Background: Molecular mechanisms controlling the late stages of endochondral ossification are unclear.
Results: Osterix functions as a downstream and transcriptional partner of Runx2 and induces MMP13 during chondrocyte differentiation.
Conclusion: Osterix is essential for late-stage endochondral ossification.
Significance: Osterix affects the ossification of cartilage matrices and matrix vesicles and might be involved in the development of osteoarthritis and related disorders.
Keywords: Cartilage, Cartilage Biology, Chondrocytes, Development, Molecular Biology, Transcription Factors, MMP13, Runx2
Abstract
Endochondral ossification is temporally and spatially regulated by several critical transcription factors, including Sox9, Runx2, and Runx3. Although the molecular mechanisms that control the late stages of endochondral ossification (e.g. calcification) are physiologically and pathologically important, these precise regulatory mechanisms remain unclear. Here, we demonstrate that Osterix is an essential transcription factor for endochondral ossification that functions downstream of Runx2. The global and conditional Osterix-deficient mice studied here exhibited a defect of cartilage-matrix ossification and matrix vesicle formation. Importantly, Osterix deficiencies caused the arrest of endochondral ossification at the hypertrophic stage. Microarray analysis revealed that matrix metallopeptidase 13 (MMP13) is an important target of Osterix. We also showed that there exists a physical interaction between Osterix and Runx2 and that these proteins function cooperatively to induce MMP13 during chondrocyte differentiation. Most interestingly, the introduction of MMP13 stimulated the calcification of matrices in Osterix-deficient mouse limb bud cells. Our results demonstrated that Osterix was essential to endochondral ossification and revealed that the physical and functional interaction between Osterix and Runx2 were necessary for the induction of MMP13 during endochondral ossification.
Introduction
Endochondral ossification is a unique and important biological event that is essential to mammalian skeletal development and tissue patterning and is involved in osteoarthritis pathogenesis (1, 2). Endochondral ossification begins with the condensation of multipotent mesenchymal cells that subsequently differentiate into chondrocytes and express chondrogenic matrix proteins, including Col2a1, Col11a2, and aggrecan (1–4). These chondrocytes further differentiate into hypertrophic chondrocytes that express Indian hedgehog (Ihh), Col10a1, and parathyroid hormone-related peptide (1, 2, 5). Cartilage tissues are subsequently replaced with bone tissues following a series of events, including apoptosis of chondrocytes, degradation of cartilage matrices, and vascular invasion into cartilage (1–3). These complementary, complex steps are sequentially coordinated during endochondral ossification.
Genetic and biochemical studies indicate that a specific transcription factor, Sox9, is essential for mesenchymal cell condensation and the initiation of chondrocyte differentiation. Mutations of the SOX9 gene in humans cause campomelic dysplasia characterized by severe chondrodysplasia (6, 7). Cartilage-specific Sox9 conditional knock-out mice showed severely impaired chondrogenesis early in the ossification process (8). In addition, Sox9 directly regulates chondrogenic genes including Col2a1, Col10a2, and aggrecan (9–13), and recent studies suggest that Sox9 inhibits chondrocyte maturation (14, 15).
In contrast, Runx2 and Runx3 are indispensable for chondrocyte hypertrophy (16, 17). In Runx2 knock-out mice, such hypertrophy was severely impaired (17); Runx2 and Runx3 double knock-out mice showed a complete lack of hypertrophic chondrocytes (17). Runx2 has also been shown to be critical for the regulation of Ihh, Col10a1, and vascular endothelial growth factor (VEGF) (17–20).
Although apoptosis of chondrocytes, degradation of cartilage matrices, and vascular invasion into cartilage are necessary components of the endochondral ossification process, the molecular network that regulates these mechanisms remains elusive. In particular, it remains unknown which transcription factors regulate calcification during endochondral ossification. As osteoarthritis and other conditions related to cartilage breakdown appear to result from a disorder of cartilage-matrix calcification (21, 22), a clear understanding of the molecular network that is active in the late stages of endochondral ossification is both biologically and clinically important.
In this study, we attempted to identify the transcription factor that functions downstream of Runx2 and regulates calcification during endochondral ossification. We found that Osterix is essential for endochondral ossification and the formation of matrix vesicles. Moreover, we demonstrated that Osterix specifically targets matrix metalloproteinase 13 (MMP13)2 during endochondral ossification. We also showed that up-regulation of MMP13 is required for there to exist a physical and functional interaction between Osterix and Runx2. Thus, we expect that our findings would contribute to the understanding of the molecular mechanisms that regulate endochondral ossification.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Infection
Limb bud cells were isolated from mouse embryos and digested with 0.1% trypsin and 0.1% collagenase. The cells were cultured in α-modified Eagle's medium containing 10% fetal calf serum (FCS). For the micromass culture, condensed limb bud cells (2 × 105 cells/well) were incubated in α-modified Eagle's medium containing 10% FCS, 0.1 mg/ml ascorbic acid, and 5 mm β-glycerophosphate (Sigma) for 7 days. After this culture period, cells were fixed with 4% formalin-phosphate-buffered saline (PBS) and then stained with alcian blue or alizarin red. Cells were infected with the adenovirus at 50 multiplicity of infection. ATDC5 and 293 cells were provided by the RIKEN cell bank and were cultured in either α-modified Eagle's medium or DMEM. Transfection was performed with FuGENE 6 (Roche Applied Science) per the manufacturer's instructions. MMP13 inhibitor (pyrimidine-4,6-dicarboxylic acid, bis-(4-fluoro-3-methyl-benzylamide)) was purchased from Merck and used at 1 μm.
Plasmids and Adenoviruses
Myc6-Osterix was generated by the subcloning of polymerase chain reaction (PCR)-amplified full-length Osterix cDNA (amino acids 2–429) into an EcoRI and XbaI site of pcDNA3 expression vector containing six tandem repeats of the Myc tag at the N-terminal portion. The sequence of Osterix cDNA was confirmed by DNA sequence analysis. 3× FLAG-Runx2 was kindly provided by Dr. Di Chen (University of Rochester, Rochester, NY). Adenoviruses were generated using an adenovirus construction kit (Takara) per the manufacturer's instructions as described previously (23, 24). DsRed-tagged-Runx2 and Venus-tagged-Osterix were generated by the subcloning of corresponding cDNA into DsRed-tagged and Venus-tagged expression vectors, respectively. The pAxCALNLhMMP13 cosmid was provided by the RIKEN gene bank (25); the Cre adenovirus was purchased from Takara. The MMP13 gene promoter containing its exon 1 and intron 1 (1.2 kb) was subcloned into a pGL4 luciferase reporter vector (Invitrogen).
Real-time RT-PCR
Total RNA was isolated using a total-RNA isolation kit (Macherey-Nagel). After the denaturation of total RNA at 70 °C for 10 min, cDNA was synthesized with an oligo dT primer and reverse transcriptase (Takara). Real-time (reverse-transcription) RT-PCR amplification was performed using the TaqMan PCR protocol and the ABI 7300 real-time PCR system (Applied Biosystems; Table 1); mRNA expression was subsequently normalized to β-action mRNA expression.
TABLE 1.
