kescape, and Fgem (13, 38).
The E7 gate pathway in SwMb is also supported strongly and independently by time-resolved x-ray crystallography measurements with mutants at the 29(B10) position. The lifetimes of CO in the Xe1 pockets of WT, L29F, and L29W SwMb are ∼2–10 μs (19), 100–300 μs (14, 18), and 1–2 ms (16), respectively, and the double mutant L29Y/H64Q SwMb shows intermediate behavior (15). If ligands moved directly to solvent from the Xe1 site, the size of the amino acid at the B10 position should have had no effect on the lifetime for ligand escape from this position. Instead, ligands appear to move back to the distal pocket, pass between the B10 side chain and the plane of the heme, and then exit through the E7 channel.
When His-64(E7) is replaced with Trp in SwMb, the calculated bimolecular rate of entry decreases from 34 to 7 μm−1 s−1, implying that up to 25% of the ligand molecules may take alternative routes to the active site (13). However, this calculation assumes that the H64W substitution completely blocks the E7 channel. Recently, Birukou et al. (52) have shown that the indole side chain in the crystal structure of H64W deoxy-Mb does not enter the E7 channel but just partially blocks the entrance, occupying a position between the heme propionates. A similar “blocked” conformation is observed in the crystal structure of Trp-63(E7) β human HbCO subunits (52). In contrast, the E7 indole side chain in the crystal structure of Trp-58(E7) deoxy-α human Hb subunits is located in the distal pocket, completely closing the E7 channel and sterically restricting access to the heme iron (52). In this completely “closed” conformation, the Trp(E7) side chain causes ≥100-fold decreases in both kNO′ and the calculated value of kentry′ for isolated mutant α subunits (49, 52). Thus, it is possible that the percentage of ligands entering through the E7 channel in Mb is significantly larger than the 75% value estimated from the rate of entry for Trp-64(E7) MbO2.
Discrepancies with MD Simulations
The mutagenesis mapping results in Figs. 3 and 4 are not in agreement with either past or recent molecular dynamics simulations, all of which suggest multiple pathways. The latest theoretical work for both CerHb and SwMb suggests 3–5 pathways with roughly equal probabilities, one of which is the E7 channel in the case of SwMb and the apolar tunnel in CerHb. Thus, the theoretical calculations predict that ∼20–33% of the ligands enter and exit through the paths identified experimentally by mutagenesis, implying that alternative routes show similar rates of entry. In contrast, the experimental data indicate that ligand movements through the E7 gate in SwMb and the apolar channel in CerHb are ∼4–10-fold faster than through the alternative routes, based on the much smaller measured bimolecular rate constants when the gate or tunnel is blocked by Trp side chains. However, these differences are relatively small on a free energy of activation scale. Thus, small inaccuracies in representing chemical and structural barriers along the individual pathways could easily affect the computed results.
One key problem is representing the barrier to apolar ligand movement through clathrate-like water clusters that cover apolar patches at the protein surface. For example, well defined crystallographic water molecules surround the highly conserved Leu-89(F4) side chain in SwMb, which is apolar and protects the iron-His-93(F8) bond from being hydrated (33). We mutated this amino acid to Gly to open a hole into the Xe1 cavity and observed that a string of 3 to 4 water molecules entered this site and were connected to well defined surface waters, forming an extended chain of “ice-like” structures (33). The rates of bimolecular ligand binding and unimolecular release were relatively unaffected by this mutation, and there was little or no effect on the total fraction of O2 geminate recombination, even though the Xe1 cavity was completely open to solvent. The only major change was the loss of the slow phase for geminate rebinding, which in WT SwMb reflects movement into and then back from the Xe1 cavity (13). These structural and computational results demonstrate that water enters into the exposed Xe1 cavity in the Gly-89(F4) SwMb mutant and forms a barrier that prevents diatomic apolar ligands from entering this site.
Another example of how the hydrophobic effect can influence ligand binding is the effect of the His-100(H11) to Phe and Trp mutations on ligand binding to CerHb (Fig. 5). In most of the published structures of WT and mutant CerHb, the imidazole side chain of His-100 is pointing directly out into the solvent (see Figs. 1B and 5, B and C). However, when this polar amino acid is replaced by Phe and Trp, the hydrophobic effect pushes the more apolar phenyl and indole rings back on the surface of the protein, which partially blocks the tunnel entrance, markedly increases Fgem,CO by trapping ligands within the CerHb tunnel, and decreases both kentry′ and kescape (Fig. 5 and Table 4).