List of sequences of Taqman probe sets for real-time RT-PCR experiments
| β-Actin | |
| Sense primer | 5′-TTAATTTCTGAATGGCCCAGGTCT-3′ |
| Anti-sense primer | 5′-ATTGGTCTCAAGTCAGTGTACAGG-3′ |
| Probe | 5′-CCTGGCTGCCTCAACACCTCAACCC-3′ |
| Osterix | |
| Sense primer | 5′-AGCGACCACTTGAGCAAACAT-3′ |
| Anti-sense primer | 5′-GCGGCTGATTGGCTTCTTCT-3′ |
| Probe | 5′-CCCGACGCTGCGACCCTCCC-3′ |
| Col2a1 | |
| Sense primer | 5′-CCTCCGTCTACTGTCCACTGA-3′ |
| Anti-sense primer | 5′-ATTGGAGCCCTGGATGAGCA-3′ |
| Probe | 5′-CTTGAGGTTGCCAGCCGCTTCGTCC-3′ |
| Col10a1 | |
| Sense primer | 5′-GCCAAGCAGTCATGCCTGAT-3′ |
| Anti-sense primer | 5′-GACACGGGCATACCTGTTACC-3′ |
| Probe | 5′-AGCACTGACAAGCGGCATCCCAGA-3′ |
| VEGF | |
| Sense primer | 5′-ACGTCAGAGAGCAACATCACC-3′ |
| Anti-sense primer | 5′-CTGTGCTGTAGGAAGCTCATCTC-3′ |
| Probe | 5′-TGCGGATCAAACCTCACCAAAGCCAGC-3′ |
| MMP13 | |
| Sense primer | 5′-GGTTATGACATTCTGGAAGGTTATCC-3′ |
| Anti-sense primer | 5′-CGTGGTTCTCAGAGAAGAAGAGG-3′ |
| Probe | 5′-CCCGTGTTCTCAAAGTGAACCGCAGCG-3′ |
| Ihh | |
| Sense primer | 5′-GACTCATTGCCTCCCAGAACTG-3′ |
| Anti-sense primer | 5′-CCAGGTAGTAGGGTCACATTGC-3′ |
| Probe | 5′-CCACAGCCAGCCTGGACATCCCGA-3′ |
| Runx2 | |
| Sense primer | 5′-CTCCTTCCAGGATGGTCCCA-3′ |
| Anti-sense primer | 5′-CTTCCGTCAGCGTCAACACC-3′ |
| Probe | 5′-CACCACCTCGAATGGCAGCACGCT-3′ |
| ALP | |
| Sense primer | 5′-ATCTTTGGTCTGGCTCCCATG-3′ |
| Anti-sense primer | 5′-TTTCCCGTTCACCGTCCAC-3′ |
| Probe | 5′-TGAGCGACACGGACAAGAAGCCCTT-3′ |
| Sox9 | |
| Sense primer | 5′-CCTTCAACCTTCCTCACTACAGC-3′ |
| Anti-sense primer | 5′-GGTGGAGTAGAGCCCTGAGC-3′ |
| Probe | 5′-CCGCCCATCACCCGCTCGCAATAC-3′ |
| PTHrPa | |
| Sense primer | 5′-GAACATCAGCTACTGCATGACAAG-3′ |
| Anti-sense primer | 5′-TCTGATTTCGGCTGTGTGGATC-3′ |
| Probe | 5′-CCATCCAAGACTTGCGCCGCCGTT-3′ |
a PTHrP, parathyroid hormone-related peptide.
Cre Transgenic Mice
CAG-Cre transgenic mice were provided by the RIKEN gene bank. Prx1-Cre, Col2-Cre, and Col11-Cre transgenic mice were used as described previously (8, 26). All experiments were performed with littermate mice under protocols approved by the Osaka University Graduate School of Dentistry's animal care committee.
Generation of Osterix-floxed Mice
A targeting vector containing a neomycin resistance gene was used to generate the floxed mice. The targeting vector was electroporated into TT2 embryonic stem (ES) cells (27). Homologous recombination was identified by Southern blotting analysis. Germ line transmission of the mutant allele was achieved using C57BL mice and confirmed by Southern blotting and genomic PCR. 5′-TCGCCTTCTTGACGAGTTCT-3′ and 5′-AGCTGGCCCTTAATTTGGTT-3′ were used as PCR primers. The accession number for Osterix-floxed mice is CDB0693K.
Generation of Osterix Global and Conditional Knock-out Mice
To generate global knock-out mice, heterozygous Osterix-floxed mice were mated with CAG-Cre transgenic mice. Deletion of the floxed allele was confirmed by genomic PCR. 5′-TGGAGTCCACCCAACTTTTC-3′ and 5′-GAACTAGGCACTGGCAAAGG-3′ were used as PCR primers. Deletion of the Cre transgene was confirmed by genomic PCR following the mating of Osterix heterozygous deficient mice with C57BL mice. To generate Osterix-conditional knock-out mice, heterozygous Osterix-floxed mice were mated with Prx1-Cre, Col2-Cre, or Col11-Cre transgenic mice. The flox/+ Cre mice were then mated with homozygous Osterix-floxed mice (flox/flox).
Histological and Immunohistochemical Analyses
For histological analyses, limbs were fixed with 4% formalin PBS. Paraffin sections of the limbs were subjected to hematoxylin-eosin (HE) staining and immunostaining with anti-annexin V (Acris), VEGFR2 (Acris), MMP13 (Abcam), or CD31 (Dianova) antibody. Von Kossa staining was performed using a Von Kossa Staining kit (Polysciences, Inc.).
Microarray Analysis
RNA was isolated from mouse limb buds using a total RNA isolation kit and subsequently analyzed using a GeneChip Mouse Gene 1.0 ST Array (Affymetrix).
Skeletal Preparation
Mice were fixed in 95% ethanol, followed by alcian blue and alizarin red staining.
in Situ Hybridization
Tibias were fixed in 4% paraformaldehyde-PBS overnight at 4 °C, embedded in paraffin, and cut into 5-μm sections. Digoxigenin-11-UTP-labeled single-stranded RNA probes were prepared using a digoxigenin RNA labeling kit (Roche Biochemical) per the manufacturer's instructions. cDNA probes for Col2a1, Col10a1, Ihh, Col1a1, Osterix, and MMP13 were provided by Dr. Toshihisa Komori (Nagasaki University, Nagasaki, Japan) and used as described previously (24).
Electron Microscopy
Limbs were fixed in 0.1 m phosphate buffer containing 2.5% glutaraldehyde and 2% paraformaldehyde, and post-fixed in 1% osmium tetroxide. Samples were stained with saturated uranyl acetate followed by lead citrate and examined with a transmission electron microscope.
Immunoprecipitation and Western Blotting
Cells were washed three times with ice-cold PBS; nuclear extracts were subsequently prepared (28). The nuclear extracts were incubated with antibodies for 4 h at 4 °C, and the protein-antibody complexes were immunoprecipitated with protein G-agarose (Roche Applied Science). Immunoprecipitates were washed five times with lysis buffer and boiled in SDS sample buffer; the supernatants were recovered as immunoprecipitate samples. Proteins in whole cell lysates or in immunoprecipitated samples were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were probed by immunoblotting using anti-Myc (Santa Cruz Biotechnology) or FLAG (Sigma) antibody.
Fluorochrome Staining
Cells transfected with Venus- or DsRed-tagged expression vectors were washed in PBS twice, fixed with 4% buffered paraformaldehyde, and treated with 0.5% Triton for 30 min; 5% BSA in PBS was used as a blocking reagent. Nucleus staining was performed with 0.1 μg/ml DAPI (Invitrogen). Slides were mounted with Vectashield (Vector, Burlingame, CA), examined with an LSM510META confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany), and analyzed with LSM5 image browser software (Carl Zeiss).
Luciferase Reporter Assay
The MMP13 gene promoter construct was transfected into ATDC5 cells or mouse limb bud cells; luciferase activity was subsequently measured by a luminometer (Promega) per the manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) Assay
We performed a ChIP assay using the MAGifty chromatin immunoprecipitation system (Invitogen) according to the manufacturer's protocol. The sonicated chromatin samples followed by immunoprecipitation with anti-Myc antibody (ChIP grade) (Abcam), and the input chromatin samples were determined by real-time PCR. Taqman probes and primers used for amplification were as follows: the 500 bp upstream of the MMP13 gene promoter (sense primer, 5′-TACCCACCTCGCTTCAAGGA-3′; antisense primer, 5′-CACTGTAGATGATTGAGTCCTGCC-3′; probe, 5′-CCCCATACCCGACTGCGGTGGTCT-3′), the 130 bp upstream of the Satb2 gene promoter (sense primer, 5′-GCGCCTCCTCTGGAGCTAG-3′; antisense primer, 5′-CTGGTTCGGAGATGGTTGTTATGA-3′; probe, 5′-AGCCAGCAGCCGCCGCAGC-3′).