A key test of the accuracy of theoretical calculations would be to examine the effects of the L89G SwMb and H100W CerHb mutations on simulated structures, starting from the WT proteins, and on ligand trajectories, carefully looking at water structure in and around the Mb Xe1 cavity near the F4 helical position and the exit aperture of the CerHb tunnel near the H11 helical position. Other valuable tests would be simulations of the effects of the F46V SwMb mutation at the CD4 helical position on movement of the His-64(E7) side chain and of the motions of the indole side chain in Trp-64(E7) Mb. The F46V mutation accelerates ligand entry and release in SwMb and causes outward movement of the distal histidine (34). The H64W mutation slows ligand entry ∼4-fold, even though only the entrance to the E7 channel is blocked (13, 52). The effects of all four of these mutations on ligand entry rates are well defined experimentally and should be replicated in any molecular dynamics simulation.
Physiological Relevance
Ironically, the fastest rates of ligand uptake and release are observed when the longer apolar tunnel in CerHb is used rather than the shorter E7 gate channel in Mb. The rates of ligand binding to CerHb are close to diffusion-controlled and very similar to model heme compounds with no distal steric hindrance or to SwMb and human Hb mutants in which the distal histidine is replaced by Gly or Ala. Thus, the Nemertean mini-Hb from C. lacteus is able to take up and release O2 roughly 10 times more rapidly than mammalian Mbs and Hbs. CerHb surrounds the sea worm's brain and axons and acts as an O2 storage protein. High rates of O2 release give the worm an advantage for high neuronal activity under hypoxic conditions in the sea floor when it hunts for prey (40).
Apolar tunnels have also evolved in the truncated HbN from Mycobacterium tuberculosis (MtTrHbN) to facilitate rapid dioxygenation of NO as part of the bacterium's defense against host macrophages (58–63). In this case, there appear to be two channels that allow rapid access to the heme iron atom, a short one between the G and H helices and a longer one leading from the solvent interface at the AB and GH loops into the distal pocket (59). The rate constants for bimolecular CO and O2 binding to MtTrHbN become similar to or greater than those for CerHb when steric restrictions due to distal pocket water are removed by apolar distal pocket mutations, and the rate of NO dioxygenation of wild-type MtTrHbN-O2 is ∼750 μm−1 s−1, which is ∼10-fold greater than that for either human HbO2 or mammalian MbO2 (61). Both results indicate that, like in CerHb, ligands can move freely into the active site of MtTrHbN. Simulations have suggested that Phe-62(E15) closes the shorter tunnel when O2 is bound, but NO can still rapidly enter the protein through the longer pathway (60). However, these ideas are controversial (62, 63) but could be verified or refuted by mutagenesis mapping approaches similar to those shown in Figs. 3 and 4.
Conclusions
The experimental mutagenesis mapping results shown in Figs. 3 and 4 are unambiguous. Ligands enter and exit CerHb through the interior apolar tunnel at least ∼90% of the time, whereas ligands enter SwMb from the opposite direction ≥75% of the time through the E7 gate. The significance of these results is 3-fold. 1) Our rational mutagenesis mapping strategy is robust and can identify interior apolar pathways. 2) Globins have evolved different routes for ligand entry, and the longer but more open apolar pathway allows more rapid rates of entry and exit. 3) The SwMb and CerHb results provide experimental benchmarks for testing the validity of molecular dynamics trajectories, one with a short pathway between the heme propionates that is gated by a polar residue and another with an internal route from the heme pocket to an opening at the opposite end of the protein.
Supplementary Material
Acknowledgments
We thank Loren Stagg, Naxhiely Martinez Ramón, Benjamin Shields, and Cambria Alpha from the Biochemistry and Cell Biology Department at Rice University for constructing some of the initial CerHb mutants as part of their 1st year rotation or undergraduate senior research projects, and Alyssa Kuschel from the Kenneth Olsen laboratory at Loyola University for helping to construct several of the new Mb mutants as part of a summer research project.
This work was supported, in whole or in part, by National Institutes of Health Grants GM035649 and HL047020. This work was also supported by Robert A. Welch Foundation Grant C-0612 (to J. S. O.), Italian Ministry of University and Scientific Research Grant FIRB Project “Biologia Strutturale,” RBLA03B3KC, the University of Milano (to M. B.), and Fund for Scientific Research Flanders Project “Hypoxia and Nerve Globins” Grant G.0331.04N (to L. M.).