RESULTS
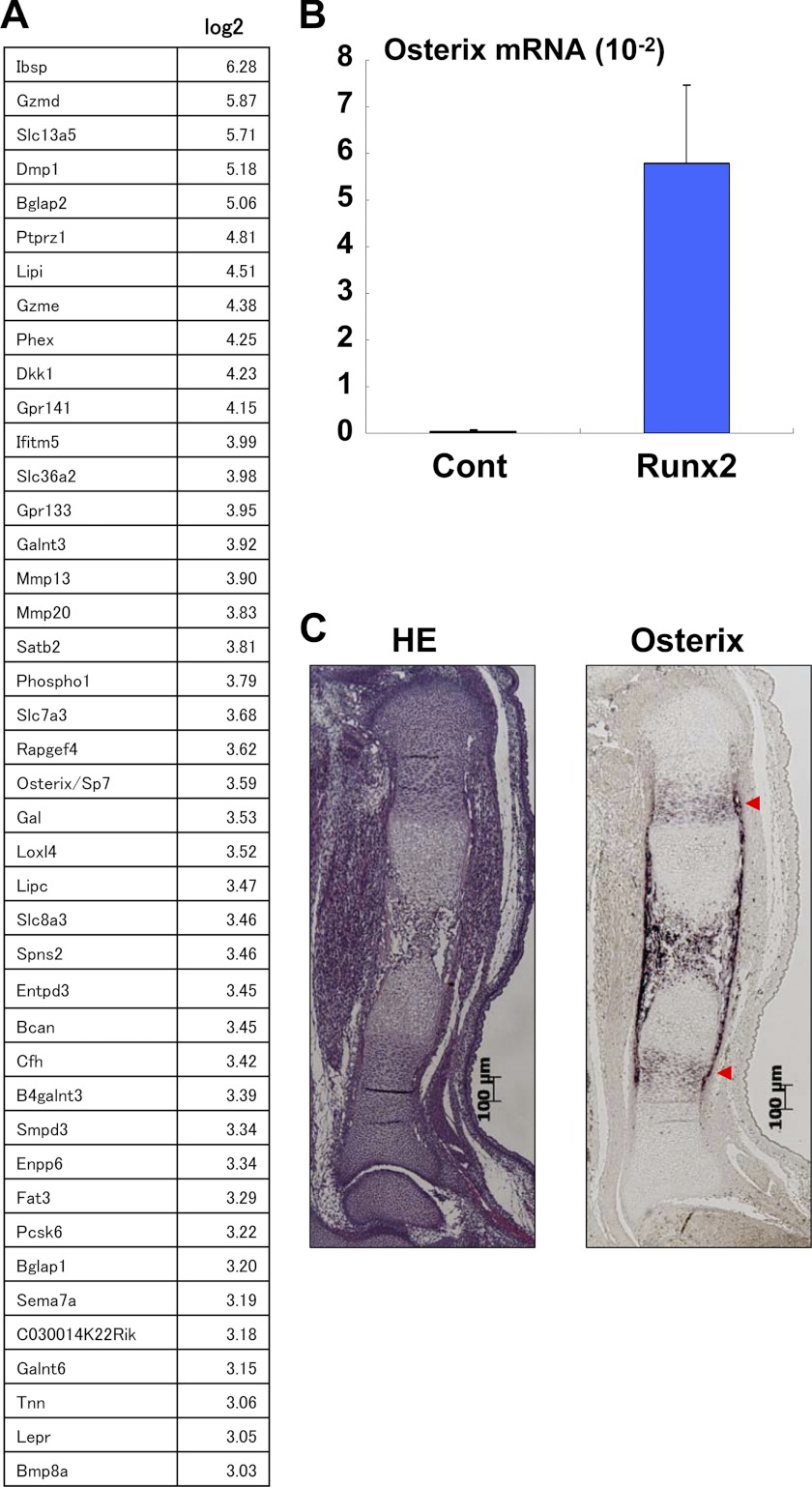
To identify the downstream targets of Runx2/3 that function during endochondral ossification, we performed microarray analyses using mouse limb bud cells that overexpressed Runx2. Subsequent microarray analyses revealed that expression of Osterix/Sp7, a transcription factor essential for osteoblastogenesis (29), was markedly up-regulated in limb bud cells that overexpressed Runx2 (Fig. 1A). Consistent with the microarray analysis, overexpression of Runx2 by infection with the Runx2 adenovirus dramatically induced Osterix expression in mouse limb bud cells (Fig. 1B). In situ hybridization analysis confirmed that Osterix was expressed in prehypertrophic chondrocytes in vivo (Fig. 1C). Consistent with a previous finding that Osterix may function as a downstream transcript of Runx2 during osteoblast development (29), our results suggested that Osterix might similarly play a role in the late stage of endochondral ossification as a downstream target of Runx2. Although the previous study showed impaired endochondral ossification and intact chondrocyte differentiation in Osterix knock-out mice (29), it remains unknown whether this impaired endochondral ossification is due to the inhibition of osteoblast development or that of chondrocyte development. Furthermore, the role of Osterix appears to be quite complex, as illustrated by a study reporting the inhibitory role of Osterix in chondrocyte differentiation (30) and another study showing increased calcification in the cartilage tissues of postnatal conditional Osterix knock-out mice (31).
FIGURE 1.
Up-regulation by Runx2 and expression in cartilage of Osterix. A, mouse limb bud cells were infected with control (Cont) or the Runx2 adenovirus, and then the RNA isolated from the cells was determined by microarray analysis. Up-regulated genes by Runx2 overexpression are listed. B, up-regulation of Osterix expression in mouse limb bud cells by Runx2 was determined using real-time PCR. Values represent means ± S.D. C, Osterix expression was determined by HE staining and in situ hybridization. Hypertrophic-positive areas are indicated by red arrows. Scale bar, 100 μm.
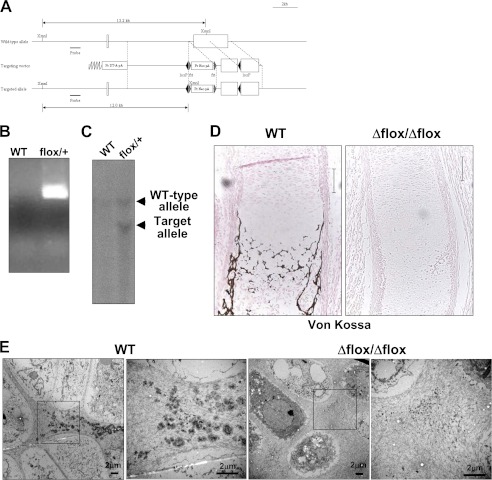
To investigate the role of Osterix in endochondral ossification, we generated mice in which LoxP sites were introduced in the coding region of the Osterix gene (Fig. 2A). The floxed allele in the mice was confirmed by PCR (Fig. 2B) and Southern blotting analyses (Fig. 2C). We first generated the global knock-out mice by mating Osterix-floxed mice with CAG-Cre transgenic mice. Deletion of the floxed allele was confirmed by PCR analysis. Osterix knock-out mice showed no bone formation (supplemental Fig. 1A), as previously demonstrated (29). Ossification of ribs, limbs, and vertebrae was not observed at E15.5 days in the Osterix knock-out mice (supplemental Fig. 1A). Ossification in fore and hind limbs was also markedly inhibited at E18.5 days, and the limbs of these embryos were significantly shorter than those of control littermates (supplemental Fig. 1B).
FIGURE 2.
Impaired ossification and matrix vesicle formation in Osterix knock-out mice. A, schematic representation of the Osterix-floxed allele. B, PCR analysis of Osterix-floxed mice. C, heterozygous Osterix-floxed mice were determined by Southern blotting analysis. D, Von Kossa staining of tibia at E16.5 days. WT, wild-type littermate: Δflox/Δflox, Osterix global knock-out mice. Scale bar, 100 μm. E, electron microscopy images of hypertrophic chondrocyte zone of WT and Osterix global knock-out mice (Δflox/Δflox). Right panels show higher magnification. Scale bar, 2 μm. Pr Neo, promoter neomycin resistant gene; Pr DT-A, promoter diphtheria toxin fragment A.
To examine the role of Osterix in cartilage development we histologically assessed cartilage in Osterix knock-out mice. Interestingly, endochondral ossification was totally stopped at the hypertrophic stage in the Osterix knock-out mice (supplemental Fig. 1C), and calcification of cartilage matrices was not observed (Fig. 2D). Importantly, matrix vesicles were not evident in the hypertrophic chondrocyte zone of Osterix knock-out mice (Fig. 2E). As Runx2- and Runx3-deficient mice show no hypertrophy of chondrocytes (17), these results suggest that Osterix functioned at a later stage of endochondral ossification than Runx2, chiefly at the calcification stage.