This article contains supplemental Tables S1–S4.
We dedicate this paper to Quentin H. Gibson who passed away at the age of 92 on March 16, 2011. Quentin was member of the United States National Academy of Sciences and the Royal Society of London. He was an inspiration to the corresponding author, both as Ph.D. advisor and then as a close colleague and friend for 40 years. His ideas and technical innovations provided the original methodology for rational mutagenesis mapping of ligand migration pathways in globins and the effects of high pressures of Xe gas.
- Mb
- myoglobin
- Hb
- hemoglobin
- Sw
- sperm whale
- Cer
- Cerebratulus lacteus
- MD
- molecular dynamics
- PDB
- Protein Data Bank. Amino acids are identified with their topological site numbers as defined in the conventional globin fold, e.g. His-64(E7) in Mb is the distal histidine located at the position 7 from the N terminus of the E helix.
REFERENCES
- 1. Perutz M. F., Mathews F. S. (1966) An x-ray study of azide methaemoglobin. J. Mol. Biol. 21, 199–202 [DOI] [PubMed] [Google Scholar]
- 2. Bolognesi M., Cannillo E., Ascenzi P., Giacometti G. M., Merli A., Brunori M. (1982) Reactivity of ferric Aplysia and sperm whale myoglobins towards imidazole. X-ray and binding study. J. Mol. Biol. 158, 305–315 [DOI] [PubMed] [Google Scholar]
- 3. Ringe D., Petsko G. A., Kerr D. E., Ortiz de Montellano P. R. (1984) Reaction of myoglobin with phenylhydrazine. A molecular doorstop. Biochemistry 23, 2–4 [DOI] [PubMed] [Google Scholar]
- 4. Tilton R. F., Jr., Kuntz I. D., Jr. (1982) Nuclear magnetic resonance studies of xenon-129 with myoglobin and hemoglobin. Biochemistry 21, 6850–6857 [DOI] [PubMed] [Google Scholar]
- 5. Tilton R. F., Jr., Kuntz I. D., Jr., Petsko G. A. (1984) Cavities in proteins. Structure of a metmyoglobin-xenon complex solved to 1.9 Å. Biochemistry 23, 2849–2857 [DOI] [PubMed] [Google Scholar]
- 6. Schoenborn B. P. (1965) Binding of xenon to horse hemoglobin. Nature 208, 760–762 [DOI] [PubMed] [Google Scholar]
- 7. Schoenborn B. P. (1969) Structure of alkaline metmyoglobin-xenon complex. J. Mol. Biol. 45, 297–303 [DOI] [PubMed] [Google Scholar]
- 8. Schoenborn B. P., Nobbs C. L. (1966) The binding of xenon of sperm whale deoxymyoglobin. Mol. Pharmacol. 2, 495–498 [PubMed] [Google Scholar]
- 9. Schoenborn B. P., Watson H. C., Kendrew J. C. (1965) Binding of xenon to sperm whale myoglobin. Nature 207, 28–30 [DOI] [PubMed] [Google Scholar]
- 10. Lim M., Jackson T. A., Anfinrud P. A. (1995) Binding of CO to myoglobin from a heme pocket docking site to form nearly linear Fe-C-O. Science 269, 962–966 [DOI] [PubMed] [Google Scholar]
- 11. Lim M., Jackson T. A., Anfinrud P. A. (1997) Ultrafast rotation and trapping of carbon monoxide dissociated from myoglobin. Nat. Struct. Biol. 4, 209–214 [DOI] [PubMed] [Google Scholar]
- 12. Scott E. E., Gibson Q. H. (1997) Ligand migration in sperm whale myoglobin. Biochemistry 36, 11909–11917 [DOI] [PubMed] [Google Scholar]
- 13. Scott E. E., Gibson Q. H., Olson J. S. (2001) Mapping the pathways for O2 entry into and exit from myoglobin. J. Biol. Chem. 276, 5177–5188 [DOI] [PubMed] [Google Scholar]
- 14. Aranda R., 4th, Levin E. J., Schotte F., Anfinrud P. A., Phillips G. N., Jr. (2006) Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin. Acta Crystallogr. D Biol. Crystallogr. 62, 776–783 [DOI] [PubMed] [Google Scholar]
- 15. Bourgeois D., Vallone B., Arcovito A., Sciara G., Schotte F., Anfinrud P. A., Brunori M. (2006) Extended subnanosecond structural dynamics of myoglobin revealed by Laue crystallography. Proc. Natl. Acad. Sci. U.S.A. 103, 4924–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt M., Nienhaus K., Pahl R., Krasselt A., Anderson S., Parak F., Nienhaus G. U., Srajer V. (2005) Ligand migration pathway and protein dynamics in myoglobin. A time-resolved crystallographic study on L29W MbCO. Proc. Natl. Acad. Sci. U.S.A. 102, 11704–11709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schotte F., Lim M., Jackson T. A., Smirnov A. V., Soman J., Olson J. S., Phillips G. N., Jr., Wulff M., Anfinrud P. A. (2003) Watching a protein as it functions with 150-ps time-resolved x-ray crystallography. Science 300, 1944–1947 [DOI] [PubMed] [Google Scholar]
- 18. Schotte F., Soman J., Olson J. S., Wulff M., Anfinrud P. A. (2004) Picosecond time-resolved x-ray crystallography. Probing protein function in real time. J. Struct. Biol. 147, 235–246 [DOI] [PubMed] [Google Scholar]
- 19. Srajer V., Ren Z., Teng T. Y., Schmidt M., Ursby T., Bourgeois D., Pradervand C., Schildkamp W., Wulff M., Moffat K. (2001) Protein conformational relaxation and ligand migration in myoglobin. A nanosecond to millisecond molecular movie from time-resolved Laue x-ray diffraction. Biochemistry 40, 13802–13815 [DOI] [PubMed] [Google Scholar]
- 20. Teng T. Y., Srajer V., Moffat K. (1997) Initial trajectory of carbon monoxide after photodissociation from myoglobin at cryogenic temperatures. Biochemistry 36, 12087–12100 [DOI] [PubMed] [Google Scholar]
- 21. Elber R., Karplus M. (1990) Enhanced sampling in molecular dynamics. Use of the time-dependent Hartree approximation for simulation of carbon monoxide diffusion through myoglobin. J. Am. Chem. Soc. 112, 9161–9175 [Google Scholar]
- 22. Elber R. (2010) Ligand diffusion in globins. Simulations versus experiment. Curr. Opin. Struct. Biol. 20, 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bossa C., Amadei A., Daidone I., Anselmi M., Vallone B., Brunori M., Di Nola A. (2005) Molecular dynamics simulation of sperm whale myoglobin. Effects of mutations and trapped CO on the structure and dynamics of cavities. Biophys. J. 89, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nutt D. R., Meuwly M. (2004) CO migration in native and mutant myoglobin. Atomistic simulations for the understanding of protein function. Proc. Natl. Acad. Sci. U.S.A. 101, 5998–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruscio J. Z., Kumar D., Shukla M., Prisant M. G., Murali T. M., Onufriev A. V. (2008) Atomic level computational identification of ligand migration pathways between solvent and binding site in myoglobin. Proc. Natl. Acad. Sci. U.S.A. 105, 9204–9209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brunori M., Bourgeois D., Vallone B. (2004) The structural dynamics of myoglobin. J. Struct. Biol. 147, 223–234 [DOI] [PubMed] [Google Scholar]
- 27. Elber R., Gibson Q. H. (2008) Toward quantitative simulations of carbon monoxide escape pathways in myoglobin. J. Phys. Chem. B 112, 6147–6154 [DOI] [PubMed] [Google Scholar]
- 28. Golden S. D., Olsen K. W. (2008) Identification of ligand-binding pathways in truncated hemoglobins using locally enhanced sampling molecular dynamics. Methods Enzymol. 437, 459–475 [DOI] [PubMed] [Google Scholar]
- 29. Mouawad L., Maréchal J. D., Perahia D. (2005) Internal cavities and ligand passageways in human hemoglobin characterized by molecular dynamics simulations. Biochim. Biophys. Acta 1724, 385–393 [DOI] [PubMed] [Google Scholar]
- 30. Orlowski S., Nowak W. (2007) Locally enhanced sampling molecular dynamics study of the dioxygen transport in human cytoglobin. J. Mol. Model. 13, 715–723 [DOI] [PubMed] [Google Scholar]
- 31. Orlowski S., Nowak W. (2008) Topology and thermodynamics of gaseous ligands diffusion paths in human neuroglobin. Biosystems 94, 263–266 [DOI] [PubMed] [Google Scholar]
- 32. Huang X., Boxer S. G. (1994) Discovery of new ligand binding pathways in myoglobin by random mutagenesis. Nat. Struct. Biol. 1, 226–229 [DOI] [PubMed] [Google Scholar]