We next examined the expression of chondrogenic marker genes by performing in situ hybridization. Although Col2a1, Col10a1, and Ihh were expressed in Osterix knock-out mice, endochondral ossification was clearly delayed (supplemental Fig. 2). Interestingly, MMP13 expression was not detected in Osterix knock-out mice (supplemental Fig. 2). Thus, it is likely that Osterix is an essential transcription factor for the late stage of endochondral ossification. However, to date, the cell autonomous function of Osterix in cartilage development has been elusive.
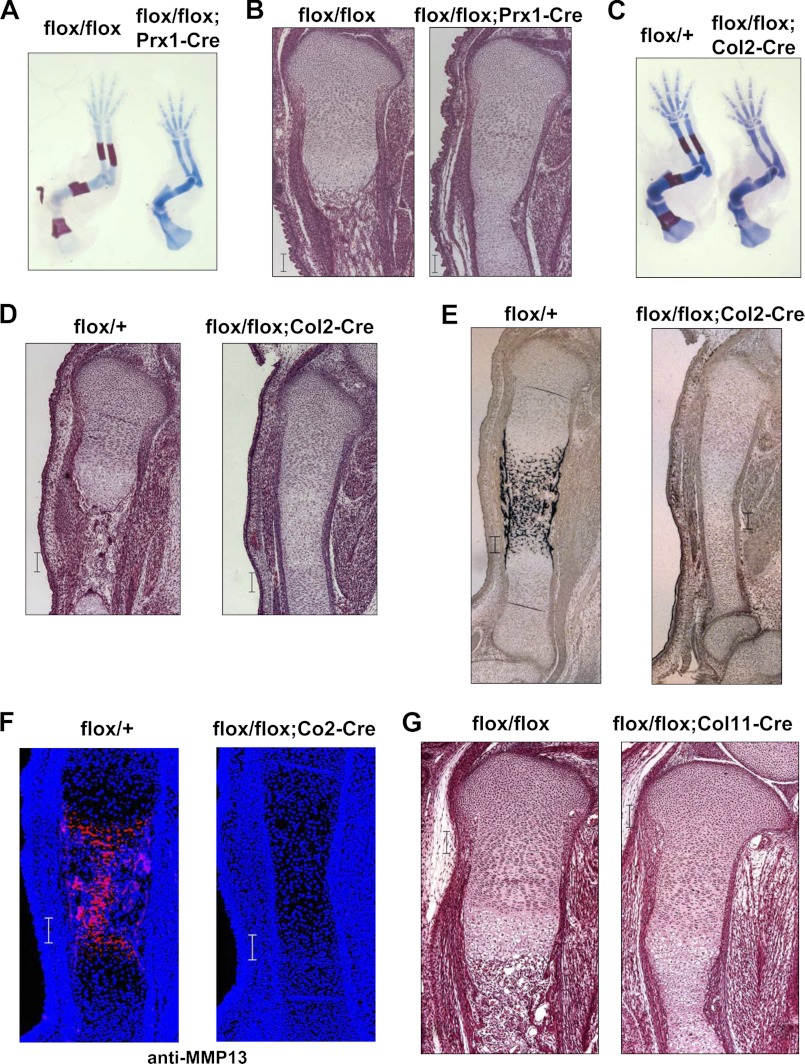
To assess whether the phenotype observed in Osterix knock-out mice was due to a lack of bone formation by osteoblasts or an impairment of chondrocyte differentiation, we generated conditional knock-out mice by mating Osterix-floxed mice with Prx1 (paired-related homeobox 1 )-Cre or Col2a1-Cre transgenic mice (8). As shown in Fig. 3A–D, limb ossification was markedly suppressed, and endochondral ossification was arrested at the hypertrophic stage in Prx1-Cre and Col2a1-Cre Osterix-conditional knock-out mice. Additionally, the calcification of cartilage matrices and MMP13 expression were also blocked in Col2-Cre Osterix-conditional knock-out mice (Fig. 3, E and F). Consistent with results of skeletal preparation (supplemental Fig. 1B), we observed modest calcification of cartilage matrices at E17.5 and E18.5 days of Col2a1-Cre Osterix-conditional knock-out mice (supplemental Fig. 3). To further assess the role of Osterix in endochondral ossification, we generated another type of Osterix-conditional knock-out mouse by mating Osterix-floxed mice with Col11a2-Cre transgenic mice (Col11Enh-Cre), allowing us to delete the target gene at a later stage than is possible with Col2a1-Cre transgenic mice (26). As expected, endochondral ossification was stopped at the hypertrophic stage in the Osterix Col11a2-Cre-conditional knock-out mice (Fig. 3G).
FIGURE 3.
Impairment of ossification in Osterix conditional mice. A and B, flox/+;Prx1-Cre transgenic mice were mated with flox/flox mice. Skeletal preparation of forelimb stained with alcian blue and alizarin red at E15.5 days. Osterix conditional knock-out (flox/flox;Prx1-Cre) and control littermate (flox/flox) (A) are shown. HE staining of tibia of Osterix-conditional knock-out and control littermate at E16.5 days are shown (B). C–E, flox/+;Col2a1(Col2)-Cre transgenic mice were mated with flox/flox mice. Skeletal preparation of forelimb stained with alcian blue and alizarin red at E16.5 days. Osterix-conditional knock-out (flox/flox;Col2-Cre) and control littermate (flox/+) are shown (C). HE staining of tibia of Osterix-conditional knock-out and control littermate at E16.5 days (D). Von Kossa staining of tibia of Osterix-conditional knock-out and control littermate at E16.5 (E). F, MMP13 immunostaining of tibia of Osterix-conditional knock-out and control littermate at E16.5 days. G, flox/+;Col11a2(Col11)-Cre transgenic mice were mated with flox/flox mice. HE staining of tibia of Osterix-conditional knock-out (flox/flox;Col11-Cre) and control littermate (flox/flox) at E16.5 days. Scale bar, 100 μm.
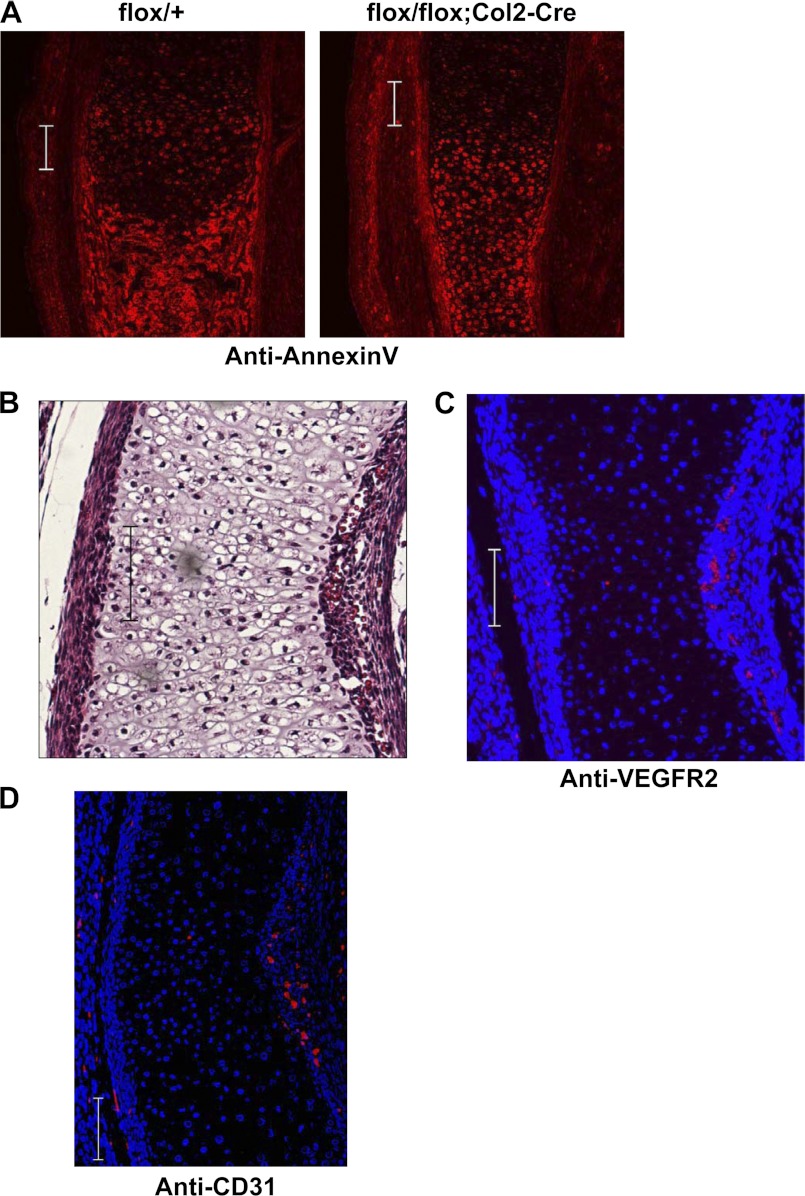
As chondrocyte apoptosis and angiogenesis are essential events in the late stages of endochondral ossification, we next examined whether Osterix is involved in chondrocyte apoptosis and/or angiogenesis during endochondral ossification. We observed that annexin V, a well known apoptosis marker, was expressed in hypertrophic chondrocytes of Col2-Cre Osterix-conditional knock-out mice as well as in control littermates (Fig. 4A), suggesting that apoptosis may proceed normally in Osterix-deficient chondrocytes. We next examined angiogenesis in Col2-Cre Osterix-conditional knock-out mice. Histological analysis indicated that angiogenesis occurred around the bone collar of the Col2-Cre Osterix-conditional knock-out mice; however, vascular invasion into cartilage was inhibited because of insufficient space in non-degraded cartilage tissues (Fig. 4B). We confirmed angiogenesis in Col2-Cre Osterix-conditional knock-out mice by performing immunostaining analyses using anti-VEGFR2 and CD31 antibodies, specific markers for vascular endothelial cells (Fig. 4, C and D). Collectively, our results indicate that Osterix plays an essential role in the calcification stage and cartilage matrix degradation process during endochondral ossification but not in the stages of apoptosis or angiogenesis.
FIGURE 4.
Apoptosis and angiogenesis in Osterix knock-out mice. A, flox/+ and flox/flox;Col2-Cre littermate mice were immunostained with anti-annexin V antibody of tibia of Osterix-conditional knock-out and control littermate at E16.5 days. B, paraffin section of Osterix-conditional knock-out mice (flox/flox;Co2-Cre) at E16.5 days was determined by HE staining. Angiogenesis around the diaphysis of the bone collar was observed in Osterix-conditional knock-out mice (flox/flox;Co2-Cre). C, histological section of Osterix-conditional knock-out mice (flox/flox;Co2-Cre) at E16.5 days was stained with anti-VEGFR2 antibody (red) and DAPI. D, histological section of Osterix-conditional knock-out mice (flox/flox;Co2-Cre) at E16.5 days was stained with anti-CD31 antibody (red) and DAPI. Scale bar, 100 μm.
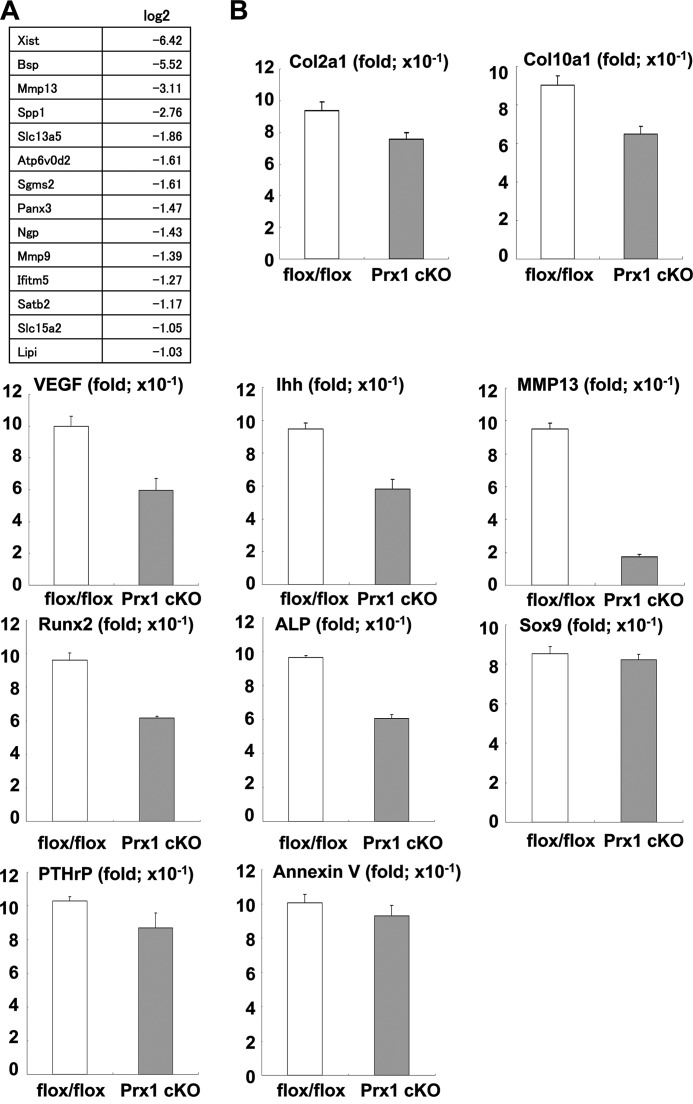
We next attempted to identify Osterix targets to study the mechanisms by which Osterix controls endochondral ossification. To address this issue, we performed microarray analysis using limb bud tissues of Prx1-Cre Osterix-conditional knock-out and control littermate mice. As shown in Fig. 5A, MMP13 expression was markedly lower in limb bud tissues from conditional knock-out mice than in those from control mice.
FIGURE 5.
Down-regulation of MMP13 expression in Osterix-conditional knock-out mice. A, microarray analysis of limb bud tissues of Osterix-conditional knock-out mice (flox/flox;Prx1-Cre) and control littermates from E16.5 days. Down-regulated genes in Osterix-conditional knock-out are listed. B, real-time PCR analyses of limb bud tissues of Osterix-conditional knock-out mice (Prx1 cKO) and control littermates (flox/flox). Values represent means ± S.D.
Consistent with our results, it has been reported that MMP13 expression was decreased in postnatal conditional Osterix knock-out mice (31). To further test this, we assessed the expression of chondrogenic marker genes in limb bud tissues from Prx1-Cre Osterix-conditional knock-out mice and control littermates using real-time RT-PCR. Col10a1, VEGF, Runx2, and ALP were moderately down-regulated in Osterix knock-out samples (Fig. 5B). Chondrocyte hypertrophy seemed intact in these animals; therefore, down-regulation of Col10a1, Runx2, and ALP might not have influenced Osterix deficiency in chondrocytes. In addition, the expression of Sox9, parathyroid hormone-related peptide, and annexin V was not altered in samples from Prx1-Cre Osterix-conditional knock-out mice (Fig. 5B). Although annexin V is reported to be involved in the formation of matrix vesicles during calcification (32), the phenotype observed in our Osterix-deficient chondrocytes did not result from down-regulation of annexin V. Consistent with in situ and microarray data (supplemental Fig. 2 and Figs. 3F and 5A), MMP13 expression was markedly reduced in limb buds from Prx1-Cre Osterix-conditional knock-out mice (Fig. 5B). Collectively, these results suggest that MMP13 was indeed a target of Osterix during endochondral ossification.