- 33. Liong E. C., Dou Y., Scott E. E., Olson J. S., Phillips G. N., Jr. (2001) Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin. J. Biol. Chem. 276, 9093–9100 [DOI] [PubMed] [Google Scholar]
- 34. Lai H. H., Li T., Lyons D. S., Phillips G. N., Jr., Olson J. S., Gibson Q. H. (1995) Phe-46(CD4) orients the distal histidine for hydrogen bonding to bound ligands in sperm whale myoglobin. Proteins 22, 322–339 [DOI] [PubMed] [Google Scholar]
- 35. Hirota S., Li T. S., Phillips G. N., Olson J. S., Kitagawa T. (1996) Perturbation of the Fe-O-2 bond by nearby residues in heme pocket. Observation of nu(Fe-O2) Raman bands for oxymyoglobin mutants. J. Am. Chem. Soc. 118, 7845–7846 [Google Scholar]
- 36. Quillin M. L., Arduini R. M., Olson J. S., Phillips G. N., Jr. (1993) High resolution crystal structures of distal histidine mutants of sperm whale myoglobin. J. Mol. Biol. 234, 140–155 [DOI] [PubMed] [Google Scholar]
- 37. Quillin M. L., Li T., Olson J. S., Phillips G. N., Jr., Dou Y., Ikeda-Saito M., Regan R., Carlson M., Gibson Q. H., Li H., et al. (1995) Structural and functional effects of apolar mutations of the distal valine in myoglobin. J. Mol. Biol. 245, 416–436 [DOI] [PubMed] [Google Scholar]
- 38. Olson J. S., Soman J., Phillips G. N., Jr. (2007) Ligand pathways in myoglobin. A review of trp cavity mutations. IUBMB Life 59, 552–562 [DOI] [PubMed] [Google Scholar]
- 39. Pesce A., Nardini M., Dewilde S., Geuens E., Yamauchi K., Ascenzi P., Riggs A. F., Moens L., Bolognesi M. (2002) The 109-residue nerve tissue minihemoglobin from Cerebratulus lacteus highlights striking structural plasticity of the α-helical globin fold. Structure 10, 725–735 [DOI] [PubMed] [Google Scholar]
- 40. Vandergon T. L., Riggs C. K., Gorr T. A., Colacino J. M., Riggs A. F. (1998) The mini-hemoglobins in neural and body wall tissue of the nemertean worm, Cerebratulus lacteus. J. Biol. Chem. 273, 16998–17011 [DOI] [PubMed] [Google Scholar]
- 41. Pesce A., Nardini M., Ascenzi P., Geuens E., Dewilde S., Moens L., Bolognesi M., Riggs A. F., Hale A., Deng P., Nienhaus G. U., Olson J. S., Nienhaus K. (2004) Thr-E11 regulates O2 affinity in Cerebratulus lacteus mini-hemoglobin. J. Biol. Chem. 279, 33662–33672 [DOI] [PubMed] [Google Scholar]
- 42. Salter M. D., Nienhaus K., Nienhaus G. U., Dewilde S., Moens L., Pesce A., Nardini M., Bolognesi M., Olson J. S. (2008) The apolar channel in Cerebratulus lacteus hemoglobin is the route for O2 entry and exit. J. Biol. Chem. 283, 35689–35702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pesce A., Nardini M., Dewilde S., Capece L., Martí M. A., Congia S., Salter M. D., Blouin G. C., Estrin D. A., Ascenzi P., Moens L., Bolognesi M., Olson J. S. (2011) Ligand migration in the apolar tunnel of Cerebratulus lacteus mini-hemoglobin. J. Biol. Chem. 286, 5347–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Orlowski S., Nowak W. (2007) Oxygen diffusion in minihemoglobin from Cerebratulus lacteus. A locally enhanced sampling study. Theor. Chem. Acc. 117, 253–258 [Google Scholar]
- 45. Springer B. A., Sligar S. G. (1987) High level expression of sperm whale myoglobin in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 84, 8961–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Springer B. A., Egeberg K. D., Sligar S. G., Rohlfs R. J., Mathews A. J., Olson J. S. (1989) Discrimination between oxygen and carbon monoxide and inhibition of autoxidation by myoglobin. Site-directed mutagenesis of the distal histidine. J. Biol. Chem. 264, 3057–3060 [PubMed] [Google Scholar]
- 47. Carver T. E., Brantley R. E., Jr., Singleton E. W., Arduini R. M., Quillin M. L., Phillips G. N., Jr., Olson J. S. (1992) A novel site-directed mutant of myoglobin with an unusually high O2 affinity and low autoxidation rate. J. Biol. Chem. 267, 14443–14450 [PubMed] [Google Scholar]
- 48. Springer B. A., Sligar S. G., Olson J. S., Phillips G. N. (1994) Mechanisms of ligand recognition in myoglobin. Chem. Rev. 94, 699–714 [Google Scholar]