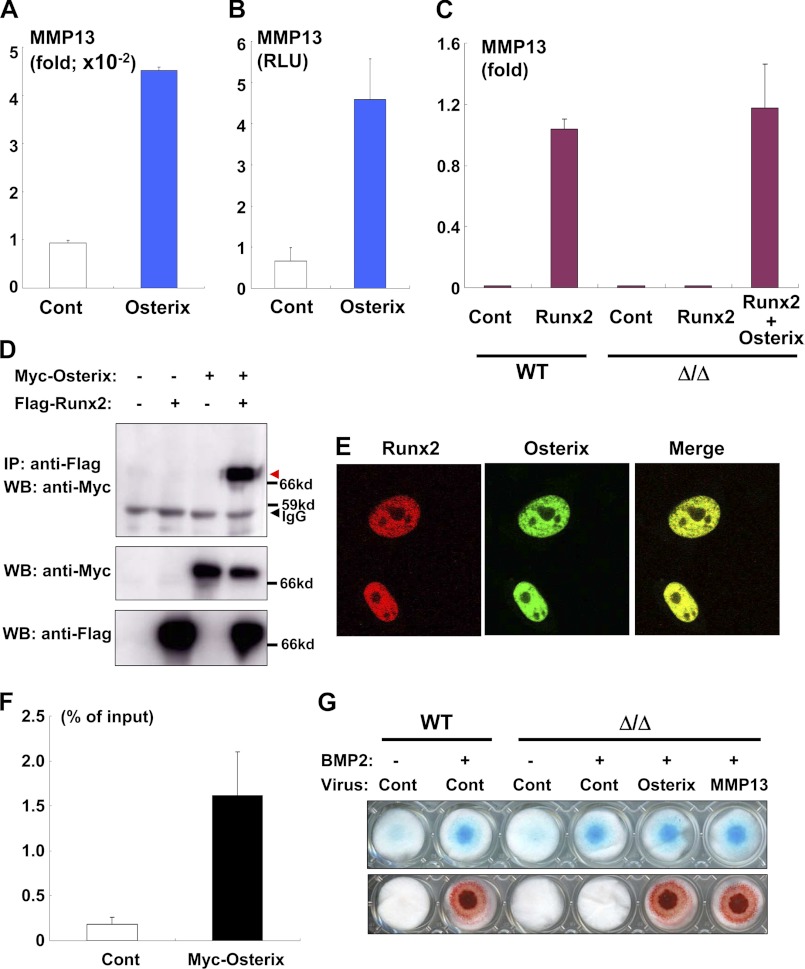
To confirm whether MMP13 was an Osterix target, we infected limb bud cells isolated from Osterix knock-out mice with Osterix adenovirus and subsequently examined MMP13 expression. Overexpression of Osterix clearly induced MMP13 expression in the Osterix-deficient background (Fig. 6A). Furthermore, overexpression of Osterix increased MMP13 gene promoter activity (Fig. 6B). As MMP13 is reportedly induced by Runx2 (16, 33), we examined the effect of Runx2 overexpression on MMP13 expression in the presence and absence of exogenous Osterix. Similar to the results of a previous study (16), we found that Runx2 overexpression markedly up-regulated MMP13 expression in the limb bud cells of wild-type mice (Fig. 6C). However, Runx2 failed to induce MMP13 expression in limb bud cells from Osterix knock-out mice (Fig. 6C). Importantly, Osterix introduction recovered Runx2-dependent induction of MMP13 (Fig. 6C). Thus, our results indicate that Osterix was required for Runx2-mediated up-regulation of MMP13; similarly, these findings raise the possibility that Osterix and Runx2 collaborate during endochondral ossification. We thus performed a series of co-immunoprecipitation experiments to more fully examine this relationship. As shown in Fig. 6D, Osterix physically associated with Runx2. In addition, Osterix and Runx2 are well colocalized in nuclei (Fig. 6E). Our ChIP assay also indicated that Osterix directly binds to the MMP13 gene promoter (Fig. 6F), and the Stab2 gene promoter (supplemental Fig. 4), which is one of the direct targets of Osterix (34). Collectively, it is likely that Osterix functioned both downstream and as a transcriptional partner of Runx2 and that Osterix regulated endochondral ossification by controlling MMP13 expression.
FIGURE 6.
Osterix and Runx2 required for up-regulation of MMP13. A, up-regulation of MMP13 by Osterix was determined by real-time PCR. Limb bud cells isolated from Osterix knock-out mice were infected with control or Osterix adenovirus, and RNA was isolated from the cells. Values represent means ± S.D. B, ATDC5 cells were transfected with the MMP13 gene promoter luciferase construct and then infected with control (Cont) or Osterix adenovirus. Luciferase activity in the cell lysates was measured. Values represent means ± S.D. C, limb bud cells of wild-type or Osterix knock-out mice (Δ/Δ) were infected with control, Runx2, and/or Osterix adenovirus, and RNA was subjected to real-time PCR analysis. Values represent means ± S.D. D, co-immunoprecipitation experiment using nuclear extracts containing 293 cells transfected with FLAG-Runx2 and/or Myc-Osterix. Co-immunoprecipitated Myc-Osterix with FLAG-Runx2 is shown by the red arrow. IP, immunoprecipitation; WB, Western blotting. E, 293 cells were transfected with DsRed-tagged-Runx2 and Venus-tagged-Osterix and then monitored under confocal microscopy. F, ATDC5 cells infected control (Cont) or Myc-Osterix adenovirus were subjected to a ChIP assay. Immunoprecipitated chromatin samples with anti-Myc antibody and input samples were determined by real-time PCR analysis using a specific Taqman probe against the ∼500 bp upstream region of the MMP13 gene promoter. G, limb bud cells of wild-type or Osterix knock-out mice (Δ/Δ) were subjected to micromass culture. The cells were infected with control, Osterix, or both MMP13 and Cre adenovirus and were subsequently cultured in the presence or absence of BMP2 for 7 days; cells were then stained with alcian blue (top panel) or alizarin red (lower panel).
To investigate the biological relevance of our findings, we examined whether the introduction of MMP13 stimulated the calcification of cartilage matrices in Osterix-deficient limb bud cells. BMP2 (bone morphogenetic protein 2) stimulated limb bud cells from wild-type mice to differentiate into chondrocytes producing alcian blue-positive and calcified matrices (Fig. 6G); this treatment also stimulated limb bud cells from Osterix knock-out mice to differentiate into alcian blue-positive chondrocytes (Fig. 6G). However, BMP2 treatment did not cause calcification in the limb bud cells of Osterix knock-out mice (Fig. 6G). These results were consistent with the phenotype seen in Osterix knock-out mice, and they strongly supported the hypothesis that Osterix is required for the differentiation of chondrocytes required to produce calcified cartilage matrices. Notably, the introduction of MMP13 by adenovirus stimulated the calcification of matrices in the limb bud cells of Osterix knock-out mice (Fig. 6G). In addition, the presence of an MMP13 inhibitor blocked Osterix-dependent calcification of the matrices (supplemental Fig. 5). These results suggest that MMP13 is a critical transcriptional target of Osterix during endochondral ossification.
DISCUSSION
It is physiologically and clinically important to understand the molecular mechanisms that regulate the calcification step of endochondral ossification. In this study, we used microarray analysis to help identify the downstream transcription factor of Runx2 during endochondral ossification. We successfully identified Osterix as the downstream transcript of Runx2 in mouse limb bud cells and found that Osterix is necessary for endochondral ossification and the formation of matrix vesicles in mice. We also successfully demonstrated that limb bud cells isolated from Osterix knock-out mice failed to calcify in vitro. Taken together, these results show that Osterix is an essential transcription factor for endochondral ossification and the formation of matrix vesicles.
Although a previous study showed a similar phenotype of Osterix knock-out mice in cartilage, the authors hypothesized that chondrogenesis could be intact and that the phenotype seen in the studied cartilage might result from the impairment of osteoblastogenesis; in fact, the authors of that study did not investigate beyond the hypertrophic stage of the endochondral ossification process (29). Furthermore, it has not been investigated whether impaired endochondral ossification in Osterix knock-out mice (29) is due to the cell autonomous function of Osterix.
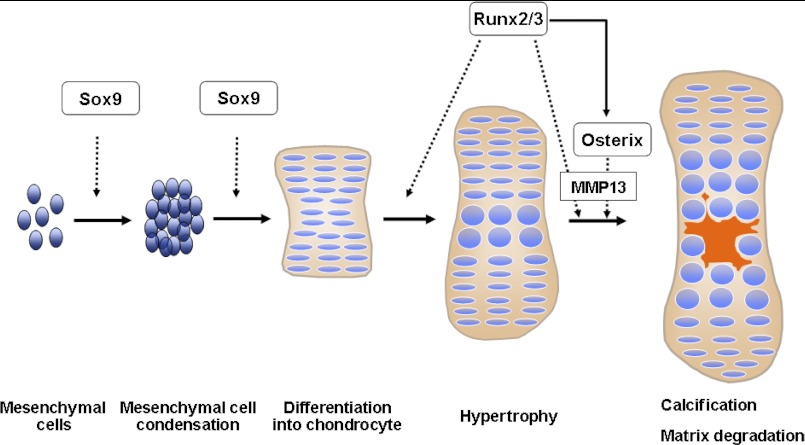
It has, however, been demonstrated previously that Sox9 and Runx2/3 are indispensable transcription factors for the initiation of chondrogenesis and the hypertrophy of chondrocytes, respectively (Fig. 7) (4, 20). Therefore, our findings suggest that a Sox9-Runx2-Osterix axis temporally and spatially mediates chondrogenesis (Fig. 7).
FIGURE 7.
Schematic diagram of endochondral ossification. Endochondral ossification is sequentially regulated by Sox9, Runx2, and Osterix. Osterix functions as both a downstream and transcriptional partner of Runx2/3 during calcification and matrix degradation in cartilage, and cooperation between Osterix and Runx2/3 are required for MMP13 expression.
A previous study reported that Osterix suppressed the differentiation and maturation of chondrocytes in vitro (30). These results are completely inconsistent with our findings. One possibility for this discrepancy is that different experimental approaches were used in the two studies, even though they each performed knockdown experiments using siRNA against Osterix. Our study took the gene-targeting approach, resulting in a consistent finding that the Osterix gene plays an essential role in chondrocyte development in vivo; we further confirmed this finding ex vivo by using limb bud cells isolated from Osterix knock-out mice. In addition, we observed the phenotype of arrest of chondrogenesis in global Osterix knock-out and cartilage-specific Osterix conditional knock-out mice. Thus, we believe that our research has proven that Osterix is truly necessary for chondrogenesis.
Although a previous study using postnatal Osterix-deficient mice suggested that MMP13 would be a target of Osterix (31), it remained unanswered whether MMP13 played a key role as Osterix target genes during endochondral ossification. Our in situ hybridization, microarray, and real-time PCR analyses collectively indicated that Osterix was required for MMP13 expression during endochondral ossification. More importantly, we demonstrated that Osterix directly regulated MMP13 expression, as shown by our discovery that overexpression of Osterix induced MMP13 expression, increased MMP13 gene promoter activity in vitro, and directly associated with the MMP13 gene promoter.
The phenotype of MMP13-deficient mice has been consistently shown to resemble that of Osterix knock-out mice (35). Thus, MMP13 is an important target of Osterix during endochondral ossification. Notably, postnatal Osterix-deficient mice showed a marked increase in calcified matrix in the growth plate (31). The postnatal Osterix-deficient mice showed a lack of resorption of mineralized cartilage and participation of osteoclasts (31). As MMP13 is required for the generation of space in cartilage (35, 36), it is therefore likely that the different phenotype in cartilage between the previous study and ours results from the timing of deficiency of the Osterix gene.
As our histological analysis suggested that vascular invasion into cartilage tissues required the degradation of cartilage matrices, we posit that Osterix is likely to be essential for the subsequent replacement of cartilage tissues with bone tissues. This idea is also supported by a previous study using MMP13 conditional knock-out mice (36). Importantly, we observed that the addition of exogenous MMP13 improved the calcification of chondrocyte matrices in Osterix-deficient limb bud cells. However, we also found that the MMP13 inhibitor suppressed Osterix-dependent calcification of the matrices. In global and chondrocyte-specific conditional MMP13-deficient mice, chondrogenic matrices accumulated in the hypertrophic zone of the growth plate (36). In particular, Col2a1 and aggrecan seem to be major substrates of MMP13 (36).
Collectively, it is possible that the degradation of cartilaginous collagen is required for the calcification of cartilage matrices or the formation of matrix vesicles in cartilage, although the precise role of MMP13 in the calcification of chondrocyte matrices remains unclear. Interestingly, MMP13 knock-out mice showed a marked increase in trabecular bones (36). It is worth noting that the global and conditional Osterix knock-out mice studied here showed no bone formation; future research into the role of Osterix in endochondral ossification might focus on the relationship that exists among Osterix, MMP13, and metabolism of trabecular bone. Yet, despite the fact that bone formation was not present in our conditional Osterix knock-out mice, we expect that MMP13 expression in osteoblasts, but not chondrocytes, might be involved in the metabolism of trabecular bone, as chondrocyte-specific MMP13-deficient mice did not demonstrate a clear increase in trabecular bone (36).
Runx2 has been demonstrated to function upstream of Osterix during osteoblast differentiation (23, 29) and to be required for the induction of MMP13 (16, 33). In this study, we found that Osterix not only plays an important role downstream of Runx2 but that it is also a transcriptional partner of Runx2. In addition to our biochemical experiments, we demonstrated that both Osterix and Runx2 are necessary for the up-regulation of MMP13 expression. It is therefore likely that the functional interaction between Osterix and Runx2 is important for endochondral ossification, especially for the calcification and degradation of chondrogenic matrices. As AP1 complex has been shown to collaborate with Runx2 to control MMP13 expression (33), we expect that a complex of Runx2, Osterix, and AP1 might be important to the regulation of MMP13 expression.
Interestingly, our experiments showed that endochondral ossification and calcification of cartilage matrices were impaired but still observed in Osterix knock-out mice at E18.5 days (supplemental Figs. 1B and 3). This observation suggested that other molecules partly compensated for the function of Osterix during endochondral ossification. We are currently attempting to identify these molecules, and plan to examine the relationship between Osterix and these molecules in a future research project.
As described above, MMP13 is essential for the chondrogenesis of growth plates (35). However, MMP13 is also integral to the pathogenesis of osteoarthritis, which is associated with heterotopic ossification of articular chondrocytes (37, 38). Thus, we believe that our findings might contribute to the understanding of the molecular mechanisms of endochondral ossification and will similarly be helpful in the development of therapeutic reagents for osteoarthritis and other cartilage disorders.
Supplementary Material
Acknowledgment
We thank the RIKEN gene bank for providing CAG-Cre transgenic mice and the pAxCALNLhMMP13 cosmid.
This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology grants-in-aid for Scientific Research (to T. Y. and R. N.); the 21st Century COE Program (to T. Y. and R. N.); the Uehara Memorial Foundation (to R. N.); and the Astellas Foundation for Research on Metabolic Disorders (to R. N.).

This article contains supplemental Figs. 1–5.
- MMP13
- matrix metallopeptidase 13
- E15
- embryonic day 15.