- 49. Birukou I., Schweers R. L., Olson J. S. (2010) Distal histidine stabilizes bound O2 and acts as a gate for ligand entry in both subunits of adult human hemoglobin. J. Biol. Chem. 285, 8840–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Olson J. S., Foley E. W., Maillett D. H., Paster E. V. (2003) Measurement of rate constants for reactions of O2, CO, and NO with hemoglobin. Methods Mol. Med. 82, 65–91 [DOI] [PubMed] [Google Scholar]
- 51. Knapp J. E., Pahl R., Cohen J., Nichols J. C., Schulten K., Gibson Q. H., Srajer V., Royer W. E., Jr. (2009) Ligand migration and cavities within Scapharca dimeric HbI. Studies by time-resolved crystallography, Xe binding, and computational analysis. Structure 17, 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Birukou I., Soman J., Olson J. S. (2011) Blocking the gate to ligand entry in human hemoglobin. J. Biol. Chem. 286, 10515–10529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deng P., Nienhaus K., Palladino P., Olson J. S., Blouin G., Moens L., Dewilde S., Geuens E., Nienhaus G. U. (2007) Transient ligand docking sites in Cerebratulus lacteus mini-hemoglobin. Gene 398, 208–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pflugrath J. W. (1999) The finer things in x-ray diffraction data collection. Acta Crystallogr. D. Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 55. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 57. Germani F., Pesce A., Venturini A., Moens L., Bolognesi M., Dewilde S., Nardini M. (2012) High resolution crystal structures of the Cerebratulus lacteus mini-Hb in the unligated and carbonmonoxy states. Int. J. Mol. Sci. 13, 8025–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Milani M., Pesce A., Ouellet H., Guertin M., Bolognesi M. (2003) Truncated hemoglobins and nitric oxide action. IUBMB Life 55, 623–627 [DOI] [PubMed] [Google Scholar]
- 59. Milani M., Pesce A., Ouellet Y., Ascenzi P., Guertin M., Bolognesi M. (2001) Mycobacterium tuberculosis hemoglobin N displays a protein tunnel suited for O2 diffusion to the heme. EMBO J. 20, 3902–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bidon-Chanal A., Martí M. A., Crespo A., Milani M., Orozco M., Bolognesi M., Luque F. J., Estrin D. A. (2006) Ligand-induced dynamical regulation of NO conversion in Mycobacterium tuberculosis truncated hemoglobin-N. Proteins 64, 457–464 [DOI] [PubMed] [Google Scholar]
- 61. Ouellet Y. H., Daigle R., Lagüe P., Dantsker D., Milani M., Bolognesi M., Friedman J. M., Guertin M. (2008) Ligand binding to truncated hemoglobin N from Mycobacterium tuberculosis is strongly modulated by the interplay between the distal heme pocket residues and internal water. J. Biol. Chem. 283, 27270–27278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Daigle R., Rousseau J. A., Guertin M., Lagüe P. (2009) Theoretical investigations of nitric oxide channeling in Mycobacterium tuberculosis truncated hemoglobin N. Biophys. J. 97, 2967–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Savard P. Y., Daigle R., Morin S., Sebilo A., Meindre F., Lagüe P., Guertin M., Gagné S. M. (2011) Structure and dynamics of Mycobacterium tuberculosis truncated hemoglobin N. Insights from NMR spectroscopy and molecular dynamics simulations. Biochemistry 50, 11121–11130 [DOI] [PubMed] [Google Scholar]
- 64. Laskowski R. A. (1995) SURFNET. A program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph. 13, 323–330 [DOI] [PubMed] [Google Scholar]
- 65. Berka K., Hanák O., Sehnal D., Banás P., Navrátilová V., Jaiswal D., Ionescu C. M., Svobodová Vareková R., Koca J., Otyepka M. (2012) MOLEonline 2.0. Interactive web-based analysis of biomacromolecular channels. Nucleic Acids Res. 40, W222–W227 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.