REFERENCES
- 1. Kronenberg H. M. (2003) Developmental regulation of the growth plate. Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 2. de Crombrugghe B., Lefebvre V., Behringer R. R., Bi W., Murakami S., Huang W. (2000) Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 19, 389–394 [DOI] [PubMed] [Google Scholar]
- 3. Komori T. (2011) Signaling networks in RUNX2-dependent bone development. J. Cell. Biochem. 112, 750–755 [DOI] [PubMed] [Google Scholar]
- 4. Nishimura R., Hata K., Ikeda F., Ichida F., Shimoyama A., Matsubara T., Wada M., Amano K., Yoneda T. (2008) Signal transduction and transcriptional regulation during mesenchymal cell differentiation. J. Bone Miner. Metab. 26, 203–212 [DOI] [PubMed] [Google Scholar]
- 5. Ornitz D. M., Marie P. J. (2002) FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 16, 1446–1465 [DOI] [PubMed] [Google Scholar]
- 6. Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F.D., Keutel J., Hustert E., Wolf U., Tommerup N., Schempp W., Scherer G. (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79, 1111–1120 [DOI] [PubMed] [Google Scholar]
- 7. Foster J. W., Dominguez-Steglich M. A., Guioli S., Kwok C., Weller P. A., Stevanović M., Weissenbach J., Mansour S., Young I. D., Goodfellow P. N. (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372, 525–530 [DOI] [PubMed] [Google Scholar]
- 8. Akiyama H., Chaboissier M. C., Martin J. F., Schedl A., de Crombrugghe B. (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ng L. J., Wheatley S., Muscat G. E., Conway-Campbell J., Bowles J., Wright E., Bell D. M., Tam P. P., Cheah K. S., Koopman P. (1997) SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev. Biol. 183, 108–121 [DOI] [PubMed] [Google Scholar]
- 10. Lefebvre V., Huang W., Harley V. R., Goodfellow P. N., de Crombrugghe B. (1997) SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro-α1(II) collagen gene. Mol. Cell. Biol. 17, 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Li H., Tanaka K., Tsumaki N., Yamada Y. (2000) Identification of an enhancer sequence within the first intron required for cartilage-specific transcription of the α2(XI) collagen gene. J. Biol. Chem. 275, 12712–12718 [DOI] [PubMed] [Google Scholar]
- 12. Takahashi I., Nuckolls G. H., Takahashi K., Tanaka O., Semba I., Dashner R., Shum L., Slavkin H. C. (1998) Compressive force promotes sox9, type II collagen, and aggrecan and inhibits IL-1β expression resulting in chondrogenesis in mouse embryonic limb bud mesenchymal cells. J. Cell Sci. 111, 2067–2076 [DOI] [PubMed] [Google Scholar]
- 13. Takigawa Y., Hata K., Muramatsu S., Amano K., Ono K., Wakabayashi M., Matsuda A., Takada K., Nishimura R., Yoneda T. (2010) The transcription factor Znf219 regulates chondrocyte differentiation by assembling a transcription factory with Sox9. J. Cell Sci. 123, 3780–3788 [DOI] [PubMed] [Google Scholar]
- 14. Amano K., Hata K., Sugita A., Takigawa Y., Ono K., Wakabayashi M., Kogo M., Nishimura R., Yoneda T. (2009) Sox9 family members negatively regulate maturation and calcification of chondrocytes through up-regulation of parathyroid hormone-related protein. Mol. Biol. Cell 20, 4541–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hattori T., Müller C., Gebhard S., Bauer E., Pausch F., Schlund B., Bösl M. R., Hess A., Surmann-Schmitt C., von der Mark H., de Crombrugghe B., von der Mark K. (2010) SOX9 is a major negative regulator of cartilage vascularization, bone marrow formation, and endochondral ossification. Development 137, 901–911 [DOI] [PubMed] [Google Scholar]
- 16. Enomoto H., Enomoto-Iwamoto M., Iwamoto M., Nomura S., Himeno M., Kitamura Y., Kishimoto T., Komori T. (2000) Cbfa1 is a positive regulatory factor in chondrocyte maturation. J. Biol. Chem. 275, 8695–8702 [DOI] [PubMed] [Google Scholar]
- 17. Yoshida C. A., Yamamoto H., Fujita T., Furuichi T., Ito K., Inoue K., Yamana K., Zanma A., Takada K., Ito Y., Komori T. (2004) Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 18, 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng Q., Zhou G., Morello R., Chen Y., Garcia-Rojas X., Lee B. (2003) Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J. Cell Biol. 162, 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwon T. G., Zhao X., Yang Q., Li Y., Ge C., Zhao G., Franceschi R. T. (2011) Physical and functional interactions between Runx2 and HIF-1α induce vascular endothelial growth factor gene expression. J. Cell. Biochem. 112, 3582–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Komori T. (2010) Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 339, 189–195 [DOI] [PubMed] [Google Scholar]
- 21. Rutges J. P., Duit R. A., Kummer J. A., Oner F. C., van Rijen M. H., Verbout A. J., Castelein R. M., Dhert W. J., Creemers L. B. (2010) Hypertrophic differentiation and calcification during intervertebral disc degeneration. Osteoarthritis Cartilage 18, 1487–1495 [DOI] [PubMed] [Google Scholar]
- 22. Kawaguchi H. (2008) Endochondral ossification signals in cartilage degradation during osteoarthritis progression in experimental mouse models. Mol. Cells 25, 1–6 [PubMed] [Google Scholar]
- 23. Matsubara T., Kida K., Yamaguchi A., Hata K., Ichida F., Meguro H., Aburatani H., Nishimura R., Yoneda T. (2008) BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J. Biol. Chem. 283, 29119–29125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hata K., Nishimura R., Muramatsu S., Matsuda A., Matsubara T., Amano K., Ikeda F., Harley V. R., Yoneda T. (2008) Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice. J. Clin. Invest. 118, 3098–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suzuki Y., Yoshitomo-Nakagawa K., Maruyama K., Suyama A., Sugano S. (1997) Construction and characterization of a full-length-enriched and a 5′-end-enriched cDNA library. Gene 200, 149–156 [DOI] [PubMed] [Google Scholar]
- 26. Ikegami D., Akiyama H., Suzuki A., Nakamura T., Nakano T., Yoshikawa H., Tsumaki N. (2011) Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development 138, 1507–1519 [DOI] [PubMed] [Google Scholar]
- 27. Yagi T., Tokunaga T., Furuta Y., Nada S., Yoshida M., Tsukada T., Saga Y., Takeda N., Ikawa Y., Aizawa S. (1993) A novel ES cell line, TT2, with high germ line-differentiating potency. Anal. Biochem. 214, 70–76 [DOI] [PubMed] [Google Scholar]
- 28. Nishimura R., Moriyama K., Yasukawa K., Mundy G. R., Yoneda T. (1998) Combination of interleukin-6 and soluble interleukin-6 receptors induces differentiation and activation of JAK-STAT and MAP kinase pathways in MG-63 human osteoblastic cells. J. Bone Miner. Res. 13, 777–785 [DOI] [PubMed] [Google Scholar]
- 29. Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29 [DOI] [PubMed] [Google Scholar]
- 30. Kaback L. A., Soung do Y., Naik A., Smith N., Schwarz E. M., O'Keefe R. J., Drissi H. (2008) Osterix/Sp7 regulates mesenchymal stem cell mediated endochondral ossification. J. Cell. Physiol. 214, 173–182 [DOI] [PubMed] [Google Scholar]
- 31. Zhou X., Zhang Z., Feng J. Q., Dusevich V.M., Sinha K., Zhang H., Darnay B. G., de Crombrugghe B. (2010) Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc. Natl. Acad. Sci. U.S.A. 107, 12919–12924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kirsch T., Wuthier R. E. (1994) Stimulation of calcification of growth plate cartilage matrix vesicles by binding to type II and X collagens. J. Biol. Chem. 269, 11462–11469 [PubMed] [Google Scholar]
- 33. Mengshol J. A, Vincenti M. P., Brinckerhoff C. E. (2001) IL-1 induces collagenase-3 (MMP-13) promoter activity in stably transfected chondrocytic cells: requirement for Runx-2 and activation by p38 MAPK and JNK pathways. Nucleic Acids Res. 29, 4361–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang W., Li Y., Osimiri L., Zhang C. (2011) Osteoblast-specific transcription factor Osterix (Osx) is an upstream regulator of Satb2 during bone formation. J. Biol. Chem. 286, 32995–33002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Inada M., Wang Y., Byrne M. H., Rahman M. U., Miyaura C., López-Otín C., Krane S. M. (2004) Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc. Natl. Acad. Sci. U.S.A. 101, 17192–17197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stickens D., Behonick D. J., Ortega N., Heyer B., Hartenstein B., Yu Y., Fosang A.J., Schorpp-Kistner M., Angel P., Werb Z. (2004) Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development 131, 5883–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. von der Mark K., Kirsch T., Nerlich A., Kuss A., Weseloh G., Glückert K., Stöss H. (1992) Type X collagen synthesis in human osteoarthritic cartilage. Indication of chondrocyte hypertrophy. Arthritis Rheum. 35, 806–811 [DOI] [PubMed] [Google Scholar]
- 38. Fuerst M., Bertrand J., Lammers L., Dreier R., Echtermeyer F., Nitschke Y., Rutsch F., Schäfer F. K., Niggemeyer O., Steinhagen J., Lohmann C.H., Pap T., Rüther W. (2009) Calcification of articular cartilage in human osteoarthritis. Arthritis Rheum. 60, 2694–2703 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.